Abstract
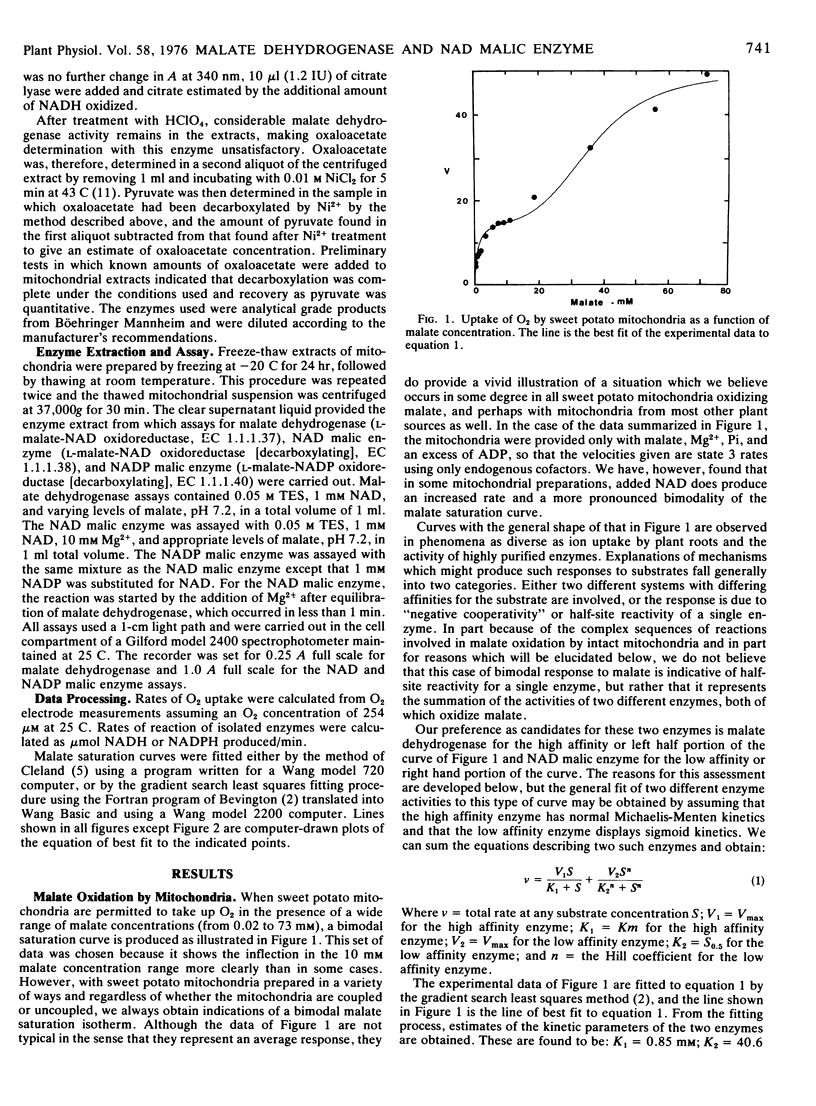
Over a range of concentrations from less than 0.1 mm to more than 70 mm, sweet potato root mitochondria display a bimodal substrate saturation isotherm for malate. The high affinity portion of the isotherm has an apparent Km for malate of 0.85 mm and fits a rectangular hyperbolic function. The low affinity portion of the isotherm is sigmoid in character and gives an apparent S0.5 of 40.6 mm and a Hill number of 3.7.
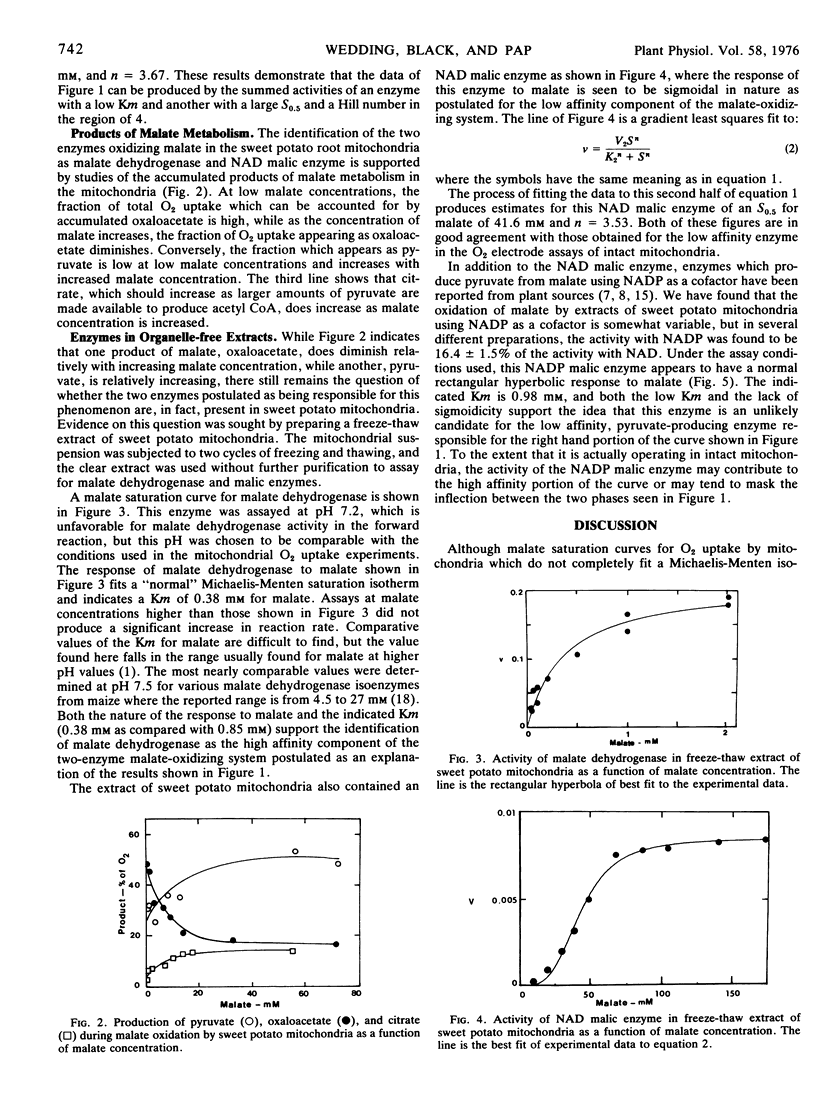
Extracts of sweet potato mitochondria contain both malate dehydrogenase and NAD malic enzyme. The malate dehydrogenase, assayed in the forward direction at pH 7.2, shows typical Michaelis-Menten kinetics with a Km for malate of 0.38 mm. The NAD malic enzyme shows pronounced sigmoidicity in response to malate with a Hill number of 3.5 and an S0.5 of 41.6 mm.
On the basis of the normal kinetics, the Km, and the fact that oxaloacetate production from malate by mitochondria appears most active at low malate concentrations, the high affinity portion of the malate isotherm with mitochondria is attributed to malate dehydrogenase. The low affinity portion of the malate isotherm with mitochondria is thought, on the basis of the similarity of S0.5 values, the Hill numbers, and the greater production of pyruvate from malate at high malate concentrations, to represent the activity of the NAD malic enzyme.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunton C. J., Palmer J. M. Pathways for the oxidation of malate and reduced pyridine nucleotide by wheat mitochondria. Eur J Biochem. 1973 Nov 1;39(1):283–291. doi: 10.1111/j.1432-1033.1973.tb03125.x. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. The statistical analysis of enzyme kinetic data. Adv Enzymol Relat Areas Mol Biol. 1967;29:1–32. doi: 10.1002/9780470122747.ch1. [DOI] [PubMed] [Google Scholar]
- Coleman J. O., Palmer J. M. The oxidation of malate by isolated plant mitochondria. Eur J Biochem. 1972 Apr 24;26(4):499–509. doi: 10.1111/j.1432-1033.1972.tb01792.x. [DOI] [PubMed] [Google Scholar]
- Davies D. D., Patil K. D. Regulation of 'malic' enzyme of Solanum tuberosum by metabolites. Biochem J. 1974 Jan;137(1):45–53. doi: 10.1042/bj1370045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R., Bonner W. D., Jr Oxalacetate control of Krebs cycle oxidations in purified plant mitochondria. Biochem Biophys Res Commun. 1972 May 12;47(3):619–624. doi: 10.1016/0006-291x(72)90923-0. [DOI] [PubMed] [Google Scholar]
- Lance C., Hobson G. E., Young R. E., Biale J. B. Metabolic processes in cytoplasmic particles of the avocado fruit. IX. The oxidation of pyruvate and malate during the climacteric cycle. Plant Physiol. 1967 Apr;42(4):471–478. doi: 10.1104/pp.42.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae A. R. Isolation and properties of a 'malic' enzyme from cauliflower bud mitochondria. Biochem J. 1971 May;122(4):495–501. doi: 10.1042/bj1220495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae A. R., Moorhouse R. The oxidation of malate by mitochondria isolated from cauliflower buds. Eur J Biochem. 1970 Sep;16(1):96–102. doi: 10.1111/j.1432-1033.1970.tb01058.x. [DOI] [PubMed] [Google Scholar]
- Yang N. S., Scandalios J. G. Purification and biochemical properties of genetically defined malate dehydrogenase in maize. Arch Biochem Biophys. 1974 Apr 2;161(2):335–353. doi: 10.1016/0003-9861(74)90314-2. [DOI] [PubMed] [Google Scholar]


