Abstract
MicroRNAs (miRs) are small non-coding RNAs that regulate protein expression through post-transcriptional mechanisms. They participate in broad aspects of biology from the control of developmental processes to tumorigenesis. Recent studies in Drosophila show that they also regulate activity-dependent and sensory-specific protein expression and support olfactory memory formation. Among the hundreds of miRs described, several have been demonstrated to be required for normal learning, memory, or for the development of neuronal circuits that support memory formation. Fly models of human diseases offer promise of identifying miRs whose expression becomes dysregulated and part of the pathological state, providing models for understanding brain disorders and drug discovery.
Keywords: Learning, memory, Drosophila, RISC, microRNA
INTRODUCTION
MicroRNAs (miRs) are small (~22 nt) non-coding RNAs that provide post-transcriptional regulation of gene expression [1,2]. They have been implicated in many different aspects of biology, from development to tumorigenesis [3–5]. Recent studies extend their influence into the biology of memory formation and memory disorders [6–8•], with miRs being offered as early biomarkers of Alzheimer disease (AD) [9] and as potential therapeutic targets [10,11].
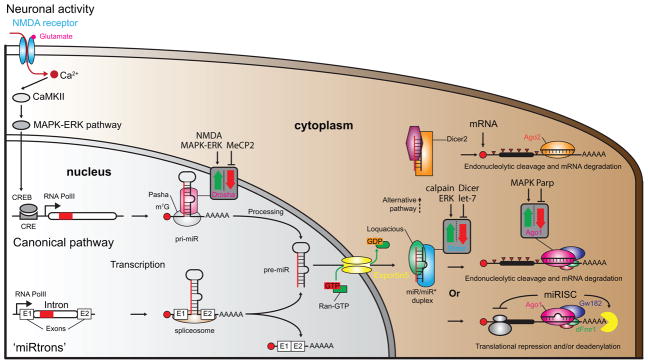
MiRs are usually transcribed from the genome as long primary miR hairpins (pri-miRs) by RNA polymerase II (Figure 1) [12]. In some cases, miRs are spliced-out from introns by the spliceosome and termed ‘miRtrons’ [13]. Pri-miRs are then processed in the nucleus by the Drosha/Pasha microprocessor complex into ~70 nt long precursor-miRs (pre-miRs). Exportin5 actively (with Ran-GTP) translocates pre-miRs to the cytoplasm where they undergo the next processing step by the Dicer/Loquatious complex to produce a mature duplex composed of a guide strand and its passenger. The guide strand is preferentially inserted in a protein complex [14] called the RNA-induced silencing complex (RISC), made up of a member of the Argonaute (Ago) family of proteins and multiple other ribonucleoproteins (RNP). RISC guides the miR to the mRNA target based on sequence complementarity between the miR recognition element (MRE) in the 3′UTR of the mRNA, and a ‘seed region’ (nt 2 to 8) at the 5′ end of the miR [15]. RISC inhibits mRNA translation or triggers degradation depending on the degree of complementarity between the miR and the mRNA [16], thereby producing post-transcriptional control over the gene expression.
Figure 1. Biogenesis of microRNAs.
MiR gene expression is regulated in ways similar to protein coding genes. Neuronal activity (depolarization, neurotrophins, sensory stimuli, etc.) can induce miR expression [12,17,82]. MiR-132, for example, possesses a CRE sequence in its promoter allowing CREB to regulate its activity. The NMDA receptor, CaMKII and the MAPK-ERK pathway are known to control miR-132 expression although different upstream signaling pathways that may regulate other miR genes [12]. There are two miR biogenesis pathways in the nucleus, the canonical and the ‘miRtrons’ pathway. In the canonical pathway, RNA polymerase II transcribes miR genes into pri-miRs that are further processed into pre-miRs by the Pasha/Drosha microprocessor complex. In the miRtron pathway, miRs are spliced from introns by the spliceosome to pre-miRs. Exportin5 along with Ran-GTP actively transports pre-miRs to the cytoplasm. Dicer/Loquacious complex further processes the pre-miRs in the cytoplasm to mature duplexes - with a guide strand (miR) and its passenger (miR*). A member of the Argonaute family of proteins (Ago1 or Ago2 depending on the pathway) loads one of the strands into the miRISC complex (lower right corner). The complex includes the Gw182 protein and other auxiliary proteins such as dFmr1. This outcome leads to mRNA degradation or translational inhibition depending on the match quality of miR/mRNA hybrid. In rare cases (upper right corner), duplexes with a very high level of complementarity between strands are sorted to an alternative pathway using Dicer2, leading to mRNA degradation by Ago2 and inducing competition between the processing pathways [83]. Neural activity, genetic manipulations or disorders have been shown to positively or negatively influence the processing enzymes involved in miRNA biogenesis (grey squares with red and green arrows) [12,82,84].
From a pure conceptual viewpoint, miRs are attractive molecular candidates for influencing memory formation [6,7,17]. On the one hand, they might quickly release sequestered “memory mRNAs” for translation, either centrally or locally, in response to relevant neural activity [6–8•,17,18]. The release of mRNAs for translation could be particularly important for modifying the function and structure of synapses tagged for memory formation [19–21]. This would enable miRs to physiologically influence the dynamics of synaptic mRNA expression for intermediate- or long-term memory formation. Given that long-term memory sparks changes in nuclear gene expression [22–25], miRs might also influence the quality or quantity of mRNAs translated at the soma to marshal the required cellular differentiation for this form of memory. On the other hand, miRs regulate nervous system development [6,26] and this provides an instructive role in building the neuronal circuits involved in memory formation [27]. Roles for miRs in neurodevelopmental [10,28,29], neurodegenerative [30], and neuropsychiatric disorders have been established [17,28]. These disorders usually present with associated symptoms of learning disability and/or memory loss [8•,10,28,29,31,32].
The fruit fly, Drosophila melanogaster, offers a facile organism to dissect the roles of miRs in memory formation. This model system provides simple and quantifiable behaviors to study memory, including olfactory classical conditioning [33] and long-term odor habituation [34,35]. The fly’s olfactory nervous system (Figure 2A), the brain region principally involved in olfactory memory, has been extensively characterized with a relatively detailed description of its neuronal circuits and constituent cell types [36–40]. Moreover, there exists very significant homology between the insect and the mammalian olfactory nervous system [41], such that conceptual insights made from the fly are easily extended to mammalian olfactory memory formation. In addition, the fly offers an extensive genetic toolset that includes genomic mutants, RNAi libraries, overexpression constructs and methodology for temporal and cell type-specific control of transgene expression [42–44]. As one example, “sponge technology”, when combined with specific Gal4 drivers, allows cell type-specific and temporal inhibition of individual miRs [45,46] to test their importance in memory formation [47••,48••,49••].
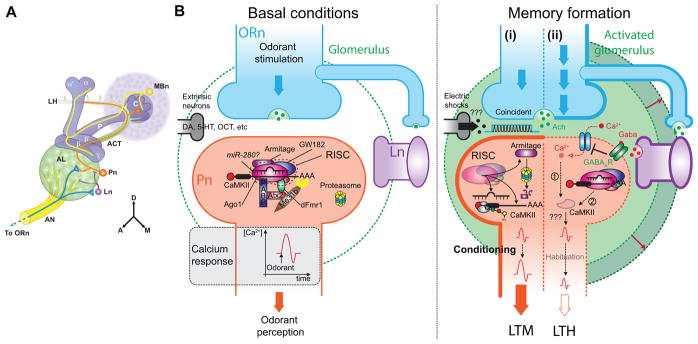
Figure 2. MiRs and RISC machinery are involved in antennal lobe-associated olfactory memory.
(A) Schematic diagram of the olfactory nervous system in the right hemisphere of the adult fly brain. Olfactory receptor neurons (ORn, blue) project their axons through the antennal nerve (AN) to terminate in antennal lobe (AL) glomeruli: discrete, spherical, neuropil structures containing synaptic connections. In the AL, the ORn pre-synaptic terminals connect with and stimulate projection neurons (Pn, orange) and local interneurons (Ln, purple) using acetylcholine as a neurotransmitter (panel B). Pn send their axons through the antennal cerebral tract (ACT) to the calyx (C) of the mushroom bodies where they stimulate the dendrites of the mushroom body neurons (MBn, yellow). The Pn axons continue on to also synapse in an area of the brain called the lateral horn (LH). The axons of the MBn bundle together to form the peduncle (P) which projects in an anterior direction into the L-shaped neuropil of MB lobes. This neuropil structure is further divided into the vertical lobes (α and α′) and the horizontal lobes (β, β′, γ). The orientation guide for the panel indicates dorsal (D), anterior (A) and medial (M).
(B, left panel) In the dendritic terminals of Pn, RISC with associated miRs (miR-280?) may be complexed under basal conditions with unidentified mRNAs (CaMKII?) required for normal memory formation. Each odorant activates specific glomeruli that can be recorded by monitoring calcium signaling [50,85].
(Right panel) During memory formation, (i) when the odorant is closely associated with mild electric shocks, probably mediated by the Ach and DA coincident activation of Pn, it induces the synthesis of CaMKII with an odorant-specific pattern and the degradation of Armitage by the proteasome following the same spatial pattern. The level of CaMKII expression seems to be crucial for the odorant-induced response in Pn. The calcium response to the odorant is increased with a glomerulus-specific pattern. Flies avoid the odorant associated with the shocks for an extended period of time. This is the behavior revealing long-term memory of the association. (ii) Repeated odorant stimulation of Pn, by Ach and GABA together, decreases the odorant-induced calcium signal in Pn and potentially diminishes CaMKII activation (1) leading to long-term reduced avoidance through unknown cellular mechanisms. An alternative hypothesis (2) is that specific mRNAs, including CaMKII, are inhibited beyond the basal level of inhibition following repeated stimulation by the RISC in specific glomeruli. As shown by Sudhakaran et al. [55•], repression is dependent on multiple proteins including Ago1, Atx2 and dFmr1. The adaptive behavior persistent for an extended period of time is called long-term habituation. RISC associated proteins are not shown on the right panel for simplicity.
There are many important questions to answer concerning the roles for miRs in Drosophila olfactory memory: (i) Which individual miRs are involved in memory formation? (ii) Where in the memory neural circuit does each miR function? (iii) When during the life cycle of the fly is each miR required? (iv) What specific phases of olfactory memory – short-, intermediate-, or long-term memory - are under the influence of individual miRs? (v) What specific aspects of neuronal physiology are affected by miR regulation? (vi) What are the target mRNAs for the miRs that are involved in olfactory memory formation? (vii) Does the dysregulation of miR expression always cause poor learning and memory, or can certain miRs be classed as memory suppressor whose normal function is to constrain memory formation? These and many related questions are tractable using the fly as the model system.
THE RISC PATHWAY IS INVOLVED IN OLFACTORY MEMORY FORMATION
The antennal lobe (AL, Figure 2A) is the first relay center for olfactory information in the fly brain [41]. At the synaptic regions of the AL – the glomeruli – the axons of olfactory receptor neurons (ORn) transmit sensory information to the dendrites of projection neurons (Pn) and local interneurons (Ln). Pn then convey olfactory information to the mushroom body neurons (MBn) and neurons of the lateral horn (LH). The AL has been well studied because of its role in processing and coding olfactory information for evaluation by higher order centers including the MB [50,51].
Ashraf et al. (2006) [52••] demonstrated the importance of miR processing proteins in the Pn for long-term olfactory memory. They showed that mutants of Armitage, a component of the RISC pathway [53], exhibit normal levels of memory performance immediately after olfactory classical conditioning but are impaired in LTM generated by spaced conditioning. They further demonstrated that the Armitage protein is rapidly degraded in certain AL glomeruli in an activity-dependent and odorant-specific way and that the level of local protein synthesis is increased. Local protein synthesis was assayed using a reporter for synaptically-localized calcium/calmodulin-dependent protein kinase II (CaMKII). Interestingly, the 3′UTR of the CaMKII mRNA contains putative binding sites for miR-280 and miR-289. These observations were combined into a model positing that neuronal activity due to odor-specific conditioning leads to degradation of RISC activity and the subsequent release of miR-dependent inhibition of synaptic protein synthesis necessary for LTM (Figure 2B).
Long-term habituation (LTH) is another form of LTM requiring plasticity in the AL between Ln and Pn [34,35] (Figure 2B). This behavior is induced by sustained exposure to an odorant resulting in a reduced sensitivity to the odor as measured by odor-avoidance in a Y-maze [39]. McCann et al. have shown that Ataxin 2 (atx2) is necessary in odorant-activated Pn for LTH and an associated growth of the glomeruli responsive to the odorant [54••]. In addition, atx2 mutation suppresses the reduced calcium signaling normally provoked by LTH in the responsive Pn. Interestingly, Atx2 associates with GW182 and Ago1, two core proteins of the miRISC-pathway. Double heterozygous mutants for atx2 and ago1 show no LTH, further indicating functional interaction between those genes. Moreover, the authors showed in mitotic clones of atx2 mutant cells that the expression of miR-dependent translational GFP reporters is released [54••]. The same group later showed that dFmr1 is also necessary in Pn for LTH and its associated reduction in calcium-response with odor application [55•]. Fmr1, when mutated, causes the Fragile X syndrome (FXS), the most common form of inherited intellectual disability (see below and [28,31]). Notably, dFmr1 has been shown to interact with Ago2, Dicer and miRs [56]. Sudhakaran et al. [55•] showed that dFmr1 also interacts with Atx2, Me31B and Ago1 and represses CaMKII expression. CaMKII mRNA is a clear target of miR-dependent translational control, however, the underlying cellular pathways controlling habituation remain elusive and still need to be explored [39]. Nevertheless, the results presented above lead us to propose the model shown in Figure 2B. In basal conditions (Figure 2B, left panel), odorant stimulation induces Ach release from ORn, producing Ca2+ response in the Pn underlying odorant perception. In these conditions, RISC, RISC-interacting proteins (Atx2, PABP and Me31b) and miRs inhibit the majority of dendritic protein synthesis in the Pn. In case of sustained odorant stimulations (right panel, ii), repeated activations of Pn by Ach and inhibition by GABA reduce calcium entry, as recorded with decreased Ca2+ signaling, and thus lead to diminished CaMKII activation. An alternative possibility is that miR-controlled translation of CaMKII is further inhibited, relative to the basal situation, by repeated exposure to the odorant. However, a reduction in the expression of CaMKII following repeated odorant exposure has not yet been shown to occur. In either case, reduced activity of CaMKII or of its expression seems to be required for LTH given that translational release of its inhibition strengthens synaptic efficacy.
Taken together, results from the research described above show that the RISC complex is involved in two distinct types of olfactory memory that require opposite forms of plasticity and behavioral outputs. LTH leads to decreased odor avoidance while LTM leads to increased odor avoidance. Interestingly, each form of plasticity seems to require proper control of CaMKII expression (Figure 2B) – controlled by the miR pathway – with elevated levels of CaMKII expression favoring increased synaptic transmission at odorant-activated synapses and reduced expression weakening the efficacy of synaptic transmission.
MiRs INVOLVED IN MEMORY FORMATION
Beyond the speculated involvement of miR-280 and its regulated mRNAs including CaMKII in the Pn, Li et al. were the first to show that an individual miRNA, miR-276a, modulates memory formation through its regulation of dopamine receptor (DopR) expression [47••]. DopR is a central actor in olfactory memory formation, currently thought to convey the signal for the unconditioned stimulus (electric shocks) to the MBn [57]. Li and co-workers showed that partial miR-276a inhibition in the MBn using the “sponge” technique produces a deficit in LTM, which was reversed by removing one genomic copy of the DopR gene. The LTM deficit was associated with an increase in DopR expression and led to the model that overexpression of DopR resulting from miR-276a inhibition impairs LTM. The authors also reported reduced odor avoidance due to miR-276a inhibition, but this effect was mapped to the ellipsoid body, outside of the MBn olfactory learning center. These results stress the possibility that a complete loss of a miR expression may produce pleiotropic effects due to regulation of different sets of mRNAs in distinct regions and the compartmentalized functions among various brain circuits [6,10,28,30].
There are more than 1000 miRs in the human and although the number is less in the fly (256 sequences with 150 of high confidence; www.miRBase.org), it is still sufficiently large given the enormous regulatory potential of this class of molecules. Moreover, miR sequences are strikingly conserved from C. elegans to human [58]. Thus, it would be extremely valuable to investigate a large set of miRs in one organism to obtain a global view of the spectrum of miRs that are involved in producing a single phenotype.
A recent genetic screen surveyed 134 miRs individually using sponge technology for their potential involvement in intermediate-term memory (ITM, [48••]). In essence, a sponge transgene for each of the 134 miRs was expressed using a pan-neuronal Gal4 driver and memory at 3h after olfactory classical conditioning was measured. Five different miRNAs (miR-9c, miR-31a, miR-305a, miR-974 and miR-980) were implicated in memory formation from the initial screen and re-screens. Two of these were analyzed for their effects on memory with expression in different subsets of neurons in the olfactory nervous system. This initial screen prompted several important questions regarding the function of individual miR players: (1) What specific neuronal populations require normal miR expression for memory formation? (2) Does each miR alter the development of the olfactory nervous system leading to memory dysfunction, or does it alter the physiology of neurons in adult animals? (3) Does each miR broadly affect all temporal phases or memory (STM, ITM, LTM), or are some miRs involved in temporally and mechanistically-distinct forms of memory? (4) Is each implicated miR involved in acquisition (learning), memory consolidation, or active forgetting? (5) What is the set of mRNAs targeted by each miR for regulating memory formation?
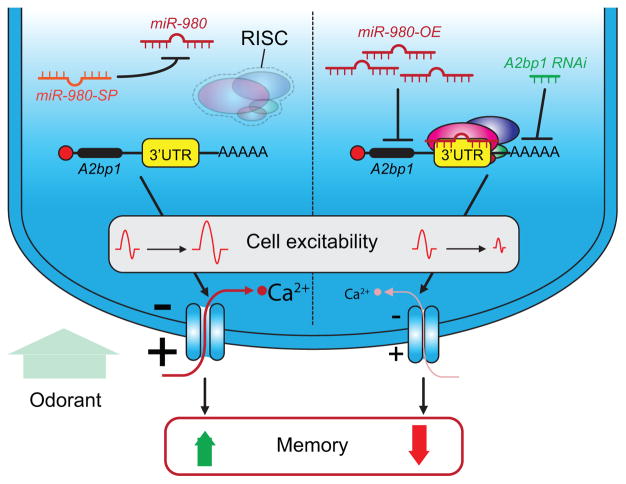
Of the five identified miRs, miR-980 captured the initial interest because its inhibition pan-neuronally increased memory performance rather than decreasing it! Similar to tumor-suppressor genes, some genes in the genome constrain memory formation and are classified as memory suppressors [49••]. Even more striking was the discovery that miR-980 inhibition in nearly all types of neurons in the olfactory nervous system increases memory performance, pointing to a shared mechanism across different set of neurons (Figure 3A). The mechanism identified was that miR-980 inhibition increases neuronal excitability. This provides an explanation for the increased memory: the hyperexcitable state of any of the neurons involved in conveying the relevant sensory information leads to increased salience of the sensory stimuli presented during conditioning. Moreover, the data argued that a primary target of miR-980 is the autism-and epilepsy-susceptibility gene, A2bp1 [59]. Four lines of evidence supported this conclusion: (1) A2bp1 mRNA has multiple MREs for miR-980 in its 3′ UTR, (2) A2bp1 protein levels vary inversely with the level of, miR-980 expression (3) the behavioral consequences of modulating A2bp1 expression varies inversely with those of miR-980 modulation, and (4) miR-980 sponge expression fails to produce the enhanced memory effects when co-expressed with A2bp1 RNAi, suggesting that A2bp1 is functionally downstream of miR-980.
Figure 3. MiR-980 impacts MB excitability to influence memory formation.
In adult flies, miR-980 controls A2bp1 expression in most of the neuronal populations involved in olfactory memory formation, including olfactory receptor neurons (ORn), projection neurons (Pn), and mushroom body neurons (MBn) (Figure 2A). MiR-980 inhibition releases A2bp1 repression producing an increase in neuronal excitability and calcium responses induced by odor presentation to the fly. The increase in A2bp1 expression leads to an increase in memory performance. MiR-980 over-expression or A2bp1 inhibition produces the opposite phenotype of memory impairment. This is associated with decreased neuronal excitability and calcium signaling.
Proper wiring of neurons involved in olfactory memory formation is required for normal behavior [60,61]. Impairments in neuronal wiring are thought to underlie neurodevelopmental disorders including intellectual disability and autism [6,8• 26,27]. Kucherenko et al. [62••] found that let-7 miR loss-of-function perturbs normal MB development and produces a learning impairment (Figure 4A). In physiological conditions, a peak of ecdysone hormone induces let-7 expression in MBn during the larval to pupae transition. Let-7 inhibits the Abrupt (Ab) transcription factor and in turn promotes the expression of the cell adhesion molecule FasII, a molecule that influences the differentiation of the αβ MBn. Insults to this pathway produce structural alterations in the αβ MBn that are critical for memory retrieval [63] explaining the STM defect (Figure 4A). Interestingly let-7 is involved in neuronal differentiation in other systems and in disease states such as Parkinson’s disease [64,65], showing the diverse roles for individual miRs depending on the cell type and time during development.
Figure 4. Individual miRs impact MB development to influence memory formation.
(A) At the larval to pupal transition during development, a peak of ecdysone leads to an increase of miR let-7 expression. Let-7 blocks the expression of the inhibitory transcription factor Abrupt (Ab). Abrupt inhibition releases the expression of the cell adhesion molecule FasII, with FasII being involved in the differentiation of αβ MBn. Let-7 loss-of-function reduces the volume of the MB αβ lobes producing an associated learning deficit of about 40%.
(B) MiR-iab8-3p is necessary in MBn during development. Its inhibition in αβ neurons induces structural alterations including cell soma hypertrophy, a volume decrease of the αβ MB lobe and a length decrease of the primary segment of the alpha axonal branch. Those alterations lead to learning disabilities and memory deficits in adult flies. The deficits may be caused by miR-iab8-3p targeting of CG4585, a gene coding for an enzyme with ceramide phosphoethanolamine synthase activity and whose inhibition strongly increases memory.
Similarly, miR-iab8-3p, a Hox miR gene involved in fertility and specification of segment identity [66–68] was recently shown to be required for proper development of the MBn [69•]. MiR-iab8-3p inhibition in αβ MBn led to cell soma hypertrophy, a decrease in volume occupied by their axonal projections and a reduced size of the α branch (Figure 4B). These structural deficits are correlated with learning deficits later leading to memory impairments. A possible mRNA target for this miR was identified as a ceramide phosphoethanolamine synthase, whose inhibition, as predicted by the model, significantly increases memory performance. This study further highlights the importance of miR regulation for proper development of neural circuits for adult cognitive functions [6,10,28,29,65,70].
FROM DISEASE MODELS TO MicroRNAs
An alternative approach for identifying miRNAs that may function in memory formation is to identify miRs that are dysregulated in fly models of human diseases linked with memory deficits. Several fly models for human disease have been developed and characterized [31,42,43,71].
Kong et al. [72•] overexpressed Aβ throughout the nervous system as a fly model of Alzheimer’s disease [73] and identified 17 miRs that were dysregulated in fly heads (8 increased/9 decreased). Performance after olfactory conditioning is impaired in this model along with lifespan and locomotion. Noteworthy among the dysregulated miRs are miR-276a (see above, [47••]) and let-7 [62••], implicated in LTM and learning, respectively, as discussed above. Bioinformatic analyses suggest that numerous biochemical pathways are likely dysregulated with such broad miR dysfunction, including the MAPK pathway, sphingolipid metabolism, and fatty acid biosynthesis. Which of these insults might be related to the measured learning impairment remains unclear. The same group later found that pan-neuronal expression of both wild-type or mutant Aβ42 resulted in reduced miR-124 expression [74•]. This is consistent with the well-documented role for miR-124 in neuronal plasticity and memory [8• 32], first described in Aplysia [75]. Kong et al. [74•] also demonstrated that miR-124 loss-of-function causes shorter lifespan, reduced climbing ability, and impaired olfactory learning. Additional experiments indicated that the dysregulated miR-124 in the Aβ overexpressing flies causes a learning deficit by regulating the Notch signaling pathway, supported by the observations that RNAi inhibition of the Notch ligand, Delta, and overexpression of miR-124 ameliorated the learning impairment in Aβ overexpressing flies. However, a number of control experiments including testing the effect of miR-124 expression itself remains to be completed.
Khanna et al. [76•] employed a similar approach to identify genes that are dysregulated in the MBn of mutants of Drosophila β-amyloid protein precursor-like (Appl), the ortholog of the human β–amyloid protein precursor through microarray studies. Surprisingly, they found that non-protein coding genes were the largest group affected by appl loss-of-function, including miR genes. They detected changes in the expression of 11 miRs genes including miR-9c, which is of particular interest because the human homolog is reported to be dysregulated in Alzheimer’s and Huntington’s disease [30], and recently found to be necessary for memory formation in the fly [48••].
Fragile X syndrome (FXS) is due to mutation of the fmr1 gene and characterized by behavioral phenotypes that include learning disability [31,77]. A fly model of FXS has been made by knocking-down dFmr1, the fly homolog of Fmr1 [31,78]. As described above, the dFMR protein has been shown to interact with Ago1, Ago2, Dicer and some specific miRs [31,56,79•], and generally thought to negatively regulate protein translation [79•]. dFmr1 potentially regulates memory formation in two separate ways. First, it causes defects in the development of the αβ MBn, which underlie learning impairment [77,79•,80]. Second, dFMR1 interacts with RISC and specific miRs to regulate protein expression at the synapse [79•]. For instance, dFMR1 regulates miR-124a expression (see above and [81]). The fact that dFMR1 is required for both the normal brain development and during adult memory formation physiologically [79•,80] adds complexity to the study of dFMR1 function with miRs and other miR-associated proteins which blurs the somewhat artificial distinction between the molecular mechanisms of development and adult physiology.
CONCLUSION
MicroRNAs provide a rapid cellular mechanism by modulating the expression of clusters of genes at post-transcriptional level. These features, along with the regulation they offer in synaptic compartments make them a particularly attractive class of molecules for modulating memory formation. Past research has clearly shown that the molecular machinery required for the biosynthesis of miRs is critical for normal LTM formation. In addition, several individual Drosophila miRs, including miR-276a, miR-980, and let-7, are required in certain brain regions for normal memory formation. Notably, their roles range from the normal development of the neural circuits that mediate memory formation, roles in the physiology of cells necessary for STM and ITM to an involvement in LTM through regulating synaptic protein synthesis. Identifying the mRNAs that are regulated by the relevant miRs and confirming their functional involvement in memory formation remains a challenge. Nevertheless, the tools and strategies to pursue these questions are available and Drosophila remains as arguably the best model system for such systematic investigations. Given the conservation of biological processes including memory formation across species, such investigations using the fly promise to identify the importance and logic of miR-mediated gene regulation in cognitive processes.
Highlights.
RISC is required for activity-dependent and sensory-specific protein translation.
RISC and associated proteins are required for olfactory long-term habituation.
MiR-276a regulates long-term memory by controlling dopamine receptor expression.
Let-7 is necessary for normal development of αβ MBn and learning in adult flies.
MiR-980, a memory suppressor, regulates memory formation through A2bp1.
Acknowledgments
Research in the authors’ laboratory was supported by grants R35NS097224 and P01NS090994 from the National Institute for Neurological Disorders and Stroke to R.L.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 5.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012;19:359–68. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- 8•.Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–9. doi: 10.1016/j.neuropharm.2014.01.026. A comprehensive review of miRs involved in memory formation based on studies with different model organisms and the implications these have for human memory disorders. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Reddy PH. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim Biophys Acta. 2016;1862:1617–1627. doi: 10.1016/j.bbadis.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunsberger JG, Austin DR, Chen G, Manji HK. MicroRNAs in mental health: from biological underpinnings to potential therapies. Neuromolecular Med. 2009;11:173–82. doi: 10.1007/s12017-009-8070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–34. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 13.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 17.Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity--a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014:369. doi: 10.1098/rstb.2013.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–9. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 19.Frey U, Morris RG. Synaptic tagging: implications for late maintenance of hippocampal long-term potentiation. Trends Neurosci. 1998;21:181–8. doi: 10.1016/s0166-2236(97)01189-2. [DOI] [PubMed] [Google Scholar]
- 20.Bailey CH, Kandel ER, Si K. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 2004;44:49–57. doi: 10.1016/j.neuron.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Rogerson T, Cai DJ, Frank A, Sano Y, Shobe J, Lopez-Aranda MF, Silva AJ. Synaptic tagging during memory allocation. Nat Rev Neurosci. 2014;15:157–69. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–59. [PubMed] [Google Scholar]
- 23.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 24.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci U S A. 1996;93:13445–52. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 26.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–70. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–31. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun E, Shi Y. MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Exp Neurol. 2015;268:46–53. doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tessier CR, Broadie K. Molecular and genetic analysis of the Drosophila model of fragile X syndrome. Results Probl Cell Differ. 2012;54:119–56. doi: 10.1007/978-3-642-21649-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez-Rapp J, Rainone S, Hébert SS. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2016 doi: 10.1016/j.pnpbp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A. 1985;157:263–77. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 34.Sachse S, Rueckert E, Keller A, Okada R, Tanaka NK, Ito K, Vosshall LB. Activity-dependent plasticity in an olfactory circuit. Neuron. 2007;56:838–50. doi: 10.1016/j.neuron.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 35.Das S, Sadanandappa MK, Dervan A, Larkin A, Lee JA, Sudhakaran IP, Priya R, Heidari R, Holohan EE, Pimentel A, et al. Plasticity of local GABAergic interneurons drives olfactory habituation. Proc Natl Acad Sci U S A. 2011;108:E646–54. doi: 10.1073/pnas.1106411108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busto GU, Cervantes-Sandoval I, Davis RL. Olfactory learning in Drosophila. Physiology (Bethesda) 2010;25:338–46. doi: 10.1152/physiol.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahsai L, Zars T. Learning and memory in Drosophila: behavior, genetics, and neural systems. Int Rev Neurobiol. 2011;99:139–67. doi: 10.1016/B978-0-12-387003-2.00006-9. [DOI] [PubMed] [Google Scholar]
- 38.Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twick I, Lee JA, Ramaswami M. Olfactory habituation in Drosophila-odor encoding and its plasticity in the antennal lobe. Prog Brain Res. 2014208:3–38. doi: 10.1016/B978-0-444-63350-7.00001-2. [DOI] [PubMed] [Google Scholar]
- 40.Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21:519–26. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis RL. Olfactory learning. Neuron. 2004;44:31–48. doi: 10.1016/j.neuron.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63:411–36. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rincon-Limas DE, Jensen K, Fernandez-Funez P. Drosophila models of proteinopathies: the little fly that could. Curr Pharm Des. 2012;18:1108–22. doi: 10.2174/138161212799315894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Androschuk A, Al-Jabri B, Bolduc FV. From Learning to Memory: What Flies Can Tell Us about Intellectual Disability Treatment. Front Psychiatry. 20156:85. doi: 10.3389/fpsyt.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, et al. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun. 2015;6:7279. doi: 10.1038/ncomms8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Li W, Cressy M, Qin H, Fulga T, Van Vactor D, Dubnau J. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J Neurosci. 2013;33:5821–33. doi: 10.1523/JNEUROSCI.4004-12.2013. The first Drosophila study implicating an individual miR, miR-276a, in memory formationMiR-276a tunes DopR expression, which conveys the unconditioned stimulus to the MBn during memory formation. This miR is also implicated in naïve odor avoidance through regulation of DopR but in the EB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48••.Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, Davis RL. microRNAs That Promote or Inhibit Memory Formation in Drosophila melanogaster. Genetics. 2015;200:569–80. doi: 10.1534/genetics.114.169623. This study employed a large-scale behavioral approach, identifying several miRs involved in memory formation due to regulating adult physiology or nervous system development. The authors identified five miRs from screening more than 130 miR genes, whose inhibition either decrease or increase memory. The final hits include miR-9c, miR-31a, miR-305, miR-974 and miR-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Guven-Ozkan T, Busto GU, Schutte SS, Cervantes-Sandoval I, O’Dowd DK, Davis RL. MiR-980 Is a Memory Suppressor MicroRNA that Regulates the Autism-Susceptibility Gene A2bp1. Cell Rep. 2016;14:1698–709. doi: 10.1016/j.celrep.2016.01.040. A role for miR-980 in memory formation was identified in this study. MiR-980 surprisingly acts as a memory suppressor miR since its inhibition increases memory. Moreover, this miR regulates neuronal excitability through A2bp1, an autism susceptibility gene, in most cell types of the olfactory system tested. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenböck G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–74. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 51.Wilson RI. Early olfactory processing in Drosophila: mechanisms and principles. Annu Rev Neurosci. 2013;36:217–41. doi: 10.1146/annurev-neuro-062111-150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. This groundbreaking study showed that the activity-induced and sensory-specific degradation of a component (Armitage) of RISC locally releases inhibition on the protein synthesis during long-term memory formation in the antennal lobe of the fly. [DOI] [PubMed] [Google Scholar]
- 53.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–41. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 54••.McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci U S A. 2011;108:E655–62. doi: 10.1073/pnas.1107198108. The authors identified several genes (including Atx2 and dFmr1) required for behavioral LTH and structural plasticity in the antennal lobe. Those genes interact together and additionally with components of the miRNA-pathway, including Ago1, to control the expression of CaMKII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hülsmeier J, Sarov M, Parker R, VijayRaghavan K, Ramaswami M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc Natl Acad Sci U S A. 2014;111:E99–E108. doi: 10.1073/pnas.1309543111. This paper followed McCann et al. and identified the role of dFmr1 in LTH and showed the translational control of CaMKII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27:7640–7. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ibáñez-Ventoso C, Vora M, Driscoll M. Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One. 2008;3:e2818. doi: 10.1371/journal.pone.0002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–13. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 60.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahin M, Sur M. Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science. 2015:350. doi: 10.1126/science.aab3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012;31:4511–23. doi: 10.1038/emboj.2012.298. This paper fully describes the induction of let-7 by ecdysone during development and its translational control of the Ab transcription factor leading to FasII regulation and to the differentiation of αβ MBn. The defects lead to brain structure and memory deficits. The authors connect miR, development and learning abilities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–3. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 64.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–41. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochem Soc Trans. 2013;41:815–20. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- 66.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–9. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Busto GU, Guven-Ozkan T, Chakraborty M, Davis RL. Developmental Inhibition of miR-iab8-3p Disrupts Mushroom Body Neuron Structure and Adult Learning Ability. Dev Biol. In press. A new role for miR-iab8-3p is described in αβ MBn development. Its inhibition leads to structural deficits associated with learning disabilities and memory deficits highlighting the crucial role for this miR during development for proper learning abilities in adult. [Google Scholar]
- 70.Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54:1103–12. doi: 10.1007/s11427-011-4249-8. [DOI] [PubMed] [Google Scholar]
- 71.Lim JY, Ott S, Crowther DC. Drosophila melanogaster as a Model for Studies on the Early Stages of Alzheimer’s Disease. Methods Mol Biol. 2016;1303:227–39. doi: 10.1007/978-1-4939-2627-5_13. [DOI] [PubMed] [Google Scholar]
- 72•.Kong Y, Wu J, Yuan L. MicroRNA expression analysis of adult-onset Drosophila Alzheimer’s disease model. Curr Alzheimer Res. 2014;11:882–91. The authors identified several miRs whose expression is modulated in a genetic model of Alzheimer disease. These miRs include miR-9c and miR-276a confirming the central role for these miRs in memory formation. [PubMed] [Google Scholar]
- 73.Crowther DC, Kinghorn KJ, Miranda E, Page R, Curry JA, Duthie FA, Gubb DC, Lomas DA. Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience. 2005;132:123–35. doi: 10.1016/j.neuroscience.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 74•.Kong Y, Wu J, Zhang D, Wan C, Yuan L. The Role of miR-124 in Drosophila Alzheimer’s Disease Model by Targeting Delta in Notch Signaling Pathway. Curr Mol Med. 2015;15:980–9. doi: 10.2174/1566524016666151123114608. This study shows a down-regulation of miR-124 in a model of Alzheimer’s disease. This miR might influence memory performance by regulating the Notch pathway. [DOI] [PubMed] [Google Scholar]
- 75.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–17. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76•.Khanna MR, Fortini ME. Transcriptomic Analysis of Drosophila Mushroom Body Neurons Lacking Amyloid-β Precursor-Like Protein Activity. J Alzheimers Dis. 2015;46:913–28. doi: 10.3233/JAD-141491. The authors of this study used an original approach to isolate MBn and identify the potential targets of APPL. Among the targets identified, non-protein coding genes including miRs represented the largest class. [DOI] [PubMed] [Google Scholar]
- 77.Siomi H, Ishizuka A, Siomi MC. RNA interference: a new mechanism by which FMRP acts in the normal brain? What can Drosophila teach us? Ment Retard Dev Disabil Res Rev. 2004;10:68–74. doi: 10.1002/mrdd.20011. [DOI] [PubMed] [Google Scholar]
- 78.Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 79•.Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–5. doi: 10.1038/nn.2175. The authors showed that dFmr1 is necessary for memory formation and interacts with Ago1. Interestingly, an excess of protein synthesis impairs memory formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu XL, Li Y, Wang F, Gao FB. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J Neurosci. 2008;28:11883–9. doi: 10.1523/JNEUROSCI.4114-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 83.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fiorenza A, Barco A. Role of Dicer and the miRNA system in neuronal plasticity and brain function. Neurobiol Learn Mem. 2016 doi: 10.1016/j.nlm.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Yu D, Ponomarev A, Davis RL. Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron. 2004;42:437–49. doi: 10.1016/s0896-6273(04)00217-x. [DOI] [PubMed] [Google Scholar]