Introduction
Genetic variation is crucial for the survival of all organisms and for crop improvement (Glaszmann et al., 2010). It is in the untapped potential of uncharacterized individuals within a population where we may find new and improved traits for plant adaptation and resilience (Riely & Martin, 2001; Saintenac et al., 2013). One of the major deterrents to crop productivity is disease. It is estimated that the actual loss of productivity due to pathogens for the major cultivated crops is close to 14% (Oerke, 2006). This loss of productivity could be reduced if the plant’s defense mechanisms against pathogens could be heightened.
Plants have intricate defense mechanisms that impede pathogen colonization and infection, and that minimize fitness costs to the infected plants. Two defense mechanisms that occur almost in parallel when a plant encounters a potential pathogen are pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Jones & Dangl, 2006). During PTI, epitopes of molecules ubiquitously present in microbes (e.g., flagellin for bacteria and chitin for fungi) are perceived by pattern-recognition receptors (PRRs), which ultimately contributes to halted microbial growth (Zipfel et al., 2004; Wan et al., 2008). How this is achieved is not currently well defined. Successful pathogens are able to dampen PTI responses, mainly by the activity of toxins and effector proteins (Melotto et al., 2006; Cheng et al., 2011; Xiang et al., 2011). However, certain plant individuals carry specific resistance (R) proteins that recognize either the presence or the activity of effectors, which ultimately triggers ETI (Grant et al., 1995; Gassmann et al., 1999). ETI usually leads to a localized programmed cell death response, the hypersensitive response, in the cells that are in contact with the pathogen, a phenomenon that is thought to limit pathogen spread. However, the boundaries distinguishing PTI and ETI are not as clear, as in nature both responses form a continuum (Thomma et al., 2011).
Phytohormones influence both PTI and ETI (Tsuda et al., 2009). Salicylic acid (SA) is a phenolic hormone involved in local defense as well as systemic acquired resistance (SAR), the latter of which protects infected plants from future pathogen colonization in uninfected tissue (Fu & Dong, 2013). Accumulation of SA increases after PTI elicitation (Tsuda et al., 2008), and pre-treatment of plants with an SA analogue potentiates several flagellin responses (Tateda et al., 2014). As for the involvement of salicylic acid in ETI, SA biosynthesis is partially required for effective ETI in some, but not all, effector-R protein pairs (Tsuda et al., 2009). Furthermore, SA accumulation increases in a biphasic manner during ETI (Malamy et al., 1990), while enzymatic depletion of SA accumulation causes ETI pathogen containment (but not cell death) to fail (Mur et al., 1997).
Until now, studies on plant resistance have focused on a limited number of natural accessions or cultivars of a plant species. A fundamental question that remains to be answered in plant–pathogen interactions is how many types of resistance mechanisms a given plant species would already possess to defend against a potential pathogen that apparently has not co-evolved with the plant. Pseudomonas syringae pv. tomato (Pst) DC3000 is a phytobacterial pathogen that has been extensively studied for its ability to infect tomato (from which it was initially isolated; Cuppels, 1986) and Arabidopsis (Whalen et al., 1991), and has been used as a model to probe plant defense responses (Xin & He, 2013). Pst DC3000 delivers more than 30 effector proteins into the plant cell using a type III secretion system (T3SS) (Wei et al., 2015), which collectively with a phytotoxin, coronatine (Melotto et al., 2006), are the two most important virulence-promoting mechanisms of this pathogen. Thus far, several studies have evaluated the variation in disease resistance to P. syringae in Arabidopsis, the largest of which examined 75 Arabidopsis accessions (Kover & Schaal, 2002; Perchepied et al., 2006; Hossain & Sultana, 2015). To address the question of how many types of resistance mechanisms a given plant species would already possess to defend against a potential pathogen that has potentially not co-evolved with the plant, we investigated the responses of over 1,000 Arabidopsis accessions to infection by Pst DC3000 and identified 14 accessions that were resistant. Further characterization separated these accessions into four defined categories: (1) two accessions were only resistant if bacteria were inoculated onto the leaf surface; (2) six accessions were able to mount an ETI-like response; (3) three accessions showed increased basal SA accumulation; and (4) three accessions did not fall into any of the previous three categories. AvrPto and HopAM1 were identified as effectors that influence the resistance in several accessions that show an ETI-like response to Pst DC3000. These results highlight the diverse mechanisms of resistance already in place in individuals of a population even before exposure to a particular pathogen strain occurs and, like in tomato, AvrPto recognition appears to play a prominent role in mediating the ETI-type interaction between A. thaliana and Pst DC3000.
Materials and Methods
Materials and methods detailing the crosses between Arabidopsis accessions, next-generation sequencing (NGS), SHOREmap mapping of resistance loci, and statistical analyses are described in the Supporting Information Methods S1–S3 and Notes S1.
Bacterial strains and antibiotics
Pseudomonas syringae van Hall strains were grown in modified LB (LM; 10 g l−1 tryptone, 6 g l−1 yeast extract, 1.5 g l−1 KH2PO4, 0.6 g NaCl, and 0.4 g MgSO4•7H2O) or King’s B medium at 30°C, while Escherichia coli (Migula) Castellani and Chalmers strains were grown in LB (Lennox) medium at 37°C (Table S1). Antibiotics were used at the following concentrations: 100–400 μg ml−1 for ampicillin, 50 μg ml−1 for kanamycin, 100 μg ml−1 for rifampicin, and 50 μg ml−1 for spectinomycin.
Plant growth conditions
Arabidopsis thaliana (L.) Heynh seeds were stratified for 6 d at 4°C before sowing. Before stratification, seeds were incubated with 1.8% sodium hypochlorite for 15 min, since two accessions, Xan-2 and Xan-5, require this treatment for even germination. Plants were grown in a growth chamber with a 12-h photoperiod and a temperature of 23°C during the day and 21°C during the night, under a partially covered transparent dome.
Construction of Pst ΔhopAM1-1ΔhopAM1-2 mutant
Pst DC3001 is a strain that has a c. 10-kb deletion in Pst DC3000 plasmid A that includes hopAM1-2 (Landgraf et al., 2006). Deletion of hopAM1-2 in Pst DC3001 was confirmed using three primers; P1, P5, and T1 (Table S2); and GoTaq® DNA polymerase (Promega), as PCR would produce amplicons of different sizes for wild-type Pst DC3000 and mutated Pst DC3001. Pst ΔhopAM1-1ΔhopAM1-2 mutant was constructed following a previously described procedure (Kvitko & Collmer, 2011). Effector hopAM1-1 was deleted from Pst DC3001 by conjugation, integration of the deletion construct, and sucrose counter-selection of double crossover strains using E. coli S17-1 pCPP5914 (pK18mobsacB::ΔhopAM1-1; Cunnac et al., 2011). After sucrose counter-selection, genomic DNA was extracted from several putative deletion strains using Gentra Puregene Yeast/Bact. kit (QIAGEN). Deletion of hopAM1-1 from Pst DC3001 was confirmed using polymerase chain reaction (PCR) with primers P2615 and P2616 and Phusion® high-fidelity DNA polymerase (Thermo Fisher Scientific).
Plasmid transformation into Pseudomonas syringae followed the protocol of Choi et al. (2006). Pseudomonas cultures were grown until they reached an absorbance at 600 nm of 0.5–0.8, washed twice with 0.5 M sucrose, and then used to electroporate the corresponding plasmids using the following parameters: 1.8 kV, 25 μF, and 200 Ω.
Screen for Arabidopsis thaliana accessions resistant to Pst DC3000
Arabidopsis accessions used in the screen included an initial set of 96 accessions from a study that evaluated genetic polymorphism in A. thaliana (Nordborg et al., 2005), and all the available accessions at the Arabidopsis Biological Resource Center (ABRC; The Ohio State University, USA) on February of 2009 (Table S3). Five-week-old plants were inoculated by dipping them into a Pst DC3000 suspension of 2 × 108 colony-forming units (CFU) ml−1 with 0.025% Silwet L-77. Plants were left covered under a transparent plastic dome for the duration of the experiment, in order to maintain high humidity. At 5 d post-inoculation, plants were evaluated for disease symptoms. Inoculated accessions lacking conspicuous disease symptoms (chlorosis, necrosis, or leaf collapse) were selected for further study.
Bacterial multiplication assays
Dip inoculation was done as described for the screen except that bacteria were resuspended in 0.25 mM MgCl2 to an inoculum of 108 CFU ml−1. For infiltration-based inoculation, leaves of 4–4.5-wk-old-Arabidopsis plants were poked with a needle and infiltrated with a bacterial suspension of 1 × 105 to 5 × 106 CFU ml−1 in 0.25 mM MgCl2 using a needleless syringe. After the infiltrated leaves dried (c. 1 h post infiltration), plants were left covered under a transparent dome for the duration of the experiment. At least three plants were inoculated per treatment to evaluate bacterial multiplication.
To evaluate if the accessions exhibited accelerated cell death in response to high bacterial inoculum, leaves were infiltrated with a bacterial suspension of 108 CFU ml−1 in 0.25 mM MgCl2 using a needleless syringe. After the infiltrated leaves dried, plants were left partially covered with a transparent dome for the duration of the experiment. Between 18–96 h post-inoculation, plants were evaluated for cell death. Individual leaves were visually categorized as having no necrosis, partial necrosis, or full cell death (Fig. S1). Alternatively, when individual effectors were expressed from Pst Δ28E strain, leaves were categorized as either showing no observable changes or chlorosis. At least four leaves from different plants were inoculated per treatment.
Hormone quantification
Hormones were extracted and quantified as described previously (Zeng et al. 2011). Approximately 50 mg of frozen leaf tissue was ground and then incubated at 4°C for 20 h in 80% methanol containing 0.1% formic acid, 0.1 g l−1 butylated hydroxytoluene (BHT), and 100 nM deuterated abscisic acid (ABA-d6, courtesy of Dr A. Daniel Jones, as an internal standard to account for hormone loss during extraction). Samples were vortexed, centrifuged and filtered using 0.2-μm PTFE filter units (Merck KGaA), and the flow through was used for hormone quantification.
Ten μl of plant extracts were injected onto an Ascentis® Express C18 column (50 × 2.1 mm, 2.7 μm; SIGMA-Aldrich) installed in the column heater (50°C) of an Acquity Ultra Performance Liquid Chromatography (UPLC) system (Waters Corp.). For UPLC separation, we used a 5-min gradient method starting with a 9 : 1 mixture (v/v) of 0.1% aqueous formic acid (solvent A) and 100% methanol (solvent B) and increasing linearly to 100% solvent B with a mobile phase flow rate of 0.4 ml min−1. After separation, samples were injected into a Quattro Premier XE mass spectrometer (Waters Corp.) equipped with an electrospray ionization (ESI) source operated in negative ion mode. Capillary voltage, cone voltage, and extractor voltage were set to 3.5 kV, 25 V, and 5 V, respectively, with the source temperature set to 120°C and the desolvation temperature set to 350°C. Desolvation gas and cone gas were set to flow rates of 600 l h−1 and 50 l h−1, respectively.
Selected ion monitoring (SIM) was performed to quantify ABA (m/z 263.1>153.1), ABA-d6 (m/z 269.1>159.1), jasmonic acid (JA; m/z 209.1>59), jasmonoyl isoleucine (JA-Ile; m/z 322.2>130.1), SA (m/z 137>93), and SA glucoside (SAG; m/z 299.1>137). The Quan-Optimize software was used to identify the parent and daughter SIM pairs, and the QuanLynx software 4.1 (Waters Corp.) was used to determine analyte responses relative to the internal standard ABA-d6. Hormones were quantified using standard curves prepared with purified hormones for each compound (hormones were purchased from SIGMA-Aldrich, except for JA-Ile, which was a kind gift from Dr Paul Staswick and SAG, for which the SA standard was used). Total SA was calculated by adding SA and SAG concentrations. Final concentrations are expressed as ng of hormone per gram of sample fresh weight (FW).
Benzothiadiazole (BTH) treatment
Arabidopsis accessions were sprayed with a solution of 0.025% Silwet L-77 with or without 100 μM BTH until the plants were thoroughly wet. Twenty-four hours after spraying, leaf tissue was collected and frozen for further analysis.
Protein extraction
Frozen leaf tissue was ground using the TissueLyser II homogenizer (QIAGEN) and 3-mm zirconium oxide beads (Glen Mills Inc.). Ground tissue was incubated for 10 min at 4°C with 3 volumes of extraction buffer (0.5% Triton X-100, 150 mM NaCl, 100 mM Tris-HCl pH 7.5, 10 mM dithiothreitol (DTT), 5 mM EDTA, and protease inhibitor cocktail for plant cell and tissue extracts; SIGMA-Aldrich) per mg of tissue to extract proteins. After removal of tissue debris, protein concentration was determined using the Bradford method (Bio-Rad protein assay), so that equivalent protein concentrations would be used for every sample.
Electrophoresis and Western blot
Polyacrylamide gel electrophoresis (PAGE) was performed using the NuPAGE® electrophoresis system (Thermo Fisher Scientific) and NuPAGE® Novex® 4–12% Bis-Tris gels. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and stained with Ponceau S stain (0.1% Ponceau S in 5% acetic acid) to confirm efficient transfer. Western blotting was done using α-PR1 (courtesy of Dr Xinnian Dong) and α-rabbit IgG-HRP (Thermo Scientific) antibodies. Protein detection used SuperSignal™ West Dura extended duration substrate (Thermo Fisher Scientific) and Blue Ultra Autoradiography films (GeneMate).
Reactive oxygen species (ROS) detection
ROS production was detected using a luminol-based assay. Four-mm-diameter-leaf discs were placed on white 96-well plates (Greiner Bio-One International) and floated overnight in water. The next day, water was removed and leaf discs floated in a 2 × 108 CFU ml−1 Pst DC3000 suspension in 0.25 mM MgCl2, 34 μg ml−1 of luminol (Sigma-Aldrich) and 10 μg ml−1 of horseradish peroxidase (type VI-A; Sigma-Aldrich). Luminescence was detected with a SpectraMax L microplate reader (Molecular Devices) using 1.5–2-s-integration intervals. Each treatment had 6–8 samples and each biological repeat was done in triplicate or quadruplicate.
Results
In order to discover as many types of A. thaliana resistance mechanisms to Pst DC3000 as possible, a large collection of 1,041 A. thaliana accessions obtained from the Arabidopsis Biological Resource Center (ABRC, The Ohio State University) (Table S3) were infected by dip-inoculation into a suspension of Pst DC3000. Fourteen accessions did not show any disease symptoms and were classified as being resistant to infection by Pst DC3000 (Fig. S2). Their geographic collection origin did not reveal any distinguishable pattern, as they were scattered throughout the native range of A. thaliana, which is restricted to Europe, the north of Africa and western Asia (Fig. S3) (Nordborg et al., 2005; 1001 Genomes Consortium, 2016).
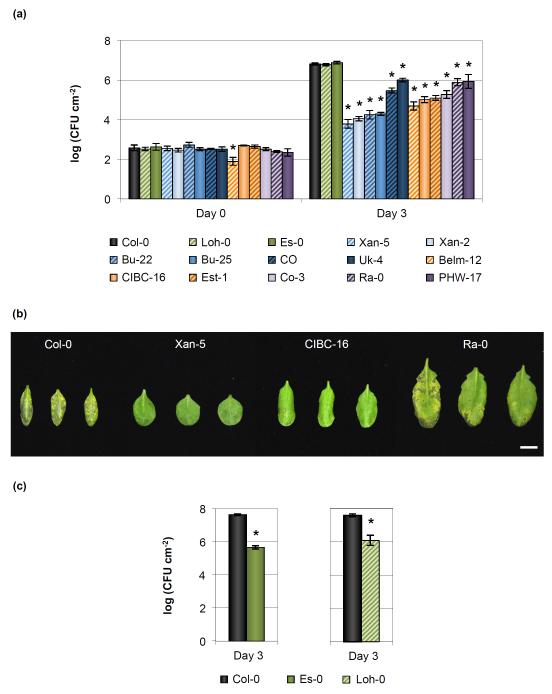
As dip-inoculation does not distinguish resistance mechanisms based on the leaf surface vs the leaf apoplast, and the lack of disease symptoms could be caused by reduced bacterial multiplication and/or disease tolerance, we further examined the 14 resistant accessions by an infiltration-based infection assay (i.e. by delivering bacterial inoculum directly into the leaf apoplast) and recorded both disease symptoms and bacterial multiplication (Figs 1a,b, S4). The 14 accessions could be categorized into two groups: 12 accessions showed heterogeneous reduction of bacterial growth and disease symptoms, from accessions like Ra-0 showing only a six-fold decrease in bacterial growth (compared to the susceptible Col-0 control) and slightly reduced symptoms, to accessions like Xan-5, in which bacterial growth was reduced more than 650-fold and no symptoms were observed. The other two accessions, Es-0 and Loh-0, were no more resistant to Pst DC3000 than Col-0 in an infiltration-based infection assay (Fig. 1a). However, when bacteria were inoculated onto the surface of these two accessions, as was done for the initial screen, it was quite evident that these accessions were resistant to Pst DC3000 (Figs 1c, S5).
Fig. 1.
Natural variation of resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 amongst Arabidopsis accessions. (a) Bacterial growth in resistant Arabidopsis thaliana accessions at 0 and 3 d after syringe-infiltration with 105 colony-forming units (CFU) ml−1 of Pst DC3000. Error bars show ± SE of the mean of 3 (for day 0) or at least 5 (day 3) biological samples. Bars are colored according to the type of resistance observed in this study: green, plant surface-mediated resistance; blue, hypersensitive-like cell death response; orange, enhanced salicylic acid defenses; and purple, unknown. The asterisk indicates accessions whose bacterial growth was significantly different when compared to the susceptible Col-0 control as determined by a Dunnett’s test (P < 0.05). Statistical analyses for each day after infection were done separately. (b) Disease symptoms in select resistant A. thaliana accessions 3 d after syringe-infiltration with 105 CFU ml−1 of Pst DC3000. Bar, 1 cm. Image was composed from accessions’ individual images from a single experiment. (c) Bacterial populations are reduced in Es-0 and Loh-0 accessions when bacteria are inoculated on the surface of plants. Bacterial growth was quantified 3 d after dip-inoculation with 108 CFU ml−1 of Pst DC3000. Error bars show ± SE of the mean of 4 biological samples. The asterisk indicates accessions whose bacterial growth was significantly different when compared to the susceptible Col-0 control as determined by a Student’s t-test (P < 0.05).
Accessions showing a hypersensitive-like cell death response to Pst DC3000
In an infection using low bacterial inoculum, recognition of effectors by R proteins manifests without any visible symptoms on the plants, as the plant cells that die due to localized programmed cell death are not visible macroscopically. However, when using higher bacterial titers, effector recognition will cause collapse of the infiltrated area by hypersensitive-like cell death that proceeds much faster (by several hours) than that which is observed due to disease (i.e. the one that would be observed in Col-0 leaves, an accession that does not carry any resistance genes against Pst DC3000; Whalen et al., 1991). In order to evaluate if resistance to Pst DC3000 in any of the accessions was due to effector recognition (i.e. ETI), accessions were infiltrated with a high bacterial titer of Pst DC3000. Six accessions showed a hypersensitive-like cell death response when compared to Col-0, reminiscent of what would be observed for ETI (Table 1; Figs 2a, S6) (Lewis et al., 2010). The hypersensitive-like cell death response was more pronounced for Bu-22, Bu-25, and Xan-5 accessions, with cell death observed for Bu-22 and Bu-25 as early as 18 h post-inoculation. The appearance of cell death in CO and Uk-4 accessions was only slightly faster than in susceptible Col-0 plants, which might reflect why these accessions do not restrict bacterial growth as much as the other 4 accessions (Fig. 1a), as they may mount weaker defense responses.
Table 1.
Several Arabidopsis accessions show an accelerated cell death response after being inoculated with high titers of Pseudomonas syringae pv. tomato (Pst) DC3000
| Accession | Plant response to Pst DC3000 infection | |||
|---|---|---|---|---|
| No | Partial | Collapsed | Total | |
| Col-0 | 31 | 3 | 2 | 36 |
| Loh-0 | 18 | 0 | 0 | 18 |
| Es-0 | 12 | 6 | 0 | 18 |
| Bu-22 | 0 | 5 | 13 | 18 |
| Bu-25 | 0 | 0 | 24 | 24 |
| CO | 13 | 11 | 0 | 24 |
| Uk-4 | 11 | 11 | 2 | 24 |
| Xan-2 | 1 | 9 | 8 | 18 |
| Xan-5 | 0 | 0 | 18 | 18 |
| Belm-12 | 18 | 0 | 0 | 18 |
| CIBC-16 | 11 | 7 | 0 | 18 |
| Est-1 | 14 | 4 | 0 | 18 |
| Co-3 | 17 | 1 | 0 | 18 |
| PHW-17 | 21 | 2 | 1 | 24 |
| Ra-0 | 16 | 2 | 0 | 18 |
Cell death after infiltration with 108 colony-forming units (CFU) ml-1 Pst DC3000 is accelerated in six Arabidopsis accessions, compared to that observed for Col-0. Cell death was evaluated 26 h after infiltration into three categories: (1) no leaf area showing necrosis symptoms; (2) partial necrosis symptoms; (3) fully collapsed leaf. Eighteen to thirty-six leaves were evaluated per accession, with three leaves being infiltrated per plant. Highlighted in bold and in italics are those accessions whose response to Pst DC3000 inoculation was different from Col-0, as determined by a Fisher’s exact test (P < 0.0036).
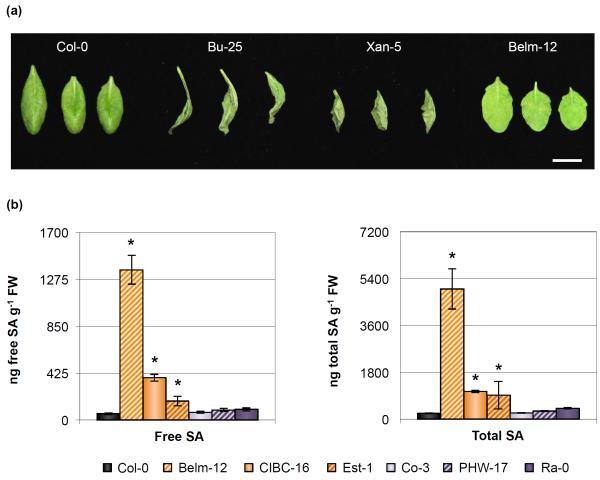
Fig. 2.
Several Arabidopsis accessions either show an accelerated cell death response reminiscent of a hypersensitive response or have higher basal levels of salicylic acid (SA) accumulation. (a) Cell death symptoms 27 h after infiltration with 108 colony-forming units (CFU) ml−1 of Pseudomonas syringae pv. tomato (Pst) DC3000 in select Pst DC3000-resistant ecotypes. Bar, 1 cm. Image was composed from accessions’ individual images from a single experiment. (b) Free SA and total SA concentration in 5-wk-old leaves of Col-0 and Pst DC3000-resistant accessions. Error bars represent ± SE of the mean from six plants. The asterisk indicates accessions whose hormone concentration was significantly different when compared to the susceptible Col-0 control as determined by a Dunnett’s test on the log10-transformed data (so that variances would be homogeneous, P < 0.05).
Several accessions have elevated basal salicylic acid accumulation
Plants that over-accumulate SA have been reported to be more resistant to infection by Pseudomonas syringae (Greenberg et al., 1994; Bowling et al., 1997; Jirage et al., 2001; Todesco et al., 2010). We measured SA accumulation in the remaining 6 Pst DC3000-resistant accessions that were resistant when bacteria were delivered directly into the apoplast but that did not display a hypersensitive-like cell death response after Pst DC3000 inoculation. Three of these accessions had higher accumulation of free and total SA when compared to Col-0 (Fig. 2b), which might explain why these plants were more resistant to Pst DC3000 infection, as these high SA concentration levels could potentially prime these accessions for enhanced defense. Under the conditions in which plants were grown, accession Est-1 showed variable accumulation of SA, even though individual plants always had higher SA accumulation than Col-0 plants and this correlated with Est-1 plants always being more resistant to Pst DC3000 infection.
In contrast to SA, ABA and JA/JA-Ile accumulation was not significantly different between these Pst DC3000-resistant accessions and Col-0 (Fig. S7).
Further characterization of selected resistant accessions with different types of resistance mechanisms
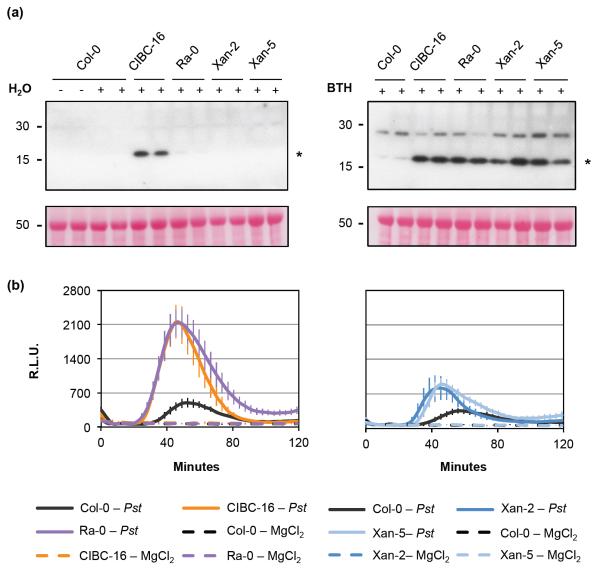
Four out of the 14 Pst DC3000-resistant Arabidopsis accessions were chosen for further characterization of their molecular and/or cellular defense responses. Accessions Xan-2 and Xan-5 had a hypersensitive-like cell death response (Table 1), CIBC-16 had an elevated basal accumulation of SA (Fig. 2b), while Ra-0 had a yet-to-be characterized mechanism of resistance. A commonly used marker for the induction of the SA defense pathway is the increase of PATHOGENESIS-RELATED GENE 1 (PR1) transcript and protein accumulation (Yalpani et al., 1991), a response that may be induced by BTH (an SA analogue) (Lawton et al., 1996), but that can also be observed during senescence and after exposure to other stressful environmental stimuli (Sharma et al., 1996; Surplus et al., 1998; Zhang et al., 2013). Consistent with the elevated basal SA accumulation detected in CIBC-16 (Fig. 2b), there was an increased basal accumulation of PR1 protein in this accession (Fig. 3a), whereas no PR1 protein was detectable for Ra-0, an accession whose SA accumulation was equivalent to that observed for Col-0. Interestingly, Ra-0, Xan-2, and Xan-5 accessions showed higher accumulation of PR1 protein after induction of SA signaling with BTH compared to Col-0 (Fig. 3a).
Fig. 3.
Several defense responses are enhanced in Pseudomonas syringae pv. tomato (Pst) DC3000-resistant Arabidopsis accessions. (a) Pathogenesis-related gene 1 (PR1) protein accumulation 24 h after H2O and 100 μM benzothiadiazole (BTH) treatment in select Pst DC3000-resistant accessions shows an enhanced response after elicitation with BTH. Two samples are shown per each treatment. Untreated (-) Col-0 is shown as a negative control. Proteins were detected using α-PR1 antibodies while the asterisk points to the expected molecular weight of PR1. Bottom image shows the Ponceau S staining of the polyvinylidene difluoride (PVDF) membranes. Twenty μg of total protein were loaded per well. (b) Reactive oxygen species (ROS) production in Col-0 and resistant accessions in response to 2 × 108 colony-forming units (CFU) ml−1 Pst DC3000 or 0.25 mM MgCl2 as the ROS elicitors. RLU, for relative light units. Error bars show the 95% confidence intervals for the means. Detection was done using SpectraMax L microplate reader (Molecular Devices).
Recognition of bacteria/PAMPs triggers rapid production of ROS, a phenomenon that is mainly due to bacterial flagellin recognition by the PRR FLS2 (FLAGELLIN-SENSING 2) in Arabidopsis (Smith & Heese, 2014). When compared to Col-0, all four resistant accessions showed an increased ROS production after elicitation with Pst DC3000 (Fig. 3b). Larger increases were observed for CIBC-16 and Ra-0, compared to Xan-2 and Xan-5, which showed modest ROS increases that were evident in only two out of the three experiments performed.
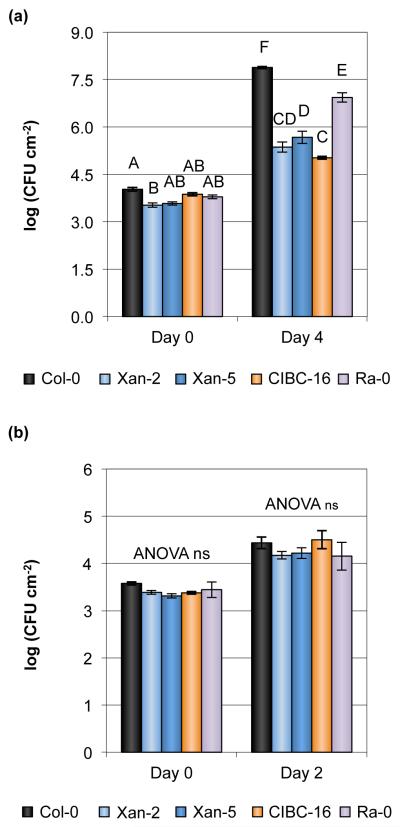
We also investigated whether the above-mentioned four accessions showed increased resistance to a different pathogenic strain of Pseudomonas, Pseudomonas cannabina pv. alisalensis ES4326R (formerly known as P. s. pv. maculicola ES4326 or CFBP 1637; Bull et al., 2010) (Figs 4a, S8a). When compared to susceptible Col-0 plants, the four accessions showed increased resistance to Pcal ES4326R, with Ra-0 being the least resistant accession of the four, similar to that which was observed for infection with Pst DC3000 (Fig. 1a). By contrast, no accession difference was observed for the growth of a non-pathogenic strain, Pst DC3000 ΔhrcC (in which a structural gene for the T3SS is deleted; Deng & Huang, 1999) (Figs 4b, S8b), suggesting that the mechanisms of resistance are associated with restricting T3SS-dependent growth.
Fig. 4.
Several Pseudomonas syringae pv. tomato (Pst) DC3000-resistant accessions are also resistant to Pseudomonas cannabina pv. alisalensis (Pcal) ES4326R. (a) Bacterial growth in Arabidopsis thaliana accessions 0 and 4 d after syringe-infiltration with Pcal ES4326R at an inoculum of 5 × 106 colony-forming units (CFU) ml−1. (b) Bacterial growth in A. thaliana accessions 0 and 2 d after syringe-infiltration with non-pathogenic Pst ΔhrcC at an inoculum of 2 × 106 CFU ml−1. Error bars show ± SE of the mean of 3 (day 0) or at least 5 (days 2 and 4) biological samples. Different letters above the bars indicate significant differences, as determined by a Tukey HSD test (P < 0.05). ns, not significant. ANOVA performed separately for each day for (b).
Segregation of the resistance to Pst DC3000 in F2 populations reveals the action of multiple loci
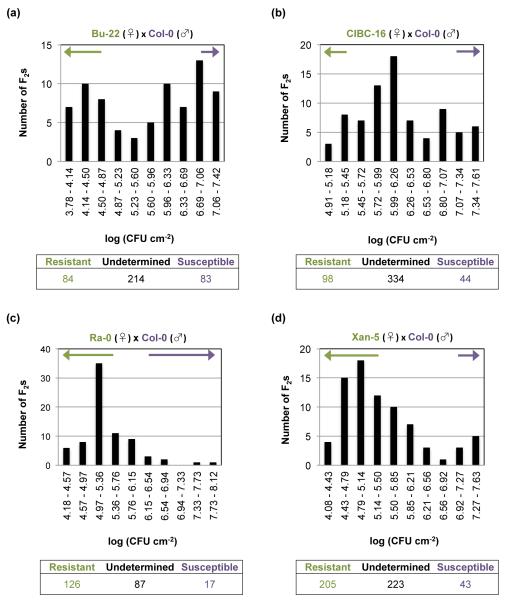
To evaluate the genetic segregation of the resistance to Pst DC3000, crosses between 3 resistant accessions, CIBC-16, Ra-0, and Xan-5, to the susceptible accession Col-0 were performed (Fig. S9b–d). The F2 population of these 3 crosses was inoculated with Pst DC3000, and the bacterial numbers within each plant determined (Fig. S10). F2 individuals were characterized as resistant if their in planta bacterial growth was lower than the highest data point for the resistant parent, and susceptible if their bacterial growth was higher than the lowest data point for Col-0. The F2 segregation of the resistance did not follow a Mendelian segregation that would be expected from the effect of 1 or 2 genes (Fig. 5b–d; Table S4), and many of the F2 individuals could neither be classified as resistant or susceptible (i.e. their values were above the highest data point for the resistant parent and below the lowest data point for Col-0). Nevertheless, we attempted to map the loci involved in the resistance of these accessions using bulked segregant analysis (BSA) and NGS. Since we expected major genes to be controlling the resistance, we used a method that identifies qualitative traits for mapping (SHOREmap; Sun & Schneeberger, 2015). Unfortunately, no association was observed for any chromosomal region in the F2 populations for any of the three accessions, suggesting a complicated polygenic nature of the resistance (Fig. S11). This quantitative nature has been observed before in Arabidopsis for resistance to P. syringae (Forsyth et al., 2010). The inability to map the loci controlling the resistance after BSA suggests that several gene combinations in the F2 individuals could create the same resistance phenotype as the one observed in the parents.
Fig. 5.
Multiple loci are involved in resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in the resistant Arabidopsis accessions. Segregation of the resistance to Pst DC3000 in the F2 generation derived from crosses of the resistant accessions (♀) (a) Bu-22, (b) CIBC-16, (c) Ra-0, and (d) Xan-5 with the susceptible parent Col-0 (♂). The segregation of the resistance of c. 80 individuals is shown in each graph. The arrows above the bars indicate those F2 individuals that were identified as either resistant (green arrow) or susceptible (purple arrow) to Pst DC3000 infection. F2 individuals were determined as resistant if their value of in planta bacterial growth was lower than the highest data point for the resistant parent, and were deemed as susceptible if that value was higher than the lowest data point for the susceptible parent. The table below the graph indicates the segregation of the resistance for all of the F2 individuals evaluated. CFU, colony-forming units.
Search for bacterial effectors responsible for the hypersensitive cell death response in Xan-5, Bu-22, and Bu-25 accessions
To further investigate whether the hypersensitive-like cell death response in the resistant accessions (Table 1) is due to effector perception by R proteins (i.e. ETI), we inoculated these accessions with Pst DC3000 mutant strains in which various effector genes were deleted in order to uncover the relevant effectors responsible for triggering the ETI-type resistance. The three accessions with the fastest cell death response, Bu-22, Bu-25, and Xan-5, were chosen for this analysis. Pst DC3000 is reported to contain 36 effector genes (Wei et al., 2015 and Table S5; two identical genes coding for HopAM1 are present in Pst DC3000 and counted only once), and a Pst DC3000 mutant strain in which 28 of these 36 effector genes are deleted (Pst Δ28E; Cunnac et al., 2011) is available. Pst Δ28E was unable to cause any visible changes when inoculated into the leaves of any of the resistant accessions (Table S6a), suggesting that one or more of the missing 28 effectors is responsible for the observed hypersensitive-like cell death response.
To identify the specific effectors that cause hypersensitive-like cell death in Bu-22, Bu-25, and Xan-5, we tested other Pst DC3000 mutant strains in which smaller subsets of effector genes were deleted. For example, 19 effector genes of Pst DC3000 are clustered in the genome and mutant strains deleted in these gene clusters are available (Kvitko et al., 2009; Table S1). However, we found that none of the 19 clustered effectors were responsible for the hypersensitive-like cell death phenotype in Bu-22, Bu-25, or Xan-5 (Table S6b; based on infection with Pst ΔIΔIIΔIVΔIXΔX, which has 15 effector genes deleted, and Pst ΔCEL, which has 4 effectors deleted). We then inoculated these 3 accessions with a strain with the avrPto and avrPtoB (hopAB2) effector genes deleted. We found that the hypersensitive-like cell death phenotype in Bu-25 was reduced (Table S6c), suggesting that one of these two effectors might be responsible for the resistance to Pst DC3000 observed in Bu-25.
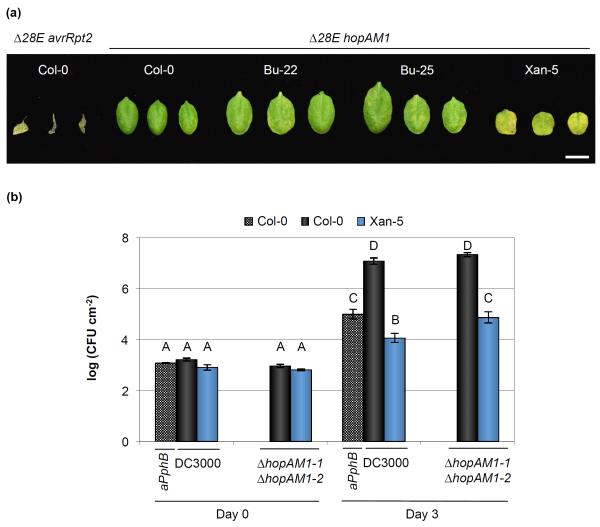
To test the potential recognition of the remaining Pst DC3000 effectors, we introduced individually each of these eight-effector genes into Pst Δ28E and tested if these strains caused any observable changes in the leaves after infiltration. As a positive control, we used a Pst Δ28E strain expressing a heterologous effector not present in Pst DC3000, avrRpt2 (which is recognized by the RPS2 resistance protein in Col-0 and therefore capable of causing tissue collapse; Bent et al., 1994) (Fig. 6a). Of the eight strains, only the Pst Δ28E strain expressing effector hopAM1 was capable of causing chlorosis on the leaves of Xan-5 (sometimes, a few chlorotic spots were observed in the other accessions), indicating that this effector could be involved in the resistance phenotype observed for Xan-5 (Table S7; Fig. 6a).
Fig. 6.
HopAM1 recognition influences resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in the Xan-5 Arabidopsis accession. (a) Cell death symptoms 4 d after inoculation with 2 × 107 colony-forming units (CFU) ml−1 of Pst Δ28E carrying pBBR:hopAM1-1 (Δ28E hopAM1) and pUCP19::avrRpt2 (Δ28 avrRpt2). Image was composed from accessions’ individual images from a single experiment. Bar, 1 cm. Pst Δ28E avrRpt2 strain was used to confirm that the strain with 28 effectors deleted was still capable of mounting a hypersensitive response (HR) response in Col-0. (b) HopAM1 recognition is partially responsible for the Pst DC3000 resistance in the Xan-5 accession. Leaves were infiltrated with Pst DC3000, Pst DC3001 ΔhopAM1-1 (ΔhopAM1-1ΔhopAM1-2), and Pst DC3000 pDSK600::avrPphB (aPphB) at an inoculum of 106 CFU ml−1. Error bars show ± SE of 3 and 6 biological samples for day 0 and day 3, respectively. Letters above each bar indicate similar groups as determined with a Tukey HSD test (P < 0.05). A slight reproducible increase in in planta bacterial growth was observed for Xan-5 when inoculated with a strain lacking hopAM1.
In planta bacterial multiplication confirms the involvement of AvrPto and HopAM1 recognition in resistance to Pst DC3000 in Bu-22, Bu-25, Xan-2, and Xan-5 accessions
As the presence of hopAM1 in Pst Δ28E caused a chlorotic response in Xan-5 leaves, we decided to delete this gene from Pst DC3000. The hopAM1 gene is present in 2 duplicated copies in Pst DC3000, one on the chromosome (hopAM1-1) and the other on the endogenous plasmid A (hopAM1-2, formerly known as avrPpiB2Pto; Buell et al., 2003). We used strain Pst DC3001, which has a 10-kb deletion that includes hopAM1-2 (Landgraf et al., 2006), to delete hopAM1-1. This strain, which was lacking both hopAM1 genes, was inoculated into Xan-5 at 106 CFU ml−1 (a titer normally used for disease assays). When compared to Pst DC3000, a reproducible increase (4- to 10-fold) in bacterial growth was observed in Xan-5 (Fig. 6b). Because another accession with a hypersensitive-like response, Xan-2, was originally collected in the same region of Azerbaijan as Xan-5, infection of Xan-2 with a strain lacking hopAM1 was performed. A similar increase in Pst ΔhopAM1-1ΔhopAM1-2 population compared to Pst DC3000 was observed in Xan-2 as had been previously identified for Xan-5 (Fig. S12), suggesting analogous recognition mechanisms for HopAM1 on both Xan-2 and Xan-5. However, deletion of both hopAM1 genes in Pst DC3000 did not fully restore virulence in Xan-2 or Xan-5, as evidenced by the lower final bacterial population when compared to that achieved in Col-0. Furthermore, the resistance of Xan-5 to a strain lacking hopAM1 was similar to that observed with a strain expressing avrPphB, an effector recognized in Col-0 by the resistance protein RPS5 (Warren et al., 1998; Fig. 6b). Therefore, there are more factors besides HopAM1 recognition that influence resistance to Pst DC3000 in the Xan-2 and Xan-5 accessions.
Next, we conducted bacterial multiplication assays to determine if the slower cell death response to inoculation by a strain lacking both avrPto and avrPtoB in Bu-25 (Table S6c) was accompanied by a loss of resistance in this accession. In contrast to Pst ΔhopAM1-1ΔhopAM1-2 infection of Xan-5, in which only a modest increase in growth was observed when compared to Pst DC3000 infection of Xan-5, Pst ΔavrPtoΔavrPtoB became fully virulent in Bu-25 (Fig. S13). We also infected Bu-22 and Xan-5 with Pst ΔavrPtoΔavrPtoB and found that, remarkably, Bu-22 was also fully susceptible to a strain lacking both effector genes, while Xan-5 maintained full resistance (Fig. S13). The minor (non-statistically significant) increase in growth of Pst ΔavrPtoΔavrPtoB observed in Bu-22 in comparison to Col-0 (also notice Fig. 7a for Pst ΔavrPto growth in these accessions) might explain why no effect was initially noticed in Bu-22 when evaluating the hypersensitive-like cell death caused by a strain lacking both effector genes, since an increased bacterial growth at earlier infection times could have caused disease-associated cell death to progress faster in Bu-22 than in Col-0.
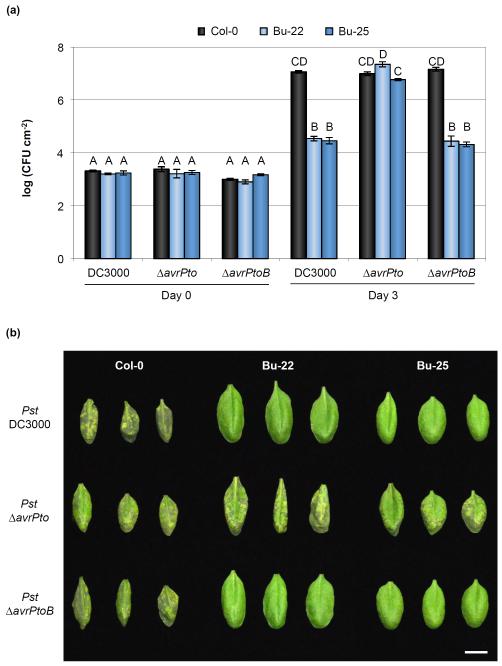
Fig. 7.
AvrPto recognition is a major factor mediating resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 in Bu-22 and Bu-25 Arabidopsis accessions. (a) In planta bacterial growth in Pst DC3000-resistant accessions after infiltration with Pst DC3000, Pst ΔavrPto, and Pst ΔavrPtoB at an inoculum of 5 × 105 colony-forming units (CFU) ml−1. Error bars show ± SE of 3 and at least 7 biological samples for days 0 and 3, respectively. Letters above each bar indicate similar groups as determined with a Tukey HSD test (P < 0.05). Notice that the absence of AvrPto allows Pst to multiply to high titers in both Bu-22 and Bu-25 accessions. (b) Disease symptoms 3 d after inoculation with 5 × 105 CFU ml−1 of Pst DC3000, Pst ΔavrPto, and Pst ΔavrPtoB. Image was composed from accessions’ individual images from a single experiment. Bar, 1 cm.
To evaluate if AvrPto alone, AvrPtoB alone, or both effectors were recognized in Bu-22 and Bu-25, bacterial strains with avrPto or avrPtoB genes individually deleted were inoculated into plants. No differences in bacterial growth were observed in Bu-22 or Bu-25 after inoculation with either Pst ΔavrPtoB or Pst DC3000, while a large increase of bacterial growth and the appearance of disease symptoms similar to those observed for Pst DC3000 in Col-0 were observed in Bu-22 and Bu-25 accessions inoculated with a strain lacking avrPto (Fig. 7a,b). Compared to Bu-22, Bu-25 had a slightly lower bacterial growth when infected with Pst ΔavrPto, suggesting that either Pst ΔavrPto is more virulent in Bu-22 due to the combined action of Pst DC3000 effectors (without AvrPto) or that there is a minor locus in Bu-25 controlling resistance against Pst DC3000. Evaluation of the F2 segregation of the resistance to Pst DC3000 in Bu-22 revealed that most likely a single locus controlled recognition of the AvrPto effector (Figs 5a, S9a; Table S4). Bu-25 was collected from the same region in Germany; it remains to be determined whether or not AvrPto recognition in this accession is similar to that of Bu-22. Either way, it seems that AvrPto recognition is the main determinant for resistance to Pst DC3000 in both Bu-22 and Bu-25 accessions.
Discussion
We have performed a large-scale screen of 1,041 A. thaliana natural accessions to address a fundamental question in plant–pathogen interactions: How many types of resistance mechanisms a plant species may already have against potential infection by a bacterial pathogen that has not apparently co-evolved with the plant species? It is well known that disease phenotypes are strongly influenced by environmental conditions. To select for robustly resistant accessions, we performed our screen under high humidity that simulates disease-conducive conditions (Xin et al., 2016). Of the 14 Pst DC3000-resistant accessions identified in this screen, two accessions, Est-1 and Ra-0, had already been shown in previous studies to be more resistant to Pst DC3000 infection (Todesco et al., 2010; Hossain & Sultana, 2015), further validating the results of our screen. Other accessions that had been observed before to be resistant to Pst DC3000 (Kover & Schaal, 2002; Perchepied et al., 2006; Hossain & Sultana, 2015) were not resistant in our screen, most likely because our disease-conducive (e.g., high humidity) conditions favor the development of disease; therefore, we only characterized accessions with a robust resistance phenotype. Further characterization of the 14 resistant accessions enabled us to classify them into four distinct categories: two accessions had a plant surface-based resistance mechanism, six accessions showed an ETI-like response, three accessions exhibited increased basal SA accumulation, while in the three remaining accessions the mechanism of resistance could not be classified (Fig. S14; Table S8). The mechanisms controlling resistance in these unclassified accessions remain to be determined and could be due to preformed antimicrobial physical barriers or compounds, and/or resistance to bacterial retrieval of nutrients and water during infection. Overall, to our knowledge, this is the first time all four types (possibly more) of pre-existing resistance mechanisms have been uncovered in different individuals of a single host population/species against the same potential pathogen in a single study.
Accessions Es-0 and Loh-0 showed Pst resistance only by surface inoculation (Fig. 1c) and do not have a mechanism that restricts Pseudomonas growth once the bacteria reach the apoplast. This is interesting because a previous genetic screen identified two Arabidopsis mutants (scord5 and scord7, in the Col-7 genetic background) that were exclusively more susceptible by surface inoculation (but not apoplastic infiltration inoculation) to a coronatine-deficient strain of Pst DC3000 (Zeng et al., 2011). Stomata are the most common entry point for foliar infecting Pseudomonads (Melotto et al., 2006), and while the wild-type Col-0 stomata close in response to the coronatine-deficient strain of Pst DC3000 as an early defense response in plants, the scord5 and scord7 mutant stomata are unable to close after bacterial inoculation. It remains to be determined whether Es-0 and Loh-0 represent natural accessions that have enhanced stomatal defense against Pst DC3000. Alternatively, the increased resistance could be caused by a more hostile epiphytic environment experienced by Pst DC3000 in these accessions, which would decrease the number of bacteria before they could enter the leaves through the stomata and reach the apoplast. Of note, epiphytic community differences between Arabidopsis accessions have been observed (Horton et al., 2014), so genetic differences among accessions could have an impact on the initial pathogen epiphytic colonization.
Arabidopsis accessions Belm-12, CIBC-16, and Est-1 showed an enhanced basal SA accumulation (Fig. 2b). In Est-1, this elevated SA accumulation is caused by a hyperactive allele of ACD6 (Rate et al., 1999; Todesco et al., 2010). The loci controlling the Pst DC3000 resistance in Belm-12 and CIBC-16 accessions await discovery. Mutagenesis experiments, done mostly in accessions Col-0 and Ler, show that mutant alleles of many Arabidopsis genes cause enhanced SA and Pst resistance, including, to name a few, CPR5, involved in nuclear pore trafficking (Bowling et al., 1997; Gu et al., 2016); CPR30, coding for an F-box protein (Guo et al., 2009); DND2 (also known as CNGC4), encoding a cyclic nucleotide gated channel (Jurkowski et al., 2004); and ACD2, coding for an enzyme involved in chlorophyll breakdown (Greenberg et al., 1994; Mach et al., 2001). Almost invariably, SA-enhancing mutants of Col-0 tend to be dwarf, possibly due to the growth-defense tradeoff (Huot et al., 2014). By contrast, Belm-12 and CIBC-16 do not exhibit any obvious dwarfism. As such, it would be interesting in the future to determine why natural accessions like Belm-12 and CIBC-16 can accumulate a high basal level of SA and have elevated pathogen resistance, but maintain apparently normal growth and development.
In addition to leaf-surface- or elevated SA-based resistance, six accessions have an ETI-like response to Pst DC3000 infection, with Bu-22, Bu-25, and Xan-5 showing a strong hypersensitive-like cell death response (Table 1). Our further characterization of these three accessions led to identification of Pst DC3000 effectors that trigger ETI in these accessions. Specifically, recognition of HopAM1 partially controlled resistance to Pst DC3000 in Xan-2 and Xan-5 accessions. When hopAM1 was expressed from an effectorless Pst DC3000 strain, instead of tissue collapse resulting from coalescing cell death (as was observed for expression of avrRpt2; Fig. 6a), only enhanced chlorosis was observed. Therefore, HopAM1 recognition in Xan-2 and Xan-5 seems to be involved in a weak ETI-like response. A quantitative nature of the cell death response after HopAM1 bacterial delivery was recently reported in 98 Arabidopsis accessions (Iakovidis et al., 2016). However, even though deletion of hopAM1 slightly increased bacterial growth in strong ETI-like accession Bur-0 (an accession evaluated in our screen that, unlike Xan-2 and Xan-5, was susceptible to Pst DC3000 infection), no correlation was found between the accelerated cell death response and disease resistance, as the same effect of increased bacterial growth for a strain lacking hopAM1 was observed after infection of both Col-0 and Bur-0 (Iakovidis et al., 2016). The observed lack of increased growth of Pst ΔhopAM1-1ΔhopAM1-2 in Col-0 plants in this study (although, a slight, non-statistically significant 2-fold growth increase was observed; Figs 6b, S12a) probably reflects the fact that bacterial populations were much higher in our experiments (c. 5 × 105 CFU cm−2 in Iakovidis et al., 2016; in comparison to 2 × 107 CFU cm−2; Fig. 6b), and, as such, could have been saturated (this same lack of growth difference in Col-0 was observed in a different previous study; Boch et al., 2002). In addition, accessions Xan-2 and Xan-5 are resistant to Pcal ES4326R (Fig. 4a), a strain that does not carry HopAM1. This further suggests that the major mechanism(s) controlling resistance to Pst DC3000 in Xan-2 and Xan-5 is likely independent of HopAM1 recognition, consistent with the polygenic nature of resistance in Xan-5 based on our analysis of F2 populations (Figs 5d, S11c).
We found that AvrPto plays a major role in conditioning Pst DC3000 resistance in Bu-22 and Bu-25 accessions. AvrPto recognition was first identified in the Solanaceae as being conferred by a cytoplasmic kinase, Pto, introgressed from S. pimpinellifolium into cultivated tomato (Martin et al., 1993). Pto-mediated resistance absolutely requires a nucleotide-binding site–leucine-rich repeat (NBS-LRR) protein (Prf, encoded in the same locus as Pto for resistance; Salmeron et al., 1996). Pto can also recognize the structurally unrelated effector AvrPtoB (Kim et al., 2002). In contrast to Pto, the factor controlling resistance in Arabidopsis accessions Bu-22 and Bu-25 recognizes only AvrPto and not AvrPtoB (Fig. 7). Guided by the Pto-AvrPto and Pto-AvrPtoB structures, Pto mutations have been made that abolish interaction with AvrPtoB but maintain AvrPto interaction (Dong et al., 2009). Also, alleles of Pto have been found in a wild tomato species (Solanum chmielewskii) that are capable of recognizing only AvrPtoB (Kraus et al., 2016). In other Solanaceae, phosphorylation of the C-terminus of AvrPto conditions resistance, a domain not involved in Pto-mediated resistance (Yeam et al., 2010). It will be exciting to discover if the R gene in Bu-22 and Bu-25 encodes a kinase like Pto or a more typical NBS-LRR like Prf, and which AvrPto regions are involved in recognition.
The mechanisms that a plant can potentially employ to defend against and evade microbes are numerous. These include, but are not limited to, ETI (Martin et al., 1993), PTI (Zipfel et al., 2004), SAR (Fu & Dong, 2013), elevated basal SA accumulation (Todesco et al., 2010), production of inhibitors of pathogen cell wall degrading enzymes (Ferrari et al., 2006), RNA silencing (Yang et al., 2004), phytoalexin and phytoanticipin production (Fan et al., 2011), toxin detoxification (Johal & Briggs, 1992), and physical barriers to entry and colonization (Melotto et al., 2006). In this study, enhanced PTI, ETI, SA accumulation and defense, and surface-mediated barriers were among the mechanisms that were identified as contributing to resistance in individuals of natural populations of Arabidopsis. These mechanisms were present even though Pst DC3000 is apparently not a native pathogen of Arabidopsis, which emphasizes how plants are able to cope with future pathogen attack even if they lack an adaptive immune system similar to that found in vertebrates. The detection of an ETI-like mechanism in some accessions suggests that these accessions are likely co-evolving with some adapted pathogen(s) in nature, and they recognize effectors that happen to be present in Pst DC3000. In this sense, ETI can be a ‘cryptic’ defense strategy against infection by emerging new pathogens that carry the same effectors. In fact, it would be advantageous for plants to recognize conserved effectors that are present in multiple pathogens. For example, a study found that avrPto was present in more than half of the evaluated P. syringae strains (Baltrus et al., 2011), highlighting how this effector recognition could have evolved in nature as a mechanism against Arabidopsis pathogens other than Pst DC3000. Remarkably, even though Pst DC3000 is not known to be a natural pathogen of A. thaliana, under laboratory conditions with dip-inoculation of a high bacterial titer, the vast majority of A. thaliana accessions developed visible disease symptoms. Only 14 accessions (c. 1.3% of all accessions) were resistant to Pst DC3000. This result suggests that, in nature, a major reason for Pst DC3000 not being a natural pathogen of A. thaliana is likely because of the high inoculum needed for infection and/or a mismatch of the environmental conditions needed for Pst DC3000 infection. The molecular basis for the need of a high inoculum and matching environmental conditions are important topics for future studies; however, plants seem to already possess a myriad of mechanisms to defend against a potential invader.
Supplementary Material
Summary.
Plants are continuously threatened by pathogen attack, and as such, they have evolved mechanisms to evade, escape, and defend themselves against pathogens. However, it is not known what types of defense mechanisms a plant would already possess to defend against a potential pathogen that has not co-evolved with the plant. We addressed this important question in a comprehensive manner by studying the responses of 1,041 accessions of Arabidopsis thaliana to the foliar pathogen Pseudomonas syringae pv. tomato (Pst) DC3000.
We characterized the interaction using a variety of established methods, including different inoculation techniques, bacterial mutant strains, and assays for the hypersensitive response, salicylic acid (SA) accumulation, and reactive oxygen production.
Fourteen accessions showed resistance to infection by Pst DC3000. Of these, two accessions had a surface-based mechanism of resistance, six accessions showed a hypersensitive-like response, while three had elevated SA levels. Interestingly, A. thaliana was discovered to have a recognition system for the effector AvrPto, and HopAM1 was found to modulate Pst DC3000 resistance in two accessions.
Our comprehensive study has significant implications for the understanding of natural disease resistance mechanisms at the species level and for the ecology and evolution of plant–pathogen interactions.
Acknowledgements
We would like to thank Dr Weiqing Zeng for his help in hormone quantification, Dr A. Daniel Jones for (ABA)-d6, Dr Paul Staswick for JA-Ile, Dr Doug Schemske for the Belm-12 accession, Dr Roger Innes for providing strain Pst DC3001, Diane Dunham and Dr Greg Martin for strain Pst ΔavrPto, and Dr Alan Collmer, Dr Hailei Wei and Dr Suma Chakravarthy for providing strain E. coli S17-1 pCPP5914 and several Pst DC3000 mutant strains. Funding was provided by the Gordon and Betty Moore Foundation (GBMF3037 to S.Y.H.), and the National Institutes of Health (GM109928 to S.Y.H.).
Footnotes
Author contributions
A.C.V. designed and performed most of the experiments, analyzed the data, and wrote the manuscript; M.O. designed and performed the screen and the initial characterization of the accessions; B.H. performed experiments and analyzed the data, S.X. performed experiments; and S.Y.H. designed and supervised the experiments, and wrote the manuscript.
Additional Supporting Information may be found online in the Supporting Information tab for this article:
References
- 1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. The Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences, USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Molecular Microbiology. 2002;44:73–88. doi: 10.1046/j.1365-2958.2002.02877.x. [DOI] [PubMed] [Google Scholar]
- Bull CT, Manceau C, Lydon J, Kong H, Vinatzer BA, Fischer-Le Saux M. Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Sutic & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Systematic and Applied Microbiology. 2010;33:105–115. doi: 10.1016/j.syapm.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, Mathieu J, Schwizer S, Wang S, Yan YB, Wang J, Martin GB, Chai J. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III Effector. Cell Host & Microbe. 2011;10:616–626. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. Journal of Microbiological Methods. 2006;64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proceedings of the National Academy of Sciences, USA. 2011;108:2975–2980. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels DA. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Applied and Environmental Microbiology. 1986;51:323–327. doi: 10.1128/aem.51.2.323-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng WL, Huang HC. Cellular locations of Pseudomonas syringae pv. syringae HrcC and HrcJ proteins, required for harpin secretion via the type III pathway. Journal of Bacteriology. 1999;181:2298–2301. doi: 10.1128/jb.181.7.2298-2301.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Xiao F, Fan F, Gu L, Cang H, Martin GB, Chai J. Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. The Plant Cell. 2009;21:1846–1859. doi: 10.1105/tpc.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C. Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science. 2011;331:1185–1188. doi: 10.1126/science.1199707. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Vairo D, Cervone F, De Lorenzo G. Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Molecular Plant–Microbe Interactions. 2006;19:931–936. doi: 10.1094/MPMI-19-0931. [DOI] [PubMed] [Google Scholar]
- Forsyth A, Mansfield JW, Grabov N, de Torres M, Sinapidou E, Grant MR. Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Molecular Plant–Microbe Interactions. 2010;23:1545–1552. doi: 10.1094/MPMI-02-10-0047. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. The Plant Journal. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Glaszmann JC, Kilian B, Upadhyaya HD, Varshney RK. Accessing genetic diversity for crop improvement. Current Opinion in Plant Biology. 2010;13:167–173. doi: 10.1016/j.pbi.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM. Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. The Plant Journal. 2009;60:757–770. doi: 10.1111/j.1365-313X.2009.03995.x. [DOI] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell. 2016;166:1526–1538. doi: 10.1016/j.cell.2016.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nature Communications. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MM, Sultana F. Genetic variation for induced and basal resistance against leaf pathogen Pseudomonas syringae pv. tomato DC3000 among Arabidopsis thaliana accessions. SpringerPlus. 2015;4:296. doi: 10.1186/s40064-015-1070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Molecular Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakovidis M, Teixeira PJPL, Exposito-Alonso M, Cowper MG, Law TF, Liu Q, Vu MC, Dang TM, Corwin JA, Weigel D, et al. Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics. 2016;204:337–353. doi: 10.1534/genetics.116.190678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J. Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. The Plant Journal. 2001;26:395–407. doi: 10.1046/j.1365-313x.2001.2641040.x. [DOI] [PubMed] [Google Scholar]
- Johal GS, Briggs SP. Reductase activity encoded by the HM1 disease resistance gene in maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK, Jr, Yu IC, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the "defense, no death" phenotype. Molecular Plant–Microbe Interactions. 2004;17:511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Kover PX, Schaal BA. Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proceedings of the National Academy of Sciences, USA. 2002;99:11270–11274. doi: 10.1073/pnas.102288999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus CM, Munkvold KR, Martin GB. Natural variation in tomato reveals differences in the recognition of AvrPto and AvrPtoB effectors from Pseudomonas syringae. Molecular Plant. 2016;9:639–649. doi: 10.1016/j.molp.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Kvitko BH, Collmer A. Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Methods in Molecular Biology. 2011;712:109–128. doi: 10.1007/978-1-61737-998-7_10. [DOI] [PubMed] [Google Scholar]
- Kvitko BH, Park DH, Velásquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathogens. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf A, Weingart H, Tsiamis G, Boch J. Different versions of Pseudomonas syringae pv. tomato DC3000 exist due to the activity of an effector transposon. Molecular Plant Pathology. 2006;7:355–364. doi: 10.1111/j.1364-3703.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. The Plant Journal. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genetics. 2010;6:e1000894. doi: 10.1371/journal.pgen.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proceedings of the National Academy of Sciences, USA. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic Acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mur LA, Bi YM, Darby RM, Firek S, Draper J. Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. The Plant Journal. 1997;12:1113–1126. doi: 10.1046/j.1365-313x.1997.12051113.x. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biology. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerke EC. Crop losses to pests. Journal of Agricultural Science. 2006;144:31–43. [Google Scholar]
- Perchepied L, Kroj T, Tronchet M, Loudet O, Roby D. Natural variation in partial resistance to Pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS One. 2006;1:e123. doi: 10.1371/journal.pone.0000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. The Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely BK, Martin GB. Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proceedings of the National Academy of Sciences, USA. 2001;98:2059–2064. doi: 10.1073/pnas.98.4.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HN, Akhunov E, Dubcovsky J. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science. 2013;341:783–786. doi: 10.1126/science.1239022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- Sharma YK, Léon J, Raskin I, Davis KR. Ozone-induced responses in Arabidopsis thaliana: the role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proceedings of the National Academy of Sciences, USA. 1996;93:5099–5104. doi: 10.1073/pnas.93.10.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Heese A. Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods. 2014;10:6. doi: 10.1186/1746-4811-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Schneeberger K. SHOREmap v3.0: fast and accurate identification of causal mutations from forward genetic screens. Methods in Molecular Biology. 2015;1284:381–395. doi: 10.1007/978-1-4939-2444-8_19. [DOI] [PubMed] [Google Scholar]
- Surplus SL, Jordan BR, Murphy AM, Carr JP, Thomas B, Mackerness SA. Ultraviolet-B-induced responses in Arabidopsis thaliana: role of salicylic acid and reactive oxygen species in the regulation of transcripts encoding photosynthetic and acidic pathogenesis-related proteins. Plant, Cell & Environment. 1998;21:685–694. [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT. Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. The Plant Cell. 2014;26:4171–4187. doi: 10.1105/tpc.114.131938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. The Plant Cell. 2011;23:4–15. doi: 10.1105/tpc.110.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F. Interplay between MAMP-triggered and SA-mediated defense responses. The Plant Journal. 2008;53:763–775. doi: 10.1111/j.1365-313X.2007.03369.x. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genetics. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. The Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. The Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H-L, Chakravarthy S, Mathieu J, Helmann TC, Stodghill P, Swingle B, Martin GB, Collmer A. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host & Microbe. 2015;17:752–762. doi: 10.1016/j.chom.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. The Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proceedings of the National Academy of Sciences, USA. 2004;101:6297–6302. doi: 10.1073/pnas.0304346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zhang J, Chen J, Chen M, Zhou JM. BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Molecular Plant–Microbe Interactions. 2011;24:100–107. doi: 10.1094/MPMI-04-10-0096. [DOI] [PubMed] [Google Scholar]
- Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annual Review of Phytopathology. 2013;51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. Bacteria establish an aqueous living space as a crucial virulence mechanism. Nature. 2016;539:524–529. doi: 10.1038/nature20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Silverman P, Wilson TM, Kleier DA, Raskin I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. The Plant Cell. 1991;3:809–818. doi: 10.1105/tpc.3.8.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeam I, Nguyen HP, Martin GB. Phosphorylation of the Pseudomonas syringae effector AvrPto is required for FLS2/BAK1-independent virulence activity and recognition by tobacco. The Plant Journal. 2010;61:16–24. doi: 10.1111/j.1365-313X.2009.04028.x. [DOI] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY. A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathogens. 2011;7:e1002291. doi: 10.1371/journal.ppat.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proceedings of the National Academy of Sciences, USA. 2013;110:14807–14812. doi: 10.1073/pnas.1302702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.