Abstract
Cyclooxygenase‐2 (COX‐2) is highly expressed in tumor cells and has been regarded as a hallmarker for cancers, but the excise regulatory mechanism of COX‐2 in tumorigenesis remains largely unknown. Here, we pulled down and identified a novel COX‐2 regulator, heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2/B1), which could specifically bind to COX‐2 core promoter and regulate tumor growth in non‐small‐cell lung cancers (NSCLCs). Knockdown of hnRNPA2/B1 by shRNA or siRNA downregulated COX‐2 expression and prostaglandin E2 (PGE2) production, and suppressed tumor cell growth in NSCLC cells in vitro and in vivo. Conversely, overexpression of hnRNPA2/B1 up‐regulated the levels of COX‐2 and PGE2 and promoted tumor cell growth. We also showed that hnRNPA2/B1 expression was positively correlated with COX‐2 expression in NSCLC cell lines and tumor tissues, and the up‐regulated expression of hnRNPA2/B1 and COX‐2 predicted worse prognosis in NSCLC patients. Furthermore, we demonstrated that the activation of COX‐2 expression by hnRNPA2/B1 was mediated through the cooperation with p300, a transcriptional co‐activator, in NSCLC cells. The hnRNPA2/B1 could interact with p300 directly and be acetylated by p300. Exogenous overexpression of p300, but not its histone acetyltransferase (HAT) domain deletion mutation, augmented the acetylation of hnRNPA2/B1 and enhanced its binding on COX‐2 promoter, thereby promoted COX‐2 expression and lung cancer cell growth. Collectively, our results demonstrate that hnRNPA2/B1 promotes tumor cell growth by activating COX‐2 signaling in NSCLC cells and imply that the hnRNPA2/B1/COX‐2 pathway may be a potential therapeutic target for human lung cancers.
Keywords: hnRNPA2/B1, COX‐2, p300, Lung cancer
Highlights
hnRNPA2/B1 is a novel COX‐2 regulator which can specifically bind to COX‐2 promoter in NSCLC cells.
hnRNPA2/B1 activates COX‐2 expression, upregulates PGE2 production, and promotes cell growth in NSCLC cells.
hnRNPA2/B1 expression is positively correlated with COX‐2 expression in NSCLC and predicts poor prognosis in lung cancer patients.
P300 interacts with and acetylates hnRNPA2/B1 protein, thereby promoting COX‐2 expression and cell growth in NSCLC cells.
1. Introduction
Non‐small‐cell lung cancer (NSCLC) patients have bad prognostic and short term survival (Belani et al., 2012; Chen et al., 2015). Besides smoking, occupational and environment factors, chronic inflammation has been extensively proved to be a common feature in NSCLCs (Hashim and Boffetta, 2014; Lee and Hashibe, 2014; Takiguchi et al., 2014; Florou et al., 2014). Recent evidence suggests that in inflammatory response heterogeneous nuclear ribonucleoproteins (hnRNPs) have ability to modulate the expression of inflammatory mediators (Tauler and Mulshine, 2009). Overexpression of hnRNPs, such as hnRNPA2/B1, can affect mRNA stability to regulate post‐transcription in lung cancer (Percipalle et al., 2009; Han et al., 2010). hnRNPs are consist of RNA and protein which present in the cell nucleus.
The inflammatory microenvironment can promote tumor formation and stimulate tumor progression. In lung cancers, bacterial infection and neutrophilia can contribute to a poor prognosis (Razmi et al., 2013; Okada, 2014). A large amount of evidences support the role of cyclooxygenase‐2 (COX‐2) in inflammation and oncogenesis. High expression of COX‐2 is significantly associated with cell apoptosis, tumor occurrence, development and invasion (Aziz et al., 2014; Norouzi et al., 2015). COX‐2 inhibition has been shown to suppress tumor growth and lymph node metastasis (Zhao et al., 2010; Masferrer et al., 2000), and, of importance, is an effective strategy for cancer treatment. Prostaglandin E2 (PGE2), a COX‐2 product, can enhance angiogenesis and lymphangiogenesis during chronic inflammation and tumor progression (Qiu et al., 2014, 2014). Therefore, the crucial role of COX‐2 in tumor progression highlights the importance of discovering and identifying novel regulators of COX‐2. In this study, we combined streptavidin‐agarose pulldown assay and mass spectrum identification criteria to pull down and discover several new COX‐2 expression regulators in NSCLC cells, and identified hnRNPA2/B1 (heterogeneous nuclear ribonucleoprotein A2/B1) as a specific COX‐2 promoter binding protein. However, the precise mechanism of hnRNPA2/B1 involved in the regulation of COX‐2 expression and lung cancer growth remains unknown.
hnRNPA2/B1 has been supposed to be overexpressed in a variety of cancers, including breast, pancreas, liver, and prostate cancer (Tauler et al., 2010; Torosyan et al., 2010; Katsimpoula et al., 2009; Turck et al., 2004). hnRNPA2/B1 is a protein which ubiquitously participates in RNA‐binding and pre‐RNA processing, and is involved in the regulation of cancer cell metabolism, migration, invasion, proliferation, survival, and apoptosis (Clower et al., 2010, 2005, 2009, 2009, 2003). It also plays an important role in epithelial–mesenchymal transition (Tauler et al., 2010). hnRNPA2/B1 is not the unique characteristic for lung cancer. It is also expressed in benign and malignant lung diseases, such as sarcoidosis, pneumonia, tuberculosis, mineral dust disease and smoker lung (Ma et al., 2009). However, whether hnRNPA2/B1 regulates lung cancer growth by modulating inflammatory mediator COX‐2 expression in lung cancer cells is still unclear.
In this study, we pulled down and identified hnRNPA2/B1 as a novel transcriptional regulator of COX‐2, and further investigated how hnRNPA2/B1 modulated COX‐2 expression thereby promoting tumor growth in NSCLC cells. We found that hnRNPA2/B1 bound to the promoter region of COX‐2 gene, and regulated its transcription and expression. We also assessed the clinical significance of the hnRNPA2/B1/COX‐2 signaling in NSCLC patients. The results from our study may provide new insights into understanding the regulatory mechanism of COX‐2 and exploring new therapeutic targets for lung cancer treatment.
2. Materials and methods
2.1. Cell lines and cell culture
H1299, H322 (NCI‐H322), HLF (HLF‐a) and HBE cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum. A549 and H460 (NCI‐H460) cells were cultured in RPMI1640 medium supplemented with 5% fetal bovine serum. The cells were grown at 37 °C in an atmosphere of 5% CO2. All the cell lines were obtained from the American Type Culture Collection (ATCC, Manassas,VA).
2.2. Mass spectrometry
Cell nuclear extracts were incubated with the biotin‐labeled COX‐2 promoter probe and streptavidin‐agarose beads. The purified COX‐2 promoter binding proteins were electrophoresised on SDS‐PAGE and silver stained. The protein bands of interests were excised from the gel and conducted liquid chromatography‐mass spectrometry (LC‐MS/MS) and proteomic analysis. The proteins which had p < 0.05 and PSMs scores ≥ 2 were considered as promising hits.
2.3. Transfection
The cells (2 × 105/ml) seeded in 6‐well plates overnight were mixed gently 2.5 ug of siRNA and 5 ul of Lipofectamine 3000 (Invitrogen) in 250 ul opti‐MEM (Gibco) and incubated at 37 °C for 48 h.
2.4. Plasmids
The plasmid pPR244‐hnRNPA2 (HM639989) and pBS‐hnRNPA2 (HM639989) were obtained from Addgene. H1299 cells were transfected with pBS‐hnRNPA2 to overexpress hnRNPA2/B1, and H460 cells were transfected with pPR244‐hnRNPA2 to inhibit hnRNPA2/B1.
2.5. Lysate preparation from tumor tissues
Lung cancerous tumors and adjacent tissues were obtained from 20 patients who underwent surgery therapy at Dalian Medical University‐First Affiliated Hospital between 2013 and 2014. The surgery and the study had been approved by Dalian Medical University‐First Affiliated Hospital medical ethics committee, the informed consent was obtained from patients in accordance with the Declaration of Helsinki and with institutional guidelines. The tissues (100 mg) were washed with PBS to remove blood, then transferred to liquid nitrogen immediately and homogenized thoroughly with RIPA buffer with protease inhibitor. After incubation on ice 30 min, the tissues were sonicated for 2–5 min at power of about 180 W. The lysates were centrifuged at 12,000 × g for 20 min at 4 °C, and the supernatants were transferred to new tubes.
2.6. Plasmid vectors and expression constructs
pcDNA3.1 (invitrogen, V790‐20) plasmid contains T7 promoters for sequencing and in vitro RNA production. To produce antisense COX‐2 5′UTR and 3′UTR RNA, a 189 bp antisense COX‐2 5′UTR full length sequence was cloned into pcDNA3.1 expression vector using Nhe I and Kpn I sites. The 2562 bp antisense COX‐2 3′UTR full length sequence was cloned into pcDNA3.1 expression vector using Kpn I and Apa I sites. The primer of COX‐2 5′UTR upstream were upstream 5′CTAgctagcGACCAATTGTCATACGACTTGCAGT3′, downstream 5′CTAgctagcGACCAATTGTCATACGACTTGCAGT3′. The primer of COX‐2 3′UTR upstream 5′GGggtaccAAGTCTAATGATCATATTTATTTATT3′, downstream5′GGggtaccAAGTCTAATGATCATATTTATTTATT3′.
2.7. In vitro RNA production and labeling
First, the constructed plasmid COX‐2 5′UTR and 3′UTR DNA were digested with enzymes sufficiently. 1 ug of linear DNA were prepared for in vitro transcription using Riboprobe in vitro Transcription Systems Kit (Promega, #P1460). 2.4 ul (100 uM) Biotin‐CTP were added in the reaction. The reaction was carried out at 37 °C for 2 h after which DNA was digested by addition of 2 units DNAse I at 37 °C for 15 min. Next, the RNA probes were purified by RNeasy Mini Kit (QIAGEN).
2.8. RNA pulldown
Briefly, 50 pmol of the target RNA probe were placed in a 400 ul mixture containing 400 ug nuclear extracts, RNase inhibitor (0.1 U/ul, 3 ul), and 50 ul Streptavidin‐Agarose beads (Sigma) incubated for 3 h at 4 °C on a rotator. RNA beads were then washed with 2 M NaCl and equilibrated in washing buffer (5 mM HEPES pH 7.9, 1 mM MgCl2, 0.8 mM magnesium acetate). The beads were subsequently pelleted by centrifugation at 3000 rpm for 3 min and washed 3 times with 1 ml of washing buffer. Heat the eluted samples for 10 min at 100 °C. Electrophorese samples on a SDS‐PAGE gel and analyzed by western blot. A fragment of non‐specific RNA probe was used as negative control followed by the same treatment.
2.9. In vitro protein IP with subsequent RNA detection
For in vitro protein RNA complex immunoprecipitation, 250 ng RNA, 400 ug nuclear extracts, and 30 μl Protein A/G agarose beads (Invitrogen) were saturated in 500 μl buffer IPB containing 40 mM Tris–HCl (pH 8.0), 150 mM sodium chloride, 0.5 mM Magnesium acetate, 1 mM DTT, 5% Glycerol, supplemented with 0.5% BSA, 0.1 U/ul RNase inhibitor and protease inhibitors (Sigma) and mixed with 3 μg anti‐hnRNPA2/B1 antibody (Santa Cruze) for 3 h with continuous rotation at 4 °C. As a control, pre‐blocked Protein A/G agarose beads lacking hnRNPA2/B1 antibody, but adding non‐specific IgG, was added to the same amount of protein‐RNA complex and processed identically to the sample tube. Three separate washes for 5 min duration each were implemented using IPB buffer. The RNA from the residual mixture containing protein‐RNA complex was extracted with phenolchloroform, and then was precipitated by ethanol for resuspension in 10 μl water in order to use for cDNA synthesis with EasyScrip one‐Step gDNA Removal and cDNA Synthesis kit (Transgen). After completion of the reverse transcription, DNA samples were subjected to PCR. The primer of COX‐2 3′UTR upstream were 5′TTGCGGAGAAAGGAGTCATAC3′, downstream 5′TTTACAGGTGATTCTACCCTA3′ (457 bp).
2.10. Animal study
Female nude mice (4–5weeks old) were obtained from Dalian Medical University and were maintained in SPF laboratory Animal Central. All animal maintenance and procedures were carried in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals, and passed through the training progress and approval by Animal Care and Ethics Committee of Dalian Medical University. Each nude mouse was injected with 2 × 105 human H460 cells which were suspended in 100 ul PBS, subcutaneously‐growing near the axillary fossa. Mice were randomly divided into 4 groups (6 mice per group): (a) Control siRNA; (b) LPS (lipopolysaccharides); (c) hnRNPA2/B1 siRNA; (d) hnRNPA2/B1 siRNA and LPS. The complex of cholesterol‐conjugated siRNA (10ug) which suspended in 100 ul saline were injected twice a week for 3weeks. The dose of LPS for each nude mouse is 10 ug/kg. LPS was injected twice a week for 2 weeks. Tumors volume and body weights were measured twice a week. Tumor volume was calculated as V = 1/2 (width2 × length). Mice were humanely sacrificed by euthanasia after treatment. The hnRNPA2/B1 siRNA and control siRNA were obtained from GenePharma (Shanghai, China). The sequence of the hnRNPA2/B1 siRNA1 was 5′‐GAAAUACCAUACCAUCAAUtt‐ 3′ (sense strand) and 5′‐AUUGAUGGUAUGGUAUUUCtt‐3′ (antisense strand). The sequence of the hnRNPA2/B1 siRNA2 was 5′‐ACAACTATGGAGGAGGAAAtt‐3′ (sense strand) and 5′‐TTTCCTCCTCCATAGTTGTCAtt‐3′ (antisense strand). Because siRNA2 had no effect on hnRNPA2/B1, we chose siRNA1 for all the experiments. The sequence of the negative control siRNA was 5′‐UUCUCCGAACGUGUCACGUtt‐3′ (sense strand) and 5′‐ACGUGACACGUUCGGAGAAtt‐3′ (antisense strand).
2.11. Immunohistochemistry
Tumors were dissected and fixed in 10% formalin overnight. Small tissues were embedded in paraffin for sectioning and incised to 4 um thick. Antigens were retrieved in hot citrate buffer (PH6.0) for 20 min. Immunohistochemistry was performed following the DAB (3, 3′‐diaminobenzidine) Kit (Origene, China). The primary antibodies hnRNPA2/B1 (Santa Cruz Tech., dilution 1:200) and COX‐2 (Abcam, dilution 1:100) were used. Sections were stained with hematoxylin to recognize nuclear.
2.12. Western blot
Protein lysates (30 ug) were separated on a 4%–12% SDS‐PAGE, transferred to PVDF membranes and immunoblotted with antibodies against hnRNPA2/B1, p300 (Santa Cruz Biotechnology, Santa Cruz,CA), β‐actin, TFIIB, COX‐2 (Cell Signaling Technology, Beverly, MA). The protein bands were detected by enhanced chemiluminescence. Densitometric analyses were performed with Image software.
2.13. RT‐PCR
Total RNA was isolated using Tri–Zol reagent according to the manufacture's instruction. RNA (A260/A280 value of 1.8–2.1) concentration was measured using the NanoDrop spectrophotometer. cDNA synthesis was performed using PrimeScriptTM RT‐PCR Kit (TaKaRa) according to the protocol described. The sequence of hnRNPA2/B1 primer was 5′‐GTTGAGCCAAAACGTGCTGT‐3′ (sense strand),5′‐ATCCCCAAATCCACGTCCAC‐3′ (antisense strand). COX‐2 (sense:5′‐TCACAGGCTTCCATTGACCAG‐3′,antisense:5′‐CCGAGGCTTTTCTACCAGA‐3′).
2.14. PGE2 assay
The cells were transfected with plasmids. After 36 h, cell culture media were collected at the same time. The amounts of PGE2 in the media were determined by Human Prostagland in E2 (PGE2) ELISA Kit (Shenzhen Kerunda Biotech).
2.15. Analysis of promoter activity
The promoter of human COX‐2 gene (−891 to +9) was fractured into 6 fragments in different length. Each fragment and a non‐specific vector were constructed into a luciferase reporter vector pGL3. Luciferase reporter assays were performed using the kit (Promega, Madison, WI).
2.16. Cell viability assay
H1299 and H460 cells were seeded into 96 well plates (2000 cells per well). H1299 cells were used to transfect the overexpression plasmid pBS‐hnRNPA2. pPR244‐hnRNPA2 was used to knock down hnRNPA2 in H460 cells. At 48 h after transfection, cell viability was measured by MTT assay.
2.17. Chromatin immunoprecipitation (ChIP)
Cells were fixed with 1% formaldehyde for 10 min and the end cross link with 0.125M glycine 10 min. Cells were washed with 1 × PBS twice, scraped and collected, resuspended in SDS lysis buffer (1% SDS, 10 mM Tris–HCL, pH 8.0), then sonicated on ice with 150 W 5 repeats of 15 s. The 1/4 volume of lysates were used as the input control. The rest of lysates were incubated with non‐immune rabbit IgG or specific antibody at 4 °C overnight. The complex of antibody‐chromatin were immunoprecipitated by protein A/G plus agarose beads. The protein/DNA complex were washed with elution buffer (20%SDS, 1M NaHCO3), then revers‐crosslinking at 65 °C and phenol/chloroform extracted DNA. The primer of PCR amplification was COX‐2 promoter (5′‐−488ATGTCAGCCTTTCTTAACCT−468‐3′, antisense:5′‐−10CCGATAGAACCTTCCTTTTT−30‐3′), (5′‐−362CATCCAAGGCGATCAGTCCA−342‐3′,antisense:5′‐−196TCGGAAACCCAGGAAGCTGC−216‐3′) (5′‐−196GCAGCTTCCTGGGTTTCCGA−176‐3′,antisense:5′‐−30AAGACTGAAAACCAAGCCCA−50‐3′)
2.18. Confocal immunofluorescence
H1299 cells were incubated on chamber slides in 6‐well plates with or without p300 transfection. The samples were fixed with 4% polyoxymethylene 30 min at room temperature and permeabilized with PBST (PBS with 0.2%Triton X‐100), then blocked with bovine serum albumin (BSA) 30 min and incubated with hnRNPA2/B1 and p300 antibodies (1:200 dilution) overnight at 4 °C. After washing 3 times, cells were incubated with the fluorescein isothiocyanate‐ and rhodamine‐conjugated secondary antibodies for 1 h. The nuclei were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI). The results were visualized by Leica DM 14000B cofocal laser scanning microscope.
2.19. Immunoprecipitation (IP)
300 ug nuclear protein was used for each IP. The proteins were pulled down with Protein A/G sepharose and detected by Western blot analysis with specific antibodies.
2.20. Migration assay
H460 cells (3 × 105 cells per well) were seeded in 6‐well plates. Five groups were divided: (a) control, (b) Celecoxib (CB) (Sigma Aldrich, St. Louis, USA, 15 uM), (c) LacZ, (d) shRNA of pPR244‐hnRNPA2 plasmid, (e) shRNA of pPR244‐hnRNPA2 plasmid and Celecoxib. After transfection for 12 h, cell layers were scratched by sterile tips and photographed at 0, 24, and 48 h.
2.21. Human tissue micro array
The human tissue micro arrays were purchased from Outdo Biotech Company (Shanghai, China). Total 150 cases from 75 lung cancer patents were arranged into two tissue array blocks. The antibodies against hnRNPA2/B1 (Santa Cruz Tech., dilution 1:200) and COX‐2 (Abcam, dilution 1:100) were used.
2.22. DNA‐protein binding by pulldown assay
hnRNPA2/B1 binding to COX‐2 a core promoter probes were verified by a streptavidin‐agarose pulldown assay as previously described (Deng et al., 2007). COX‐2 promoter probe is the biotin‐labeled double‐stranded oligonucleotide probe. Biotin labeled in the 5'end. The 3 fragments of COX‐2 promoter sequence were synthesized by Invitrogen Company. The sequence of non‐specific probe (NSP) is 5′‐AGAGTGGTCACTACCCCCTCTG‐3′. The COX‐2 promoter probes were synthesized by PCR. The biotin‐labeled primer sequences were: sense:5′‐−891GGCCATCGCCGCTTCCTTTG−871‐3′, antisense:5′‐−30AAGACTGAAAACCAAGCCCA−50‐3′; sense: 5′‐−459GGCGGCGGGAGAGGGGATTC−439‐3′, antisense: 5′‐−30AAGACTGAAAACCAAGCCCA−50‐3′; sense: 5′‐−362CATCCAAGGCGATCAGTCCA−342‐3′, antisense: 5′‐−30AAGACTGAAAACCAAGCCCA−50‐3′. All the probes were biotin‐labeled.
2.23. Statistical analysis
The SPSS 19.0 was used to analyze statistics. The Student's t‐test, Fisher's exact test Kaplan–Meier and Spearman's correlation test were used as deemed appropriate. Statistical significance was assumed when p < 0.05.
3. Results
3.1. hnRNPA2/B1 was identified as a COX‐2 promoter‐binding protein
COX‐2 promoter region has many functional binding sites. To discover the novel and specific COX‐2 promoter regulators in NSCLC cell lines, we incubated the cell nuclear extracts with streptavidin‐agarose beads and the biotin‐labeled DNA probe corresponding to the COX‐2 promoter (−891 to −30) (Figure 1A). The pulled down and purified protein factors bound to the COX‐2 promoter probe were separated by SDS/PAGE electrophoresis and then visualized by silver staining (Figure 1B). Compared to the differential binding capabilities on COX‐2 promoter between human normal cell lines (HLF), immortalized HBE cells and NSCLC cell lines (A549, H1299), we selected the protein bands of interests for further analysis using mass spectrum and proteomic techniques. One protein band with molecular weight about 36 kDa was identified as hnRNPA2/B1 (Figure 1B).
Figure 1.
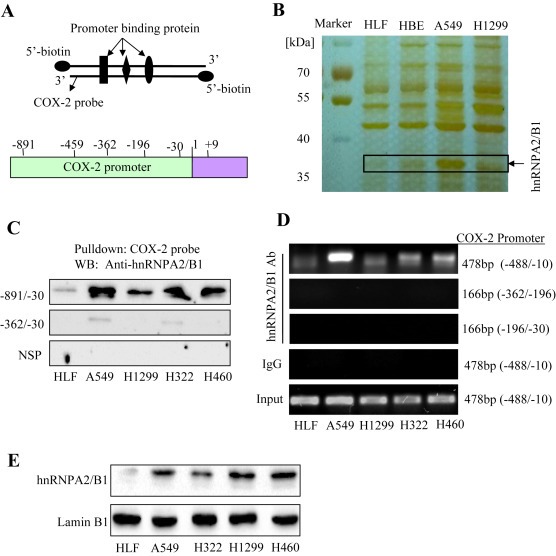
Binding of hnRNPA2/B1 to the COX‐2 promoter. (A) The COX‐2 promoter probe and structure. (B) SDS‐PAGE and silver staining of COX‐2 promoter binding proteins. (C) Binding of hnRNPA2/B1 to the biotinylated COX‐2 promoter probe. The anti‐hnRNPA2/B1 antibody was used to validate hnRNPA2/B1 protein in COX‐2 probe‐streptavidin bead complex by Western blot. NSP, a non‐specific DNA probe. (D) Binding of hnRNPA2/B1 to the COX‐2 promoter in chromatin structure by ChIP assay. The complex of hnRNPA2/B1 antibody‐chromatin were immunoprecipitated, and the PCR products were amplified using the different primers of COX‐2 promoter. IgG, a negative control for ChIP assay. (E) Detection of hnRNPA2/B1 expression in cell nuclear extracts. LaminB1, a loading control for nuclear proteins.
To further validate the binding of hnRNPA2/B1 to the COX‐2 promoter, we used western blot assay to detect the proteins which were pulled down in the streptavidin‐biotinylated COX‐2 promoter probe complex in various NSCLC cell lines. As shown in Figure 1C, all four NSCLC cell lines (A549, H1299, H322 and H460) showed high binding of hnRNPA2/B1 on the COX‐2 promoter probe corresponding to the sequence −891 to −30, whereas a very light binding of hnRNPA2/B1 was detected on the COX‐2 promoter probe corresponding to the sequence −360 to −30. By contrast, hnRNPA2/B1 did not bind to the irrelevant non‐specific DNA sequence (NSP) (Figure 1C). To further verify the binding of hnRNPA2/B1 on COX‐2 promoter region in the chromatin structure in living cells, ChIP assay was performed. The results showed that the DNA immunoprecipitated from the NSCLC cell lines (A549, H1299, H322 and H460) by antibody against hnRNPA2/B1 were amplified by PCR using the primers corresponding to the COX‐2 promoter region (−488 to −10). However, the DNA was not amplified by the primers corresponding to the COX‐2 promoter regions (−362 to −196) and (−362 to −30), indicating that hnRNPA2/B1 did not bind to these two COX‐2 promoter regions (Figure 1D). We also did not detect the amplification of COX‐2 promoter using the negative IgG control for the ChIP assay (Figure 1D). These results demonstrated the binding ability of hnRNPA2/B1 on COX‐2 promoter in lung cancer cells.
We also performed RNA‐Pulldown experiments to test the binding between hnRNPA2/B1 and COX‐2 5′‐ and 3′‐UTR. A very light binding of hnRNPA2/B1 was detected on the COX‐2 3′‐UTR probe and nearly no binding was seen on the COX‐2 5′‐UTR (Supplementary Figure 3A). Similarly, in vitro protein IP assay indicated that COX‐2 3′‐UTR could be detected in the immunoprecipitated complexes formed by hnRNPA2/B1 and its antibody, but 5′‐UTR was hardly detected (Supplementary Figure 3B). These data demonstrated the binding of hnRNPA2/B1 at COX‐2 3′‐UTR, though such binding is not so strong compared to its binding at COX‐2 promoter sequence.
To see whether the differential binding of hnRNPA2/B1 to COX‐2 promoter is due to the differential expression between lung normal and cancer cells, we next detected the hnRNPA2/B1 proteins in the nuclear extracts by Western blot. As shown in Figure 1E, the comparatively high levels of hnRNPA2/B1 proteins in nuclear extracts were detected in the four NSCLC cell lines compared to the normal cell line HLF.
3.2. hnRNPA2/B1 upregulated COX‐2 expression and PGE2 production
To see whether hnRNPA2/B1 could mediate the regulation of COX‐2 in NSCLC cells, we determined the effect of hnRNPA2/B1 on the protein expression and promoter activity of COX‐2 in 3 NSCLC cell lines. Knockdown of hnRNPA2/B1 by transfecting H460 and H322 cells with hnRNPA2/B1 shRNA (pPR244‐hnRNP plasmid) or hnRNPA2/B1 siRNA1 effectively inhibited COX‐2 expression at protein and mRNA levels (Figure 2A) in H460 and H322 cells. We found that the expression levels of COX‐2 was comparatively low in H1299 cell line. Therefore, we used H1299 cell line for the overexpression experiment. Conversely, overexpression of hnRNPA2/B1 by transfecting H1299, H460 and H322 cells with hnRNPA2/B1 expressing vector (hnRNP) dramatically up‐regulated COX‐2 protein expression (Figure 2D).
Figure 2.
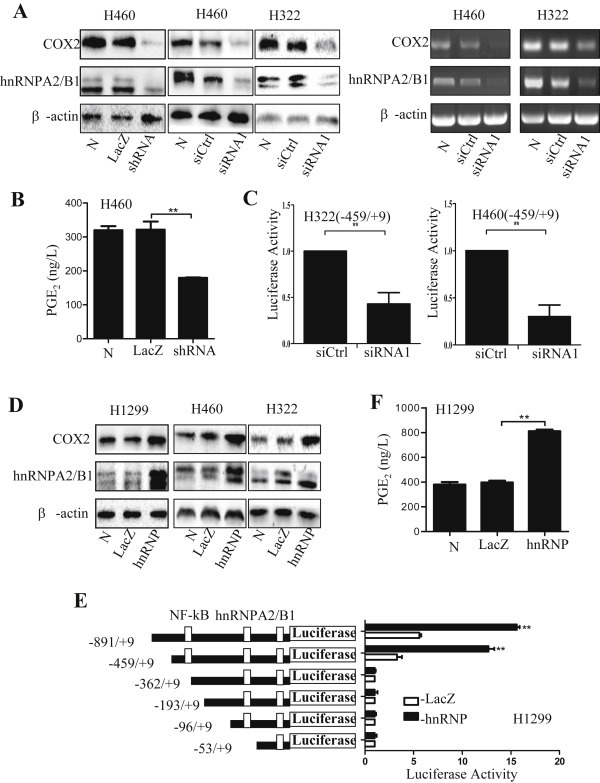
Regulation of COX‐2 and PGE2 by hnRNPA2/B1. (A–C) H460 and H322 cells were transfected with hnRNPA2/B1 siRNA or shRNA (pPR244‐hnRNPA2 plasmids). At 48 h after transfection, the expression of COX‐2 and hnRNPA2/B1 proteins and mRNA was detected by Western blot and RT‐PCR (A), the PGE2 in cell culture media (B) and COX‐2 promoter activity (C) were also determined. (D–F) The H1299, H322 and H460 cells were transfected with pBS‐hnRNPA2 plasmid (hnRNP) to overexpress hnRNPA2/B1. H1299 cells were induced by pyromellitic acid (PMA, 100 nM) for 8 h. At 48 h after transfection, the expression of COX‐2 and hnRNPA2/B1 proteins (D) was detected by Western blot, and the PGE2 in cell culture media (E) as well as the activities of COX‐2 promoter for 6 different fragments (F) were determined. Ctrl, the control shRNA; siCtrl, the control siRNA; N, no treatment. The data were normalized and represent the mean ± SD of three independent experiments. **p < 0.05 represents significant difference.
PGE2 is a downstream product of COX‐2 (Obermajer et al., 2011). The COX‐2/PGE2 signaling is associated with the initiation and promotion of lung tumorigenesis and tumor metastasis (Castellone et al., 2005; Solomon et al., 2005). We next quantitatively analyzed the regulation of hnRNPA2/B1 on PGE2 production in cell culture media. Consistent with the effect of hnRNPA2/B1 on COX‐2 expression, knockdown of hnRNPA2/B1 by shRNA significantly inhibited PGE2 production in H460 cells (Figure 2B). Similarly, overexpression of hnRNPA2/B1 increased the level of PGE2 in H1299 cells (Figure 2E). These results indicate that hnRNPA2/B1 positively up‐regulates the COX‐2/PGE2 signaling in NSCLC cells.
3.3. hnRNPA2/B1 promoted transcriptional activation of COX‐2 promoter
Since hnRNPA2/B1 specifically bound to COX‐2 promoter in lung cancer cells, it might play a crucial role in the regulation of COX‐2 transcription. We next tested the effect of hnRNPA2/B1 on COX‐2 promoter activity in various NSCLC cell lines. Knockdown of hnRNPA2/B1 by siRNA significantly inhibited COX‐2 promoter activity in H460 and H322 cell lines (Figure 2C). We next constructed six luciferase report gene vectors containing full length promoter region of COX‐2 (−891 to +9) and its deletion mutants: COX‐2 (−459 to +9), COX‐2 (−362 to +9), COX‐2 (−193 to +9), COX‐2 (−96 to +9). As shown in Figure 2F, overexpression of hnRNPA2 in H1299 cells almost had no effect on the expression of report gene luciferase driven by the COX‐2 promoter mutants COX‐2 (−362 to +9), COX‐2 (−193 to +9) and COX‐2 (−96 to +9), while significantly up‐regulated luciferase expression driven by the promoters COX‐2 (−459 to +9) and COX‐2 (−891 to +9). These results indicate that hnRNPA2/B1 may bind on the region of COX‐2 promoter region (−459 to −362) and regulate COX‐2 expression.
3.4. hnRNPA2/B1 and COX‐2 were highly expressed in NSCLC cell lines and tumor tissues
We next analyzed the expression of hnRNPA2/B1 and COX‐2 in NSCLC cell lines and tumor tissues. High expression of hnRNPA2/B1 at protein (Figure 3A) and mRNA (Figure 3B) levels were detected in NSCLC cells (A549, H1299, H322, H460) and immortalized cell line (HBE), but not in the lung normal cell line (HLF). The COX‐2 protein (Figure 3A) and mRNA (Figure 3B) were also highly expressed in A549, H460 and HBE cells but not in HLF cells. Consistent with the previous reports, we detected a lower levels of COX‐2 expression in H1299 and H322 cells (Yang et al., 2014). In addition, we also determined the expression of hnRNPA2/B1 and COX‐2 in tumor tissue samples and their adjacent tissues from 20 NSCLC patients by Western blot analysis (Figure 3C) and immunohistochemical staining (Figure 3D). We found that 16 out of 20 tumor samples had high expression of both hnRNPA2/B1 and COX‐2 by comparison with their adjacent tissue samples, indicating that the expression of COX‐2 is positively correlated with hnRNPA2/B1 levels in NSCLC cells.
Figure 3.
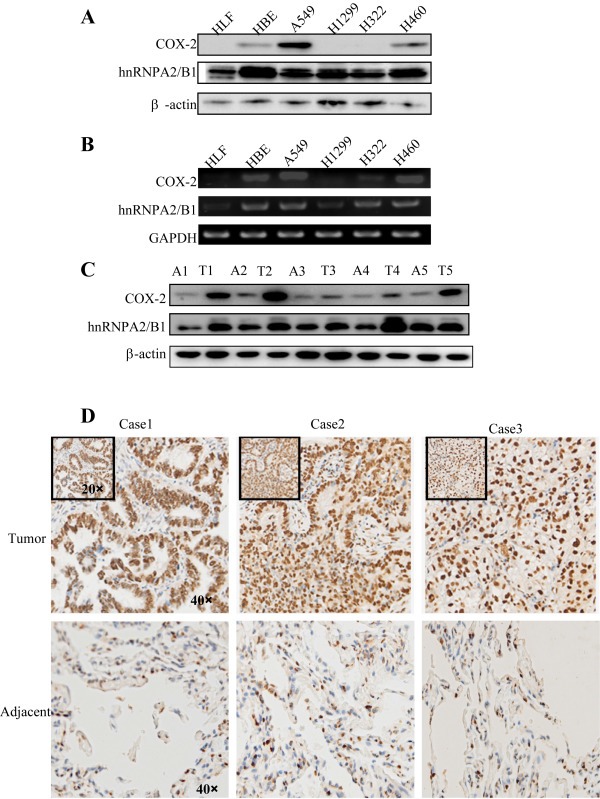
High expression of hnRNPA2/B1 and COX‐2 in NSCLC cell lines and NSCLC tumor tissues. (A) The proteins of hnRNPA2/B1 and COX‐2 in various NSCLC cell lines were detected by Western blot. (B) The levels of hnRNPA2/B1 and COX‐2 mRNA in NSCLC cell lines were determined by RT‐PCR. (C) The proteins of hnRNPA2/B1 and COX‐2 in NSCLC tumor tissues (T) and their adjacent tissues (A) were detected by Western blot. (D) hnRNPA2/B1 protein in NSCLC tumor tissues and their adjacent tissues were detected by immunohistochemical staining.
3.5. Knockdown of hnRNPA2/B1 inhibited NSCLC cell migration and proliferation
We next determined the effect of hnRNPA2/B1 on cell migration and proliferation by scratch and MTT assays respectively. The results showed that transfection of H460 cells with hnRNPA2/B1 shRNA considerably suppressed cell migration (Figure 4A) and cell proliferation and viability (Figure 4B,C). Celecoxib (CB) is known as a COX‐2‐selective inhibitor which has been used to inhibit tumor growth (Yang et al., 2014). The combined treatment with hnRNPA2/B1 shRNA and celecoxib (CB) did not significantly enhance the cell viability inhibition (Figure 4C). By contrast, overexpression of hnRNPA2/B1 in H1299 cells by transfecting cells with its expressing vector (hnRNP) markedly increased cell proliferation, and the addition of celecoxib (CB) reduced the hnRNPA2/B1‐mediated promotion of cell proliferation (Figure 4D).
Figure 4.
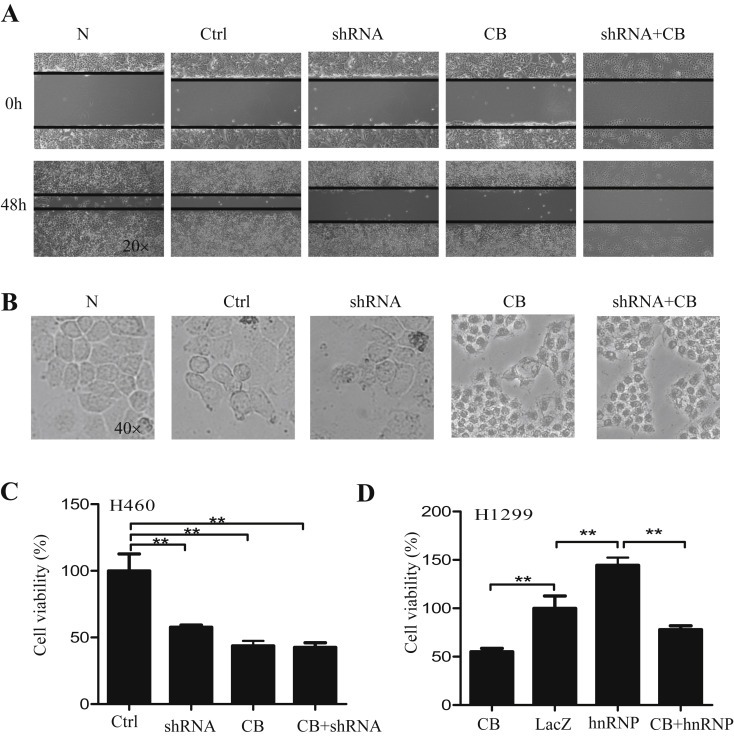
Regulation of cell migration and proliferation by hnRNPA2/B1. (A–C) H460 cells were transfected with hnRNPA2/B1 shRNA (pPR244‐hnRNPA2 plasmid) or control shRNA (Ctrl) and treated with or without celecoxib (CB). After 48 h, cell migration (A), cell growth (B) and cell viability (C) were determined. (D) H1299 cells were transfected with hnRNPA2 overexpressing plasmids and treated with or without celecoxib (CB). After 48 h, cell viability (D) was determined. The LacZ was used a control. The data represent the mean ± SD of three independent experiments. p < 0.05 represents significant difference.
3.6. Knockdown of hnRNPA2/B1 suppressed tumor growth in a xenograft mouse model
We also validated the hnRNPA2/B1‐mediated regulation of tumor growth via COX‐2 signaling in a mouse model. H460 cells were injected into nude mice (flank). Knockdown of hnRNPA2/B1 by siRNA significantly inhibited tumor volume (Figure 5A,B,C,E) and tumor weight (Figure 5D). However, treatment with LPS, a COX‐2 inducer, effectively rescued the inhibitions of tumor growth caused by hnRNPA2/B1 silencing (Figure 5A–E). The results from Western blot also showed that the hnRNPA2/B1 knockdown considerably suppressed COX‐2 protein expression in the mouse groups treated with hnRNPA2/B1 siRNA compared to the control siRNA groups (Figure 5F). Moreover, immunohistochemical staining also confirmed that the expression of hnRNPA2/B1 was positively correlated with COX‐2 expression (Figure 5G). These data further demonstrate that the hnRNPA2/B1‐mediated regulation of tumor cell growth is in part realized through the modulation of COX‐2 signaling in NSCLC cells.
Figure 5.
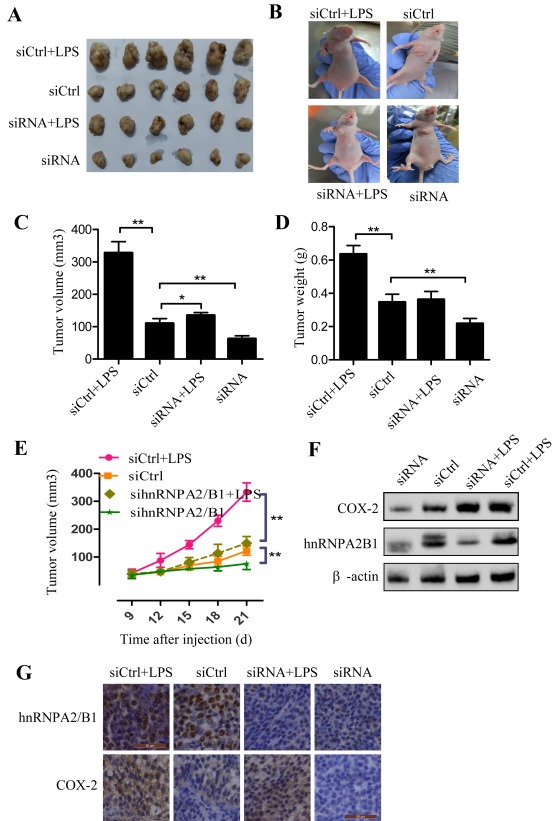
Inhibition of tumor growth and COX‐2 expression by hnRNPA2/B1 siRNA in a xenograft mouse model. Human H460 cell lines were inject into right flank of nude mice to evaluate the effects of different treatments on tumor growth. The four treatment groups were used: (1) Control siRNA (siCtrl); (2) siCtrl + LPS; (3) hnRNPA2/B1 siRNA (si‐hnRNPA2/B1); (4) si‐hnRNPA2/B1 + LPS. Each group included 6 mice. (A) Tumors from mice. (B) Xenografts mice. (C) Tumor volume. (D) Tumor weight. (E) Dynamic development of tumor volume. (F) Western blot analysis of tumor tissues of mice. (G) Immunohistochemical analysis of hnRNPA2/B1 and COX‐2 from tumor xenografts. Hematoxylin was used to stain nuclear. The data were the mean ± SD of 6 mice. p < 0.05 represents significant difference.
3.7. hnRNPA2/B1 expression was positively correlated with COX‐2 expression in NSCLC patients
We next evaluated the expression of hnRNPA2/B1 and COX‐2 in tumor tissues and their adjacent samples in 150 tumor samples from 75 NSCLC patients by tissue microarray and immunohistochemical assay. We found that the expression level of COX‐2 positively correlated with hnRNPA2/B1 expression (p = 0.011) (Figure 6A,B,C), and the overall survival in these patients was significantly poorer than in those with low expression of hnRNPA2/B1 (p = 0.014) (Supplementary Figure 1A). Cumulative hazard function in the patients with high expression of hnRNPA2/B1 was higher than those with low expression of hnRNPA2/B1 (Supplementary Figure 1B). In addition, the worse statues of clinical TNM stage was significantly correlated with the high expression of hnRNPA2/B1 (HR = 0.306, p = 0.003) (Figure 6C).
Figure 6.
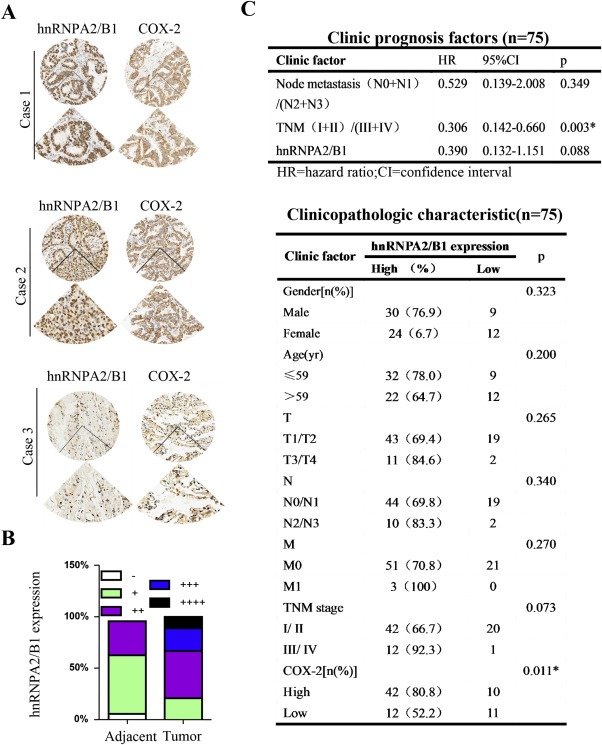
Clinical data analysis for hnRNPA2/B1 and COX‐2 in human tissue microarrays from NSCLC patients. (A) Expression of hnRNPA2/B1 and COX‐2 in tumor tissues from NSCLC patients detected by immunohistochemical staining. (B) Expression of hnRNPA2/B1 in tumor tissues and their adjacent non‐malignant lung tissues from NSCLC patients. (C) Clinical prognosis factors and clinicopathologic characteristics of 75 NSCLC patients.
P300 interacted with and acetylated hnRNPA2/B1 to regulate COX‐2 expression.
P300 is an important co‐activator for COX‐2 transcription and has been shown to acetylate COX‐2 promoter‐bound transactivators (Deng et al., 2003; Subbaramaiah et al., 2001). To determine whether p300 plays a role in the hnRNPA2/B1‐mediated regulation of COX‐2 transcription, we first determined the interaction of p300 with hnRNPA2/B1. As shown in Figure 7A, direct interaction between hnRNPA2/B1 and p300 were observed in A549, H1299 and H460 cell lines by immunoprecipitation experiments. To further confirm the interaction between p300 and hnRNPA2/B1, we also analyzed the subcellular co‐localization of p300 and hnRNPA2/B1 in H460, H322, H1299 cells by immunofluorescence confocal assay, and found that most of hnRNPA2/B1 and p300 proteins colocalized in the cell nuclei of H460 cells (Figure 7B) and H322, H1299 cells (Supplementary Figure 4), which are consistent with the previous study (Mizuno et al., 2012).
Figure 7.
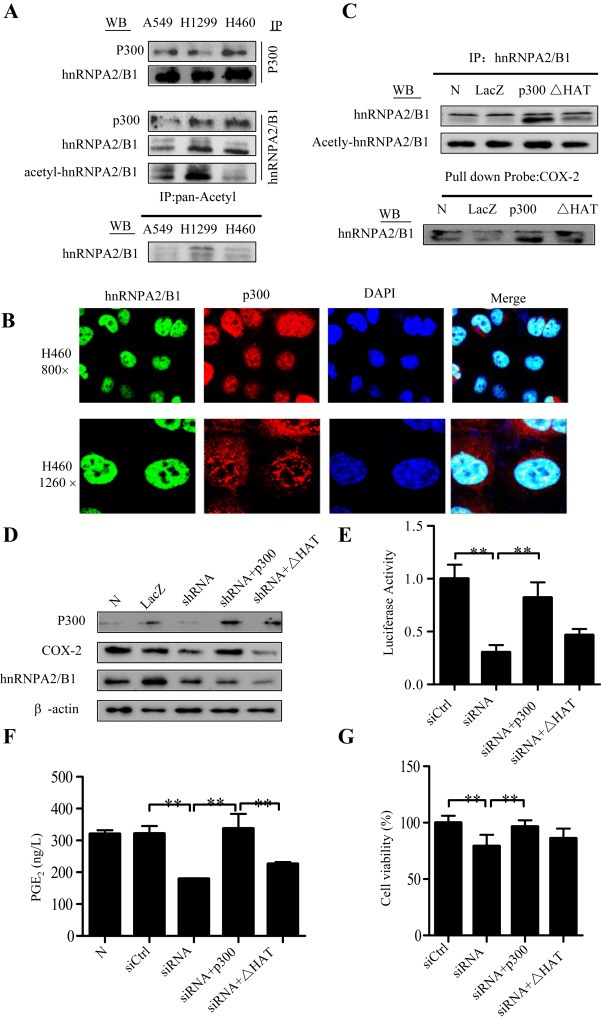
Acetylation of hnRNPA2/B1 by p300. (A) Cell lysates were subjected to immunoprecipitation (IP) assays with anti‐p300, anti‐hnRNPA2/B1, or anti‐pan‐Acetyl antibody. Western blot (WB) assays were performed the antibodies against anti‐hnRNPA2/B1 and anti‐p300. (B) Cellular co‐localization of p300 and hnRNPA2/B1 in H460 cells was detected by confocal immunofluorescence assay. (C) H1299 cells were transfected with the expressing plasmids of p300 or its HAT deletion mutant. LacZ was used for the control. After 48 h, the nuclear extracts were used for IP assay using anti‐hnRNPA2/B1 antibody following by WB assay using anti‐hnRNPA2/B1 or anti‐pan‐Acetyl antibody, and for DNA‐binding protein pulldown assay using biotin‐labeled COX‐2 promoter probe following by Western blot assay using anti‐hnRNPA2/B1. (D–G) H460 cells were transfected with hnRNPA2/B1 shRNA combining with the expressing plasmids of p300 or its HAT deletion mutant. After 48 h, the effects of the shRNA on COX‐2 protein (D), COX‐2 promoter (−459/+9) activity (E), PGE2 production in cell culture media (F), and cell viability (G) were determined, respectively. The data is the mean ± SD of three independent experiments. p < 0.05 represents significant difference.
To see whether hnRNPA2/B1‐induced COX‐2 expression is mediated by its acetylation, we used hnRNPA2/B1 antibody to immunoprecipitate the total hnRNPA2/B1, and used the pan‐acetylated antibody to detected acetylated‐hnRNPA2/B1 (Figure 7A). The results demonstrated the acetylation of hnRNPA2/B1 in lung cancer cells tested.
P300 has HAT domain and can acetylate multiple transactivators, such as NF‐kB, which binds on COX‐2 promoter thereby activating COX‐2 expression. To illustrate whether p300 could also acetylate hnRNPA2/B1 and promote its binding activity, we evaluated the effect of overexpression of p300 and its HAT deletion mutant (ΔHAT) on hnRNPA2/B1 acetylation and its binding on COX‐2 promoter. The results showed that overexpression of p300, but not its HAT deletion mutant (ΔHAT), augmented hnRNPA2/B1 acetylation (Figure 7C) and enhanced its binding on COX‐2 promoter (Figure 7C). We also further validate whether p300 HAT is important for the hnRNPA2/B1‐mediated activation of COX‐2 expression. As expected, overexpression of p300, but not its HAT deletion mutant (ΔHAT), rescued the shRNPA2/B1 shRNA‐mediated inhibitions of COX‐2 protein expression (Figure 7D), promoter activity (Figure 7E), PGE2 production (Figure 7F), as well as cell proliferation (Figure 7G) in H460 cells.
4. Discussion
The data presented here has demonstrated that hnRNPA2/B1 functioned as a COX‐2 regulator in human NSCLC cells. The up‐regulated expression of COX‐2 and hnRNPA2/B1 has been documented in various cancer types (Markkula et al., 2014; Rizzo, 2011; Yan‐Sanders et al., 2002; Matsuyama et al., 2000). Our data suggest hnRNPA2/B1 plays a crucial role in the regulation of COX‐2 transactivation. Inhibition of hnRNPA2/B1 resulted in the decrease of COX‐2 expression and PGE2 production. In contrast, overexpression of hnRNPA2/B1 promoted COX‐2 expression and PGE2 production. Knockdown of hnRNPA2/B1 in NSCLC cells also inhibited cell migration and proliferation.
In this study, we chose the cell lines H1299, H460 and H322 as the cell models in the in vitro experiments. We found that the expression levels of COX‐2 in H460 and H322 cell lines were higher, but comparatively low in H1299 cell line. Therefore, we used H1299 cell line for the overexpression experiments and used H460 and H322 cell lines for siRNA knockdown experiments. We also confirmed that transfection with hnRNPA2/B1 siRNA inhibited tumor growth and COX‐2 expression in a mouse model in vivo. More importantly, in human tissue arrays, overexpression of hnRNPA2/B1 indicated a worse prognosis in patients with NSCLCs. Consistent with our results, recent reports also have shown that high expression of hnRNPA2/B1 promoted tumor development, and the patients with high expression of hnRNPA2/B1 can ultimately develop into lung cancer (Katsimpoula et al., 2009; Pino et al., 2003).
In this study, we analyzed the levels of hnRNPA2/B1 in tumors and adjacent normal tissues to demonstrate the higher expression of hnRNPA2/B1 in tumor tissues than in adjacent normal tissues, and found that hnRNPA2/B1 had only lower level in adjacent normal tissues (Supplementay Figure 2A). Although in the previous report overexpression of hnRNPB1 protein was observed in 100% of stage I lung cancer tissues but not in normal bronchial epithelium (Sueoka et al., 2001), the hnRNPA2/B1 protein was indeed observed in adjacent normal tissue in our research. Moreover, another report also showed that hnRNPB1 was widely expressed in a range of lung carcinomas. The expression of hnRNPB1 was seen in benign bronchial epithelial cells and inflammatory cells, and its expression in background bronchial epithelial cells appeared to be higher in malignant than in benign lung disease (Snead et al., 2003). Thus, it is feasible that this biomarker may be of use in the detection of early lung cancer.
We have also performed the statistic analysis for the COX‐2 expression in different tumor stages (Supplement Figure 2B). We observed more percentage of patients had high expression of COX‐2 in T4 stage than any other tumor stage, and found that there was a significant positive correlation between the co‐expression of hnRNPA2/B1 and COX‐2 in lung cancer (p = 0.011). In this study, we used 150 tumor samples from 75 NSCLC samples. We will conduct a larger cohort Meta‐Analysis research about the correlation of hnRNPA2/B1 with COX‐2 in lung cancers according to Evidence‐Based Medicine (EBM) in the future research.
hnRNPA2/B1 was reported to be able to bind in AU‐rich elements to then regulate gene post‐transcriptional expression. Our study also indicated its weak binding at 3′‐UTR of COX‐2 gene, suggesting its possible involvement in COX‐2 gene regulation at post‐transcriptional level. Much deeper studies deserve to be done to confirm its underlying molecular mechanisms about the slicing function of hnRNPA2/B1 in the post‐transcriptional regulation of COX‐2 in our future study.
As a common transcriptional coactivator, p300 has been proved to be involved in the regulation of COX‐2 gene expression. We therefore proposed the possible association of p300 with hnRNPA2/B1 and their synergizing effect in co‐regulating COX‐2 gene expression. We have confirmed our hypothesis that p300 interacted with and acetylated hnRNPA2/B1 to augment the binding of hnRNPA2/B1 to the COX‐2 promoter, thereby activating COX‐2 expression. Moreover, the elevated p300 could rescue the si‐hnRNPA2/B1‐mediated inhibition of COX‐2 expression and cell proliferation, indicating the vital role of p300 in hnRNPA2/B1‐mediated regulation of COX‐2 expression in NSCLC cells.
In summary, we have demonstrated an important mechanism of hnRNPA2/B1 in the regulation of COX‐2 expression and tumor growth in NSCLC cells. High expression of hnRNPA2/B1 contributes to a poor prognosis in human NSCLCs. Our results therefore suggest that the hnRNPA2/B1/COX‐2 pathway is a potential new therapeutic target for the treatment of lung cancers.
Conflict interest
The authors declare no conflict of interest.
Author contributions
XY, JW, LB, JL, CY, ZL, WY, TX, WY and XX carried out the molecular and cellular biology studies. ZT, RT and WG carried out the immunoassays. ML participated in the mass spectrum analysis. SM, WH, QL and YC participated in the design of the study and performed the statistical analysis. WD, XC and WG participated the design of the study and help draft the manuscript. All authors read and approved the final manuscript.
Supporting information
The following are the supplementary data related to this article:
Supplemantary Figure 1 Kaplan‐Meier analysis for overall survival (A) and cumulative hazard function (B) in NSCLCs with expression of hnRNPA2/B1. p < 0.05 represents significant difference.
Supplementary Figure 2 (A). Immunohistochemical staining showed low expression of hnRNPA2/B1 in adjacent normal tissues of NSCLC patients. (B). COX‐2 protein expression in 75 lung cancer patients had no significant difference in different T stages.
Supplementary Figure 3 The binding of hnRNPA2/B1 to COX‐2 3′UTR. (A). RNA pulldown assay identified the binding of hnRNPA2/B to the COX‐2 3′UTR. Non‐specific RNA probe (NC) was used a negative control. (B). Protein IP with subsequent RNA detection assay identified the binding of hnRNPA2/B1 to the COX‐2 3′UTR. Non‐specific IgG was used a negative control (NC).
Supplementary Figure 4 Colocalization of hnRNPA2/B1 with p300 proteins in the nuclei of H322 and H1299 cells. Images, 400×.
Acknowledgment
This work was supported by the funds from the National Natural Science Foundation of China (81173615 XC, 81470337 YC, 81472178 WD, 81372133 XX); the State “973 Program” of China (2014CB542005); the State “863 Program” of China (SS2012AA020403); the Education Department of Liaoning Province in China (the “Program for Pan‐Deng Scholars”); and the Natural Science Foundation of Liaoning Province in China (2014023009).
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.11.010.
Xuan Yang, Wang Jingshu, Ban Liying, Lu Jian-Jun, Yi Canhui, Li Zhenglin, Yu Wendan, Li Mei, Xu Tingting, Yang Wenjing, Tang Zhipeng, Tang Ranran, Xiao Xiangsheng, Meng Songshu, Chen Yiming, Liu Quentin, Huang Wenlin, Guo Wei, Cui Xiaonan, Deng Wuguo, (2016), hnRNPA2/B1 activates cyclooxygenase‐2 and promotes tumor growth in human lung cancers, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.11.010.
Contributor Information
Wei Guo, Email: cxn23@sina.com.
Xiaonan Cui, Email: wei1015@dmu.edu.cn.
Wuguo Deng, Email: dengwg@sysucc.org.cn.
References
- Aziz, F. , Yang, X. , Wang, X. , Yan, Q. , 2015. Anti-LeY antibody enhances therapeutic efficacy of celecoxib against gastric cancer by downregulation of MAPKs/COX-2 signaling pathway: correlation with clinical study. J. Cancer Res. Clin. Oncol. 141, 1221–1235. [DOI] [PubMed] [Google Scholar]
- Belani, C.P. , Goss, G. , Blumenschein, G. , 2012. Recent clinical developments and rationale for combining targeted agents in non-small cell lung cancer (NSCLC). Cancer Treat. Rev. 38, 173–184. [DOI] [PubMed] [Google Scholar]
- Castellone, M.D. , Teramoto, H. , Williams, B.O. , Druey, K.M. , Gutkind, J.S. , 2005. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 310, 1504–1510. [DOI] [PubMed] [Google Scholar]
- Chen, B. , Hu, B. , Li, W. , Xue, J. , 2015. Lancet Oncol. 16, e309–311. [DOI] [PubMed] [Google Scholar]
- Clower, C.V. , Chatterjee, D. , Wang, Z. , Cantley, L.C. , Vander Heiden, M.G. , Krainer, A.R. , 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. U. S. A. 107, 1894–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W.G. , Montero, A.J. , Wu, K.K. , 2007. Interferon-gamma suppresses cyclooxygenase-2 promoter activity by inhibiting C-jun and C/EBPbeta binding. Arterioscler. Thromb. Vasc. Biol. 27, 1752–1759. [DOI] [PubMed] [Google Scholar]
- Deng, W.G. , Zhu, Y. , Wu, K.K. , 2003. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J. Biol. Chem. 278, 4770–4777. [DOI] [PubMed] [Google Scholar]
- Florou, A.N. , Gkiozos, I.C. , Tsagouli, S.K. , Souliotis, K.N. , Syrigos, K.N. , 2014. Clinical significance of smoking cessation in subjects with cancer: a 30-year review. Respir. Care. 59, 1924–1936. [DOI] [PubMed] [Google Scholar]
- Han, S.P. , Tang, Y.H. , Smith, R. , 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430, 379–392. [DOI] [PubMed] [Google Scholar]
- Hashim, D. , Boffetta, P. , 2014. Occupational and environmental exposures and cancers in developing countries. Ann. Glob. Health. 80, 393–411. [DOI] [PubMed] [Google Scholar]
- He, Y. , Brown, M.A. , Rothnagel, J.A. , Saunders, N.A. , Smith, R. , 2005. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J. Cel. Sci. 118, 3173–3183. [DOI] [PubMed] [Google Scholar]
- He, Y. , Rothnagel, J.A. , Epis, M.R. , Leedman, P.J. , Smith, R. , 2009. Downstream targets of heterogeneous nuclear ribonucleoprotein A2 mediate cell proliferation. Mol. Carcinog. 48, 167–179. [DOI] [PubMed] [Google Scholar]
- Katsimpoula, S. , Patrinou-Georgoula, M. , Makrilia, N. , Dimakou, K. , Guialis, A. , Orfanidou, D. , Syrigos, K.N. , 2009. Overexpression of hnRNPA2/B1 in bronchoscopic specimens: a potential early detection marker in lung cancer. Anticancer Res. 29, 1373–1382. [PubMed] [Google Scholar]
- Lee, Y.C.A. , Hashibe, M. , 2014. Tobacco, alcohol, and cancer in low and high income countries. Ann. Glob. Health. 80, 378–383. [DOI] [PubMed] [Google Scholar]
- Li, Q.L. , Liu, L.X. , Zhang, Q.Y. , Liu, S. , Ge, D.X. , You, Z.B. , 2014. Interleukin-17 indirectly promotes M2 macrophage differentiation through stimulation of COX-2/PGE2 pathway in the cancer cells. Cancer Res. Treat. 46, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, K.W. , Au, S.W. , Waye, M.M. , 2009. Over-expression of SUMO-1 induces the up-regulation of heterogeneous nuclear ribonucleoprotein A2/B1 isoform B1 (hnRNPA2/B1 isoform B1) and uracil DNA glycosylase (UDG) in hepG2 cells. Cell Biochem. Funct. 27, 228–237. [DOI] [PubMed] [Google Scholar]
- Markkula, A. , Simonsson, M. , Rosendahl, A.H. , Gaber, A. , Ingvar, C. , Rose, C. , Jernstrom, H. , 2014. Impact of COX2 genotype, ER status and body constitution on risk of early events in different treatment groups of breast cancer patients. Int. J. Cancer J. Int. du Cancer. 135, 1898–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masferrer, J.L. , Leahy, K.M. , Koki, A.T. , Zweifel, B.S. , Settle, S.L. , Woerner, B.M. , Edwards, D.A. , Flickinger, A.G. , Moore, R.J. , Seibert, K. , 2000. Antiangiogenic and antitumor activities of cyclooxygenase-2 inhibitors. Cancer Res. 60, 1306–1311. [PubMed] [Google Scholar]
- Matsuyama, S. , Goto, Y. , Sueoka, N. , Ohkura, Y. , Tanaka, Y. , Nakachi, K. , Sueoka, E. , 2000. Heterogeneous nuclear ribonucleoprotein B1 expressed in esophageal squamous cell carcinomas as a new biomarker for diagnosis. Jpn. J. Cancer Res. Gann. 91, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, H. , Honda, M. , Shirasaki, T. , Yamashita, T. , Yamashita, T. , Mizukoshi, E. , Kaneko, S. , 2012. Heterogeneous nuclear ribonucleoprotein A2/B1 in association with hTERT is a potential biomarker for hepatocellular carcinoma. Liver Int. 32, 1146–1155. [DOI] [PubMed] [Google Scholar]
- Moran-Jones, K. , Grindlay, J. , Jones, M. , Smith, R. , Norman, J.C. , 2009. hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res. 69, 9219–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzi, M. , Norouzi, S. , Amini, M. , Amanzadeh, A. , Irian, S. , Salimi, M. , 2015. Apoptotic effects of two COX-2 inhibitors on breast adenocarcinoma cells through COX-2 independent pathway. J. Cell Biochem. 116, 81–90. [DOI] [PubMed] [Google Scholar]
- Obermajer, N. , Muthuswamy, R. , Odunsi, K. , Edwards, R.P. , Kalinski, P. , 2011. PGE(2)-induced CXCL12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 71, 7463–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, F. , 2014. Inflammation-related carcinogenesis: current findings in epidemiological trends, causes and mechanisms. Yonago Acta Med. 57, 65–72. [PMC free article] [PubMed] [Google Scholar]
- Patry, C. , Bouchard, L. , Labrecque, P. , Gendron, D. , Lemieux, B. , Toutant, J. , Lapointe, E. , Wellinger, R. , Chabot, B. , 2003. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 63, 7679–7688. [PubMed] [Google Scholar]
- Percipalle, P. , Raju, C.S. , Fukuda, N. , 2009. Actin-associated hnRNP proteins as transacting factors in the control of mRNA transport and localization. RNA Biol. 6, 171–174. [DOI] [PubMed] [Google Scholar]
- Pino, I. , Pio, R. , Toledo, G. , Zabalegui, N. , Vicent, S. , Rey, N. , Lozano, M.D. , Torre, W. , Garcia-Foncillas, J. , Montuenga, L.M. , 2003. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 41, 131–143. [DOI] [PubMed] [Google Scholar]
- Qiu, X. , Cheng, J.C. , Chang, H.M. , Leung, P.C. , 2014. COX2 and PGE2 mediate EGF-induced E-cadherin-independent human ovarian cancer cell invasion. Endocr. Relat. Cancer. 21, 533–543. [DOI] [PubMed] [Google Scholar]
- Razmi, A. , Zarghi, A. , Arfaee, S. , Naderi, N. , Faizi, M. , 2013. Evaluation of anti-nociceptive and anti-inflammatory activities of novel chalcone derivatives. Iran J. Pharm. Res. 12, 149–155. [PMC free article] [PubMed] [Google Scholar]
- Rizzo, M.T. , 2011. Cyclooxygenase-2 in oncogenesis. Clin. Chim. Acta Int. J. Clin. Chem. 412, 671–687. [DOI] [PubMed] [Google Scholar]
- Snead, D.R. , Perunovic, B. , Cullen, N. , Needham, M. , Dhillon, D.P. , Satoh, H. , Kamma, H. , 2003. hnRNP B1 expression in benign and malignant lung disease. J. Pathol. 200, 88–94. [DOI] [PubMed] [Google Scholar]
- Solomon, S.D. , McMurray, J.J. , Pfeffer, M.A. , Wittes, J. , Fowler, R. , Finn, P. , Anderson, W.F. , Zauber, A. , Hawk, E. , Bertagnolli, M. , Adenoma Prevention with Celecoxib Study, I.2005. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. New Engl. J. Med. 352, 1071–1080. [DOI] [PubMed] [Google Scholar]
- SubbaPR ramaiah, K. , Lin, D.T. , Hart, J.C. , Dannenberg, A.J. , 2001. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J. Biol. Chem. 276, 12440–12448. [DOI] [PubMed] [Google Scholar]
- Sueoka, E. , Sueoka, N. , Goto, Y. , Matsuyama, S. , Nishimura, H. , Sato, M. , Fujimura, S. , Chiba, H. , Fujiki, H. , 2001. Heterogeneous nuclear ribonucleoprotein B1 as early cancer biomarker for occult cancer of human lungs and bronchial dysplasia. Cancer Res. 61, 1896–1902. [PubMed] [Google Scholar]
- Takiguchi, Y. , Sekine, I. , Iwasawa, S. , Kurimoto, R. , Tatsumi, K. , 2014. Chronic obstructive pulmonary disease as a risk factor for lung cancer. World J. Clin. Oncol. 5, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauler, J. , Mulshine, J.L. , 2009. Lung cancer and inflammation: interaction of chemokines and hnRNPs. Curr. Opin. Pharmacol. 9, 384–388. [DOI] [PubMed] [Google Scholar]
- Tauler, J. , Zudaire, E. , Liu, H.T. , Shih, J. , Mulshine, J.L. , 2010. hnRNPA2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 70, 7137–7147. [DOI] [PubMed] [Google Scholar]
- Torosyan, Y. , Dobi, A. , Glasman, M. , Mezhevaya, K. , Naga, S. , Huang, W. , Paweletz, C. , Leighton, X. , Pollard, H.B. , Srivastava, M. , 2010. Role of multi-hnRNP nuclear complex in regulation of tumor suppressor ANXA7 in prostate cancer cells. Oncogene. 29, 2457–2466. [DOI] [PubMed] [Google Scholar]
- Turck, N. , Richert, S. , Gendry, P. , Stutzmann, J. , Kedinger, M. , Leize, E. , Simon-Assmann, P. , Van Dorsselaer, A. , Launay, J.F. , 2004. Proteomic analysis of nuclear proteins from proliferative and differentiated human colonic intestinal epithelial cells. Proteomics. 4, 93–105. [DOI] [PubMed] [Google Scholar]
- Yan-Sanders, Y. , Hammons, G.J. , Lyn-Cook, B.D. , 2002. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 183, 215–220. [DOI] [PubMed] [Google Scholar]
- Yang, P.Y. , Cartwright, C. , Chan, D.N. , Ding, J.B. , Felix, E. , Pan, Y. , Pang, J.H. , Rhea, P. , Block, K. , Fischer, S.M. , Newman, R.A. , 2014. Anticancer activity of fish oils against human lung cancer is associated with changes in formation of PGE(2) and PGE(3) and alteration of Akt phosphorylation. Mol. Carcinog. 53, 566–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.A. , Hao, Y.B. , Ji, H.G. , Fang, Y.Y. , Guo, Y.H. , Sha, W. , Zhou, Y.F. , Pang, X.W. , Southerland, W.M. , Califano, J.A. , Gu, X.B. , 2010. Combination effects of salvianolic acid B with low-dose celecoxib on inhibition of head and neck squamous cell carcinoma growth in vitro and in vivo. Cancer Prev. Res. 3, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplemantary Figure 1 Kaplan‐Meier analysis for overall survival (A) and cumulative hazard function (B) in NSCLCs with expression of hnRNPA2/B1. p < 0.05 represents significant difference.
Supplementary Figure 2 (A). Immunohistochemical staining showed low expression of hnRNPA2/B1 in adjacent normal tissues of NSCLC patients. (B). COX‐2 protein expression in 75 lung cancer patients had no significant difference in different T stages.
Supplementary Figure 3 The binding of hnRNPA2/B1 to COX‐2 3′UTR. (A). RNA pulldown assay identified the binding of hnRNPA2/B to the COX‐2 3′UTR. Non‐specific RNA probe (NC) was used a negative control. (B). Protein IP with subsequent RNA detection assay identified the binding of hnRNPA2/B1 to the COX‐2 3′UTR. Non‐specific IgG was used a negative control (NC).
Supplementary Figure 4 Colocalization of hnRNPA2/B1 with p300 proteins in the nuclei of H322 and H1299 cells. Images, 400×.


