Abstract
Type II germ cell tumors arise after puberty from a germ cell that was incorrectly programmed during fetal life. Failure of testicular germ cells to properly differentiate can lead to the formation of germ cell neoplasia in situ of the testis; this precursor cell invariably gives rise to germ cell cancer after puberty. The Nodal co‐receptor Cripto is expressed transiently during normal germ cell development and is ectopically expressed in non‐seminomas that arise from germ cell neoplasia in situ, suggesting that its aberrant expression may underlie germ cell dysregulation and hence germ cell cancer. Here we investigated methylation of the Cripto promoter in mouse germ cells and human germ cell cancer and correlated this with the level of CRIPTO protein expression. We found hypomethylation of the CRIPTO promoter in undifferentiated fetal germ cells, embryonal carcinoma and seminomas, but hypermethylation in differentiated fetal germ cells and the differentiated types of non‐seminomas. CRIPTO protein was strongly expressed in germ cell neoplasia in situ along with embryonal carcinoma, yolk sac tumor and seminomas. Further, cleaved CRIPTO was detected in media from seminoma and embryonal carcinoma cell lines, suggesting that cleaved CRIPTO may provide diagnostic indication of germ cell cancer. Accordingly, CRIPTO was detectable in serum from 6/15 patients with embryonal carcinoma, 5/15 patients with seminoma, 4/5 patients with germ cell neoplasia in situ cells only and in 1/15 control patients. These findings suggest that CRIPTO expression may be a useful serological marker for diagnostic and/or prognostic purposes during germ cell cancer management.
Keywords: Cripto, Testicular germ cell cancer, Methylation, Diagnostic
Highlights
Promoter methylation of CRIPTO is differentially regulated between normal germ cell development and in germ cell tumors.
CRIPTO is strongly expressed in the germ cell precursor cell GCNIS, EC, YST and SE.
CRIPTO protein is cleaved from the cell surface and detectable in serum of patients with GCNIS‐only, EC, YST and SE.
Abbreviations
- GCNIS
Germ cell neoplasia In Situ
- CH
choriocarcinoma
- dcp
days post coitum
- ELISA
enzyme-linked immunosorbent assay
- FACS
fluorescence activated cell sorting
- GCC
germ cell cancer
- NS
non-seminoma
- SE
seminoma
- TE
teratoma
- YST
yolk sac tumor
1. Introduction
Testicular germ cell cancers (GCCs), also known as Type II germ cell tumors (Oosterhuis and Looijenga, 2005), account for ∼60% of all malignancies in men aged 20–40 (Adami et al., 1994; van de Geijn et al., 2009). The cell of origin or ‘cancer stem cell’, germ cell neoplasia in situ (GCNIS, according to the newest WHO classification, 2016), previously known as carcinoma in situ (CIS) and intratubular germ cell neoplasia unclassified (IGCNU), is considered to be an embryonic germ cell that has failed to differentiate into a pre‐spermatogonium during development (Skakkebaek et al., 1987). Although GCNIS may be present before birth, it does not transform into GCC until after puberty when tumor pathology is classified into seminoma (SE) and non‐seminoma (NS) (Sonne et al., 2009; van de Geijn et al., 2009). SE is characterized by fetal germ cell‐like expression profile, and NS comprises both highly pluripotent/undifferentiated tumors (embryonal carcinoma; EC) and differentiated tumors: yolk‐sac tumor (YST); choriocarcinoma (CH); teratoma (TE) and combinations of these.
The ‘fetal origins hypothesis’ of GCNIS predicts developmental pathways that control fetal germ cell pluripotency/differentiation contribute to their malignant potential. We recently discovered that the TGFβ signaling molecule Nodal and its obligate receptor Cripto are expressed at a critical point during fetal XY germ cell development in mice and that Nodal/Cripto signaling is active, apparently acting to maintain pluripotency and oppose differentiation (Spiller et al., 2012). We also found that Nodal/Cripto signaling is ectopically activated in NS and we therefore hypothesize that ectopic activation of Nodal signaling, or failure to silence it, contributes to GCC formation (Spiller et al., 2013).
Nodal, a member of the TGFβ family, signals by binding to Activin receptors in the presence of the receptor Cripto (also known as teratocarcinoma derived growth factor 1; Tdgf1). Nodal signaling is absent in normal adult tissues, but is critical for patterning events during embryogenesis (Shen, 2007). Cripto is also essential during embryogenesis, and plays additional roles in stem cell self‐renewal and pluripotency in human embryonic stem cells (Bianco et al., 2010; Wei et al., 2005). Its continuous activation is associated with initiation or progression of cancer in many tissues including skin, pancreas, intestine, breast, bladder and brain (Klauzinska et al., 2014). As a cell‐surface receptor for Nodal, Cripto must remain tethered to the cell membrane via its glycosylphosphatidylinositol (GPI) anchor at its carboxy terminal (Watanabe et al., 2007b). Cleavage of Cripto at the GPI anchor by GPI‐phospholipase D produces a shorter, biologically active form of Cripto that can promote endothelial cell migration, independent of Nodal signaling (Watanabe et al., 2007a). Detection of cleaved Cripto in serum has been identified as a promising diagnostic for breast, colon and brain cancer (Bianco et al., 2006; Pilgaard et al., 2014).
Hypomethylation of oncogenes and hypermethylation of tumor‐suppressor genes are commonly seen in cancer, therefore it is possible that dysregulation of Cripto expression in GCC may reflect aberrant methylation of regulatory sequences. In this study we investigated the methylation status of the Cripto promoter during normal fetal germ cell development in mice and contrasted this to human GCC. We also assessed Cripto protein expression in GCNIS and GCC of different histologies. Lastly we used ELISA to quantitate levels of Cripto protein present in conditioned media from GCC cell lines and serum from patients with GCC.
2. Materials and methods
2.1. Mouse strains
Protocols and use of animals in these experiments were approved by the Animal Ethics Committee of the University of Queensland. Embryos were collected from timed matings of the CD1 strain and Oct4ΔPE:eGFP (OG2) strain (Szabo et al., 2002), with noon of the day on which the mating plug was observed designated 0.5 days post coitum (dpc).
2.2. Gonadal collection, germ cell isolation
Gonads without mesonephros were dissected at 11.5–17.5 dpc from CD1 embryos. At 11.5 dpc embryos were genotyped by PCR for genetic sex (McFarlane et al., 2013); later stages were sexed visually by gonad morphology. For each stage three pools of ≥4 gonads were processed for qRT‐PCR. For germ cell isolation, sexed gonadal tissue was collected from Oct4ΔPE:eGFP matings. Tissue was dissociated and germ cell (GFP+) populations isolated by fluorescence‐activated cell sorting (FACS) using a FACSAria Cell Sorter (BD Biosciences). For each sex and timepoint approximately 100,000 cells were collected and processed for methylation profiling.
2.3. Cell line culture and 5‐azacytidine treatment
TCam‐2 and JKT‐1 were maintained in RPMI (Life Technologies), and NT2 and TERA‐1 cells in DMEM (Life Technologies) with the addition of 10% fetal calf serum (GE Healthcare Life Sciences, HyClone Laboratories), 100 U/ml penicillin and 100 ug/ml streptomycin in a humidified 5% CO2, 37 °C incubator. Treatment with 5‐azacytidine (AG scientific) was used at a final concentration of 10 μM (Wermann et al., 2010).
2.4. Human GCC samples
Use of tissue samples for scientific reasons was approved by an institutional review board (MEC 02.981 and CCR2041) in The Netherlands and by the Human Research Ethics Committee at the University of Queensland. Samples were used according to the “Code for Proper Secondary Use of Human Tissue in The Netherlands” developed by the Dutch Federation of Medical and Scientific Societies (FMWV) (Version 2, update 2011). Tissues were collected as described (Looijenga et al., 2003) and diagnosed according to WHO standards by an experienced pathologist (J.W Oosterhuis, Erasmus MC, Rotterdam, The Netherlands). The testicular parenchyma samples investigated have been described previously (Mosselman et al., 1996) and comprised of tissue micro‐arrays as well as single biopsies. Samples (paraffin blocks, gDNA, RNA and serum) were extracted at Erasmus MC, Rotterdam, The Netherlands and selected to exemplify different histological subtypes.
2.5. Quantitative RT‐PCR
Total RNA was extracted (RNeasy kit; Qiagen) and cDNA generated (High capacity cDNA Reverse Transcription kit; Applied Biosystems) according to manufacturers' instructions. Taqman probes (Applied Biosystems) used were: mouse (Tbp; Mm00446973_m1, Cripto/Tdgf1; Mm00783944_g1) and human (TBP; Hs00427621_m1, CRIPTO/TDGF1; Hs02339499_g1, NODAL; Hs00415443_m1, LEFTY1; Hs00764128_s1, LEFTY2; Hs00745761_s1, SOX2; Hs01053049_s1, SOX17; Hs00751752_s1, GFD3; Hs00220998, DNMT3L; Hs01081364_m1). Relative transcript abundance was calculated using the 2−ΔCT method. Error bars represent S.E.M calculated from independent biological replicates.
2.6. Methylation profiling – mouse germ cells
DNA was extracted (Qiagen, DNeasy kit) from freshly isolated germ cells and bisulfite‐treated (Zymo Research, EZ DNA Methylation Kit). Between 2 and 4 ng of bisulfite‐treated and untreated (control) DNA was subjected to two rounds of PCR amplification with Hot‐start Taq DNA Polymerase (NEB) (conditions described in Supplementary Table 1). The first amplification of 35 cycles used flanking primers in a 25 μl reaction. Subsequently 1 μl of this reaction was subjected to a further 35 cycles using nested primers. PCR products were gel purified, cloned into pGEM‐T Easy (Promega) and 10–15 clones sequenced using T7 and T3 primers at the Australian Genome Research Facility (Brisbane, Australia).
2.7. Methylation profiling – GCC cell lines and tumor samples
DNA was extracted from cell lines and tumor biopsies according to manufacturer's instructions (Qiagen, DNeasy kit). DNA was bisulfite‐treated (Zymo Research, EZ DNA Methylation Kit) and 2.5 μg of bisulfite treated and untreated (control) DNA was subjected to one round of PCR amplification of 40 cycles with Zymo Taq DNA Polymerase (Zymo Research) (conditions described in Supplementary Table 1). PCR reactions were subjected to ExoSAP‐IT PCR clean‐up protocol (Affymetrix). For direct sequencing, 5 μl of the purified PCR product was used with the ABI Prism BigDye Terminator sequencing kit (Applied Biosciences, v1.1) with forward and reverse primers. For colony sequencing, 1 μl of the PCR product was cloned into the TOPO‐TA vector (Life Technologies) and 10–15 clones sequenced using the ABI Prism BigDye Terminator sequencing kit (Applied Biosciences, v1.1) with M13 primer.
2.8. Immunohistochemistry
Paraffin sections (3 μm) of previously characterized tumors were deparaffinised and heated under pressure to 1.2 bar in 0.01 M Tris, 0.001 M EGTA, pH 9.0. Slides were blocked with Avidin and Biotin solution (Vector Laboratories) and incubated with monoclonal anti‐CRIPTO antibody (1:10000; commissioned from Abmart; raised against epitope sequence FPQAFLPG) in 0.2%BSA/PBS overnight at 4 °C. Biotinylated Rabbit anti‐mouse antibody (1:150; Dako E413) was incubated for 30 min at RT followed by incubation with Avidin‐Biotin‐HRP complex (Vector Laboratories) for 30 min and staining visualized with DAB reagents (Vector Laboratories). Slides were counterstained with haematoxylin and mounted.
2.9. Western blotting
NT2 cells were lysed in RIPA buffer and sonicated, run on a 10% SDS‐PAGE gel, transferred to PVDF membrane then blocked/probed in 5% skim milk/PBS. Primary antibodies (anti‐CRIPTO; Abmart 8295; 1:10000, TUBA1A; Sigma T5168; 1:1000) were incubated overnight at 4 °C and secondary antibodies (mouse‐HRP; Sigma a8924; 1:2000) for 1 h at RT. Detection was performed using ECL (BioRad) and imaged using BioRad ChemiDoc.
2.10. Cripto ELISA
Human Cripto‐1 DuoSet ELISA Development kit (DY145; R&D Systems) was used as per manufacturer's instructions. Cell culture media was diluted 1:5 with 0.1% BSA/PBS. Complete media diluted 1:5 in 0.1% BSA/PBS with standards was used as the control to generate the standard curve for interpolation. For serum analysis, whole serum was diluted 1:2 in 0.1% BSA/PBS and standards spiked into control serum diluted 1:2 in 0.1% BSA/PBS to generate the standard curve for interpolation. All standards and unknowns were assayed in duplicate. Optical density was determined using a microplate reader (Bio‐Rad) with the 570 nm reading subtracted from the 450 nm reading. Duplicate readings for each sample were averaged and the zero standard optical density subtracted from this value. Prism software was used to generate a four‐parameter logistic curve‐fit and unknown values were interpolated to give the final CRIPTO concentration. For assessment of our commissioned CRIPTO antibody (Abmart) the ELISA assay was performed exactly as per manufacturer's instructions with the exception that the R&D Systems capture antibody was replaced with the Abmart CRIPTO antibody at a concentration of 5 ug/ml. Recombinant CIRPTO standard range was 0–4000 ng/ml.
3. Results
3.1. Cripto promoter methylation in mouse fetal germ cells
Because we have shown that Cripto is expressed for a short time in fetal germ cells but is ectopically expressed in GCC (Spiller et al., 2012), we wanted to investigate the mechanism by which Cripto expression is properly silenced under normal conditions using the mouse model. We began by investigating methylation of CpG dinucleotides in the promoter region, since male germ cells are known to undergo genome‐wide remethylation from 13.5 to 18.5 days post coitum (dpc) in mice (La Salle et al., 2004; Western et al., 2010). Using Methyl Primer Express software (v1.0, Applied Biosystems) we identified two regions in the mouse Cripto promoter with high CpG site‐content. Region 1 is 1383 bp upstream of the transcription start site (15 CpG sites) and Region 2 spans the transcription start site (18 CpG sites) (Figure 1A).
Figure 1.
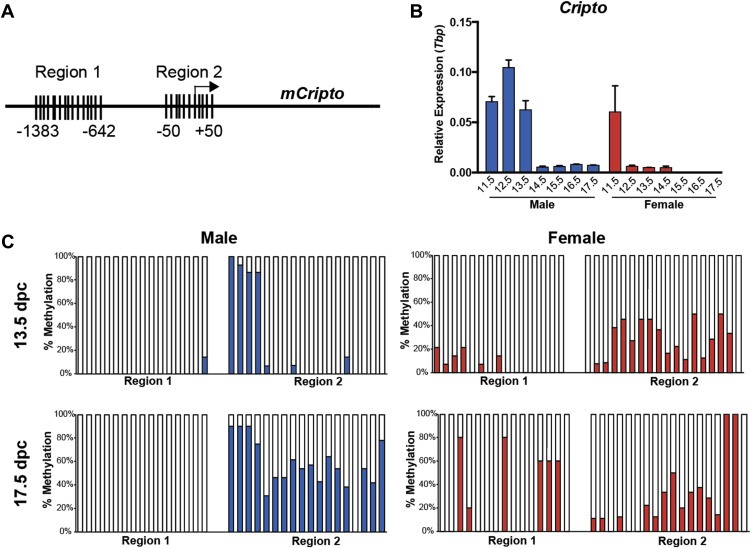
Bisulfite analysis of CpG methylation in the promoter region of Cripto in 13.5 and 17.5 dpc germ cells. (A) The mouse Cripto promoter has two CpG islands in the proximal promoter region: Region 1 at position ‐1383 – 642 contains 15 CpG sites. Region 2 at position −50 – +50 contains 18 CpG sites. (B)Cripto is expressed maximally in male gonads at 12.5 dpc and has declined by 14.5 dpc. Expression is similar in male and female gonads at 11.5 dpc before becoming downregulated by 12.5 dpc in the female. qRT‐PCR of pooled gonad‐only male and female samples at 11.5–17.5 dpc. Expression normalized to Tbp; error bars represent ±S.E.M; n = 3. (C) Methylation (expressed as % methylation) for each CpG site are shown for purified male and female germ cells at 13.5 and 17.5 dpc. For each sample ≥10 clones were sequenced.
To assess methylation specifically in germ cells, we isolated male and female (control) germ cells from Oct4ΔPE:eGFP mouse embryos (Szabo et al., 2002) using FACS at 13.5 dpc (Cripto is expressed in the male) and at 17.5 dpc (expression has declined in the male; Figure 1B, Supplementary Figure 1). Bisulfite‐treated DNA was subjected to PCR amplification of the two CpG regions before cloning and sequence analysis. We found that XX (female) germ cells displayed increased methylation in Region 1 from 13.5 to 17.5 dpc (6%–24% average methylation) while methylation of Region 2 remained unchanged (27% both timepoints; Figure 1C). Conversely, XY (male) germ cells displayed an increase in methylation of Region 2 from 13.5 to 17.5 dpc (22%–56%), whilst CpG sites in Region 1 remained unmethylated at both timepoints (1% and 0%) (Figure 1C). These data indicate that methylation of CpG sites in Region 2 correlates with declining Cripto expression male germ cells.
3.2. CRIPTO promoter methylation in human GCC
We next investigated whether methylation of the human CRIPTO promoter might also correlate with levels of gene expression in the various GCC specimens, using a combination of direct‐ and colony‐bisulfite sequencing. The human CRIPTO proximal promoter contains two CpG regions that contain 14 and 7 CpG sites, respectively (Figure 2A). These regions have previously been shown in the EC cell line NT2/D1 to be differentially methylated according to expression level and potential malignancy of these cells (Watanabe et al., 2010).
Figure 2.

Bisulfite analysis of GpG methylation in the promoter region of CRIPTO in GCC pathologies. (A) The human CRIPTO promoter has two CpG islands in the proximal promoter region: Region 1 at position −800 to −662 contains 13 CpG sites. Region 2 at position −225 to −84 contains 7 CpG sites. (B) qRT‐PCR analysis of CRIPTO expression (Log2 scale) in normal testis (control) and NS and SE samples; n = 5, 4, 9, 10, 7, 10. Methylation for each CpG site is shown for CH (C), TE (D), EC (E), YST (F) SE (G) and JKT‐1 control cell line (H). For each sample, graphs represent colony sequencing of ≥10 clones of one sample, each bar representing one CpG site (expressed at % methylation). Direct sequencing of 2–4 different tumor samples is presented as circles at each CpG site. Open circles represent unmethylated site, closed circles represent a methylated site and half‐filled circles represent heterogeneity of methylation at this site within a given sample.
Gene expression of CRIPTO was highest in EC and YST, with low transcript expression detectable in SE, TE and CH samples (Figure 2B). By colony sequencing, CH and TE (low CRIPTO expression) displayed high levels of methylation (average 75% and 72%, respectively) in both regions (Figure 2C,D). Conversely, in EC, which displays high levels of CRIPTO expression, methylation levels were relatively low (20%; Figure 2E). In contrast, despite high expression of CRIPTO in YST, both regions remained moderately‐to‐highly methylated (67%; Figure 2F). Further, in SE, which expresses low levels of CRIPTO, promoter methylation was relatively low (24%; Figure 2G). These results were confirmed by direct sequencing (Figure 2C–G). As a negative control we included the JKT‐1 cell line that is hypothesized to originate from a more mature germ cell (spermatocyte) (de Jong et al., 2007) and is negative for CRIPTO expression (Supplementary Figure 2A). In accordance with the lack of CRIPTO expression, JKT‐1 cells were found to be highly methylated using colony sequencing (80%; Figure 2H). Together, these data indicate that hypermethylation of the Cripto promoter is associated with low gene expression in CH and TE and hypomethylation is associated with high gene expression in EC. This correlation does not hold true for SE and YST, which presumably utilize other methods (or methylated regions) for gene regulation.
We also included several GCC cell lines in the methylation analysis. CRIPTO was expressed most strongly at the mRNA level in the TERA‐1 and NCCIT cells, at lower levels in NT2, TCam2 and 833 KE cells, and was undetectable in control JKT‐1 cells (Supplementary Figure 2A). The EC (833 KE, NT2, NCCIT, TERA‐1) and SE (TCam‐2) cell lines, which express CRIPTO at varying levels, all showed low methylation by direct sequencing, although generally higher methylation in Region 2 compared to Region 1 (Supplementary Figure 2B).
Lastly, we performed in vitro demethylation of the genome by 5‐azacytidine treatment of NT2 and TCam2 cell lines to observe the effect on CRIPTO expression. Although both cell lines display low levels of CRIPTO promoter methylation originally, upon 5‐azacytadine treatment methylation was reduced even further in both regions, assessed by direct sequencing (Supplementary Figure 3A). Interestingly, qRT‐PCR analysis revealed that CRIPTO mRNA expression was slightly reduced 48 h post treatment in both NT2 and TCam2 cells lines (0.5 and 0.2 fold, respectively; Supplementary Figure 3B). Because many genes have altered expression under such artificial conditions we also investigated expression of NODAL and the negative regulators of NODAL/CRIPTO signaling: LEFTY1 and LEFTY2 (Supplementary Figure 3B). We found NODAL expression to be greatly induced by demethylation in NT2 cells, and slightly reduced in TCam2 cells. Expression of both inhibitors LEFTY1 and LEFTY2 were induced robustly in both cell lines. We also assessed expression of pluripotency markers SOX2, SOX17 and GDF3 (Supplementary Figure 3B). Concordant with other studies, SOX17 expression was not significantly altered under the demethylating conditions in either cell line (Wermann et al., 2010) and SOX2 and GDF3 were decreased in NT2 cells (Biswal et al., 2012). The DNA methyltransferase DNMT3L, was induced in both cell lines under these conditions, as also reported previously in TCam2 cells (Wermann et al., 2010). These results suggest that under the artificial condition of induced genome‐wide demethylation, CRIPTO expression follows expression of other key pluripotency markers, consistent with its role in stem cell self‐renewal and maintenance (Bianco et al., 2010; Biswal et al., 2012; Wei et al., 2005).
3.3. CRIPTO expression in GCNIS and GCC
We next investigated CRIPTO protein expression in the same tissues and cell lines as investigated for methylation and mRNA expression. Based on qRT‐PCR data (Figure 2A) we expected strong expression of CRIPTO in EC and YST, and low or no expression in SE and TE biopsies. We commissioned the production of a mouse monoclonal antibody raised against part of the signal peptide of CRIPTO and verified that this antibody recognizes a single band in NT2 cells at the predicted molecular weight of 26 kDa (Supplementary Figure 4A). Further, this antibody outperformed the commercial R&D Systems capture antibody in recognizing recombinant CRIPTO in a sandwich ELISA assay (Supplementary Figure 4B). By immunohistochemistry we detected strong reactivity in EC and YST (Figure 3A,B). Despite a lower transcript abundance, we detected CRIPTO protein uniformly, but at lower intensity, in SE (lymphocytes were negative) (Figure 3C). In the differentiated GCC histology TE we found positive staining in small regions of differentiated cells, although large areas of tumor were negative for CRIPTO (Figure 3D).
Figure 3.
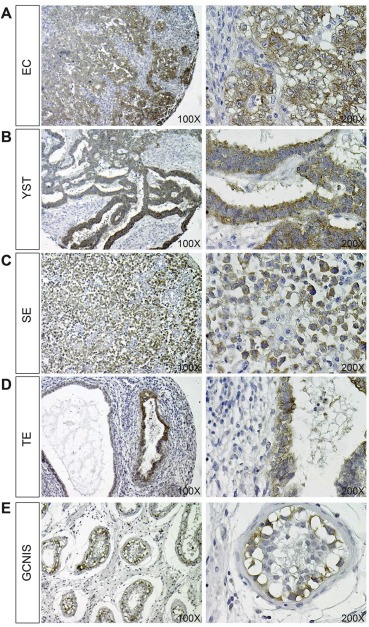
Immunohistochemistry for CRIPTO expression (brown staining) in EC (A), YST (B), SE (C), TE (D) and GCNIS (E) pathologies, counterstained with haematoxylin. Images are representative of ≥8 biopsies analyzed for each subtype at a magnification of both 100 and 200× (left and right images, respectively).
We also investigated whether CRIPTO was expressed in GCNIS cells. Consistent with previous results showing elevated CRIPTO expression in testis biopsies containing GCNIS (Spiller et al., 2012), strong expression of membrane‐bound CRIPTO was detected in GCNIS cells (verified by Pathologist J.W. Oosterhuis; Figure 3E). In normal testicular tissue, membranous CRIPTO was detectable only in spermatogonial stem cells (verified by J.W. Oosterhuis; Supplementary Figure 5A). Some positive staining was detected in nuclei of spermatocytes: we presume that this represents non‐specific binding as is seen for various antibodies (unpublished observations). Spermatids and all interstitium were negative for CRIPTO. Negative controls omitting primary antibody showed no staining (Supplementary Figure 5B).
3.4. Soluble CRIPTO detection in GCC cell line media
Although GPI‐anchored to the cell membrane, CRIPTO can be cleaved at this site and signal as a soluble ligand (Watanabe et al., 2007, 2007). The presence of soluble CRIPTO has been reported in the serum of patients with breast cancer, colon cancer and glioblastoma multiforme brain tumors (Bianco et al., 2006; Pilgaard et al., 2014) which suggests that it may be detectable, similarly, in serum of patients with GCC. Initially we investigated this possibility in GCC cell lines. By immunohistochemistry we found CRIPTO expressed in TCam2, TERA‐1 and NT2 cell lines, but not JKT‐1 control cells (Figure 4A). Although TERA‐1 and NT2 cells uniformly expressed CRIPTO, individual TCam2 cells displayed variable expression (absent to strong). We next evaluated the concentration of CRIPTO present in the media of each of these cell lines using sandwich ELISA. CRIPTO was undetectable in JKT‐1 culture media (Figure 4B), consistent with the negative immunohistochemistry and qRT‐PCR data (Supplementary Figure 1A). Highest average levels of cleaved CRIPTO were detected in media of the EC cell lines NT2 (11.7 ng/ml) and TERA‐1 (8.7 ng/ml), and lower levels in SE cell line TCam2 (1.4 ng/ml) (Figure 4B). Levels of expression varied depending on confluency of the cells (ranging 10–100%), which is reflected in the range of values detected.
Figure 4.
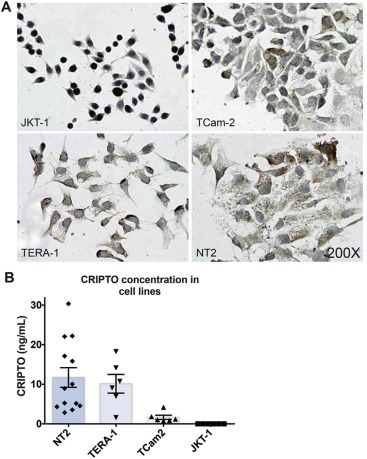
(A) Immunohistochemistry for CRIPTO expression in JKT‐1, TCam‐2, TERA‐1 and NT2 cell lines (B) CRIPTO concentration (ng/mL) assessed by ELISA in cell culture media collected from cell lines NT2 (range: 2.85–30.35), TERA‐1 (range: 1.58–18.28), TCam2 (range: 0.78–4.19) and JKT‐1 (range: 0.0–0.0) at varying confluences (ranging 10–100%). Error bars represent ±S.E.M; n = 13,6,6,6.
3.5. Soluble CRIPTO detection in GCC patient serum
Having found CRIPTO detectable in GCC by immunohistochemistry (Figure 3) we next assessed serum of patients with various GCC pathologies (Figure 5 and Table 1). Using ELISA, only one of 15 control patients (6.6%) generated detectable CRIPTO in serum (Control patients had no testicular malignancy at biopsy, but presented with some testicular abnormality such as testicular torsion or epididymitis. Inclusion in this ‘control’ group did not exclude the possibility that other somatic malignancies may have been present without our knowledge at the time of serum collection). Highest levels of CRIPTO were detected in SE samples, of which 5 of 15 (33%) were positive and had an average value of 7.6 ± 24.2 ng/ml. The large standard deviation in this group is accounted for by one sample, of unknown clinical stage, that generated an unusually high reading of 94.36 ng/ml of CRIPTO. In EC serum samples, CRIPTO was detected in 6 of 15 (33%) at an average concentration of 3.6 ± 11.6 ng/ml. One of 3 YST samples (33%) was positive (average 1.5 ± 2.6 ng/ml). Lower levels of CRIPTO were detected in 4 of the 5 GCNIS‐only samples (80%). The GCNIS‐only group had an average concentration of 0.6 ± 0.7 ng/ml and was distinct, in a statistically significant manner, from the control group. The 5 TE and 1 CH serum samples were negative for CRIPTO expression in this assay. When these data were analyzed against the control group omitting the outlying sample (high CRIPTO concentration of 11.5 ng/ml), we found that EC, SE and GCNIS‐only groups were significantly distinct from control (Table 1).
Figure 5.
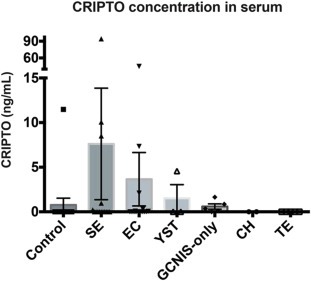
CRIPTO concentration (ng/mL) assessed by ELISA in patient serum of control (no testicular malignancy), SE, EC, YST, GCNIS‐only, CH and TE pathologies. Error bars represent ±S.E.M; n = 15,15,15,3,5,1,5.
Table 1.
CRIPTO concentration in serum.
| Patient group | No. Positive samples/total | CRIPTO concentration (ng/mL) | Statistics | ||
|---|---|---|---|---|---|
| Mean (SD) | Range | Pa | Pb | ||
| Control | 1/15 | 0.77 (2.97) | 0.0–11.48 | – | – |
| SE | 5/15 | 7.61 (24.22) | 0.0–94.36 | 0.1433 | 0.0437 |
| EC | 6/15 | 3.66 (11.59) | 0.0–44.98 | 0.0695 | 0.0178 |
| YST | 1/3 | 1.52 (2.63) | 0.0–4.56 | 0.3137 | 0.1875 |
| GCNIS‐only | 4/5 | 0.59 (0.67) | 0.0–1.63 | 0.0049 | 0.0016 |
| TE | 0/5 | – | – | – | – |
| CH | 0/1 | – | – | – | – |
Mann–Whitney U test of patient group versus control.
Mann–Whitney U test of patient group versus control excluding positive outlier.
4. Discussion
Previous studies in mice have established a role for Nodal/Cripto signaling in the maintenance of germ cell pluripotency during a short window of germ cell development (Spiller et al., 2012). This period of development is believed to correspond to a window of vulnerability for germ cell transformation into GCNIS, the common precursor of all GCC. The finding that CRIPTO is relatively highly expressed in GCC (Baldassarre et al., 2001; Spiller et al., 2012) suggests that embryonic CRIPTO expression may fail to extinguish during malignant transformation. In the present study we found that epigenetic factors may be responsible for this aberrant expression during adult life. We also provide proof‐of‐principle that detection of cleaved CRIPTO protein in serum might be a useful avenue for prognostic and/or diagnostic application to GCC.
As expected, we found that methylation was highest in mouse germ cell populations where Cripto expression is low. Of the two regions analyzed in fetal germ cells, it appears Region 2, spanning the transcription start site, is more important: CpG sites in Region 2 became hypermethylated in XY germ cells as expression declined, while the more distal CpG sites (Region 1) remained hypomethylated at both timepoints. There was little change to methylation in XX germ cells in either region over time, likely because they do not undergo genome‐wide remethylation but, rather, enter meiosis (La Salle et al., 2004; McLaren, 2003; Western et al., 2010). We speculate that the hypomethylation of Region 1 is responsible for allowing the transient expression of Cripto in XY germ cells, and further, that hypermethylation of this site may be a meiosis‐specific methylation pattern in XX germ cells that helps prevent expression of Cripto that might otherwise disrupt meiosis. Methylation analysis of sexed, 11.5 dpc germ cells before sex‐specific gene expression begins, although technically challenging, would shed light on these possibilities.
Similarly to mouse, the human CRIPTO promoter has two regions enriched for CpG sites (Bianco et al., 2013; Watanabe et al., 2010). In our cell line analysis, we find both regions generally hypomethylated in SE and NS cell lines that express CRIPTO, regardless of relative expression levels, and both regions hypermethylated in the control cell line that is negative for CRIPTO expression. These results concur with recent findings using DNA methylation PCR for Region 2 of CRIPTO in which <10% of this promoter region was found to be methylated (Bianco et al., 2013). In order to link methylation of these regions to CRIPTO gene expression, Watanabe et al. (2010) performed retinoic acid‐induced DNA methylation in the EC cell line NT2/D1 and showed that time‐dependent methylation of the same CpG regions we investigated, correspond to reduction in CRIPTO expression. Further, they showed that differences in methylation of CpG Region 1 correspond to gene expression in this cell line, supporting the claim that methylation is partially responsible for CRIPTO gene regulation (Watanabe et al., 2010). Here we tested the opposite approach of in vitro genome wide demethylation using 5‐azacytadine in NT2 and TCam2 cells and found that under these conditions CRIPTO expression was slightly decreased post‐treatment. Although somewhat surprising, this could be due to a combination of factors, including that these regions are already sufficiently hypomethylated or that upregulation of negative inhibitors of CRIPTO/NODAL signaling such as LEFTY1, ‐2 (and many others that we did not test for) could have negative feedback on CRIPTO expression. In addition to these possibilities it is also possible, and likely, that CRIPTO falls within the complement of classic ‘pluripotency’ factors that become downregulated under the artificial in vitro conditions of global demethylation (along with NANOG, SOX2, GDF3 and MYC target genes; (Biswal et al., 2012)).
In the GCC samples, we found a clear correlation between CRIPTO promoter hypermethylation and low CRIPTO expression in TE and CH tumor subtypes, and vice versa for EC. These correlations were not found for YST and SE. Instead, in YST samples (which display high levels of CRIPTO expression), we found high levels of methylation in Region 1 of the CRIPTO promoter, while Region 2 showed moderate levels of methylation. However, as YST is known to display high levels of global methylation (Amatruda et al., 2013; Furukawa et al., 2009), it is possible that the moderate level of methylation in Region 2 is sufficient for gene transcription, although it is likely that additional mechanisms are involved. Similarly, for SE, the hypomethylation observed in our findings is seemingly contrary to low CRIPTO expression, but is consistent with the global hypomethylation that characterizes this GCC subtype (Netto et al., 2008; Smiraglia et al., 2002; Wermann et al., 2010). It is possible that modest methylation of Region 1 is sufficient to suppress gene transcription in this genomic environment of global hypomethylation.
Exactly how a GCNIS cell transforms into the two distinct tumor types, SE and NS, which are characterized by hypo‐ and hypermethylation, respectively, is unknown. This issue is of interest because of the different sensitivity of the SE and NS elements to irradiation and cisplatin‐based chemotherapy. GCNIS cells, like SE, exhibit global hypomethylation, akin to their fetal germ cell precursors, so de novo methylation is thought to be largely responsible for establishment of the methylation profile in the overt tumor (Almstrup et al., 2010; Netto et al., 2008; Wermann et al., 2010). Based on our fetal germ cell data, we might expect CRIPTO Region 1 to be completely hypomethylated and Region 2 to have variable methylation in a given GCNIS cell. Indeed it has been shown that differing levels of CRIPTO expression and methylation in the NT2 human EC cell line correlate with malignancy potential of these cells (Watanabe et al., 2010). It will be interesting to investigate whether individual GCNIS cells express CRIPTO at varying levels, and whether methylation of Region 1 may be predictive of a GCNIS cell that will give rise to EC (hypomethylation) or SE (moderate levels of methylation).
We found in our cell line analysis that protein expression of CRIPTO, detected by immunohistochemistry, was comparable between NT2 and TERA‐1 cells, despite lower gene expression in NT2 cells. This discrepancy is likely the result of different confluency of the cells when assayed for RNA and protein, as confluency was seen to greatly affect levels of soluble CRIPTO detected in media using the ELISA assay. This is consistent with a report from Watanabe et al. (2010) that see CRIPTO expression decline in NT2/D1 cells at confluency. Similarly, although we expected strong protein expression of CRIPTO in EC and YST, based on other reports and our own gene expression data, we were surprised to find that it could also be detected in SE and small regions of TE biopsies, where gene expression is overall low. However, expression of CRIPTO in GCNIS makes it extremely likely that it would also be expressed in SE, as these two cell types share similar expression profiles (Almstrup et al., 2005). We don't consider it likely that our antibody is detecting an unspecific target in SE based on the Western blot results and the fact that our immunohistochemistry data was corroborated by the independently‐tested antibodies in the sandwich ELISA, which detected CRIPTO protein in the SE cell line TCam2 and in serum of patients with SE. This surprising finding of low gene expression with high protein detection is not unique, and for example has also reported for GDF3, in which the transcript is predominantly expressed in EC and YST but the protein is also detected in GCNIS and SE (Gopalan et al., 2009; Korkola et al., 2006). This similarity is of interest as GDF3 is a known ligand for CRIPTO and is one of the stem cell genes in the 200 kb region of chromosome 12p12.31 commonly over‐represented in both SE and NS (Roelofs et al., 2000; Zafarana et al., 2003). The overlapping expression patterns of CRIPTO and GDF3 in GCNIS and the GCC subtypes (Figure 6) suggests that GDF3 (alone, or in combination with NODAL) could trigger CRIPTO activation. In this context it is of interest to mention that gain of 12p is related to progression from GCNIS to either SE or NS (Rosenberg et al., 2000). It will be interesting to determine how the specific combinations of CRIPTO/GDF3/NODAL contribute to the malignancy potential of each subtype, a question that we are now investigating.
Figure 6.
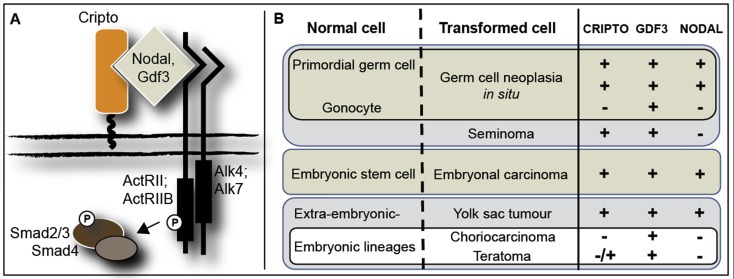
Schematic of CRIPTO/NODAL/GDF3 signaling (A) and the combinations of receptor/ligand expression in normal germ cells, embryonic stem cells and the subtypes of GCC (B). Symbols represent positive expression (+), negative expression (−) and heterogeneous expression (−/+).
Screening using serum markers is minimally invasive, rapid and cost‐effective. Detection of CRIPTO for diagnostic and prognostic purposes has been assessed for breast, colon and brain cancer (Bianco et al., 2006; Pilgaard et al., 2014); both studies similarly utilized sandwich ELISA for CRIPTO detection. In our analysis we detected highest average CRIPTO readings for SE followed by EC, YST then GCNIS‐only samples, of which % sensitivity was 33%, 40%, 33%, 33% and 80% respectively. Our false positive rate was 6%, however it is possible that the presence of CRIPTO in this one sample is attributable to another undetected malignancy at the time of serum collection (CRIPTO is expressed in a wide range of epithelial cancers (Normanno et al., 2001)). A larger sample size would reduce the effect such outliers have on the data set. Our background reading of CRIPTO expression was 0 ng/ml for 14 of 15 control samples. Conversely, Bianco et al. (2006) detected CRIPTO levels in plasma of above 0.07 ng/ml for all samples (n = 21) and Pilgaard et al. (2014) above 0.3 ng/ml for all samples (n = 27). Discrepancies between our basal levels of CRIPTO and those from the previous studies can most likely be attributable to differences in the ELISA assays (Bianco et al., 2006) and serum dilution factor differences (Bianco et al., 2006; Pilgaard et al., 2014). We found a 1:2 dilution factor necessary to consistently achieve good linearity‐of‐dilution and spike‐and‐recovery efficiencies when optimizing our assay (data not shown). It may be possible to increase the sensitivity of our assay by increasing the capture and/or detection antibody concentrations.
An alternative and more promising approach that we are currently pursuing is detection of CRIPTO using ELISA in semen samples. As the CRIPTO‐expressing GCNIS and GCC cells are in close proximity with semen, it is feasible that this will prove highly sensitive for our diagnostic and prognostic purposes. So far, CRIPTO is undetectable in 6/6 controls (malignancy‐free, normozoospermic) in our assay optimized for semen (data not shown). A great advantage of this approach, in addition to a likely increase in sensitivity, is that positivity for CRIPTO in semen would presumably be a specific indicator of the precursor of GCC, unlike positivity in serum, as outlined above.
In conclusion, our studies have highlighted regions of the CRIPTO promoter that appear to be differentially methylated in fetal germ cells and GCC derived from incorrect germ cell differentiation. We have also demonstrated for the first time the potential suitability of CRIPTO as a serological marker for diagnostic and/or prognostic purposes for GCC. We are now investigating whether detection in semen may offer an alternative and potentially more sensitive means for GCC diagnosis.
Disclosure/conflicts of interest
No conflicts of interest are declared.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgements
We thank Prof Sohei Kitazawa and Prof Riko Kitazawa from the Department of Molecular Pathology, Ehime University Graduate School of Medicine, Japan, for providing TCam2 cells. Flow cytometry was performed with the assistance of the Queensland Brain Institute Flow Cytometry Facility. This work was supported by the National Health and Medical Research Council [APP1030146] and Cancer Council Queensland [APP1012325]. CS is a University of Queensland Postdoctoral Fellow. PK is an NHMRC Senior Principal Research Fellow.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.11.003.
Spiller Cassy M., Gillis Ad J.M., Burnet Guillaume, Stoop Hans, Koopman Peter, Bowles Josephine, Looijenga Leendert H.J., (2016), Cripto: Expression, epigenetic regulation and potential diagnostic use in testicular germ cell tumors, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.11.003.
References
- Adami, H.O. , Bergstrom, R. , Mohner, M. , Zatonski, W. , Storm, H. , Ekbom, A. , Tretli, S. , Teppo, L. , Ziegler, H. , Rahu, M. , 1994. Testicular cancer in nine northern European countries. Int. J. Cancer. 59, 33–38. [DOI] [PubMed] [Google Scholar]
- Almstrup, K. , Hoei-Hansen, C.E. , Nielsen, J.E. , Wirkner, U. , Ansorge, W. , Skakkebaek, N.E. , Rajpert-De Meyts, E. , Leffers, H. , 2005. Genome-wide gene expression profiling of testicular carcinoma in situ progression into overt tumours. Br. J. Cancer. 92, 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almstrup, K. , Nielsen, J.E. , Mlynarska, O. , Jansen, M.T. , Jorgensen, A. , Skakkebaek, N.E. , Rajpert-De Meyts, E. , 2010. Carcinoma in situ testis displays permissive chromatin modifications similar to immature foetal germ cells. Br. J. Cancer. 103, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda, J.F. , Ross, J.A. , Christensen, B. , Fustino, N.J. , Chen, K.S. , Hooten, A.J. , Nelson, H. , Kuriger, J.K. , Rakheja, D. , Frazier, A.L. , Poynter, J.N. , 2013. DNA methylation analysis reveals distinct methylation signatures in pediatric germ cell tumors. BMC Cancer. 13, 313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre, G. , Tucci, M. , Lembo, G. , Pacifico, F.M. , Dono, R. , Lago, C.T. , Barra, A. , Bianco, C. , Viglietto, G. , Salomon, D. , Persico, M.G. , 2001. A truncated form of teratocarcinoma-derived growth factor-1 (cripto-1) mRNA expressed in human colon carcinoma cell lines and tumors. Tumour Biol. 22, 286–293. [DOI] [PubMed] [Google Scholar]
- Bianco, C. , Castro, N.P. , Baraty, C. , Rollman, K. , Held, N. , Rangel, M.C. , Karasawa, H. , Gonzales, M. , Strizzi, L. , Salomon, D.S. , 2013. Regulation of human Cripto-1 expression by nuclear receptors and DNA promoter methylation in human embryonal and breast cancer cells. J. Cell Physiol. 228, 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, C. , Rangel, M.C. , Castro, N.P. , Nagaoka, T. , Rollman, K. , Gonzales, M. , Salomon, D.S. , 2010. Role of Cripto-1 in stem cell maintenance and malignant progression. Am. J. Pathol. 177, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco, C. , Strizzi, L. , Mancino, M. , Rehman, A. , Hamada, S. , Watanabe, K. , De Luca, A. , Jones, B. , Balogh, G. , Russo, J. , Mailo, D. , Palaia, R. , D'Aiuto, G. , Botti, G. , Perrone, F. , Salomon, D.S. , Normanno, N. , 2006. Identification of cripto-1 as a novel serologic marker for breast and colon cancer. Clin. Cancer Res. 12, 5158–5164. [DOI] [PubMed] [Google Scholar]
- Biswal, B.K. , Beyrouthy, M.J. , Hever-Jardine, M.P. , Armstrong, D. , Tomlinson, C.R. , Christensen, B.C. , Marsit, C.J. , Spinella, M.J. , 2012. Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA damage-associated p53 activation, anti-pluripotency and DNA demethylation. PLoS One. 7, e53003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, J. , Stoop, H. , Gillis, A.J. , van Gurp, R.J. , van Drunen, E. , Beverloo, H.B. , Lau, Y.F. , Schneider, D.T. , Sherlock, J.K. , Baeten, J. , Hatakeyama, S. , Ohyama, C. , Oosterhuis, J.W. , Looijenga, L.H. , 2007. JKT-1 is not a human seminoma cell line. Int. J. Androl. 30, 350–365. [DOI] [PubMed] [Google Scholar]
- Furukawa, S. , Haruta, M. , Arai, Y. , Honda, S. , Ohshima, J. , Sugawara, W. , Kageyama, Y. , Higashi, Y. , Nishida, K. , Tsunematsu, Y. , Nakadate, H. , Ishii, M. , Kaneko, Y. , 2009. Yolk sac tumor but not seminoma or teratoma is associated with abnormal epigenetic reprogramming pathway and shows frequent hypermethylation of various tumor suppressor genes. Cancer Sci. 100, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan, A. , Dhall, D. , Olgac, S. , Fine, S.W. , Korkola, J.E. , Houldsworth, J. , Chaganti, R.S. , Bosl, G.J. , Reuter, V.E. , Tickoo, S.K. , 2009. Testicular mixed germ cell tumors: a morphological and immunohistochemical study using stem cell markers, OCT3/4, SOX2 and GDF3, with emphasis on morphologically difficult-to-classify areas. Mod. Pathol. 22, 1066–1074. [DOI] [PubMed] [Google Scholar]
- Klauzinska, M. , Castro, N.P. , Rangel, M.C. , Spike, B.T. , Gray, P.C. , Bertolette, D. , Cuttitta, F. , Salomon, D. , 2014. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin. Cancer Biol. 29, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkola, J.E. , Houldsworth, J. , Chadalavada, R.S. , Olshen, A.B. , Dobrzynski, D. , Reuter, V.E. , Bosl, G.J. , Chaganti, R.S. , 2006. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 66, 820–827. [DOI] [PubMed] [Google Scholar]
- La Salle, S. , Mertineit, C. , Taketo, T. , Moens, P.B. , Bestor, T.H. , Trasler, J.M. , 2004. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev. Biol. 268, 403–415. [DOI] [PubMed] [Google Scholar]
- Looijenga, L.H. , de Leeuw, H. , van Oorschot, M. , van Gurp, R.J. , Stoop, H. , Gillis, A.J. , de Gouveia Brazao, C.A. , Weber, R.F. , Kirkels, W.J. , van Dijk, T. , von Lindern, M. , Valk, P. , Lajos, G. , Olah, E. , Nesland, J.M. , Fossa, S.D. , Oosterhuis, J.W. , 2003. Stem cell factor receptor (c-KIT) codon 816 mutations predict development of bilateral testicular germ-cell tumors. Cancer Res. 63, 7674–7678. [PubMed] [Google Scholar]
- McFarlane, L. , Truong, V. , Palmer, J.S. , Wilhelm, D. , 2013. Novel PCR assay for determining the genetic sex of mice. Sex Dev. 7, 207–211. [DOI] [PubMed] [Google Scholar]
- McLaren, A. , 2003. Primordial germ cells in the mouse. Dev. Biol. 262, 1–15. [DOI] [PubMed] [Google Scholar]
- Mosselman, S. , Looijenga, L.H. , Gillis, A.J. , van Rooijen, M.A. , Kraft, H.J. , van Zoelen, E.J. , Oosterhuis, J.W. , 1996. Aberrant platelet-derived growth factor alpha-receptor transcript as a diagnostic marker for early human germ cell tumors of the adult testis. Proc. Natl. Acad. Sci. U. S. A. 93, 2884–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto, G.J. , Nakai, Y. , Nakayama, M. , Jadallah, S. , Toubaji, A. , Nonomura, N. , Albadine, R. , Hicks, J.L. , Epstein, J.I. , Yegnasubramanian, S. , Nelson, W.G. , De Marzo, A.M. , 2008. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod. Pathol. 21, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno, N. , Bianco, C. , De Luca, A. , Salomon, D.S. , 2001. The role of EGF-related peptides in tumor growth. Front Biosci. 6, D685–D707. [DOI] [PubMed] [Google Scholar]
- Oosterhuis, J.W. , Looijenga, L.H. , 2005. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 5, 210–222. [DOI] [PubMed] [Google Scholar]
- Pilgaard, L. , Mortensen, J.H. , Henriksen, M. , Olesen, P. , Sorensen, P. , Laursen, R. , Vyberg, M. , Agger, R. , Zachar, V. , Moos, T. , Duroux, M. , 2014. Cripto-1 expression in glioblastoma multiforme. Brain Pathol. 24, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs, H. , Mostert, M.C. , Pompe, K. , Zafarana, G. , van Oorschot, M. , van Gurp, R.J. , Gillis, A.J. , Stoop, H. , Beverloo, B. , Oosterhuis, J.W. , Bokemeyer, C. , Looijenga, L.H. , 2000. Restricted 12p amplification and RAS mutation in human germ cell tumors of the adult testis. Am. J. Pathol. 157, 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, C. , Van Gurp, R.J. , Geelen, E. , Oosterhuis, J.W. , Looijenga, L.H. , 2000. Overrepresentation of the short arm of chromosome 12 is related to invasive growth of human testicular seminomas and nonseminomas. Oncogene. 19, 5858–5862. [DOI] [PubMed] [Google Scholar]
- Shen, M.M. , 2007. Nodal signaling: developmental roles and regulation. Development. 134, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Skakkebaek, N.E. , Berthelsen, J.G. , Giwercman, A. , Muller, J. , 1987. Carcinoma-in-situ of the testis: possible origin from gonocytes and precursor of all types of germ cell tumours except spermatocytoma. Int. J. Androl. 10, 19–28. [DOI] [PubMed] [Google Scholar]
- Smiraglia, D.J. , Szymanska, J. , Kraggerud, S.M. , Lothe, R.A. , Peltomaki, P. , Plass, C. , 2002. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 21, 3909–3916. [DOI] [PubMed] [Google Scholar]
- Sonne, S.B. , Almstrup, K. , Dalgaard, M. , Juncker, A.S. , Edsgard, D. , Ruban, L. , Harrison, N.J. , Schwager, C. , Abdollahi, A. , Huber, P.E. , Brunak, S. , Gjerdrum, L.M. , Moore, H.D. , Andrews, P.W. , Skakkebaek, N.E. , Rajpert-De Meyts, E. , Leffers, H. , 2009. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res. 69, 5241–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller, C.M. , Bowles, J. , Koopman, P. , 2013. Nodal/Cripto signaling in fetal male germ cell development: implications for testicular germ cell tumors. Int. J. Dev. Biol. 57, 211–219. [DOI] [PubMed] [Google Scholar]
- Spiller, C.M. , Feng, C.W. , Jackson, A. , Gillis, A.J. , Rolland, A.D. , Looijenga, L.H. , Koopman, P. , Bowles, J. , 2012. Endogenous Nodal signaling regulates germ cell potency during mammalian testis development. Development. 139, 4123–4132. [DOI] [PubMed] [Google Scholar]
- Szabo, P.E. , Hubner, K. , Scholer, H. , Mann, J.R. , 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115, 157–160. [DOI] [PubMed] [Google Scholar]
- van de Geijn, G.J. , Hersmus, R. , Looijenga, L.H. , 2009. Recent developments in testicular germ cell tumor research. Birth Defects Res. C Embryo Today. 87, 96–113. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Bianco, C. , Strizzi, L. , Hamada, S. , Mancino, M. , Bailly, V. , Mo, W. , Wen, D. , Miatkowski, K. , Gonzales, M. , Sanicola, M. , Seno, M. , Salomon, D.S. , 2007. Growth factor induction of Cripto-1 shedding by glycosylphosphatidylinositol-phospholipase D and enhancement of endothelial cell migration. J. Biol. Chem. 282, 31643–31655. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Hamada, S. , Bianco, C. , Mancino, M. , Nagaoka, T. , Gonzales, M. , Bailly, V. , Strizzi, L. , Salomon, D.S. , 2007. Requirement of glycosylphosphatidylinositol anchor of Cripto-1 for trans activity as a Nodal co-receptor. J. Biol. Chem. 282, 35772–35786. [DOI] [PubMed] [Google Scholar]
- Watanabe, K. , Meyer, M.J. , Strizzi, L. , Lee, J.M. , Gonzales, M. , Bianco, C. , Nagaoka, T. , Farid, S.S. , Margaryan, N. , Hendrix, M.J. , Vonderhaar, B.K. , Salomon, D.S. , 2010. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells. 28, 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.L. , Miura, T. , Robson, P. , Lim, S.K. , Xu, X.Q. , Lee, M.Y. , Gupta, S. , Stanton, L. , Luo, Y. , Schmitt, J. , Thies, S. , Wang, W. , Khrebtukova, I. , Zhou, D. , Liu, E.T. , Ruan, Y.J. , Rao, M. , Lim, B. , 2005. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 23, 166–185. [DOI] [PubMed] [Google Scholar]
- Wermann, H. , Stoop, H. , Gillis, A.J. , Honecker, F. , van Gurp, R.J. , Ammerpohl, O. , Richter, J. , Oosterhuis, J.W. , Bokemeyer, C. , Looijenga, L.H. , 2010. Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J. Pathol. 221, 433–442. [DOI] [PubMed] [Google Scholar]
- Western, P.S. , van den Bergen, J.A. , Miles, D.C. , Sinclair, A.H. , 2010. Male fetal germ cell differentiation involves complex repression of the regulatory network controlling pluripotency. FASEB J. 24, 3026–3035. [DOI] [PubMed] [Google Scholar]
- Zafarana, G. , Grygalewicz, B. , Gillis, A.J. , Vissers, L.E. , van de Vliet, W. , van Gurp, R.J. , Stoop, H. , Debiec-Rychter, M. , Oosterhuis, J.W. , van Kessel, A.G. , Schoenmakers, E.F. , Looijenga, L.H. , Veltman, J.A. , 2003. 12p-amplicon structure analysis in testicular germ cell tumors of adolescents and adults by array CGH. Oncogene. 22, 7695–7701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


