Abstract
Suppressor of cytokine signaling (SOCS) proteins are negative feedback regulators of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway. Dysregulation of SOCS protein expression in cancers can be one of the mechanisms that maintain STAT activation, but this mechanism is still poorly understood in oral squamous cell carcinoma (OSCC). Here, we report that SOCS2 protein is significantly downregulated in OSCC patients and its levels are inversely correlated with miR‐424‐5p expression. We identified the SOCS2 protein, which modulates STAT5 activity, as a direct target of miR‐424‐5p. The miR‐424‐5p‐induced STAT5 phosphorylation, matrix metalloproteinases (MMPs) expression, and cell migration and invasion were blocked by SOCS2 restoration, suggesting that miR‐424‐5p exhibits its oncogenic activity through negatively regulating SOCS2 levels. Furthermore, miR‐424‐5p expression could be induced by the cytokine IL‐8 primarily through enhancing STAT5 transcriptional activity rather than NF‐κB signaling. Antagomir‐mediated inactivation of miR‐424‐5p prevented the IL‐8‐induced cell migration and invasion, indicating that miR‐424‐5p is required for IL‐8‐induced cellular invasiveness. Taken together, these data indicate that STAT5‐dependent expression of miR‐424‐5p plays an important role in mediating IL‐8/STAT5/SOCS2 feedback loop, and scavenging miR‐424‐5p function using antagomir may have therapeutic potential for the treatment of OSCC.
Keywords: microRNA, SOCS2, IL‐8, Invasion, Oral squamous cell carcinoma
Highlights
miR‐424‐5p is overexpressed in OSCC.
miR‐424‐5p directly targets SOCS2, leading to increased cell migration and invasion.
STAT5 activation is required for IL‐8‐mediated miR‐424‐5p transcription.
miR‐424‐5p plays an important role in mediating IL‐8/STAT5/SOCS2 feedback loop.
Abbreviations
- 3′-UTR
3′-untranslated region
- EGFR
epidermal growth factor receptor
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- miRNA
microRNA
- MMP
matrix metalloproteinase
- OSCC
Oral squamous cell carcinoma
- PI3K
phosphoinositide 3-kinase
- qRT-PCR
quantitative real-time polymerase chain reaction
- SH2
Src homology
- SOCS
suppressors of cytokine signaling
- STAT
signal transducer and activator of transcription
1. Introduction
Oral squamous cell carcinoma (OSCC) is a common head and neck cancer form with relatively low 5‐year survival rate (Forastiere et al., 2001; Jemal et al., 2007) and ranks the fourth most common cancer in Taiwan's male population with growing incidence and mortality. Due to their frequent loco‐regional recurrences and highly metastatic neck lymph node properties (Jerjes et al., 2010), there is a need for a better understanding of the molecular basis underlying the aggressive growth of OSCC to improve therapeutic efficacy, as well as to design more effective treatment strategies. Several cellular and molecular factors that cause the occurrence and progression of cancer have been reported in OSCC, including p53 (Ahomadegbe et al., 1995), NOTCH (Agrawal et al., 2011), cyclin D1 (Callender et al., 1994), mitogen‐activated protein kinase (MAPK) (Mishima et al., 1998), phosphoinositide 3‐kinase (PI3K) (Lui et al., 2013), epidermal growth factor receptor (EGFR) (Maxwell et al., 1989) and signal transducer and activator of transcription (STAT) (Grandis et al., 2000).
The STAT family members play a critical role in regulating physiological responses to stimulation by cytokines, hormones and growth factors involved in hematopoiesis, immune regulation, growth and embryogenesis (Benekli et al., 2003; Siavash et al., 2004). Activated STAT proteins conduct signals by forming dimer complexes and subsequently translocating into the nucleus to induce transcription of specific target genes (Darnell, 1997). The activity of STAT proteins is under tight regulation by the induction of suppressors of cytokine signaling (SOCS) proteins, which silence the STAT pathways by competing binding with Janus kinase, by binding to the receptor to prevent STAT interaction, or by targeting proteins for proteasomal degradation (Siavash et al., 2004; Tamiya et al., 2011). The SOCS family consists of 8 members (SOCS1‐7 and CIS) and is characterized by a variable length N‐terminal domain, a central Src homology (SH2) domain for protein–protein interaction and a conserved C‐terminus SOCS box domain for ubiquitin‐mediated proteasomal degradation (Palmer and Restifo, 2009). Many cytokines induce the expression of SOCS, which act in a negative feedback loop to prevent further STAT signal transduction (Croker et al., 2003; Vlotides et al., 2004). However, aberrant activation of STAT pathways, particularly of STAT1, STAT3, and STAT5, has been reported in hematologic malignancies (Xia et al., 1998) and solid tumors, including OSCC (Grandis et al., 1998; Leong et al., 2002). In OSCC, there is compelling evidence that constitutive activation of STAT proteins is linked to cancer initiation and progression (Grandis et al., 2000; Song and Grandis, 2000). Therefore, dysregulation of SOCS proteins could be one of the important mechanisms of abnormal STAT activation (Inagaki‐Ohara et al., 2013).
Several mechanisms that lead to abnormal expression of SOCS proteins in cancer have been reported, such as abnormal subcellular localization (Rossa et al., 2012), genetic variations (Zhang et al., 2014), epigenetic modifications (Sutherland et al., 2004; Zhang et al., 2013), and microRNA (miRNA) regulation (Huang et al., 2013; Ru et al., 2011). miRNAs are short non‐coding RNAs that can post‐transcriptionally silence gene expression either by decreasing protein translation rate or by increasing degradation of the mRNA. Numerous studies have documented that miRNAs play vital roles in various biological functions, including cell growth, differentiation, apoptosis, development and homeostasis (Inui et al., 2010). Alterations in miRNA expression can cause various human diseases such as cancer (Calin and Croce, 2006). In fact, miRNA has been shown to regulate JAK/STAT signal transduction through control of SOCS expression. For example, miR‐155 has been shown to target SOCS1 and promote cell proliferation and colony formation in breast cancer cells (Jiang et al., 2010). MiR‐19a has been found to target SOCS3 together with increased activation of STAT3 (Collins et al., 2013). In addition, miR‐9 and miR‐98 that target SOCS3 and SOCS4, respectively, have also been reported (Hu et al., 2010; Zhuang et al., 2012). These results suggest that a significant role for miRNAs in the regulation of the STAT/SOCS pathway. However, little is known about the role of miRNAs involved in STAT/SOCS regulation in oral cancer.
In the current study, we found that miR‐424‐5p was upregulated in OSCC tissue. We also identified SOCS2 as a novel target of miR‐424‐5p. Overexpression of miR‐424‐5p in oral cancer cells promoted cell migration and invasion through the repression of SOCS2. Furthermore, miR‐424‐5p expression in OSCC cells could be induced by IL‐8 through increasing STAT5 transcriptional activity. Taken together, we discovered a feedback loop that is triggered by IL‐8/STAT5‐mediated induction of miR‐424‐5p and repression of SOCS2, which cause constitutive activation of STAT5 and promote migration and invasion in OSCC.
2. Materials and methods
2.1. Clinical samples and pathological characteristics
Oral squamous cell carcinoma tissues and their adjacent non‐tumorous epithelia were derived from 40 patients who received curative surgery from 1999 to 2010 at the National Cheng Kung University Hospital. Tissues were obtained before chemotherapy or radiation therapy and were immediately snap‐frozen in liquid nitrogen for RNA extraction. Total RNA (including miRNA) was isolated using the miRNeasy Mini Kit (Qiagen, #217004) according to the manufacturer's protocol and stored at −80 °C for further study. Clinical parameters, including age, sex, social history, pathological features, and TMN stage were retrospectively collected by reviewing patients' charts. The study protocol was reviewed and approved by the Institutional Human Experiment and Ethic Committee. Informed consent was obtained from each patient.
2.2. Cell lines and culture condition
OSCC cells, including DOK, FaDu, HSC‐3, OEC‐M1, SCC‐4, SCC‐9, SCC‐15, SCC‐25 and Tu183, were routinely cultured as previously described (Yen et al., 2014). All cells were cultured at 37 °C in a 5% CO2 atmosphere and maintained in 10% Fetal bovine serum (FBS, Kibbutz BeitHaemek, Israel) within 3 months of resuscitation from the frozen aliquots, with lower than 20 passages for each experiment.
2.3. RNA extraction and reverse‐transcriptase PCR (RT‐PCR)
Total RNA was isolated from OSCC cell lines with the use of TRIzol reagent (Life Technologies, Gaithersburg, MD) and cDNA was synthesized using random hexamer primers and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) to produce a template suitable for PCR. The PCR reactions were run on a Biometra T3000 thermocycler (Biometra GmbH, Germany). The amplification consisted of a 5 min denaturation at 95 °C, followed by 35–40 cycles of denaturation at 94 °C for 30 s, annealing at 53–55 °C for 30 s, extension at 72 °C for 30 s and one cycle of a final extension step for 10 min at 72 °C. PCR products were subjected to electrophoresis on 1% agarose gel and visualized on UVP GDS‐8000 Bioimaging System (UVP, 11th Street Upland, CA, USA). The level of glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was determined as an internal control. The primer sequences were used as followed: 5′‐TTAAA AGAGG CACCA GAAGG AAC‐3′ (forward) and 5′‐AGTCG ATCAG ATGAA CCACA CT‐3′ (reverse) for SOCS2 expression, 5′‐CAG TGG TTT GAC GGG GTG AT‐3′ (forward) and 5′‐GTC GTG GGC CTG TTG CTT AT‐3′ (reverse) for STAT5 expression, 5′‐CAC TGA GGG CCG CAC GGA T‐3′ (forward) and 5′‐CTT GAT GTC ATC CTG GGA CA‐3′ (reverse) for matrix metalloproteinase (MMP)‐2 expression, 5′‐CAA CA T CAC CTA TTG GAT CC‐3′ (forward) and 5′‐GGG TGT AGA GTC TCT CGC TG‐3′ (reverse) for MMP‐9 expression, 5′‐GAA GGT GAA GGT CGG AGT‐3′ (forward) and 5′‐GAA GAT GGT GAT GGG ATT TC‐3′ (reverse) for GAPDH expression.
2.4. Quantitative real‐time PCR (qRT‐PCR)
For mature miRNA detection, RNA samples extracted from tissues or cells were performed with miRNA‐specific primers and TaqMan Universal PCR Master Mix (Applied Biosystems) with RUN44 small nuclear RNA as an internal normalized reference. For MMP‐2, MMP‐9, SOCS2 mRNA and primary miRNA detection, reverse‐transcription reactions were performed using miScript II RT kit (Qiagen, Hilden, Germany) with GAPDH as an internal normalized reference. The primer sequences were used for PCR reaction as followed: 5′‐CCG CAG TGA CGG AAA GAT GT‐3′ (forward) and 5′‐GCC CCA CTT GCG GTC AT‐3′ (reverse) for MMP‐2 expression, 5′‐GGA CGA TGC CTG CAA CGT‐3′ (forward) and 5′‐CAG TAC TTC CCA TCC TTG AAC AAA‐3′ (reverse) for MMP‐9 expression, 5′‐TTT ATT CAC CCG CAG GTA CCC C‐3′ (forward) and 5′‐GCA GAC CCC ACC TTC TAC CT‐3′ (reverse) for primary miR‐424 expression, 5′‐CAG ATG TGC AAG GAT AAG CGG‐3′ (forward) and 5′‐GCG GTT TGG TCA GAT AAA GGT G‐3′ (reverse) for SOCS2 expression, 5′‐GAA GGT GAA GGT CGG AGT‐3′ (forward) and 5′‐GAA GAT GGT GAT GGG ATT TC‐3′ (reverse) for GAPDH expression. All qRT‐PCR reactions were run on an Applied Biosystems StepOne Plus real‐time PCR system. The expression level was defined based on the threshold cycle, and relative expression levels were calculated as △△Ct after normalization with reference control.
2.5. Plasmids
For gene expression, a 597‐base pair (bp) fragment of SOCS2 was amplified by PCR and cloned into the BamHI and EcoRI sites of the pcDNA3.1+ vector (Invitrogen, Gaithersburg, MD), using the SOCS2 cloning primers: 5′‐GGG GAT CCA TGA CCC TGC GGT GCC‐3′ (forward) and 5′‐GGG AAT TCT TAT ACC TGG AAT TAT ATT CTT CCA A‐3′ (reverse). For gene knockdown experiments, the shRNA clones for SOCS2, STAT5 and their control pLKO_TRC vector (NS) were obtained from the National RNAi Core Facility (Academia Sinica, Taiwan). For luciferase reporter assay, the SOCS2 3′‐UTR region was generated by subcloning PCR‐amplified full‐length human SOCS2 cDNA (1545 bp from the stop codon respectively) into the Nhe1/Sal1 sites of pmiRGLO firefly luciferase‐expressing vector (Promega, WI, USA), using the primers: 5′‐GGG CTA GCA TGT TTC TCT TTT TTT AAA C‐3′ (forward) and 5′‐GGG TCG ACA CAA TTT ATA ACT GAA TTT TCT CAT TC‐3′ (reverse). The miRNA binding sites mutation reporters were constructed by using Site‐Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with primers 5′‐ACT GTA ATT TGA TAG GTT GGA TGA TCA GAG TCT CTT GGG CAT TTT ATA TTT TG‐3′ (forward) and 5′‐CAT CCA ACC TAT CAA ATT ACA GTG AGG CCT GTG TCA GCT TGG TT‐3′ (reverse) for site 1, 5′‐TTT AAA CTG TGT ATG TAC TAC TAT TAC TAT TTG ATT AGA ATG TAT TAA ATA AAA AAA ACC TG‐3′ (forward) and 5′‐TAA TAG TAG TAC ATA CAC AGT TTA AAA AAC AGG CAG GTA TCC CCC TGT‐3′ (reverse) for site 2.
2.6. Transfection experiments
MiR‐424‐5p mimics (PM), miR‐424‐5p inhibitors (AM) and the respective scrambled controls (NC) were obtained from Dharmacon (Lafayette, USA). The transfection was performed using Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer's instructions. For transfection of the other plasmids, cells were transiently transfected with 2 μg of plasmids using Lipofectamine 2000 from Invitrogen (CA, USA) according to the manufacturer's protocol.
2.7. Protein extraction and western blot analysis
Total proteins were extracted from cultured cells in a lysis buffer containing 50 mM Tris–HCl, 1% NP‐40, 150 mM NaCl, 0.1% SDS, 1 mM PMSF, 1 mM Na3VO4 and 1 μl protease inhibitor cocktail (Sigma‐Aldrich, Inc.). Protein concentrations were then determined by the BCA assay kit (Thermo, USA) with bovine serum albumin as standard. Equal amount of protein lysates were subjected to 10–12% SDS polyacrylamide gels and transferred to poly‐vinylidene fluoride (PVDF) membrane (Pall Life Sciences, Glen Cove, NY). The membranes were probed with specific antibodies against SOCS2, STAT5 (GeneTex, USA), STAT1, phospho‐STAT1 (pY701) (Epitomics Inc., Burlingame, CA), STAT3, phospho‐STAT3 (pY705), phospho‐STAT5 (pY694) (Cell signaling, USA) and α‐tubulin (Thermo, USA) as an internal control. Signals from HRP‐coupled secondary antibodies were visualized by enhanced chemiluminescence (ECL) detection system (PerkinElmer, Waltham, MA) and chemiluminescence was exposed onto Kodak X‐Omat film (Kodak, Chalon/Paris, France). Protein levels were determined as the integrated area (pixels) of the band intensities by densitometry analysis with Image J software (Bethesda, MD, USA). The numerical values for protein band intensities were corrected with the values for the α‐tubulin bands.
2.8. Luciferase reporter assays
DOK and SCC‐15 cells were cultured in 24‐well plates and co‐transfected with 300 ng of SOCS2 3′‐untranslated region (UTR) wild‐type or mutant‐type pmirGLO reporter plasmid and with 25 nM of miR‐424‐5p mimics (PM) or control oligonucleotide (NC) with Lipofectamine 2000 according to the manufacturer's instructions. The luciferase assay was performed 48 h post transfection with Dual Luciferase Reporter Assay System (Promega, USA) as described by the manufacturer's protocol. Luminometry readings were obtained using an Orion L luminometer (Berthold).
2.9. Cell migration and invasion assay
The migration and invasion assays were performed using 24‐well Fluoro‐Blok insert‐based assays system (8 μm pore size membrane; BD Biosciences, Franklin Lakes, NJ). The culture insert was coated to a density of 40 μg/insert of Matrigel Basement Membrane Matrix (BD Biosciences) for invasion assay, but was coated nothing for migration assay. Subsequently, 5 × 104 cells were seeded in the upper chamber in culture medium containing 10% NuSerum. After incubating 24 h for invasion assay and 12 h for migration assay at 37 °C, the cells that passed through the Fluoro‐Blok membrane were fixed with 95% methanol and stained with propidium iodine. The fluorescence images were then counted with Analytical Imaging Station software package (Imaging Research, Ontario, Canada).
2.10. Cytokines treatment
Cells were seed at a density of 5 × 104 cells/well in 6‐well plates and maintained in culture medium containing 10% FBS for 24 h. Then, cells were treated with serum free medium including 10 ng/ml indicated cytokines, such as IL‐6 (R&D Systems, USA), IL‐8, IL‐10 or oncostatin M (OSM) (Peprotech, Rocky Hill, NJ, USA) for 0–48 h. After washed with PBS, the treated cells were lysed for quantitative real‐time PCR or western blot analysis.
2.11. Immunohistochemistry
Multiple head and neck cancer tissue array (# HN803b, US Biomax Inc., Rockville, MD) sections were deparaffinized and rehydrated through graded alcohols and rinsed with phosphate‐buffered saline solution. For heat‐induced antigen retrieval, slides were soaked in citric acid buffer (pH 6.0) and heated till 95 °C for 20 min. After quenching endogenous peroxidase activity with Peroxidase 1 (Biocare, CA, USA), sections were blocked and then incubated with anti‐human SOCS2 polyclonal antibody (Ab74533, 1:100) (Abcam, Cambridge, MA) at 4 °C overnight. Specific signals were then developed with LSAB+ kit (DakoCytomation, CA, USA) using diaminobenzidine (Biocare) as chromogen. Sections were then counterstained with hematoxylin and observed under light microscope. This multiple head and neck carcinoma tissue array contains 22 cases of tongue squamous cell carcinoma, 31 cases of larynx squamous cell carcinoma, 7 cases of nose squamous cell carcinoma, 1 case of carcinoma sarcomatodes, 8 cases of metastatic carcinoma, 9 cases of normal tongue tissue and 2 cases of adjacent normal tissue. We used 9 cases of normal tongue tissue and 22 cases of tongue squamous cell carcinoma to represent the oral cavity specimens. Tumor SOCS2 level was scored according to SOCS2 staining intensity as follows: 0 = negative, 1 = mild, 2 = moderate and 3 = intense. Two pathologists independently assessed all the scorings. Patients were then further subdivided into the low‐SOCS2 expression group (score 0, 1) and the high‐SOCS2 expression group (score 2, 3) according to their tumor SOCS2 scores.
2.12. Drug treatment
Cells were seed at a density of 2 × 105 cells/well in 6‐well plates. After 16 h, the cells were treated with 10 μg/ml of cycloheximide (cat# C7698, Sigma, MO, USA) or 10 μM of MG132 (cat# M7449, Sigma, MO, USA) for indicated times. Total proteins extraction was performed as described in “Materials and Methods” Section 2.7.
2.13. Statistical analysis
All data are expressed as the mean ± standard error (SE) from at least three independent experiments. Linear correlation and Pearson correlations were used to evaluate the correlation between 2 variants. Differences with the various treatment groups were assessed using the Student's t‐test or one‐way analysis of variance (ANOVA) analysis with multiple comparisons in cases in which more than 2 conditions were compared on the same set. Calculations were performed using Graph Pad Prism Ver. 4.01 (San Diego, CA). A p‐value < 0.05 was considered as significant.
3. Results
3.1. Downregulation of SOCS2 in human OSCC in comparison with adjacent non‐tumor tissues
In order to examine the expression profile of SOCS2 in OSCC patients, we analyzed microarray data generated from 40 pairs of OSCC specimens and their corresponding non‐tumorous epithelia (Shiah et al., 2014). The data showed that SOCS2 levels were significantly downregulated in tumors compared with their corresponding normal samples (p < 0.0001), but there was no significant correlation between SOCS2 expression and stages of the tumor (Figure 1A). We next analyzed the expression profile of SOCS2 in different tumors employing publicly available microarray datasets. From NCBI GEO database, SOCS2 expression in 22 matched pairs of squamous cell carcinoma was downregulated in tumors compared with their adjacent corresponding normal tissues (Supplementary Figure 1A). From another Oncomin database, the SOCS2 expression was also significantly decreased in other types of cancer, including breast, colon and lung tumors (Supplementary Figure 1B–D). Using qRT‐PCR to validate the mRNA expression pattern in tumors versus their normal adjacent tissues, similar results were observed for another OSCC cohort samples (Supplementary Figure 1E). To confirm this trend in SOCS2 protein level, we assessed SOCS2 expression by immunohistochemistry in normal tissue and OSCC biopsies from individual cases using tissue arrays (Figure 1B). We found that SOCS2 expression levels were high in 88.9% of normal samples (staining intensity: score 0 = 0%, score 1 = 11.1%, score 2 = 22.2%, score 3 = 66.7%), whereas only 13.6% showed a high stain signal in tumor samples (staining intensity: score 0 = 36.4%, score 1 = 50.0%, score 2 = 9.1%, score 3 = 4.5%) (Figure 1C), and these data were consistent with our qRT‐PCR results. Findings from patient specimens suggest that the decrease of SOCS2 is correlated with OSCC tumor progression.
Figure 1.
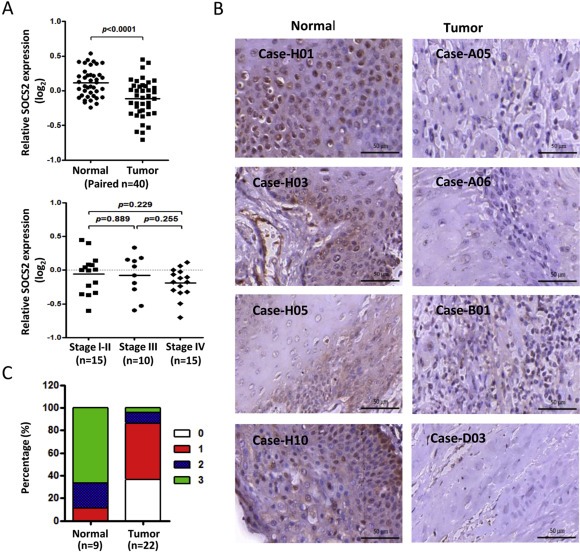
The aberrant expression of SOCS2 in OSCC. A: Microarray analysis of SOCS2 expression level in OSCC tumors (n = 40) compared with their own adjacent normal tissues or compared with patients' stage (one‐way ANOVA, p = 0.3779). SOCS2 expression levels are expressed as the log2 ratios. B: Immunohistochemical analysis of SOCS2 in oral cancer tissue microarray. Bars in the right lower corners of all photos are equivalent to 50 μm. C: Bar charts show the percentage of SOCS2 staining score for the oral specimens in the tissue microarray, including 9 cases of normal tissue and 22 cases of OSCC. The SOCS2 staining intensity was scored as follows: 0 = negative, 1 = mild, 2 = moderate and 3 = intense.
3.2. SOCS2 mediates cellular functions through STAT5 inhibition
To test whether SOCS2 contributes to the mechanisms that control OSCC tumor progression, we then evaluated the functional role of SOCS2 on the invasiveness of oral cancer cells. We first ectopically expressed SOCS2 in OEC‐M1 and SCC‐9 cells, which exhibit low levels of endogenous SOCS2 expression. The results showed that SOCS2 protein expression was significantly elevated in OEC‐M1 and SCC‐9 cells, and the expression of SOCS2 strongly reduced STAT5 phosphorylation (Figure 2A) and its transcriptional activity (Figure 2B). Complementarily, we silenced SOCS2 in DOK and SCC‐15 cells using SOCS2‐specific lentiviral shRNA construct (shSOCS2) and found that shRNA‐mediated silencing of ectopic SOCS2 increased STAT5 protein level and its phosphorylation (Figure 2C). However, overexpression of SOCS2 did not change the protein levels and phosphorylation of STAT1 and STAT3 (Figure 2A), indicating that STAT5 activity is primarily subject to negative regulation by SOCS2 in OSCC cells. To determine whether SOCS2 modulates OSCC cell invasion, we performed a Boyden Chamber invasion assay using matrigel as the basement membrane. We observed a significant decrease in the invasive activity of OEC‐M1 cells with SOCS2 overexpression as well as with STAT5 silencing (shSTAT5) (Figure 2D). Because SOCS2 and STAT5 have been reported to modulate MMP‐2 transcription in association with invasive behavior (Gutzman et al., 2007; Sato et al., 2014), we examined the effect of SOCS2 overexpression and STAT5 silencing on MMP‐2 transcription. As shown in Figure 2E and Supplementary Figure 2A, both SOCS2 overexpression and STAT5 silencing resulted in significant decreases in MMP‐2 transcripts in OEC‐M1 and SCC‐9 cells. Collectively, these observations suggested that overexpression of SOCS2 suppresses STAT5 activity and MMP‐2 expression and consequently decreases the invasion properties in OSCC cells.
Figure 2.
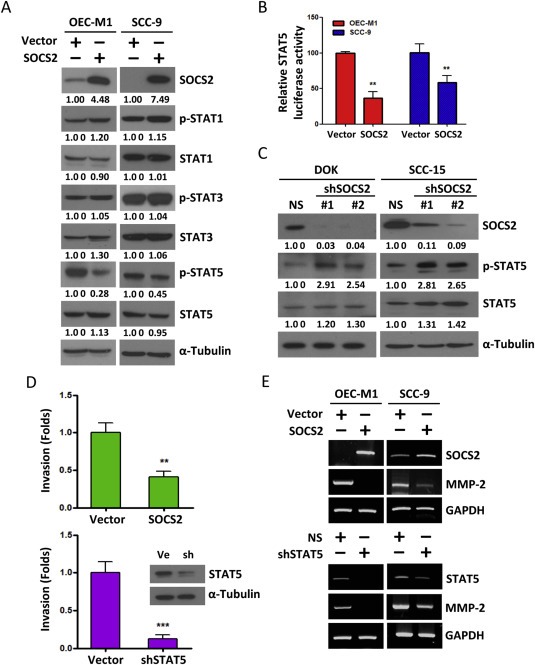
The effects of dysregulated SOCS2 in STAT5 activity and invasion ability. A: Western blot analysis of the STAT1, 3, 5 and their phosphorylation form after transfection of SOCS2 for 48 h in OEC‐M1 and SCC‐9 cells. α‐Tubulin was used as protein loading control. Numerical values for protein band intensities are shown below the gels. The values were quantitated by densitometry and normalized to α‐tubulin. B: The effect of SOCS2 on the transcriptional activity of the construct (pGL3) containing the STAT5 binding sequence. The relative luciferase activities are the ratios of Renilla luciferase normalized to the control mimics. The data are represented as mean ± SD; **, p < 0.01 versus vector control. C: Whole cell lysates from DOK and SCC‐15 cells transfected with plasmids expressing two target‐specific SOCS2 shRNA (#1 or #2, respectively) or non‐targeting shRNA plasmid (NS) were analyzed for STAT5 and phosphor‐STAT5 protein for 48 h by western blot. α‐Tubulin was used as protein loading control. The numerical values for protein band intensities were corrected with the values for the loading control α‐tubulin bands. D: Relative invasion ability of OEC‐M1 cells transfected with SOCS2 or shSTAT5 plasmids for 24 h. The data are represented as mean ± SE; **, p < 0.01; ***, p < 0.001 versus vector control. Western blot data showed that STAT5 protein level in OEC‐M1 cells after transfecting the shSTAT5 plasmid (sh) or control vector (Ve). E: RT‐PCR analysis of MMP‐2 transcript after transfection of SOCS2 or shSTAT5 plasmids for 48 h in OEC‐M1 and SCC‐9 cells. GAPDH was used as an internal control.
3.3. SOCS2 is a direct target of miR‐424‐5p
To test whether SOCS2 was targeted by miRNAs, we used targeting algorithms (miRNAmap and microRNA.org) combined with OSCC patients' miRNA microarray data (Shiah et al., 2014) to search for putative miRNAs that might bind to SOCS2 mRNA. Based on the computational screening (Figure 3A, left), we identified one miRNA, miR‐424‐5p, which was significantly upregulated in OSCC tumors compared with their corresponding normal samples (p < 0.0001) (Figure 3B) and could potentially target SOCS2 with two predicted miRNA binding sites in the 3′‐UTR of SOCS2 (Figure 3A, right). We next analyzed another OSCC testing cohort using qRT‐PCR approach and found a significant inverse correlation between the expression levels of SOCS2 and miR‐424‐5p (r = −0.3999, p < 0.001) (Figure 3C). To determine whether miR‐424‐5p could directly target SOCS2, we performed luciferase reporter assay in DOK and SCC‐15 cells. The Renilla luciferase activity of the reporter that contained SOCS2 3′‐UTR was significantly suppressed by miR‐424‐5p mimic (PM) transfection, but the activity of the reporter that contained SOCS2 mutant 3′‐UTR had no significant change (Figure 3D). Furthermore, transfection of miR‐424‐5p mimics (PM) in DOK and SCC‐15 cells not only decreased SOCS2 protein expressions but also increased STAT5 protein expression and phosphorylation in a dose‐dependent manner (Figure 3E and F). By contrast, transfection of miR‐424‐5p inhibitors (AM) increased SOCS2 protein in OEC‐M1 and SCC‐9 cells. In addition, miR‐424‐5p inhibitors (AM) decreased STAT5 protein expression and phosphorylation (Figure 3G and H). Nonetheless, due to the small sample size, we found no significant correlation between miR‐424‐5p expression and stages of the OSCC tumor (Supplementary Figure 3).
Figure 3.
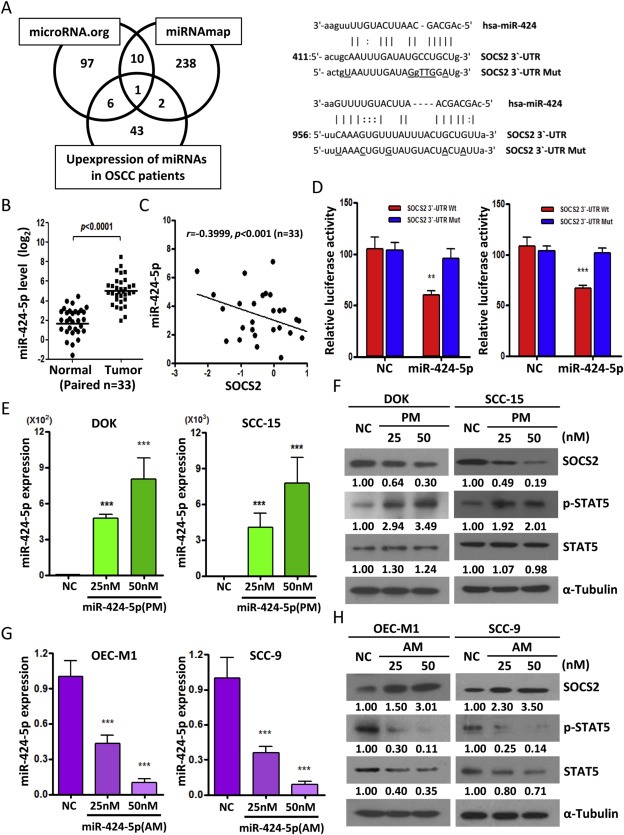
SOCS2 is a direct target of miR‐424‐5p. A: Left, Venn diagram of predicted miRNA which targets SOCS2 3′‐UTR in silico using two independent algorithms (microRNA.org and miRNAmap) combined with our patients' miRNA array data (GSE45238). Right, Schematic representation of the putative miR‐424‐5p binding sequence in the 3′‐UTR of SOCS2 with wild‐type form (SOCS2 3′‐UTR Wt) and mutant form (SOCS2 3′‐UTR Mut). The mutated nucleotides are labeled with underline. B: qRT‐PCR analysis to validate the miR‐424‐5p expression pattern in tumors versus their normal adjacent tissues for another OSCC cohort samples (n = 33). C: Correlation analysis of miR‐424‐5p and SOCS2 in human OSCC patients (n = 33) by qRT‐PCR analysis. D: The effect of miR‐424‐5p mimics (PM, 25 nM) on the luciferase activities of the constructs containing the wild‐type or mutant‐type 3′‐UTR in DOK (left) and SCC‐15 (right) cells. The relative luciferase activity of each sample is measured at 48 h after transfection and normalized to Renilla luciferase activity. E: qRT–PCR analysis showing the expression level of miR‐424‐5p in DOK and SCC‐15 cell lines after transfection with 25 nM or 50 nM of miR‐424‐5p mimics (PM) for 48 h. F: Western blot analysis of the SOCS2, STAT5 and phosphor‐STAT5 after transfection of miR‐424‐5p mimics (PM) with 25 nM or 50 nM for 48 h in DOK and SCC‐15 cells. G: qRT–PCR analysis showing the expression level of miR‐424‐5p in OEC‐M1 and SCC‐9 cells after transfection with indicated concentration of miR‐424‐5p inhibitors (AM) for 48 h. H: Western blot analysis of the SOCS2, STAT5 and phosphor‐STAT5 after transfection of miR‐424‐5p inhibitors (AM) with indicated concentration for 48 h OEC‐M1 and SCC‐9 cells. All data are presented as mean ± SE; **, p < 0.01; ***, p < 0.001 versus scramble control (NC). α‐Tubulin was used as protein loading control. Numerical values for protein band intensities are shown below the gels. The values were quantitated by densitometry and normalized to α‐tubulin.
However, miR‐424‐5p may regulate SOCS2 protein level through changing the protein degradation rate. To address this question, first, we examined the effect of proteasome inhibition on the stability of SOCS2. SCC‐15 (high expression of SOCS2) and OEC‐M1 (low expression of SOCS2) cells were treated with or without proteasome inhibitor MG132 and collected protein lysate at 12 h. The protein level of SOCS2 was accumulated in the presence of MG132 (Supplementary Figure 4A), indicating that the proteasome is required for degradation of SOCS2. Next, SCC‐15 and OEC‐M1 cells were treated with cycloheximide, an inhibitor of protein synthesis. As shown in Supplementary Figure 4B, the half‐life of SOCS2 protein was about 0.5–1 h and was no difference in SCC‐15 and OEC‐M1 cells, suggesting that the differential SOCS2 protein levels are not caused by protein degradation activity. Furthermore, SCC‐15 cells were transfected with miR‐424‐5p mimics or scramble control (NC) for 24 h and then treated with cycloheximide. We found that the half‐life of SOCS2 protein just slight shifted forward about 15 min by miR‐424‐5P‐p treatment, suggested that miR‐424‐5p does not significant affect the SOCS2 protein stability.
To consolidate our findings, we also correlated the expression level of miR‐424‐5p (Supplementary Figure 5A) to SOCS2 mRNA (Supplementary Figure 5B) using OSCC cell lines and found a strong inverse correlation (r = −0.82, p = 0.0068) between the expression levels of miR‐424‐5p and SOCS2 (Supplementary Figure 5C). Taken together, our results demonstrate that SOCS2 is a direct target of miR‐424‐5p. Overexpression of miR‐424‐5p in OSCC cells decreases SOCS2 protein expression and increases STAT5 activity and protein level, suggesting a direct interaction between miR‐424‐5p and STAT5 signaling pathway.
3.4. miR‐424‐5p promotes OSCC cell invasion and migration through direct inhibition of SOCS2 expression
Next, we attempted to investigate whether miR‐424‐5p‐induced SOCS2 downregulation plays a major role in enhancing STAT5 signaling. For this purpose, we generated a vector‐based SOCS2 expression plasmid lacking 3′‐UTR sequence, and thus the SOCS2 is not targeted by miR‐424‐5p. Cells transfected with SOCS2 plasmid without 3′‐UTR were able to reduce STAT5 phosphorylation and protein level in the presence of miR‐424‐5p mimics (Figure 4A). Moreover, SOCS2 restoration was able to reduce the expression of canonical STAT5‐transcriptional targets, such as MMP‐2 and MMP‐9 (Figure 4B and Supplementary Figure 2B). Because MMP‐2 and MMP‐9 are important executors in cell migration and invasion, we further assessed the effect of SOCS2 on the in vitro invasion potential of SCC15 cells by Matrigel‐based transwell assay. As expected, the restoration of SOCS2 potently suppressed the migration and invasion activities induced by miR‐424‐5p overexpression (Figure 4C and D). These data demonstrate that miR‐424‐5p‐mediated SOCS2 functions are crucial in regulating STAT5 activity, MMPs secretion, and the migration and invasive potential in OSCC cells.
Figure 4.
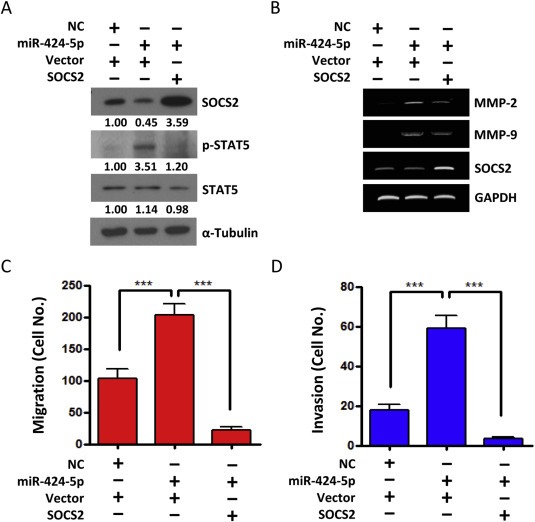
miR‐424‐5p promotes invasion and migration through SOCS2. SCC‐15 cells were transfected with miR‐424‐5p mimics or scramble control (NC) for 24 h and then transfected with SOCS2 expression vector (without 3′‐UTR) or control vector for another 24 h. A: Western blot analysis of the SOCS2, STAT5 and phosphor‐STAT5. α‐Tubulin was used as protein loading control. Numerical values for protein band intensities are shown below the gels. The values were quantitated by densitometry and normalized to α‐tubulin. B: RT‐PCR analysis of SOCS2, MMP‐2 and MMP‐9. GAPDH was used as an internal control. C: Migration assay and D: Invasion assay using Boyden Chamber system. All data are presented as mean ± SE; ***, p < 0.001, one‐way ANOVA p < 0.0001.
3.5. IL‐8 induce miR‐424‐5p expression through activation of STAT5
The JAK/STAT pathway is activated upon binding of cytokines to their receptors. Growing evidence suggests that STAT5 can be activated by IL‐6, IL‐8, IL‐10 and Oncostatin M (OSM) (Britschgi et al., 2012; Klausen et al., 2000; Nishioka et al., 2014). Since miRNAs are often reported to be components of feedback loops in cytokines mediated signaling (Collins et al., 2013; Rokavec et al., 2014), we hypothesized that miR‐424‐5p may involve in the cytokine‐induced STAT5/SOCS2 pathway. To test this hypothesis, we compared the abilities of IL‐6, IL‐8, IL‐10 and OSM to induce the miR‐424‐5p expression in OSCC cells. We found that only IL‐8 significantly induced miR‐424‐5p expression in SCC‐15 (Figure 5A) and DOK (Supplementary Figure 6A) cells. Under IL‐8 treatment, a decrease of SOCS2 protein and an increase of phosphor‐STAT5 were also detected at 24 h and up to 48 h (Figure 5B and Supplementary Figure 6B). Furthermore, IL‐8 suppressed the SOCS2 activity in a dual luciferase reporter assay when SCC‐15 cells were transfected with SOCS2 3′‐UTR reporter constructs, whereas the effect was abrogated when the mutant construct lacking the miR‐424‐5p binding sites was employed (Figure 5C and Supplementary Figure 6C). To further determine whether IL‐8‐induced miR‐424‐5p transcription is involved in the activation of STAT5, we analyzed primary miR‐424‐5p expression in SCC‐15 cells in response to IL‐8 stimulation and found a time‐dependent increase in the expression of primary miR‐424 with IL‐8 exposure (Figure 5D). More importantly, shRNA‐mediated downregulation of STAT5 prevented the induction of primary miR‐424 (Figure 5E) and mature miR‐424‐5p expression (Figure 5F) after IL‐8 exposure, demonstrating that STAT5 mediates the IL‐8‐induced transcription of miR‐424. Similar results were also observed in the DOK cells (Supplementary Figure 6D–F). Moreover, nuclear factor‐κB (NF‐κB) is known to be one of the principal downstream transcriptional factors of IL‐8 signaling (Manna and Ramesh, 2005). After treatment of SCC‐15 cells with NF‐κB inhibitor or STAT5 inhibitor blocked the IL‐8‐induced primary miR‐424 expression (Figure 5G). Interestingly, suppression of NF‐κB activity only resulted in a moderate decrease of IL‐8‐induced miR‐424‐5p expression, however, suppression of STAT5 activity dramatically reduced the IL‐8‐induced miR‐424‐5p expression (Figure 5H), suggesting that STAT5 is a major downstream effector for mature miR‐424‐5p formation. Taken together, these data demonstrated that SOCS2 mRNA is directly targeted by miR‐424‐5p upon IL‐8 treatment. They also indicate that STAT5 activation is required for IL‐8‐mediated miR‐424‐5p expression, from which the suppression of SOCS2 expression and subsequent activation of STAT5 are maintained.
Figure 5.
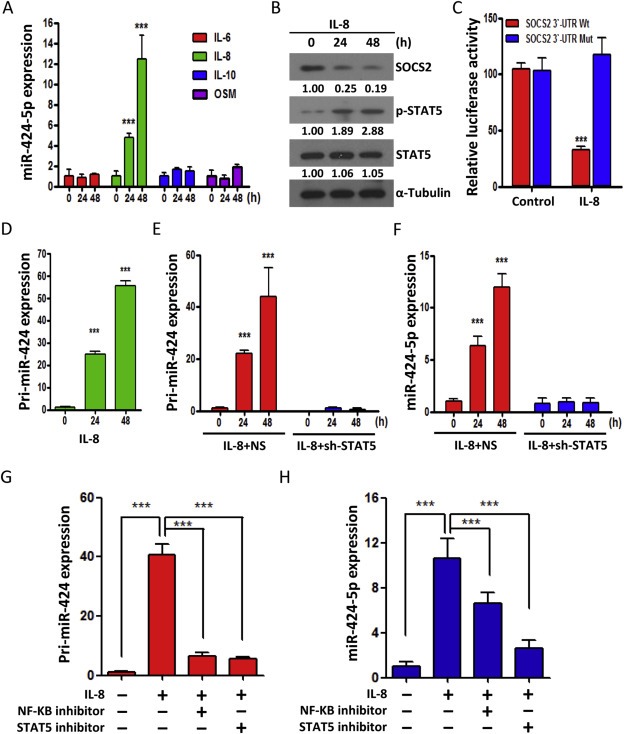
miR‐424‐5p disrupts IL‐8/STAT5/SOCS2 feedback loop. A: qRT–PCR analysis showing the expression level of miR‐424‐5p in SCC‐15 cells treated with various cytokines, including IL‐6, IL‐8, IL‐10 and OSM (each for 10 ng/ml) for indicated time periods. B: Western blot analysis of the SOCS2, STAT5 and phosphor‐STAT5 in SCC‐15 cells after addition of IL‐8 (10 ng/ml) for 24 and 48 h. The numerical values for protein band intensities were corrected with the values for the loading control α‐Tubulin bands. C: The effect of IL‐8 (10 ng/ml) on the luciferase activities of the constructs containing the wild‐type or mutant‐type SOCS2 3′‐UTR in SCC‐15 cells. The relative luciferase activity of each sample is measured at 48 h after transfection and normalized to Renilla luciferase activity. D: qRT–PCR analysis of primary miR‐424 (pri‐miR‐424) expression in SCC‐15 cells treated with IL‐8 (10 ng/ml). E–F: qRT–PCR analysis of primary miR‐424 (pri‐miR‐424) (E) and mature miR‐424 (miR‐424‐5p) (F) in SCC‐15 cells transfected with control (NS) or STAT5 shRNAs for 24 h and subsequently treated with IL‐8 for 24 or 48 h. G–H: qRT–PCR analysis of primary miR‐424 (pri‐miR‐424) (G) and mature miR‐424 (miR‐424‐5p) (H) in SCC‐15 cells treated with NF‐kB inhibitor or STAT inhibitor for 16 h and subsequently treated with IL‐8 (10 ng/ml) for 48 h. All data are presented as mean ± SE; ***, p < 0.001, one‐way ANOVA p < 0.0001.
3.6. IL‐8 promotes OSCC cell invasion and migration through direct induction of miR‐424‐5p
To determine whether miR‐424‐5p is required for IL‐8‐mediated cell migration and invasion, the endogenous miR‐424‐5p was downregulated in SCC‐15 and DOK cells with miR‐424‐5p inhibitor (AM424) followed by IL‐8 treatment. The results showed that miR‐424‐5p inhibitor not only restored the SOCS2 protein levels that suppressed by IL‐8 but also decreased the IL‐8‐induced STAT5 protein expression and phosphorylation (Figure 6A). MiR‐424‐5p inhibitor also reduced the IL‐8‐induced MMP‐2 expression (Figure 6B and Supplementary Figure 2C). Furthermore, the IL‐8‐induced tumor cell migration and invasion were prevented when miR‐424‐5p was specifically inhibited by an antagomir (AM424) (Figure 6C–D), indicating that induction of miR‐424‐5p is essential for IL‐8‐induced cellular invasiveness.
Figure 6.
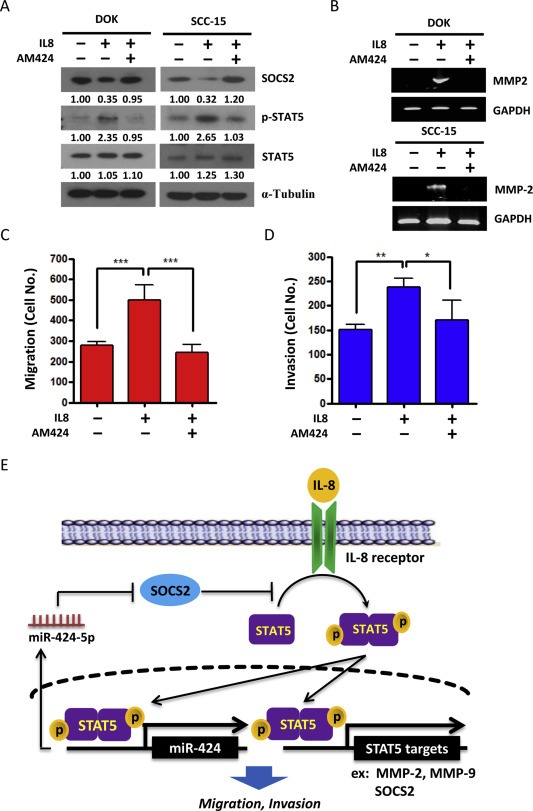
IL‐8 induces invasion and migration of OSCC cells through induction of miR‐424‐5p. DOK and SCC‐15 cells were transfected with miR‐424‐5p specific inhibitors (AM424) for 24 h, followed by addition of IL‐8 (10 ng/ml) for 48 h. A: Western blot analysis of the SOCS2, STAT5 and phosphor‐STAT5. α‐Tubulin was used as protein loading control. The numerical values for protein band intensities were corrected with the values for the loading control α‐tubulin bands. B: RT‐PCR analysis of MMP‐2 expression. GAPDH was used as an internal control. C: Migration assay using Boyden Chamber system. The data are represented as mean ± SE; ***, p < 0.001 versus vector control, one‐way ANOVA p = 0.0067. D: Invasion assay using Boyden Chamber system. The data are presented as mean ± SE; *, p < 0.05; **, p < 0.01; one‐way ANOVA p = 0.0022. E: Schematic representation of the proposed hypothesis that miR‐424‐5p disrupts IL‐8/STAT5/SOCS2 feedback loop in OSCC model.
4. Discussion
In general, cytokine binding to cognate trans‐membrane receptors results in activation of intracellular JAK/STAT signaling cascade, which culminates in appropriate cellular responses such as growth, development and immune reactions (Croker et al., 2008). However, the JAK/STAT pathway requires precise cellular control and loss of the tightly regulation can promote the inflammatory disease and tumorigenesis. In normal physiological condition, the expression of SOCS proteins can be induced by cytokine stimulation and to inhibit the JAK/STAT signaling in a negative feedback loop mechanism. In pathological or cancer disease states, dysregulation of the JAK/STAT negative feedback loop in OSCC may be due to two molecular events: i) constitutive activation by cytokine stimulation (Grandis et al., 2000; Song and Grandis, 2000); ii) loss of SOCS proteins expression (Huang et al., 2013; Zhang et al., 2013). IL‐8 is an important cytokine involved in tumor growth and metastasis, and could thus contribute to the pathogenesis in oral cancer (Chen et al., 1999). Therefore, IL‐8 is not only a prognostic marker but also a factor that may contribute to a poor prognosis for OSCC (Watanabe et al., 2002; St John et al., 2004). However, the detail mechanisms of the IL‐8 involved in oral carcinogenesis remain to be elucidated.
In this study, we found that miR‐424‐5p functions as a regulator of the IL‐8/STAT5/SOCS2 feedback loop and, as such, controls the tumor progression in OSCC cells. As shown in Figure 6E, IL‐8 binding to its receptor triggers the STAT5 activation, which then increases the expression of downstream target genes, including MMP‐2, MMP‐9 and SOCS2. SOCS2 acts as a STAT5 inhibitor, which can attenuate the IL‐8‐induced STAT5 activity. However, in cancer cells, STAT5 activation simultaneously induces the expression of miR‐424‐5p, which directly targets to SOCS2. The miR‐424‐5p‐mediated reduction of SOCS2 then leads to continuous enhancement of STAT5 activation and its target gene expression, ultimately leading to malignant cellular behavior.
SOCS2 expression has been found to be downregulated with diverse cancers (Trengove and Ward, 2013) and low expression of SOCS2 protein is associated with a decreased cell proliferation rate and tumor progression (Farabegoli et al., 2005; Iglesias‐Gato et al., 2014). Functional studies have shown that transgenic mice with SOCS2 alleles disruption exert an increase in the spontaneous development of tumors (Newton et al., 2010), strongly suggesting the tumor suppressive activity of SOCS2. Here we demonstrate that SOCS2 expression is significantly downregulated in OSCC patient samples compared with their corresponding normal tissues (p < 0.0001). As expected, overexpression of SOCS2 inhibits MMP‐2 expression in OSCC cells as well as their invasion ability, confirming that SOCS2 acts as a tumor suppressor. However, we failed to detect significant changes in SOCS2 expression between different OSCC stages. Similar results were observed in another OSCC cohort samples which suggest that SOCS2 may not be an appropriate prognostic biomarker, but could be a potential diagnostic biomarker for OSCC patients.
SOCS proteins function as classic negative regulators of JAK/STAT feedback loop downstream of cytokine signaling (Croker et al., 2003; Vlotides et al., 2004). SOCS2 overexpression has been reported to inhibit STAT3 and STAT5 activity (Yang et al., 2012). In our experiments, overexpression of SOCS2 protein did inhibit STAT5 phosphorylation but not STAT1 or STAT3 phosphorylation. Furthermore, SOCS2 overexpression not only reduced the STAT5 transcriptional activity but also the MMP‐2 expression and invasive ability, suggesting that SOCS2‐modulated STAT5 signaling plays an import role in OSCC invasion.
Otherwise, STAT5 is an oncogene as well as a key transcriptional factor for SOCS2 gene expression (Vidal et al., 2007). STAT5 activity is known to be correlated with SOCS2 expression in a cytokine‐dependent manner (Quentmeier et al., 2008). However, SOCS2 levels are significantly lowered in most cancers (Trengove and Ward, 2013), indicating the existence of unknown mechanisms that impede SOCS2 expression in OSCC tumorigenesis. One possibility is that SOCS2 gene is epigenetic silencing by hypermethylation of CpG dinucleotides in its promoter region in various cancer cells and cancer cell lines (Fiegl et al., 2004; Liu et al., 2008; Sutherland et al., 2004). Treatment of colon cancer cells with 5‐aza‐dC (a DNA methyltransferase inhibitor) has been shown to increase the basal SOCS2 expression, demonstrating that the DNA methylation is responsible for the reduced SOCS2 expression (Letellier et al., 2014). In the present study, however, we could not evaluate SOCS2 methylation in our OSCC patients' samples. We identified SOCS2 as a direct target of miR‐424‐5p which could be another pivotal mechanism causing the reduced expression of SOCS2 in OSCC.
The findings of recent studies on miR‐424‐5p expression and functions in cancers are somewhat diverse. For example, miR‐424‐5p was found to be significantly upregulated and is positively correlated with cell migration, invasion, and chemoresistance in pancreatic cancer, ovary cancer and non‐small cell lung cancers (Donnem et al., 2012; Park et al., 2013; Wu et al., 2013). On the other hand, this miRNA was reported to act as a tumor suppressor, which is downregulated and is correlated with cell proliferation and invasion and tumor progression in cervical cancer and endometrial cancer (Li et al., 2015; Xu et al., 2013). In line with these results, the distribution of miR‐424‐5p seems to be tissue type‐specific. Nevertheless, the expression and function of miR‐424‐5p in OSCC is unclear and remains to be investigated. Our data showed that miR‐424‐5p was upregulated in OSCC samples compared with their corresponding adjacent normal tissues (p < 0.0001). In the cell line studies, overexpression of miR‐424‐5p significantly induced OSCC cell invasion and migration, demonstrating that miR‐424 may act as an oncogenic miRNA and may contribute to the progression of OSCC. Furthermore, restoration of SOCS2 expression could block miR‐424‐5p‐induced migration and invasion in oral cancer cells, indicating that miR‐424‐5p‐mediated tumor malignancy is SOCS2 dependent.
We also tested the effects of IL‐6, IL‐8, IL‐10 and OSM on miR‐424‐5p expression. Interestingly, only IL‐8 caused a significant induction of miR‐424‐5p expression in primary and mature miRNA forms. Moreover, shRNA‐mediated downregulation of STAT5 prevented the induction of primary and mature miR‐424 after IL‐8 treatment, demonstrating that STAT5 modulates the transcription of miR‐424 under IL‐8 stimulation. NF‐κB is one of the major downstream transcriptional factors in IL‐8 signaling (Manna and Ramesh, 2005) and NF‐κB binding sites have been identified in the promoter region of the miR‐424 gene (Zhou et al., 2013). Thus, we also tested the possibility that NF‐κB is involved in the IL‐8‐mediated miR‐424 expression. The data showed that the NF‐κB inhibitor only caused a small decrease in IL‐8‐induced mature miR‐424‐5p formation compared with the STAT5 inhibitor, indicating that the blockade of NF‐κB activity cannot interrupt the miR424 processing to mature miR‐424‐5p. The detail mechanisms remain to be elucidated.
In summary, this study is the first to show that miR‐424‐5p is a part of the positive feedback loop of IL‐8/STAT5/SOCS2 signaling in OSCC. Through STAT5 activation, IL‐8 was found to induce miR‐424‐5p expression which in turn targets SOCS2. This reduction of SOCS2 enhances STAT5 activity and the expression of STAT5 targeting genes, such as MMP‐2 and MMP‐9, and thereby contributes to OSCC tumorigenesis. Furthermore, antagomir‐mediated inactivation of miR‐424‐5p prevented the IL‐8‐induced cell migration and invasion, demonstrating the essential role of miR‐424‐5p in IL‐8‐induced cellular invasiveness. Besides IL‐8 and STAT5, which are well established targets for cancer treatment, here we propose that scavenging miR‐424‐5p expression and function using antagomir may have therapeutic potential for the treatment of invasive OSCC.
Funding support
This work was supported by National Health Research Institutes (NHRI) grants from Taiwan (NHRI‐CA104‐PP‐03), the Department of Health (DOH), and the Ministry of Health and Welfare (MOHW) and phase II Cancer Center Support Grant (CCSG) Program from Taiwan (MOHW103‐TDU‐212‐114‐005, MOHW104‐TDU‐B‐212‐124‐008).
Conflict of interest disclosure
The authors declare no conflict of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
The authors are grateful for support with the clinical samples from the Tissue Bank, Research Center of Clinical Medicine, National Cheng Kung University Hospital. We also thank the Taiwan Mouse Clinic (MOST 104‐2325‐B‐001‐011) which is funded by the National Research Program for Biopharmaceuticals (NRPB) at the Ministry of Science and Technology (MOST) of Taiwan for technical support in immunohistochemistry experiment for optical image capture.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.03.001.
Peng Hsuan-Yu, Jiang Shih-Sheng, Hsiao Jenn-Ren, Hsiao Michael, Hsu Yuan-Ming, Wu Guan-Hsun, Chang Wei-Min, Chang Jang-Yang, Shiow-Lian Catherine Jin, Shiah Shine-Gwo, (2016), IL‐8 induces miR‐424‐5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.03.001.
References
- Agrawal, N. , Frederick, M.J. , Pickering, C.R. , Bettegowda, C. , Chang, K. , Li, R.J. , Fakhry, C. , Xie, T.X. , Zhang, J. , Wang, J. , Zhang, N. , El-Naggar, A.K. , Jasser, S.A. , Weinstein, J.N. , Trevino, L. , Drummond, J.A. , Muzny, D.M. , Wu, Y. , Wood, L.D. , Hruban, R.H. , Westra, W.H. , Koch, W.M. , Califano, J.A. , Gibbs, R.A. , Sidransky, D. , Vogelstein, B. , Velculescu, V.E. , Papadopoulos, N. , Wheeler, D.A. , Kinzler, K.W. , Myers, J.N. , 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333, 1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahomadegbe, J.C. , Barrois, M. , Fogel, S. , Le Bihan, M.L. , Douc-Rasy, S. , Duvillard, P. , Armand, J.P. , Riou, G. , 1995. High incidence of p53 alterations (mutation, deletion, overexpression) in head and neck primary tumors and metastases; absence of correlation with clinical outcome. Frequent protein overexpression in normal epithelium and in early non-invasive lesions. Oncogene. 10, 1217–1227. [PubMed] [Google Scholar]
- Benekli, M. , Baer, M.R. , Baumann, H. , Wetzler, M. , 2003. Signal transducer and activator of transcription proteins in leukemias. Blood. 101, 2940–2954. [DOI] [PubMed] [Google Scholar]
- Britschgi, A. , Andraos, R. , Brinkhaus, H. , Klebba, I. , Romanet, V. , Muller, U. , Murakami, M. , Radimerski, T. , Bentires-Alj, M. , 2012. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer cell. 22, 796–811. [DOI] [PubMed] [Google Scholar]
- Calin, G.A. , Croce, C.M. , 2006. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 66, 7390–7394. [DOI] [PubMed] [Google Scholar]
- Callender, T. , el-Naggar, A.K. , Lee, M.S. , Frankenthaler, R. , Luna, M.A. , Batsakis, J.G. , 1994. PRAD-1 (CCND1)/cyclin D1 oncogene amplification in primary head and neck squamous cell carcinoma. Cancer. 74, 152–158. [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Malhotra, P.S. , Thomas, G.R. , Ondrey, F.G. , Duffey, D.C. , Smith, C.W. , Enamorado, I. , Yeh, N.T. , Kroog, G.S. , Rudy, S. , McCullagh, L. , Mousa, S. , Quezado, M. , Herscher, L.L. , Van Waes, C. , 1999. Expression of proinflammatory and proangiogenic cytokines in patients with head and neck cancer. Clin. Cancer Res. 5, 1369–1379. [PubMed] [Google Scholar]
- Collins, A.S. , McCoy, C.E. , Lloyd, A.T. , O'Farrelly, C. , Stevenson, N.J. , 2013. miR-19a: an effective regulator of SOCS3 and enhancer of JAK-STAT signalling. PloS One. 8, e69090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker, B.A. , Krebs, D.L. , Zhang, J.G. , Wormald, S. , Willson, T.A. , Stanley, E.G. , Robb, L. , Greenhalgh, C.J. , Forster, I. , Clausen, B.E. , Nicola, N.A. , Metcalf, D. , Hilton, D.J. , Roberts, A.W. , Alexander, W.S. , 2003. SOCS3 negatively regulates IL-6 signaling in vivo. Nat. Immunol. 4, 540–545. [DOI] [PubMed] [Google Scholar]
- Croker, B.A. , Kiu, H. , Nicholson, S.E. , 2008. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cel. Dev. Biol. 19, 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell, J.E. , 1997. STATs and gene regulation. Science. 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- Donnem, T. , Fenton, C.G. , Lonvik, K. , Berg, T. , Eklo, K. , Andersen, S. , Stenvold, H. , Al-Shibli, K. , Al-Saad, S. , Bremnes, R.M. , Busund, L.T. , 2012. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PloS One. 7, e29671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabegoli, F. , Ceccarelli, C. , Santini, D. , Taffurelli, M. , 2005. Suppressor of cytokine signalling 2 (SOCS-2) expression in breast carcinoma. J. Clin. Pathol. 58, 1046–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegl, H. , Gattringer, C. , Widschwendter, A. , Schneitter, A. , Ramoni, A. , Sarlay, D. , Gaugg, I. , Goebel, G. , Muller, H.M. , Mueller-Holzner, E. , Marth, C. , Widschwendter, M. , 2004. Methylated DNA collected by tampons – a new tool to detect endometrial cancer. Cancer Epidemiol. Biomarkers Prev. 13, 882–888. [PubMed] [Google Scholar]
- Forastiere, A. , Koch, W. , Trotti, A. , Sidransky, D. , 2001. Head and neck cancer. New Engl. J. Med. 345, 1890–1900. [DOI] [PubMed] [Google Scholar]
- Grandis, J.R. , Drenning, S.D. , Chakraborty, A. , Zhou, M.Y. , Zeng, Q. , Pitt, A.S. , Tweardy, D.J. , 1998. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J. Clin. Invest. 102, 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis, J.R. , Drenning, S.D. , Zeng, Q. , Watkins, S.C. , Melhem, M.F. , Endo, S. , Johnson, D.E. , Huang, L. , He, Y. , Kim, J.D. , 2000. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 97, 4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzman, J.H. , Rugowski, D.E. , Nikolai, S.E. , Schuler, L.A. , 2007. Stat5 activation inhibits prolactin-induced AP-1 activity: distinct prolactin-initiated signals in tumorigenesis dependent on cell context. Oncogene. 26, 6341–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G. , Zhou, R. , Liu, J. , Gong, A.Y. , Chen, X.M. , 2010. MicroRNA-98 and let-7 regulate expression of suppressor of cytokine signaling 4 in biliary epithelial cells in response to Cryptosporidium parvum infection. J. Infect. Dis. 202, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Li, H. , Wu, W. , Jiang, T. , Qiu, Z. , 2013. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol. Rep. 30, 1223–1230. [DOI] [PubMed] [Google Scholar]
- Iglesias-Gato, D. , Chuan, Y.C. , Wikstrom, P. , Augsten, S. , Jiang, N. , Niu, Y. , Seipel, A. , Danneman, D. , Vermeij, M. , Fernandez-Perez, L. , Jenster, G. , Egevad, L. , Norstedt, G. , Flores-Morales, A. , 2014. SOCS2 mediates the cross talk between androgen and growth hormone signaling in prostate cancer. Carcinogenesis. 35, 24–33. [DOI] [PubMed] [Google Scholar]
- Inagaki-Ohara, K. , Kondo, T. , Ito, M. , Yoshimura, A. , 2013. SOCS, inflammation, and cancer. JAKSTAT. 2, e24053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui, M. , Martello, G. , Piccolo, S. , 2010. MicroRNA control of signal transduction. Nat. Rev. Mol. Cel. Biol. 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Jemal, A. , Siegel, R. , Ward, E. , Murray, T. , Xu, J. , Thun, M.J. , 2007. Cancer statistics, 2007. CA Cancer J. Clin. 57, 43–66. [DOI] [PubMed] [Google Scholar]
- Jerjes, W. , Upile, T. , Petrie, A. , Riskalla, A. , Hamdoon, Z. , Vourvachis, M. , Karavidas, K. , Jay, A. , Sandison, A. , Thomas, G.J. , Kalavrezos, N. , Hopper, C. , 2010. Clinicopathological parameters, recurrence, locoregional and distant metastasis in 115 T1-T2 oral squamous cell carcinoma patients. Head Neck Oncol. 2, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Zhang, H.W. , Lu, M.H. , He, X.H. , Li, Y. , Gu, H. , Liu, M.F. , Wang, E.D. , 2010. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70, 3119–3127. [DOI] [PubMed] [Google Scholar]
- Klausen, P. , Pedersen, L. , Jurlander, J. , Baumann, H. , 2000. Oncostatin M and interleukin 6 inhibit cell cycle progression by prevention of p27kip1 degradation in HepG2 cells. Oncogene. 19, 3675–3683. [DOI] [PubMed] [Google Scholar]
- Leong, P.L. , Xi, S. , Drenning, S.D. , Dyer, K.F. , Wentzel, A.L. , Lerner, E.C. , Smithgall, T.E. , Grandis, J.R. , 2002. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 21, 2846–2853. [DOI] [PubMed] [Google Scholar]
- Letellier, E. , Schmitz, M. , Baig, K. , Beaume, N. , Schwartz, C. , Frasquilho, S. , Antunes, L. , Marcon, N. , Nazarov, P.V. , Vallar, L. , Even, J. , Haan, S. , 2014. Identification of SOCS2 and SOCS6 as biomarkers in human colorectal cancer. Br. J. Cancer. 111, 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Qiu, X.M. , Li, Q.H. , Wang, X.Y. , Li, L. , Xu, M. , Dong, M. , Xiao, Y.B. , 2015. MicroRNA-424 may function as a tumor suppressor in endometrial carcinoma cells by targeting E2F7. Oncol. Rep. 33, 2354–2360. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Ren, S. , Howell, P. , Fodstad, O. , Riker, A.I. , 2008. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res. 21, 545–558. [DOI] [PubMed] [Google Scholar]
- Lui, V.W. , Hedberg, M.L. , Li, H. , Vangara, B.S. , Pendleton, K. , Zeng, Y. , Lu, Y. , Zhang, Q. , Du, Y. , Gilbert, B.R. , Freilino, M. , Sauerwein, S. , Peyser, N.D. , Xiao, D. , Diergaarde, B. , Wang, L. , Chiosea, S. , Seethala, R. , Johnson, J.T. , Kim, S. , Duvvuri, U. , Ferris, R.L. , Romkes, M. , Nukui, T. , Kwok-Shing Ng, P. , Garraway, L.A. , Hammerman, P.S. , Mills, G.B. , Grandis, J.R. , 2013. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 3, 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna, S.K. , Ramesh, G.T. , 2005. Interleukin-8 induces nuclear transcription factor-kappaB through a TRAF6-dependent pathway. J. Biol. Chem. 280, 7010–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, S.A. , Sacks, P.G. , Gutterman, J.U. , Gallick, G.E. , 1989. Epidermal growth factor receptor protein-tyrosine kinase activity in human cell lines established from squamous carcinomas of the head and neck. Cancer Res. 49, 1130–1137. [PubMed] [Google Scholar]
- Mishima, K. , Yamada, E. , Masui, K. , Shimokawara, T. , Takayama, K. , Sugimura, M. , Ichijima, K. , 1998. Overexpression of the ERK/MAP kinases in oral squamous cell carcinoma. Mod. Pathol. 11, 886–891. [PubMed] [Google Scholar]
- Newton, V.A. , Ramocki, N.M. , Scull, B.P. , Simmons, J.G. , McNaughton, K. , Lund, P.K. , 2010. Suppressor of cytokine signaling-2 gene disruption promotes Apc(Min/+) tumorigenesis and activator protein-1 activation. Am. J. Pathol. 176, 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka, C. , Ikezoe, T. , Yang, J. , Nobumoto, A. , Kataoka, S. , Tsuda, M. , Udaka, K. , Yokoyama, A. , 2014. CD82 regulates STAT5/IL-10 and supports survival of acute myelogenous leukemia cells. Int. J. Cancer. 134, 55–64. [DOI] [PubMed] [Google Scholar]
- Palmer, D.C. , Restifo, N.P. , 2009. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y.T. , Jeong, J.Y. , Lee, M.J. , Kim, K.I. , Kim, T.H. , Kwon, Y.D. , Lee, C. , Kim, O.J. , An, H.J. , 2013. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J. Ovarian Res. 6, 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentmeier, H. , Geffers, R. , Jost, E. , Macleod, R.A. , Nagel, S. , Rohrs, S. , Romani, J. , Scherr, M. , Zaborski, M. , Drexler, H.G. , 2008. SOCS2: inhibitor of JAK2V617F-mediated signal transduction. Leukemia. 22, 2169–2175. [DOI] [PubMed] [Google Scholar]
- Rokavec, M. , Oner, M.G. , Li, H. , Jackstadt, R. , Jiang, L. , Lodygin, D. , Kaller, M. , Horst, D. , Ziegler, P.K. , Schwitalla, S. , Slotta-Huspenina, J. , Bader, F.G. , Greten, F.R. , Hermeking, H. , 2014. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Invest. 124, 1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossa, C. , Sommer, G. , Spolidorio, L.C. , Rosenzweig, S.A. , Watson, D.K. , Kirkwood, K.L. , 2012. Loss of expression and function of SOCS3 is an early event in HNSCC: altered subcellular localization as a possible mechanism involved in proliferation, migration and invasion. PloS One. 7, e45197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru, P. , Steele, R. , Hsueh, E.C. , Ray, R.B. , 2011. Anti-miR-203 upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2, 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, B.L. , Sugawara, A. , Ward, M.A. , Collier, A.C. , 2014. Single blastomere removal from murine embryos is associated with activation of matrix metalloproteinases and Janus kinase/signal transducers and activators of transcription pathways of placental inflammation. Mol. Hum. Reprod. 20, 1247–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiah, S.G. , Hsiao, J.R. , Chang, W.M. , Chen, Y.W. , Jin, Y.T. , Wong, T.Y. , Huang, J.S. , Tsai, S.T. , Hsu, Y.M. , Chou, S.T. , Yen, Y.C. , Jiang, S.S. , Shieh, Y.S. , Chang, I.S. , Hsiao, M. , Chang, J.Y. , 2014. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 74, 7560–7572. [DOI] [PubMed] [Google Scholar]
- Siavash, H. , Nikitakis, N.G. , Sauk, J.J. , 2004. Signal transducers and activators of transcription: insights into the molecular basis of oral cancer. Crit. Rev. Oral Biol. Med. 15, 298–307. [DOI] [PubMed] [Google Scholar]
- Song, J.I. , Grandis, J.R. , 2000. STAT signaling in head and neck cancer. Oncogene. 19, 2489–2495. [DOI] [PubMed] [Google Scholar]
- St John, M.A. , Li, Y. , Zhou, X. , Denny, P. , Ho, C.M. , Montemagno, C. , Shi, W. , Qi, F. , Wu, B. , Sinha, U. , Jordan, R. , Wolinsky, L. , Park, N.H. , Liu, H. , Abemayor, E. , Wong, D.T. , 2004. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 130, 929–935. [DOI] [PubMed] [Google Scholar]
- Sutherland, K.D. , Lindeman, G.J. , Choong, D.Y. , Wittlin, S. , Brentzell, L. , Phillips, W. , Campbell, I.G. , Visvader, J.E. , 2004. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 23, 7726–7733. [DOI] [PubMed] [Google Scholar]
- Tamiya, T. , Kashiwagi, I. , Takahashi, R. , Yasukawa, H. , Yoshimura, A. , 2011. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler. Thromb. Vasc. Biol. 31, 980–985. [DOI] [PubMed] [Google Scholar]
- Trengove, M.C. , Ward, A.C. , 2013. SOCS proteins in development and disease. Am. J. Clin. Exp. Immunol. 2, 1–29. [PMC free article] [PubMed] [Google Scholar]
- Vidal, O.M. , Merino, R. , Rico-Bautista, E. , Fernandez-Perez, L. , Chia, D.J. , Woelfle, J. , Ono, M. , Lenhard, B. , Norstedt, G. , Rotwein, P. , Flores-Morales, A. , 2007. In vivo transcript profiling and phylogenetic analysis identifies suppressor of cytokine signaling 2 as a direct signal transducer and activator of transcription 5b target in liver. Mol. Endocrinol. 21, 293–311. [DOI] [PubMed] [Google Scholar]
- Vlotides, G. , Sorensen, A.S. , Kopp, F. , Zitzmann, K. , Cengic, N. , Brand, S. , Zachoval, R. , Auernhammer, C.J. , 2004. SOCS-1 and SOCS-3 inhibit IFN-alpha-induced expression of the antiviral proteins 2,5-OAS and MxA. Biochem. Biophys. Res. Commun. 320, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Watanabe, H. , Iwase, M. , Ohashi, M. , Nagumo, M. , 2002. Role of interleukin-8 secreted from human oral squamous cell carcinoma cell lines. Oral Oncol. 38, 670–679. [DOI] [PubMed] [Google Scholar]
- Wu, K. , Hu, G. , He, X. , Zhou, P. , Li, J. , He, B. , Sun, W. , 2013. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol. Oncol. Res. 19, 739–748. [DOI] [PubMed] [Google Scholar]
- Xia, Z. , Baer, M.R. , Block, A.W. , Baumann, H. , Wetzler, M. , 1998. Expression of signal transducers and activators of transcription proteins in acute myeloid leukemia blasts. Cancer Res. 58, 3173–3180. [PubMed] [Google Scholar]
- Xu, J. , Li, Y. , Wang, F. , Wang, X. , Cheng, B. , Ye, F. , Xie, X. , Zhou, C. , Lu, W. , 2013. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 32, 976–987. [DOI] [PubMed] [Google Scholar]
- Yang, H.L. , Sun, C. , Sun, C. , Qi, R.L. , 2012. Effect of suppressor of cytokine signaling 2 (SOCS2) on fat metabolism induced by growth hormone (GH) in porcine primary adipocyte. Mol. Biol. Rep. 39, 9113–9122. [DOI] [PubMed] [Google Scholar]
- Yen, Y.C. , Shiah, S.G. , Chu, H.C. , Hsu, Y.M. , Hsiao, J.R. , Chang, J.Y. , Huang, W.C. , Liao, C.T. , Cheng, A.J. , Lu, Y.C. , Chen, Y.W. , 2014. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol. Cancer. 13, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Y. , Fung, T.K. , Chen, F.Y. , Chim, C.S. , 2013. Methylation profiling of SOCS1, SOCS2, SOCS3, CISH and SHP1 in Philadelphia-negative myeloproliferative neoplasm. J. Cell. Mol. Med. 17, 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Li, F. , Li, N. , Zhu, Q. , Yang, C. , Han, Q. , Chen, J. , Lv, Y. , Yu, L. , Wei, P. , Liu, Z. , 2014. Genetic variations of SOCS1 are associated with chronic hepatitis B virus infection. Hum. Immunol. 75, 709–714. [DOI] [PubMed] [Google Scholar]
- Zhou, R. , Gong, A.Y. , Chen, D. , Miller, R.E. , Eischeid, A.N. , Chen, X.M. , 2013. Histone deacetylases and NF-kB signaling coordinate expression of CX3CL1 in epithelial cells in response to microbial challenge by suppressing miR-424 and miR-503. PloS One. 8, e65153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, G. , Wu, X. , Jiang, Z. , Kasman, I. , Yao, J. , Guan, Y. , Oeh, J. , Modrusan, Z. , Bais, C. , Sampath, D. , Ferrara, N. , 2012. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 31, 3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


