Abstract
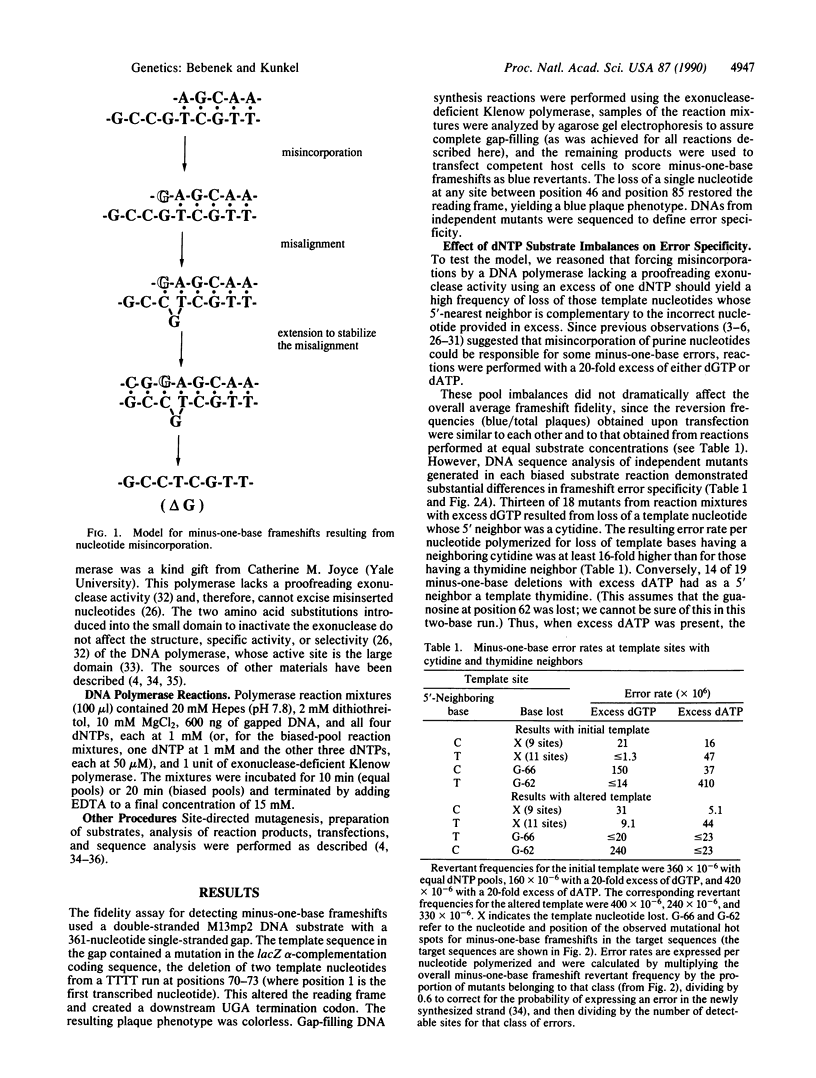
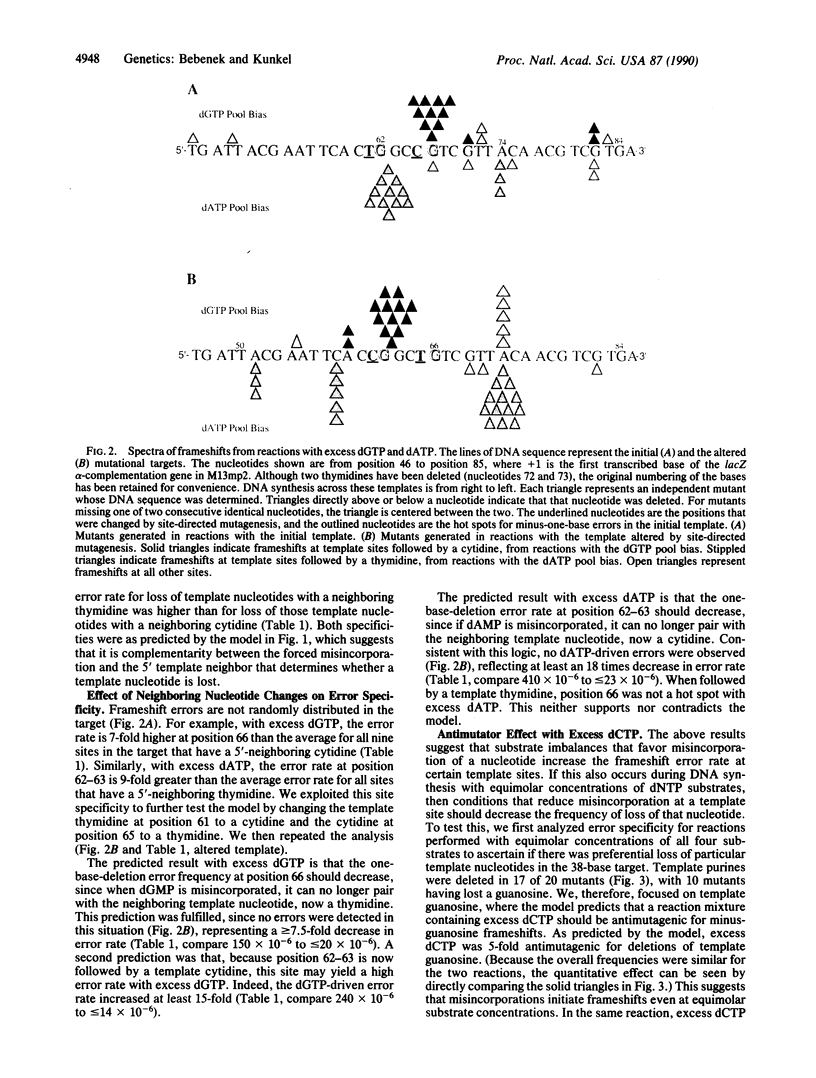
Studies presented here on the fidelity of DNA synthesis in vitro support the hypothesis that a classical base-substitution intermediate (i.e., a misincorporated nucleotide) can yield a frameshift mutation. By using a fidelity assay specifically designed to detect minus-one-base errors, nucleotide substrate pool imbalances that have previously been shown to increase the rate of misincorporation are now shown to also increase minus-one-base frameshift error rates. Examination of the specificity of the errors produced in reactions with various dNTP pool imbalances and various DNA templates revealed that template nucleotides were preferentially lost when they had as a 5' neighbor a nucleotide complementary to the dNTP provided in excess. This suggests that when a misincorporated nucleotide is complementary to the next nucleotide in the template, a misaligned intermediate containing a correct terminal base pair can form and be extended by a DNA polymerase, leading to a frameshift mutation. We present evidence that the proposed mechanism may operate in vivo and discuss the implications of this model for frameshift mutations induced by DNA damage.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashman C. R., Davidson R. L. Sequence analysis of spontaneous mutations in a shuttle vector gene integrated into mammalian chromosomal DNA. Proc Natl Acad Sci U S A. 1987 May;84(10):3354–3358. doi: 10.1073/pnas.84.10.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C. A., Randall S. K., Rayssiguier C., Radman M., Eritja R., Kaplan B. E., McEntee K., Goodman M. F. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988 Dec 15;263(35):18946–18952. [PubMed] [Google Scholar]
- Boosalis M. S., Mosbaugh D. W., Hamatake R., Sugino A., Kunkel T. A., Goodman M. F. Kinetic analysis of base substitution mutagenesis by transient misalignment of DNA and by miscoding. J Biol Chem. 1989 Jul 5;264(19):11360–11366. [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Ollis D. L., Steitz T. A., Joyce C. M. A domain of the Klenow fragment of Escherichia coli DNA polymerase I has polymerase but no exonuclease activity. Proteins. 1986 Sep;1(1):66–73. doi: 10.1002/prot.340010111. [DOI] [PubMed] [Google Scholar]
- Giroux C. N., Mis J. R., Pierce M. K., Kohalmi S. E., Kunz B. A. DNA sequence analysis of spontaneous mutations in the SUP4-o gene of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Feb;8(2):978–981. doi: 10.1128/mcb.8.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Extrahelical adenosine stacks into right-handed DNA: solution conformation of the d(C-G-C-A-G-A-G-C-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7456–7464. doi: 10.1021/bi00371a030. [DOI] [PubMed] [Google Scholar]
- Ikehata H., Akagi T., Kimura H., Akasaka S., Kato T. Spectrum of spontaneous mutations in a cDNA of the human hprt gene integrated in chromosomal DNA. Mol Gen Genet. 1989 Nov;219(3):349–358. doi: 10.1007/BF00259606. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L., Rabinovich D., Hope H., Frolow F., Appella E., Sussman J. L. The three-dimensional structure of a DNA duplex containing looped-out bases. Nature. 1988 Jul 7;334(6177):82–84. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- Joyce C. M. How DNA travels between the separate polymerase and 3'-5'-exonuclease sites of DNA polymerase I (Klenow fragment). J Biol Chem. 1989 Jun 25;264(18):10858–10866. [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988 Nov 10;951(1):1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- Kunkel T. A., Hamatake R. K., Motto-Fox J., Fitzgerald M. P., Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol Cell Biol. 1989 Oct;9(10):4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Exonucleolytic proofreading enhances the fidelity of DNA synthesis by chick embryo DNA polymerase-gamma. J Biol Chem. 1988 Mar 25;263(9):4450–4459. [PubMed] [Google Scholar]
- Kunkel T. A., Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988 Oct 15;263(29):14784–14789. [PubMed] [Google Scholar]
- Kunkel T. A. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem. 1985 May 10;260(9):5787–5796. [PubMed] [Google Scholar]
- Lee G. S., Savage E. A., Ritzel R. G., von Borstel R. C. The base-alteration spectrum of spontaneous and ultraviolet radiation-induced forward mutations in the URA3 locus of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Nov;214(3):396–404. doi: 10.1007/BF00330472. [DOI] [PubMed] [Google Scholar]
- Mendelman L. V., Boosalis M. S., Petruska J., Goodman M. F. Nearest neighbor influences on DNA polymerase insertion fidelity. J Biol Chem. 1989 Aug 25;264(24):14415–14423. [PubMed] [Google Scholar]
- Mendelman L. V., Petruska J., Goodman M. F. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J Biol Chem. 1990 Feb 5;265(4):2338–2346. [PubMed] [Google Scholar]
- Miller M., Harrison R. W., Wlodawer A., Appella E., Sussman J. L. Crystal structure of 15-mer DNA duplex containing unpaired bases. Nature. 1988 Jul 7;334(6177):85–86. doi: 10.1038/334085a0. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou C., Ripley L. S. Polymerase-specific differences in the DNA intermediates of frameshift mutagenesis. In vitro synthesis errors of Escherichia coli DNA polymerase I and its large fragment derivative. J Mol Biol. 1989 May 20;207(2):335–353. doi: 10.1016/0022-2836(89)90258-1. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q Rev Biophys. 1987 Aug;20(1-2):35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Loeb L. A. Differential extension of 3' mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989 Feb 15;264(5):2898–2905. [PubMed] [Google Scholar]
- Phear G., Armstrong W., Meuth M. Molecular basis of spontaneous mutation at the aprt locus of hamster cells. J Mol Biol. 1989 Oct 20;209(4):577–582. doi: 10.1016/0022-2836(89)90595-0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Clark A., deBoer J. G. Spectrum of spontaneous frameshift mutations. Sequences of bacteriophage T4 rII gene frameshifts. J Mol Biol. 1986 Oct 20;191(4):601–613. doi: 10.1016/0022-2836(86)90448-1. [DOI] [PubMed] [Google Scholar]
- Roberts J. D., Kunkel T. A. Fidelity of a human cell DNA replication complex. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7064–7068. doi: 10.1073/pnas.85.19.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Preston B. D., Johnston L. A., Soni A., Loeb L. A., Kunkel T. A. Fidelity of two retroviral reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989 Feb;9(2):469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Sklenar V., Appella E., Cohen J. S. Conformational perturbation due to an extra adenosine in a self-complementary oligodeoxynucleotide duplex. Biopolymers. 1987 Dec;26(12):2041–2052. doi: 10.1002/bip.360261206. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Danforth B. N., Glickman B. W. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986 May 20;189(2):273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985 Apr;109(4):633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S. A., Crothers D. M. Structural model for an oligonucleotide containing a bulged guanosine by NMR and energy minimization. Biochemistry. 1988 May 3;27(9):3130–3141. doi: 10.1021/bi00409a004. [DOI] [PubMed] [Google Scholar]
- de Boer J. G., Ripley L. S. An in vitro assay for frameshift mutations: hotspots for deletions of 1 bp by Klenow-fragment polymerase share a consensus DNA sequence. Genetics. 1988 Feb;118(2):181–191. doi: 10.1093/genetics/118.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]