ABSTRACT
Transcriptional interference (TI) is increasingly recognized as a widespread mechanism of gene control, particularly given the pervasive nature of transcription, both sense and antisense, across all kingdoms of life. Here, we discuss how transcription factor binding kinetics strongly influence the ability of a transcription factor to relieve or induce TI.
KEYWORDS: convergent promoters, dislodgement, roadblock, DNA-binding kinetics, transcriptional interference
Transcriptional interference (TI) is defined as the direct suppressive influence of one transcriptional process on another, in cis.1,2 TI operates through a variety of mechanisms by which an interfering promoter (Pinterfering) can inhibit the activity of a target promoter (Ptarget), with the specific combination of mechanisms at play being dependent upon the relative arrangement of the two promoters. The six possible orientations of Pinterfering relative to Ptarget each defines a set of potential TI mechanisms (Fig. 1A). An elongating RNA polymerase (RNAP) complex initiated from the interfering promoter may compromise the activity of the target promoter by inhibiting target promoter RNAPs or transcription factors (TFs) required for target promoter activity. For convergent promoters, which allow for the largest number of possible TI mechanisms, interfering RNAPs can inhibit the binding of RNAP (occlusion TI) or the transcription initiation process (“sitting duck” TI), impede the progress of elongating RNAPs (collision TI), or dislodge activatory TFs (Fig. 1B).
Figure 1.
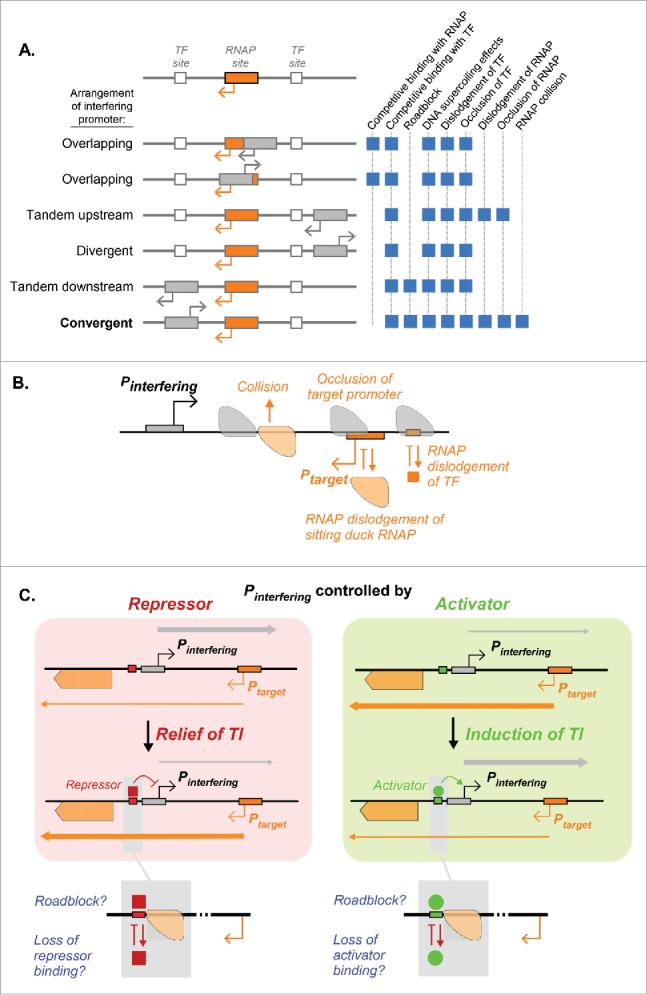
Transcriptional interference and its modulation. (A) Mechanisms of TI operating for different arrangements of the Ptarget promoter (orange) and the Pinterfering promoter (gray). (B) Mechanisms of TI at play for two convergent, non-overlapping promoters, where RNAPs from each promoter elongate over the other promoter. Elongating RNAPs can remove transcription factors or promoter-bound RNAPs from the DNA (dislodgement), or can block their binding (occlusion). Head-to-head “collisions” between elongating RNAPs can cause termination of one or both RNAPs. (C) The function of the transcription factor determines its effect on TI. If the interfering promoter is controlled by a repressor (left panel), the target promoter will experience relief from TI.5 If the TF is an activator (right panel), the target promoter will experience induced TI.
Modulation of TI by regulation of the interfering promoter opens up additional layers of transcriptional control and has been shown to operate in a variety of systems. TI provides a simple way to “invert” the function of a TF, such that a bona fide repressor can be converted to an activator by repressing the Pinterfering to relieve TI, while a bona fide activator can be converted to a repressor by activating the Pinterfering to induce TI (Fig. 1C). For example, in temperate bacteriophages λ and 186, relief of TI by repression of the strong lytic promoter appears to be a crucial step toward lysogenic development.3-5 In bacteria, such as L. monocytogenes, transcription of the mogR gene from the inducible P1 promoter brings about TI on three flagellin transcripts expressed from the opposite strand of DNA, and reduces cell mobility.6 In the commensal bacterium E. faecalis, modulation of TI through convergent prgX/prgQ gene promoters controls its competency.7
Modulation of TI as a gene control mechanism has also been observed in eukaryotic systems. In budding yeast S. cerevisiae, induction of TI by activating a promoter upstream of SER3 reduces serine biosynthesis in rich medium.8 On the other hand, relief of TI controls entry into meiosis by the a1/α2 repressor in diploid cells by repressing a convergent promoter downstream of the IME4 gene,9 and by repressing an activator for a promoter upstream of the IME1 gene.10 TI is also responsible for the establishment of mosaic expression of homeobox gene Ubx during embryonic development in D. melanogaster,11 while in mammals, the promoter for the Airn long non-coding RNA exerts cis-acting repression on the downstream Igf2r promoter by transcriptional overlap on the paternal allele, but is relieved by DNA methylation-associated repression of the Airn promoter on the maternal allele.12 TI can also operate in a multi-layered manner. For example, the yeast FLO11 transcript is controlled by the upstream ICR1 promoter, which itself is regulated by the convergent PWR1 promoter. Activation or repression of PWR1 induces or relieves TI on ICR1, causing stimulation or inhibition of FLO11 transcription.13
Using a TF to induce or relieve TI is relatively straightforward when Pinterfering is upstream of Ptarget. However, when the promoters are convergent, the situation becomes more complex (Fig. 1C). First, TI can be reciprocal, with the activity of the target promoter exerting TI on RNAP at the interfering promoter. Second, there are potential interactions between the elongating RNAPs from Ptarget and the TF in their path bound at Pinterfering. The elongating RNAP from Ptarget may dislodge the transcription factor, altering its regulation of Pinterfering.14 If the bound transcription factor is not dislodged, it may alternatively act as a roadblock to elongating RNAP, blocking downstream transcription,15,16 which may be important if the biologic function of Ptarget requires its transcription to pass Pinterfering (e.g., to express a gene beyond Pinterfering, as in Fig. 1C).
In a recent study,5 we combined mathematical modeling with in vivo experiments on the well-characterized bacteriophage λ to examine how repression of Pinterfering (in this case, the λ PR promoter) can give relief of TI on convergent Ptarget (the λ PRE promoter).5 We began by applying stochastic simulations to explore the key parameters that drive this “relief of TI” mode of regulation, and showed that for each of the three major TI mechanisms mentioned above, the magnitude of relief of TI was strongly dependent on the properties of the transcription factor and its binding site. These properties determine whether the TF acts as a roadblock to transcription from Ptarget and whether its occupancy of its binding site is sensitive to dislodgement by RNAPs from Ptarget (Fig. 1C). Roadblocking by a TF requires that the TF–DNA complex presents a strong barrier to the elongating RNAP (i.e., it is not easily dislodged) and that its spontaneous unbinding kinetics are slow relative to the RNAP elongation rate (Fig. 2A). The occupancy of the TF-binding site is most sensitive to elongating RNAP when it easily dislodged (weak barrier) and has a low rate of spontaneous unbinding, such that the increased removal due to the RNAP significantly perturbs its binding equilibrium (Fig. 2B). Modeling thus predicted that relief of TI is readily tunable, maximized by rapid repressor-binding kinetics, but can be compromised by repressors with slow-binding kinetics, as a result of roadblocking as well as loss of repression by repressor dislodgement (Fig. 2C).
Figure 2.
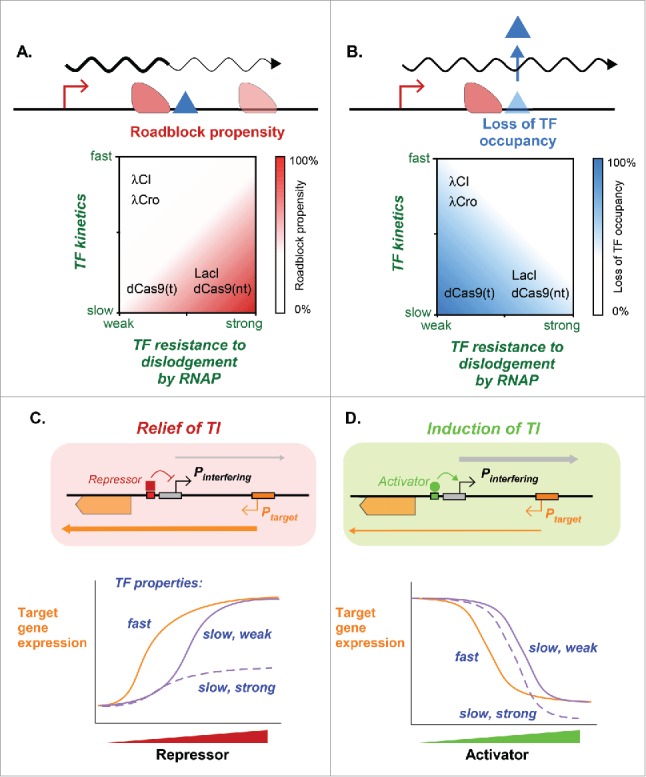
The kinetic properties of a TF determine its effect on modulating TI between convergent promoters. (A, B) TF-binding kinetics and the ability of the TF to resist dislodgement by an elongating RNAP both contribute to the overall roadblock effect (A) and to the overall occupation of the TF-binding site (B). The kinetic and resistance properties of several DNA-binding proteins (λ CI, λ Cro, LacI and dCas9 bound either to the template strand (t) or non-template strand (nt)) are classified, as determined in Hao et al.5 (C, D) TF-binding kinetics influence how gene expression from the target promoter responds to changes in TF concentration. (C) Schematic of the simulated change in target gene expression upon relief of TI by repressors with different properties (slow/fast kinetics, strong/weak barrier to elongating RNAPs).5 (D) Schematic of simulated induction of TI by activators with different properties. Higher concentrations of slow-binding TFs are needed to relieve or induce TI because their activity is reduced by elongating RNAPs from the target promoter. Slow-binding TFs, that are strong barriers, have a high roadblocking propensity, which can inhibit relief of TI by repressors or can aid induction of TI by activators.
We then validated our model predictions experimentally using the λ PR − PRE promoter pair. Repression of the strong lytic promoter PR by the λ CI or Cro repressors very efficiently relieved TI on the convergent lysogenic promoter PRE. We showed that these repressors did not roadblock RNAP and that CI repression of PR was not sensitive to PRE transcription, implying fast DNA-binding kinetics for these repressors.
Conversely, repression of the same PR promoter with a DNA-cleavage-defective Cas9 protein (dCas9) gave sub-optimal relief of TI, providing an important counter-example for TI regulation by a repressor with slow-binding kinetics.5 Interestingly, the binding orientation of dCas9, determined by the strand to which the guide RNA hybridizes,17,18 is known to influence its strength as a barrier to elongating RNAPs. This property allowed testing of a single repressor with the same binding kinetics but differing roadblock propensities. Consistent with the model prediction, our results clearly demonstrated that the effect of dCas9 on the relief of TI depends on its binding orientation: In the strong-barrier orientation, the barrier effect of dCas9 makes it incapable of relieving TI, whereas in the weak-barrier orientation, dCas9 does relieve TI but poorly, since its repression of PR was reduced due to its dislodgement by RNAPs from PRE5.
As well as demonstrating that relief of TI is dependent on TF-binding properties, the results demonstrate a novel method for estimating the in vivo DNA-binding kinetics of a repressor, based on the sensitivity or insensitivity of repression to dislodgement of the repressor by elongating RNAPs. This approach should enhance our ability to study TF binding in vivo, as current methodologies require the use of imaging techniques that rely on fluorescent tagging of proteins and do not measure specific, functional DNA binding.
As an extension of our published study,5 we used stochastic modeling of protein traffic on DNA to ask how different TI mechanisms and different TF properties might combine to influence the activity of a target promoter, Ptarget, when the TF is an activator, rather than a repressor, of the interfering promoter Pinterfering (Fig. 1C). Again, we looked at four different categories of activators: activators that are either a strong barrier or a weak barrier to elongating RNAPs, and activators with either fast or slow DNA-binding kinetics.
Simulations were performed as previously reported,5 where promoter firing is simulated as a two-step process. We simulated promoter activator with the simple assumption that the presence of bound activator accelerates RNAP loading at Pinterfering by 200-fold, and thereby Pinterfering is effectively proportional to the occupancy of the activator-binding site.
As might be expected, activation of Pinterfering with each of the classes of activator induces TI on the convergent Ptarget promoter and reduces its activity (Fig. 2D).
Strong induction of TI was seen in all TI scenarios, but the response curves varied depending on the properties of the activators. An activator with fast-binding kinetics gave a strong induction of TI at lower activator concentrations than were required by an activator with slow kinetics (Fig. 2D). This is because occupation of its binding site by the slow-binding activator is reduced due to dislodgement by elongating RNAPs, which, in turn, suppresses Pinterfering activation. In contrast, an activator with fast kinetics naturally binds and unbinds more frequently, so that a dislodged activator is quickly replaced.
Induction of TI by slow-binding activators was dependent on their ability to resist dislodgement by RNAP. An activator with slow kinetics but strong barrier properties showed a more potent induction of TI, when compared with an activator that is a weak barrier but has otherwise identical kinetics (Fig. 2D). A strong barrier activity helps induce TI by two mechanisms: it provides some level of protection against dislodgement by RNAPs, making such a TF a more effective activator of the interfering promoter; and roadblocking by the activator contributes directly to inhibition of transcription from the target promoter.
Thus, while induction of TI by an activator and relief of TI by a repressor for convergent promoters are both made more responsive by fast TF kinetics, their responses to the transcription barrier properties of the TF are reversed. A repressor that acts as a roadblock inhibits relief of TI, while an activator that acts as a roadblock augments induction of TI.
These results underscore the importance of the kinetic properties of transcription factors in their interaction with genomic traffic, and may prove useful in the field of synthetic biology for the design of artificial circuits which exploit TI.19,20
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported with supercomputing resources provided by the Phoenix HPC service at the University of Adelaide and by the Australian Research Council via a Discovery Early Career Researcher Award to N.H. [DE150100091] and a Discovery Grant [DP150103009 to K.E.S. and I.B.D.], and by the NHMRC [APP1100653].
References
- [1].Palmer AC, Egan JB, Shearwin KE. Transcriptional interference by RNA polymerase pausing and dislodgement of transcription factors. Transcription 2011; 2:9-14; PMID:21326903;http://dx.doi.org/ 10.4161/trns.2.1.13511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet 2005; 21:339-345; PMID:15922833;http://dx.doi.org/ 10.1016/j.tig.2005.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell 2004; 14:647-656; PMID:15175159; http://dx.doi.org/ 10.1016/j.molcel.2004.05.010 [DOI] [PubMed] [Google Scholar]
- [4].Dodd IB, Shearwin KE, Sneppen K. Modelling transcriptional interference and DNA looping in gene regulation. J Mol Biol 2007; 369:1200-1213; PMID:17498740; http://dx.doi.org/ 10.1016/j.jmb.2007.04.041 [DOI] [PubMed] [Google Scholar]
- [5].Hao N, Palmer AC, Ahlgren-Berg A, Shearwin KE, Dodd IB. The role of repressor kinetics in relief of transcriptional interference between convergent promoters. Nucleic Acids Res 2016; 44:6625-6638; PMID:27378773; http://dx.doi.org/ 10.1093/nar/gkw600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K et al. . The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009; 459:950-956; PMID:19448609; http://dx.doi.org/ 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- [7].Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 2011; 108:9721-9726;PMID:21606359; http://dx.doi.org/ 10.1073/pnas.1101569108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev 2005; 19:2695-2704; PMID:16291644; http://dx.doi.org/17110333 10.1101/gad.1367605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 2006; 127:735-745; PMID:17110333; http://dx.doi.org/ 10.1016/j.cell.2006.09.038 [DOI] [PubMed] [Google Scholar]
- [10].van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 2012; 150:1170-1181; PMID:22959267; http://dx.doi.org/ 10.1016/j.cell.2012.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 2006; 127:1209-1221;PMID:17174895;http://dx.doi.org/ 10.1016/j.cell.2006.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE et al. . Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012; 338:1469-1472; PMID:23239737; http://dx.doi.org/ 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- [13].Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A 2009; 106:18321-18326; PMID:19805129; http://dx.doi.org/ 10.1073/pnas.0909641106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nakanishi H, Mitarai N, Sneppen K. Dynamical analysis on gene activity in the presence of repressors and an interfering promoter. Biophys J 2008; 95:4228-4240; PMID:18658208; http://dx.doi.org/ 10.1529/biophysj.108.132894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. Transcription through the roadblocks: the role of RNA polymerase cooperation. Embo J 2003; 22:4719-4727; PMID:12970184; http://dx.doi.org/ 10.1093/emboj/cdg452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hao N, Krishna S, Ahlgren-Berg A, Cutts EE, Shearwin KE, Dodd IB. Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res 2014; 42:8861-8872; PMID:25034688; http://dx.doi.org/ 10.1093/nar/gku627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013; 152:11731183; PMID:23452860;http://dx.doi.org/ 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 2013; 41:74297437; PMID:23761437; http://dx.doi.org/ 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brophy JA, Voigt CA. Antisense transcription as a tool to tune gene expression. Mol Systems Biol 2016; 12:854; PMID:26769567; http://dx.doi.org/ 10.15252/msb.20156540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hoffmann SA, Kruse SM, Arndt KM. Long-range transcriptional interference in E. coli used to construct a dual positive selection system for genetic switches. Nucleic Acids Res 2016; 44:e95; PMID:26932362 [DOI] [PMC free article] [PubMed] [Google Scholar]


