Abstract
The E.coli single stranded DNA binding protein (SSB) is essential to all aspects of DNA metabolism. Here, it has two seemingly disparate but equally important roles: it binds rapidly and cooperatively to single stranded DNA (ssDNA) and it binds to partner proteins that constitute the SSB interactome. These two roles are not disparate but are instead, intimately linked. A model is presented wherein the intrinsically disordered linker (IDL) is directly responsible for mediating protein-protein interactions. It does this by binding, via PXXP motifs, to the OB-fold (aka SH3 domain) of a nearby protein. When the nearby protein is another SSB tetramer, this leads to a highly efficient ssDNA binding reaction that rapidly and cooperatively covers and protects the exposed nucleic acid from degradation. Alternatively, when the nearby protein is a member of the SSB interactome, loading of the enzyme onto the DNA takes places.
1. Introduction
The Escherichia coli single stranded DNA binding protein (SSB) is essential to all aspects of DNA metabolism where it has two essential roles. In the first, it stabilizes single-stranded DNA (ssDNA) intermediates generated during DNA processing (Chase and Williams, 1986; Kowalczykowski et al., 1994; Lohman and Ferrari, 1994; Meyer and Laine, 1990; Shereda et al., 2008). In the second, it mediates binding to as many as fourteen DNA-binding proteins that constitute the SSB interactome (Costes et al., 2010; Shereda et al., 2008).
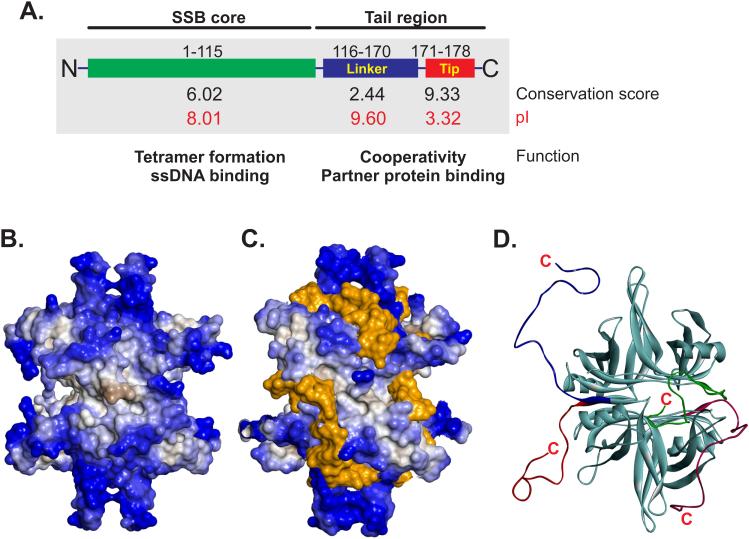
The protein, like the majority of eubacterial SSBs, exists as a stable homo-tetramer with a monomer MW of 18, 843 Da and a pI of 5.44 (Figure 1A B and (Sancar et al., 1981)). Each 178 amino acid length monomer can be divided into two domains defined by proteolytic cleavage: an N-terminal portion comprising the first 115 residues and a C-terminal domain that includes residues 116 to 178 (Figure 1A and (Williams et al., 1983)). Sequence analyses reveal that the first 115 residues are well conserved, with a conservation score of 6 out of 10 and in addition, the pI of this domain is 8.01 (Simossis and Heringa, 2005; Wilkins et al., 1999). The C-terminal domain can be further subdivided into two sub-domains: a sequence of approximately 54 amino acids comprising residues 116 to 170, that is known as the intrinsically disordered linker (IDL) and, the last 8 residues which are known as the acidic tip or C-peptide (Kozlov et al., 2015; Shereda et al., 2008). The IDL has a conservation score of 2.44 and a pI of 9.6 whereas the acidic tip has a score of 9.33 and a pI of 3.32 (Figure 1A).
Figure 1. Organization of the E. coli SSB protein.
(A). The protein is shown in schematic form divided into the N-terminal core and C-terminal tail regions by proteolytic cleavage. The tail can be further sub-divided into the IDL and acidic tip, labelled as linker and tip, respectively. The conservation scores for each region were calculated from alignments using Praline (Simossis and Heringa, 2005). The pI of each region is shown in red and was calculated using the ProtParam tool of Expasy (Wilkins et al., 1999). The pI of the intact protein is 5.44. (B). SSB exists as a tetramer. Only the core domain is clearly visible in the majority of structures due to the disordered C-terminal tail. The SSB tetramer is presented as a Connolly surface and is coloured according to hydrophobicity (Connolly, 1983). (C). Single-stranded DNA binding to the N-terminal core domain of each monomer within the tetramer occurs via wrapping. The protein and DNA are represented as Connolly surfaces and are coloured according to hydrophobicity and orange respectively, to enable clear visualization. (D). The C-terminal tail is intrinsically disordered and could adopt a variety of configurations. Images were generated using BIOVIA Discovery Studio Visualizer.
The well conserved N-terminal or core domain contains elements critical to tetramer formation and the oligonucleotide/oligosaccharide binding-fold (OB-fold) critical to the binding of ssDNA (Raghunathan et al., 2000; Williams et al., 1983). DNA binding by this domain is non-specific in nature and occurs via the wrapping of ssDNA around the SSB tetramer using an extensive network of electrostatic and base-stacking interactions with the phosphodiester backbone and nucleotide bases, respectively (Figure 1C and (Chrysogelos and Griffith, 1982; Kuznetsov et al., 2006; Raghunathan et al., 2000)).
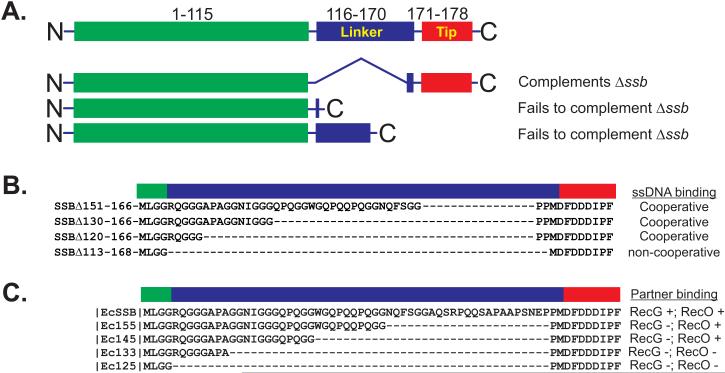
Initially, the C-terminus was proposed to be a non-essential part of the protein, as an ssb gene containing a deletion of residues 116-167 was able to complement an ssb deletion (Figure 2A and (Curth et al., 1996)). Subsequently, it was shown that truncated ssb genes terminating at residues 117 or 152 were unable to complement the same ssb deletion. Later, in vitro studies showed that deletion of the C-terminal 15-20 residues eliminated interactions with the psi and chi subunits of the DNA polymerase III complex (Kelman et al., 1998; Witte et al., 2003). More recently, studies both in vivo and in vitro using the ssbΔC8 gene and SSBΔC8 protein respectively, have suggested that the last 8 residues are important for mediating target protein binding (Buss et al., 2008; Cadman and McGlynn, 2004; Pottinger et al., 2016; Shereda et al., 2007; Tan et al., 2016; Yu et al., 2016). However, and as explained below, while removal of the last 8 residues of SSB does have a negative impact on target protein binding, this does not necessarily mean that it is the protein-protein interaction domain per se.
Figure 2. The effects of C-terminal deletions on SSB function.
(A). Analysis of the effects of deletions in vivo. A schematic of the wild type protein is shown at the top for comparison and colouring is the same as that in Figure 1A. Details of the deletions are discussed in the text and are taken from (Curth et al., 1996). (B). Summary of the effects of IDL deletions on ssDNA binding. These mutant proteins were published in (Kozlov et al., 2015). (C). Summary of the effects of IDL deletions on interactome partner binding. These mutants were studied in (Pottinger et al., 2016).
Early biochemical experiments with C-terminally truncated E. coli SSB led to a model with the acidic tip bound to the OB-fold in the N-terminal domain (Curth et al., 1996; Kozlov et al., 2010; Mason et al., 2013; Williams et al., 1983). As ssDNA and C-peptide binding by the OB-fold are competitive, this model proposed that in solution, the C-termini of SSB are bound to the tetramer and are released upon ssDNA binding, making them available for interactions with target proteins. At low pH, the SSB tetramer dissociates into monomers and this was taken advantage of to show direct binding between the C-peptide and valines 29 and 58 of the OB-fold (Shishmarev et al., 2014). While this region interacts with the N-terminal domain when SSB is monomeric, this interaction is transient in the tetramer. In fact, in solution, the acidic tip spends the majority of its time away from the tetramer core making itself available for protein-protein interactions.(Su et al., 2014). This was confirmed in vivo where binding of SSB in the absence of DNA to the fork rescue helicases RecG and PriA was observed (Yu et al., 2016). Furthermore, binding in vivo required the presence of the last 8 residues of SSB, a finding that is consistent with our work and that of others suggesting that this region of SSB is responsible for mediating interactions with key proteins involved in DNA metabolism known as the SSB interactome (Buss et al., 2008; Cadman and McGlynn, 2004; Costes et al., 2010; Genschel et al., 2000; Glover and McHenry, 1998; Handa et al., 2001; Kantake et al., 2002; Pottinger et al., 2016; Shereda et al., 2007; Suski and Marians, 2008; Umezu and Kolodner, 1994; Witte et al., 2003). However, this may not be correct as removal of the acidic tip has profound effects on SSB. First, removal virtually eliminates binding to target proteins (Pottinger et al., 2016; Yu et al., 2016). Second, it affects cooperative binding to ssDNA (Kozlov et al., 2015). Finally, it affects the stability of the SSB tetramer. In vitro in 10mM ammonium acetate, SSBΔC8 tetramers exchange subunits with 15N-SSBΔC8 tetramers at a rate twice that of wild type (Mason et al., 2013). Collectively, these studies imply that the last eight residues of the C-terminus have a regulatory role in SSB function.
If this is correct, how does SSB bind to target proteins? This leaves the least well-studied region of SSB that is, the intrinsically discorded linker as being responsible. The C-terminus comprising the IDL and acidic tip or C-peptide, is highly disordered even when the protein is bound to ssDNA and could adopt different conformations (Figure 1D and (Matsumoto et al., 2000; Savvides et al., 2004)). As a result, little interest has been shown in this region of the protein until recently. The first study from the Lohman group, used a series of deletions of varying sizes to demonstrate that this region of SSB is critical to the cooperative binding of single stranded DNA (Figure 2B and (Kozlov et al., 2015)). Cooperativity is critical to SSB function enabling rapid ssDNA binding resulting in its protection and/or conversion into a more suitable substrate for subsequent processing (Bianco et al., 1998; Lohman and Ferrari, 1994). In fact, mutation of a single residue within the conserved N-terminal domain, W54, results in defects in DNA replication and repair and these defects can be attributed to a loss of cooperativity (Ferrari et al., 1997). A second study from the Bianco laboratory used similar deletions of the IDL in an effort to understand how binding to the SSB interactome partners RecG (76kDa) and RecO (27kDa) occurred (Figure 2C and (Pottinger et al., 2016)). The results showed that the smallest deletion of the IDL producing SSB155 or SSBΔ146-167, eliminated binding to the larger RecG. In contrast, larger deletions to produce SSB133 (SSBΔ124-167) and SSB125 (SSBΔ115-167) were required to eliminate binding to the smaller RecO. Importantly, in both studies, each deletion mutant contained the acidic tip. Thus the residues comprising the acidic tip cannot be the sole determinant in protein-protein interactions. Therefore, while the IDL may not have been thought of as essential to protein function it is essential to two critical aspects of SSB function: cooperative ssDNA binding and interactions with partner proteins.
In this review, a model is proposed for the mechanism of SSB function. In this model, the IDL and key components thereof, mediate protein-protein interactions. These interactions are critical to cooperative binding of ssDNA as well as to SSB interactome function. In addition, and consistent with the work of others, the model proposes that the acidic tip is not involved in protein binding per se but is instead a regulatory element (Mason et al., 2013; Su et al., 2014).
2. Analysis of the IDL
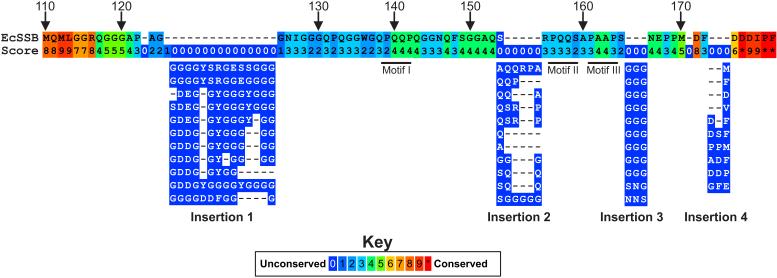
In alignments of eubacterial SSB sequences, homology is restricted to the N-terminal 1~120 residues and the C-terminal 8-10 amino acids (Kozlov et al., 2015; Shereda et al., 2008). Little homology is observed in the IDL. Therefore, to begin to understand which components, if any may be important in protein function, an alignment was done using the C-termini of 251 Proteobacterial SSB sequences. In this alignment, short ssb genes (e.g., those from Geobacter) and those which do not have the acidic tip at the extreme C-terminus were excluded. A summary of the alignment beginning at residue 110 (E. coli numbering) is shown in Figure 3 and the complete alignment in Supplementary Figure 1.
Figure 3. The IDL is not well conserved.
A summary of the alignment of the C-terminal tails of 251 Proteobacterial SSB proteins is presented with only the E. coli sequence shown. The alignment was done using PRALINE (Simossis and Heringa, 2005). The numbering above the sequence corresponds to residue numbering and the values below each amino acid correspond to the alignment score for residues in that position. The positions of the PXXP motifs are indicated by the thick black lines just below positions 140, 156 and 162. Immediately below the E. coli sequence, are representative sequences corresponding to insertions 1-4 (details are in the text). In several cases, the same sequence appeared more than once. Consequently, only one is shown for clarity. The complete alignment is available in Supplementary Figure 1.
This analysis shows that for the entire C-terminal domain, sequences are 40% identical. As expected, the homology is highest within the first seven and last 5 residues and D171, corresponding to the C-terminal end of the core domain and acidic tip, respectively. Between these conserved ends, it initially appears that there is little homology with conservation scores ranging from 0 to 3. However, there are regions of homology which have scores of 4 to 6 out of a possible 10 (green boxes, Fig. 3). These are interrupted by four insertions: the first after residue 124; the second at position 154, the third after amino acid 165 and the final one after F172. A summary of the types of insertions observed at each position are indicated below the E. coli sequence. What is evident is that for each insertion there is strong sequence commonality and for insertion1, the predominant residue inserted is glycine. The same can be said for insertion 3. In contrast, for insertion 2, the predominant residues are glutamine, serine and glycine. Finally, insertion 4 varies in length from 1 to 3 residues, with the insertions containing large hydrophobic or charged residues. Those bacteria the comprise insertion1 are predominantly Bulkholderia sp.
The nature of the residues inserted at each position suggests that for those bacteria, both longer and more flexible C-termini may be required for optimal SSB function. This may be reflected in the larger size and greater range in size, of interactome partners to which each SSB must bind. In addition to the insertions, there are several notable regions that are reasonably well conserved in some, but not all of the aligned sequences. These include the PXXP motifs occurring at residues 139, 156 and 161. The first PXXP motif, PQQP, is invariant in 27% of the 251 sequences. This is particularly evident when the first 23 sequences in the alignment are examined (Supplementary Figure 1). The second, PQQS, appears less well conserved while motif III, PAAP has an intermediate level of conservation. For the latter PXXP sequences, the lower level of homology arises due to insertions and deletions, relative to the E.coli sequence, resulting in their misalignment as well as to changes. The second motif, PQQS occurs in 22% of the 251 proteins either as PQQS or PQQY. Motif III is invariant in 10% of sequences and is replaced by PSAP, PQQS, PAAS, or PAAQ. When taken as a group, all of the motif III variants occur in 30% of the 251 sequences. Collectively, the presence of these motifs suggests they may play an important role in the function of SSB protein.
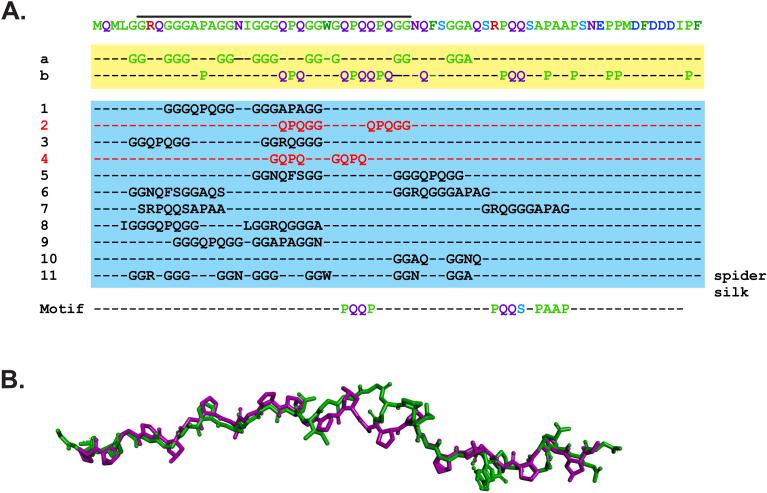
As the E.coli SSB is perhaps the most well-studied protein in this family, a more detailed analysis of its IDL was undertaken to further understand which elements may be critical for function (Pottinger et al., 2016). First, an analysis of the primary amino acid sequence of residues 110-178 shows that this region is over-represented for Gly 24.6%, Gln 17.4%, Pro 14.5%, and to a lesser extent, Ala and Ser (8.7 and 5.8%, respectively; Fig. 4A, residues highlighted in the yellow box). The presence and spacing of these over-represented residues in the N-terminal half of this region, ending at residues 148 and 149 (Phe and Ser respectively), is consistent with the formation of a polyproline II helix (PPII) found in proteins such as collagen (Adzhubei et al., 2013). However, the spacing of the proline residues in collagen occurs at every fourth position which is not the case for SSB. Instead, we observed that glycine, glutamine and arginine occur in those positions in SSB. A recent study has shown that these amino acids can in fact substitute for proline without disrupting a model PPII helix (Brown and Zondlo, 2012). As one strand of a collagen fiber is essentially a PPII helix, we wanted to determine whether this region of SSB could be modeled on a collagen-like peptide (Biasini et al., 2014; Schwede et al., 2003). Indeed, the SSB sequence can adopt a PPII helix that superimposes well with the peptide, with an RMSD = 0.8 Angstroms for the backbone atoms (Figure 4B). In contrast, the residues downstream of F148/S149 could not be modelled on the peptide using the same approach (data not shown).
Figure 4. The IDL contains sequence elements critical to its function.
(A). The IDL contains repetitive elements consisting mainly of glycine, proline and glutamine. The sequence of the C-terminal 69 residues of E.coli SSB is presented in the first line, with the most over-represented residues highlighted in lines a and b. Sequence analysis of the protein was done using REPRO at http://www.ibi.vu.nl/programs/ to determine the presence of repetitive elements. Lines 1-11 show a subset of the repeated sequence elements identified in this analysis (George and Heringa, 2000). The analysis of the IDL sequence also revealed the presence of three PXXP motifs indicated in the Motif line. (B). The core-proximal region of SSB can be modelled on a PPII helix. The amino acid sequence indicated by the black line in panel A (residues 114-147), was modelled onto a single collagen strand from PDB file 1GAC (Bella et al., 1994). Modelling was done using Swiss-Model at http://swissmodel.expasy.org/. The superimposed structures are presented with collagen coloured purple and SSB in green.
What then is the significance of the proposed PPII helix? This helix is an unusual structure with the prolines forming a continuous hydrophobic strip around the surface of the helix, while the backbone carbonyls present ideal hydrogen bonding sites, being both conformationally restricted (and therefore poorly hydrated) and electron-rich (Hongo et al., 2005; Nagarajan et al., 1998; Okuyama et al., 1981). Consequently, this helix would present an easily accessible hydrophobic surface, as well as a good hydrogen-bonding site (Williamson, 1994). The accessibility of PPII helices is greatly enhanced by the fact that they are frequently found either at the amino- or carboxyl termini of proteins where they form extended structures that have been described as ‘sticky arms’ (Adzhubei and Sternberg, 1993; Kay et al., 2000; Matsushima et al., 1990). In addition, PPII-helices are flexible, and this has been suggested to be the major feature underlying its function in protein interactions and in fact, these helices are often involved in protein-protein interactions (Adzhubei et al., 2013; Bochicchio et al., 2004). While the analysis suggests that this region of SSB could adopt a PPII helix, further experimentation is required to determine whether this is in fact the case. Regardless, removal of this region of SSB or alteration of its position relative to the tetramer core eliminates target protein binding (Pottinger et al., 2016; Tan et al., 2016).
Second, and as the gly, pro and gln residues appear in clusters in the SSB C-terminal domain (Fig. 3 and 4A, lines a and b), we analyzed the entire 69 residue region for repeats using REPRO, which identifies protein sequence repeats using a graph-based iterative clustering procedure (George and Heringa, 2000). Eleven of the identified repeats are shown in Figure 4A (lines 1-11; blue box). Some are these are identical (numbers 2 and 4), while the remaining identified repeats possess a high degree of similarity with one another (quasi-repeats), e.g., repeats 1 and 10. We also noticed the presence of seven hydrophilic GGX repeats (line 11). In addition, of the first ten repeats, nine cluster in the N-terminal half of the IDL sequence upstream of F148. Likewise, six of the seven GGX repeats also occur in this region of the protein.
Similar repeats containing G, P and Q have been identified in several eukaryotic proteins such as spider silk, the X-type HMW subunit of wheat gluten and ω-protein where they have been shown to confer important structural features and special functions in addition to conferring elastomeric properties to that region of the protein (Matsushima et al., 2008). The over-representation of gly, gln, pro, and ser residues and the presence of the repeats, may impart similar elastomeric properties to the C-terminus of SSB, enabling it to interact with a large range of partners of different sizes. In addition, these sequence elements may impart tensile strength, which may be a key component in cooperative ssDNA binding where interactions between neighboring SSB tetramers would be critical to rapidly protecting exposed DNA. Once bound to the DNA, the strength afforded by interactions between IDLs of one tetramer with that of a neighbor, would result in a very stable protein-DNA complex that can resist displacement by many potentially harmful nucleases.
Finally, our analysis also identified PXXP motifs in the C-terminus of SSB, the first one starting at residue 138 (PQQP), possibly a second starting at residue 156 (PQQS) and a third at amino acid 161 (PAAP). As alluded to above, they are reasonably well conserved in the Proteobacterial sequences. How might they contribute to SSB function? PXXP motifs are most well known for their ability to bind structurally conserved Src homology 3 (SH3) domains. These domains are ~50 residue modules that are ubiquitous in biological systems and which often occur in signaling and cytoskeletal proteins in eukaryotes (Dalgarno et al., 1997; Kay et al., 2000; Ponting et al., 1999; Sudol, 1998). The SH3 domain has a characteristic fold which consists of five or six beta-strands arranged as two tightly packed anti-parallel beta sheets. SH3 domains are similar in structure to the OB-fold present in many single-strand DNA binding proteins including SSB (Agrawal and Kishan, 2001). This suggests that interactions between either one or more PXXP motifs and the OB-fold (i.e., SH3 domain) in the adjacent tetramer, may play a critical role in SSB-ssDNA complex formation.
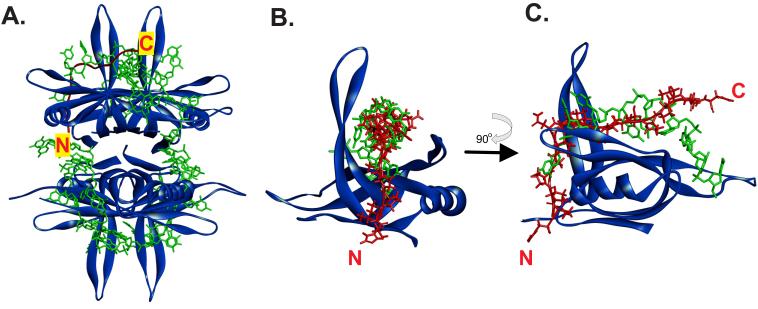
To determine whether this could occur, we superimposed an SH3 domain bound to its peptide ligand onto an SSB tetramer using TM-align (Zhang and Skolnick, 2005). For this analysis, we selected PDB file 2KXC as the SH3 domain as the ligand contains PXXP motifs in close juxtaposition similar to that observed for motifs II and III in SSB (Aitio et al., 2010). Then, we used PDB file 1EYG which contains the N-terminal domain of SSB bound to ssDNA (Raghunathan et al., 2000). Once the alignment process was complete, we removed the SH3 domain so that the position of the ligand on SSB relative to the ssDNA binding site could be readily visualized. The aligned structures are shown in Supplementary Figure 2 and the SSB-ligand-ssDNA images are presented in Figure 5. The SH3 domain and OB-fold align with an RMSD = 2.68 angstroms. This alignment positions the ligand in the ssDNA binding pocket of the OB-fold. When viewed in this way, it suggests that the region of the IDL of SSB spanning the PXXP motifs could bind to the OB-fold of either another monomer within the same tetramer or, to that of another tetramer.
Figure 5. The PXXP motifs of the IDL can bind to the OB-fold in the same position as ssDNA.
(A). Position of a PXXP ligand (i.e., the IDL) bound to one subunit of the SSB tetramer. (B) and (C). The ligand and ssDNA may bind to the same position in the OB-fold. In each panel, SSB is represented as a ribbon diagram and is coloured blue. ssDNA is shown as a stick model in green and the PXXP ligand is coloured in red. In panel A, the ligand is represented as a schematic and in panels B and C as a stick model. The positions of the N- and C-termini of the ligand are indicated. The image in C is rotated 90° relative to that in B as shown by the arrow. The PDB files used in this analysis are 1EYG for SSB and 2KXC for the ligand (Aitio et al., 2010; Raghunathan et al., 2000). The 2KXC structure was used in this analysis as the ligand contains tandem PXXP motifs similar to that in the SSB IDL. To generate the images shown, 2KXC was aligned onto file 1EYG using TM-align (Zhang and Skolnick, 2005). Then, the SH3 domain was removed so that clear visualization of the ligand (IDL) bound to the OB-fold in one monomer can be made.
3. Mechanism of SSB-ssDNA complex formation
Our model for the mechanism of protein-DNA complex formation by SSB is as follows. When the protein is in solution, the IDL adopts a variety of conformations with the acidic tip making only transient interactions with the N-terminal core (Matsumoto et al., 2000; Savvides et al., 2004; Su et al., 2014). The presence of the acidic tip is critical as it may prevent an IDL from binding to the tetramer from which it emanates. Once an SSB tetramer binds to DNA under conditions where cooperative binding is critical (SSB35 mode in low salt), the IDL from one tetramer interacts with a vacant OB-fold of a monomer within a nearby SSB. The inherent flexibility in this region of the protein as suggested by our sequence analysis may then facilitate sliding of the second tetramer relative to the first so that they occupy immediately adjacent positions on the ssDNA. There is evidence for tetramer sliding on ssDNA (Su et al., 2014). Continued IDL-OB-fold interactions would then facilitate rapid and complete binding of the ssDNA in highly cooperative fashion. Cooperativity is critical to SSB function enabling rapid single stranded DNA binding resulting in its protection and/or conversion into a more suitable substrate (Bianco et al., 1998; Lohman and Bujalowski, 1994).
4. Implications of the model for the SSB interactome
The presence of the PXXP motifs prompted us to more closely examine members of the SSB interactome for the presence of SH3 domains. Even though SH3 domains are thought of as playing important roles in eukaryotic systems, they have been identified in several E. coli proteins, including Exonuclease I, an SSB-interactome partner (Breyer and Matthews, 2000; Genschel et al., 2000). With this information in hand, we examined the structures of each of the known interactome partners and found OB-folds in each (P. Bianco, unpublished). The presence of an OB-fold was initially determined by visual examination and confirmed by submitting the region to the DALI server (Holm and Rosenstrom, 2010).
In the case of RecG a known interactome partner, we overlaid the SH3 domain in PDB file 2KXC onto the OB-fold in the wedge domain of the helicase (Bianco, 2016). Once this process was complete we removed the SH3 domain only, leaving the ligand in place to illustrate the potential binding location of the SSB IDL on RecG. Here, the ligand adopts a position on the wedge domain of RecG that is incompatible with fork binding and places the OB-folds of the SSB tetramer in a position to bind ssDNA, thereby facilitating the loading of RecG as shown previously (Sun et al., 2015). This interaction would also provide an explanation for SSB-mediated remodeling of the helicase, which prevents it from docking at a fork and instead, facilitating thermal sliding. The interaction between the PXXP-containing ligand and RecG implies a significant interface for binding which is consistent with the stability of the RecG-SSB complex, which can be purified through several chromatography steps (Yu et al., 2016).
5. Summary
The analysis presented herein has led to a model whereby the C-terminal domain of SSB plays essential roles in the function of the protein. These are the cooperative binding to ssDNA and also in the interaction with partner proteins of the SSB interactome. In the model presented herein, the intrinsically disordered linker (IDL) is directly responsible for mediating protein-protein interactions. These occur between the region spanning the PXXP motifs and the OB-fold of the partner, either another SSB or an interactome member such as RecG. In addition, and as we propose that the IDL is responsible for binding, we also suggest a regulatory role for the acidic tip, rather than a direct role in protein binding.
In our view, this makes sense. The C-peptide is the most highly conserved region of the protein – each SSB has it, although it is not always positioned at the extreme C-terminus. When the acidic tip is deleted as in the SSBΔC8 protein, binding to ssDNA as assayed by site-size and salt dependence is largely the same as that of that of wild type (Liu et al., 2011). Cooperative DNA binding is however negatively impacted (Kozlov et al., 2015). Further, a recent study shows that subunit exchange between tetramers is significantly enhanced when the acidic tip is removed, consistent with it having a stabilizing function (Mason et al., 2013). It is clear that the presence of the last 8 residues of SSB are required for binding to target proteins. When they are removed, binding is essentially eliminated. This could be interpreted to mean that the acidic tip binds directly to target proteins. However, a failure of the IDL of SSBΔC8 to interact with the target proteins can also explain the data. When the C-peptide is present, it prevents the IDL from binding either to itself, another IDL of the same protein or to an OB-fold of the SSB tetramer from which it emanates. In its absence, the IDL can bind to any of the sites listed above instead of the target protein and thereby eliminating complex formation.
As each eubacterial genome is unique and to allow for species specificity, the binding site in our view, should be unique. When sequence alignments are performed, they reveal that the IDL, while having common features, is unique to each species. We propose that binding occurs between a sub-domain within the IDL, spanning the PXXP motifs. These motifs then direct binding to the OB-fold of the target protein – either another SSB tetramer or a member of the interactome. The model predicts that if these motifs are mutated, or if OB-folds in the target protein are deleted, protein-protein interactions would be eliminated. Preliminary data suggests that this is indeed the case (P.Bianco, in preparation). The model also predicts that mutation of the PXXP motifs would have a negative impact on cooperative ssDNA binding and also on partner protein binding. DNA binding studies are underway and separately, preliminary data suggests that mutation of the PXXP motifs negatively impacts interactome partner binding (P.Bianco, in preparation).
6. Conclusion
The IDL of the C-terminus of SSB plays acritical role in the function of the protein. It is responsible for mediating protein-protein interactions. When these occur between SSB tetramers, cooperative binding to ssDNA results. When they occur between SSB and an interactome partner, loading of that partner onto the DNA occurs.
Supplementary Material
Acknowledgement
Work in the Bianco laboratory is supported by NIH Grant GM100156 to PRB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei AA, Sternberg MJ. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993;229:472–93. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Adzhubei AA, Sternberg MJ, Makarov AA. Polyproline-II helix in proteins: structure and function. J Mol Biol. 2013;425:2100–32. doi: 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Agrawal V, Kishan RK. Functional evolution of two subtly different (similar) folds. BMC Struct Biol. 2001;1:5. doi: 10.1186/1472-6807-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitio O, Hellman M, Kazlauskas A, Vingadassalom DF, Leong JM, Saksela K, Permi P. Recognition of tandem PxxP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly. Proc Natl Acad Sci U S A. 2010;107:21743–8. doi: 10.1073/pnas.1010243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- Bianco PR. RecG and SSB: an intimate association to rescue a stalled replication fork. Genes. 2016 doi: 10.1002/pro.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–8. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochicchio B, Ait-Ali A, Tamburro AM, Alix AJ. Spectroscopic evidence revealing polyproline II structure in hydrophobic, putatively elastomeric sequences encoded by specific exons of human tropoelastin. Biopolymers. 2004;73:484–93. doi: 10.1002/bip.10552. [DOI] [PubMed] [Google Scholar]
- Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol. 2000;7:1125–8. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- Brown AM, Zondlo NJ. A propensity scale for type II polyproline helices (PPII): aromatic amino acids in proline-rich sequences strongly disfavor PPII due to proline-aromatic interactions. Biochemistry. 2012;51:5041–51. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- Buss J, Kimura Y, Bianco P. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 2008;36:7029–42. doi: 10.1093/nar/gkn795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Research. 2004;32:6378–87. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl. Acad. Sci. USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–13. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 2010;6:e1001238. doi: 10.1371/journal.pgen.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–11. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno DC, Botfield MC, Rickles RJ. SH3 domains and drug design: ligands, structure, and biological function. Biopolymers. 1997;43:383–400. doi: 10.1002/(SICI)1097-0282(1997)43:5<383::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Ferrari ME, Fang J, Lohman TM. A mutation in E. coli SSB protein (W54S) alters intra-tetramer negative cooperativity and inter-tetramer positive cooperativity for single-stranded DNA binding. Biophys Chem. 1997;64:235–51. doi: 10.1016/s0301-4622(96)02223-5. [DOI] [PubMed] [Google Scholar]
- Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biological Chemistry. 2000;381:183–92. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- George RA, Heringa J. The REPRO server: finding protein internal sequence repeats through the Web. Trends Biochem Sci. 2000;25:515–7. doi: 10.1016/s0968-0004(00)01643-1. [DOI] [PubMed] [Google Scholar]
- Glover B, McHenry C. The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem. 1998;273:23476–84. doi: 10.1074/jbc.273.36.23476. [DOI] [PubMed] [Google Scholar]
- Handa P, Acharya N, Varshney U. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. Journal of Biological Chemistry. 2001;276:16992–7. doi: 10.1074/jbc.M100393200. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo C, Noguchi K, Okuyama K, Tanaka Y, Nishino N. Repetitive interactions observed in the crystal structure of a collagen-model peptide, [(Pro-Pro-Gly)9]3. J Biochem. 2005;138:135–44. doi: 10.1093/jb/mvi108. [DOI] [PubMed] [Google Scholar]
- Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15327–32. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–41. [PubMed] [Google Scholar]
- Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–49. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Cox MM, Lohman TM. Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J Biol Chem. 2010;285:17246–52. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Weiland E, Mittal A, Waldman V, Antony E, Fazio N, Pappu RV, Lohman TM. Intrinsically disordered C-terminal tails of E. coli single-stranded DNA binding protein regulate cooperative binding to single-stranded DNA. J Mol Biol. 2015;427:763–74. doi: 10.1016/j.jmb.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S, Kozlov A, Lohman T, Ansari A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Liu J, Choi M, Stanenas AG, Byrd AK, Raney KD, Cohan C, Bianco PR. Novel, fluorescent, SSB protein chimeras with broad utility. Protein Sci. 2011;20:1005–20. doi: 10.1002/pro.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T, Ferrari M. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Bujalowski W. Effects of base composition on the negative cooperativity and binding mode transitions of Escherichia coli SSB-single-stranded DNA complexes. Biochemistry. 1994;33:6167–76. doi: 10.1021/bi00186a016. [DOI] [PubMed] [Google Scholar]
- Mason CE, Jergic S, Lo AT, Wang Y, Dixon NE, Beck JL. Escherichia coli single-stranded DNA-binding protein: nanoESI-MS studies of salt-modulated subunit exchange and DNA binding transactions. J Am Soc Mass Spectrom. 2013;24:274–85. doi: 10.1007/s13361-012-0552-2. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Morimoto Y, Shibata N, Kinebuchi T, Shimamoto N, Tsukihara T, Yasuoka N. Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis. J Biochem. 2000;127:329–35. doi: 10.1093/oxfordjournals.jbchem.a022611. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Creutz CE, Kretsinger RH. Polyproline, beta-turn helices. Novel secondary structures proposed for the tandem repeats within rhodopsin, synaptophysin, synexin, gliadin, RNA polymerase II, hordein, and gluten. Proteins. 1990;7:125–55. doi: 10.1002/prot.340070204. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Yoshida H, Kumaki Y, Kamiya M, Tanaka T, Izumi Y, Kretsinger RH. Flexible structures and ligand interactions of tandem repeats consisting of proline, glycine, asparagine, serine, and/or threonine rich oligopeptides in proteins. Curr Protein Pept Sci. 2008;9:591–610. doi: 10.2174/138920308786733886. [DOI] [PubMed] [Google Scholar]
- Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–80. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan V, Kamitori S, Okuyama K. Crystal structure analysis of collagen model peptide (Pro-pro-Gly)10. J Biochem. 1998;124:1117–23. doi: 10.1093/oxfordjournals.jbchem.a022229. [DOI] [PubMed] [Google Scholar]
- Okuyama K, Okuyama K, Arnott S, Takayanagi M, Kakudo M. Crystal and molecular structure of a collagen-like polypeptide (Pro-Pro-Gly)10. J Mol Biol. 1981;152:427–43. doi: 10.1016/0022-2836(81)90252-7. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. Journal of Molecular Biology. 1999;289:729–45. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- Pottinger S, Tan HY, Nguyenduc T, Rex K, Varshney U, Bianco PR. Insight into the function of the E.coli SSB C-terminal tail: I. The length and composition of the linker is critical to target protein binding in vivo. 2016 submitted. [Google Scholar]
- Raghunathan S, Kozlov A, Lohman T, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- Sancar A, Williams KR, Chase JW, Rupp WD. Sequences of the ssb gene and protein. Proc. Natl. Acad. Sci. USA. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Science. 2004;13:1942–7. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. Journal of Biological Chemistry. 2007;282:19247–58. doi: 10.1074/jbc.M608011200. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishmarev D, Wang Y, Mason CE, Su XC, Oakley AJ, Graham B, Huber T, Dixon NE, Otting G. Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy. Nucleic Acids Res. 2014;42:2750–7. doi: 10.1093/nar/gkt1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–94. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XC, Wang Y, Yagi H, Shishmarev D, Mason CE, Smith PJ, Vandevenne M, Dixon NE, Otting G. Bound or free: interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry. 2014;53:1925–34. doi: 10.1021/bi5001867. [DOI] [PubMed] [Google Scholar]
- Sudol M. From Src Homology domains to other signaling modules: proposal of the 'protein recognition code'. Oncogene. 1998;17:1469–74. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- Sun Z, Tan HY, Bianco PR, Lyubchenko YL. Remodeling of RecG Helicase at the DNA Replication Fork by SSB Protein. Sci Rep. 2015;5:9625. doi: 10.1038/srep09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–89. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HY, Pottinger S, Yu C, Nguyen T, Bianco PR. Insight into the function of E.coli SSB C-terminal tail: II. Linker length is critical to dissociation from single stranded DNA submitted. 2016 [Google Scholar]
- Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–13. [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–52. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein: Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J. Biol. Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–60. doi: 10.1042/bj2970249. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Research. 2003;31:4434–40. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Tan HY, Choi M, Stanenas AJ, Byrd AK, Cohan CS, Bianco PR. SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells. 2016;21:163–84. doi: 10.1111/gtc.12334. K, D.R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.