Abstract
Interstrand DNA-DNA cross-links are highly toxic lesions that are important in medicinal chemistry, toxicology, and endogenous biology. In current models of replication-dependent repair, stalling of a replication fork activates the Fanconi anemia pathway and cross-links are “unhooked” by the action of structure-specific endonucleases such as XPF-ERCC1 that make incisions flanking the cross-link. This process generates a double-strand break, which must be subsequently repaired by homologous recombination. Recent work provided evidence for a new, incision-independent unhooking mechanism involving intrusion of a base excision repair (BER) enzyme, NEIL3, into the world of cross-link repair. The evidence suggests that the glycosylase action of NEIL3 unhooks interstrand cross-links derived from an abasic site or the psoralen derivative trioxsalen. If the incision-independent NEIL3 pathway is blocked, repair reverts to the incision-dependent route. In light of the new model invoking participation of NEIL3 in cross-link repair, we consider the possibility that various BER glycosylases or other DNA-processing enzymes might participate in the unhooking of chemically diverse interstrand DNA cross-links.
Keywords: DNA cross-link, cross-link repair, Fanconi anemia, homologous recombination, base excision repair, XPF-ERCC1, NEIL, abasic site, psoralen
Graphical abstract
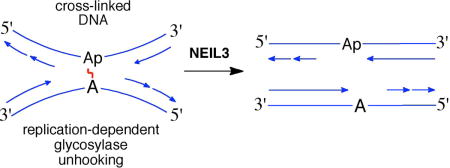
1. Introduction. DNA Interstrand cross-links in biology and medicine
Reading and replicating the genetic information encoded in the sequence of nucleobases in DNA requires cellular operations that separate the two strands of the double helix [1–3]. Interstrand cross-links introduced by bifunctional alkylating agents such as nitrogen mustard anticancer drugs[4–7] prevent strand separation and compromise critical cellular functions of duplex DNA [8, 9]. As a result, DNA cross-links are highly toxic to cells [10, 11]. A wide range of organisms, from bacteria to mammals, possess elaborate systems for the repair of interstrand cross-links [12]. Over twenty proteins are required for cross-link repair in vertebrates [12–16]. The conserved nature of such resource-intensive repair systems suggests that the formation of interstrand cross-links in cellular DNA is an inevitable fact of life. Endogenous processes or unavoidable exposure to environmental agents have the potential to generate interstrand cross-links in cellular DNA [8, 17–27], but the chemical nature of the selection pressures driving the evolution and retention of cross-link repair systems across all walks of life remains uncertain. In medicine, the repair of interstrand cross-links helps define response and resistance to drugs such as cisplatin, carmustine (BCNU), and nitrogen mustards that are commonly used in the treatment of human cancers [28–33].
2. Replication-dependent cross-link repair
When DNA replication is stalled by an interstrand cross-link, complex repair processes are activated [34]. There are excellent comprehensive reviews of replication-dependent interstrand cross-link repair [12–16] and we will summarize briefly here. Advancement of a replication fork is stalled when the Cdc45-MCM-GINS (CMG) helicase complex [2, 35, 36] encounters an interstrand cross-link. Due to the size of this complex, DNA polymerization halts between −40 and −20 nucleotides (nt) from the 3′-side of the cross-link on the leading strand (Figure 1) [34, 37]. Stalling of a replication fork leads to activation of the Fanconi anemia pathway and association of the FANCD2/FANCI heterodimer with the stalled replication fork leads to ubiquitination of this protein complex [34, 38–45]. An emerging view holds that cross-link repair is initiated when two replication forks converge at the lesion [37]. The CMG helicases then dissociate in a process that involves BRCA1-BARD1 and ubiquitin signaling [46]. Dissociation of the helicase complex [2, 35] enables DNA polymerization to progress to the −1 position immediately preceding the cross-link on the leading strand (Figure 1). The activated FANCD2/FANCI complex recruits endonucleases that make incisions on either side of the cross-link [38, 41, 45]. In this process, the SLX4 protein serves as a scaffold that coordinates the action of several structure-specific endonucleases including XPF-ERCC1, MUS81-EME1, and SLX1 [45, 47, 48]. The involvement of XPF-ERCC1 has been clearly demonstrated[49, 50] and cells deficient in these proteins are acutely hypersensitive to interstrand cross-linking agents [44, 51–54]. The identity of other endonucleases that make incisions during cross-link repair, and the circumstances under which they become involved, remains under investigation [45, 48, 51, 52, 55–58]. Incisions by structure-specific endonucleases “unhook” the cross-link and generate a double strand break (Figure 1). The unhooked cross-link remnant may be trimmed [34] to a smaller adduct (possibly by the exonuclease activity of SNM1A) [51, 59] to facilitate lesion bypass by translesion synthesis (TLS) polymerases such as Polκ (kappa), Polη (eta), and Polν (nu) [60–63]. Extension may then be carried out by a complex containing REV1 and Polζ (zeta) [34, 64, 65]. Indeed, cells deficient in REV1 and Polζ are hypersensitive to cross-linking agents.[64, 66] It remains uncertain whether REV1/Polζ cooperate with other TLS polymerases as seen in the bypass of pyrimidine dimers.[67] Homologous recombination (HR) using the newly synthesized duplex on the leading strand as the homology repair partner is required to repair the double-strand break [68–71]. Finally, nucleotide excision repair (NER) may remove the unhooked cross-link remnant to regenerate a native DNA duplex [70]. There is much evidence supporting the general pathway shown in Figure 1; however, it is worth noting that a distinct pathway involving replication traverse of a psoralen-derived cross-link without repair has been suggested [72, 73].
Figure 1.
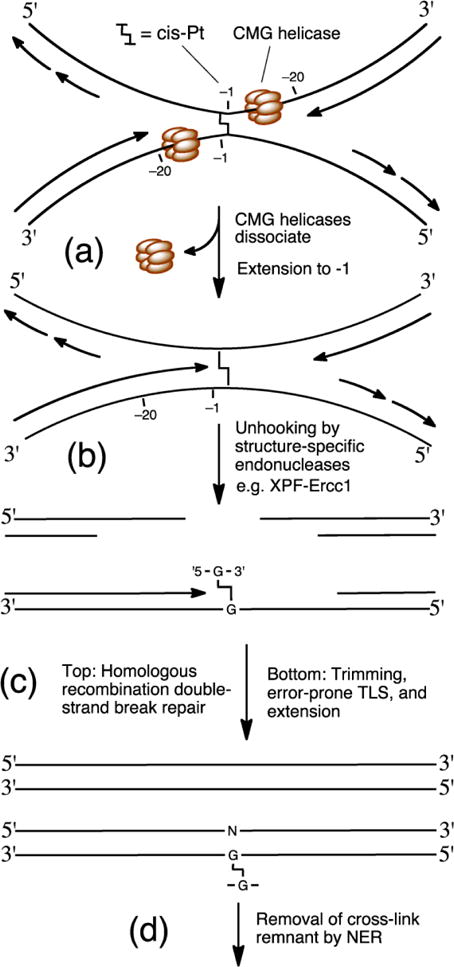
Model for replication-dependent interstrand cross-link repair. (a) CMG helicase collides with cross-link and dissociates. (b) Structure-specific endonucleases unhook the cross-link. (c) Homologous recombination repairs double-strand break (top duplex). Possible nuclease trimming of the oligonucleotides flanking the adduct, TLS, and extension past the adduct remnant (bottom duplex). (d) Cross-link remnant may be removed by NER.
3. Base excision repair DNA glycosylases and their involvement in cross-link repair
The base excision repair (BER) pathway is critical for the removal of damaged, misincorporated, and mispaired DNA bases [74–81]. BER typically begins with DNA glycosylase enzymes that recognize a damaged or mispaired nucleobase (B* in Figure 2A) and catalyze hydrolysis of the glycosidic bond holding the base to the deoxyribose backbone [82]. Various BER glycosylases remove a wide array of damaged and mispaired nucleobases [74–80]. Importantly, substrate recognition and catalysis by glycosylases typically involve “flipping out” (extrusion of) the target base from the double helix – a process that is presumably precluded for bases involved in an interstrand cross-link (Figure 2B) [81–83]. Monofunctional BER enzymes catalyze hydrolytic removal of a nucleobase leaving an intact abasic (Ap) site in the DNA (Figure 2A), while bifunctional DNA glycosylases remove the base and also catalyze a subsequent elimination (lyase) reaction that generates a strand break with a 5′-phosphoryl end group and 3′-deoxyribose phosphate sugar remnant [74, 76, 78, 79, 82, 84–86].
Figure 2.
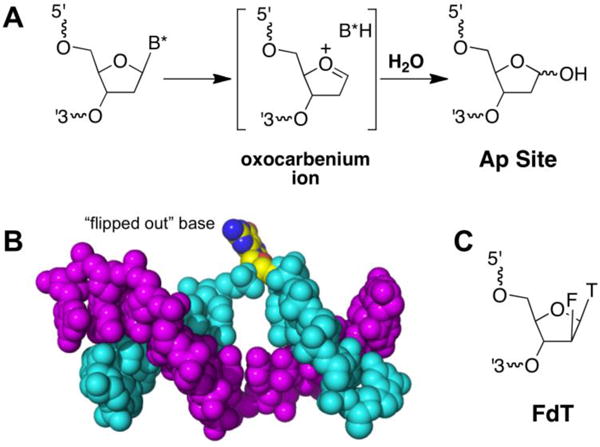
Base excision repair enzymes remove damaged or mispaired nucleobases from DNA. Panel A: The mechanism of base removal by BER enzymes involves hydrolysis of the glycosidic bond. Shown here is a dissociative hydrolysis mechanism involving an oxocarbenium ion intermediate. Panel B: BER enzymes typically act on DNA substrates in which the damaged nucleobase is extruded from the double helix. The image shows the DNA substrate in a DNA-hOGG1 complex (protein structure omitted) in which the 8-oxo-G base is extruded from the duplex (image prepared from pdb entry 3ktu). Panel C: Damaged bases attached to 2′-deoxy-2′-fluoroarabinofuranosyl residues are resistant to hydrolysis by BER enzymes because the electron-withdrawing fluoro substituent on the sugar destabilizes the oxocarbenium ion-like transition state of the glycosylase reaction.
Over the years, there have been reports that BER proteins can interact with cross-link repair pathways [87]. For example, there is evidence that murine 3-methyladenine glycosylase (Aag) plays a role in cellular resistance to psoralen-derived cross-links [88]. However, in vitro studies showed that human AAG did not bind or process a 21 base pair duplex containing a psoralen-derived cross-link, leading the authors to conclude that “Aag’s role in conferring protection against psoralen-induced (cross-links) is either indirect or involves a DNA substrate that is an intermediate of the repair process” [88]. In a separate example, the glycosylase NEIL1 was shown to accumulate at psoralen-derived interstrand cross-links in cells and inhibit repair of these lesions [89]. In vitro gel electrophoretic mobility shift assays demonstrated that NEIL1 bound to a 21 base pair cross-linked duplex, with an affinity at least 4-fold greater than that for a native control duplex [89]. In another set of studies, it was found that the deletion of the BER proteins Pol β and uracil DNA glycosylase (UNG) conferred resistance to cisplatin toxicity [90]. This led to a proposal that UNGs remove uracil residues generated by deamination of cytosine residues near cisplatin cross-links, thus inhibiting subsequent repair [91]. In a final early example where BER may influence the repair of cross-links, Couvé et al. showed that NEIL1 can excise an unhooked, psoralen-derived cross-link remnant from a three-stranded structure (e.g. bottom duplex in Figure 3) [92, 93]. This type of glycosylase activity would not provide initial unhooking of the cross-link, but could provide an alternative to NER for the final “clean-up” of the psoralen cross-link remnant (last step in Figure 3).
Figure 3.
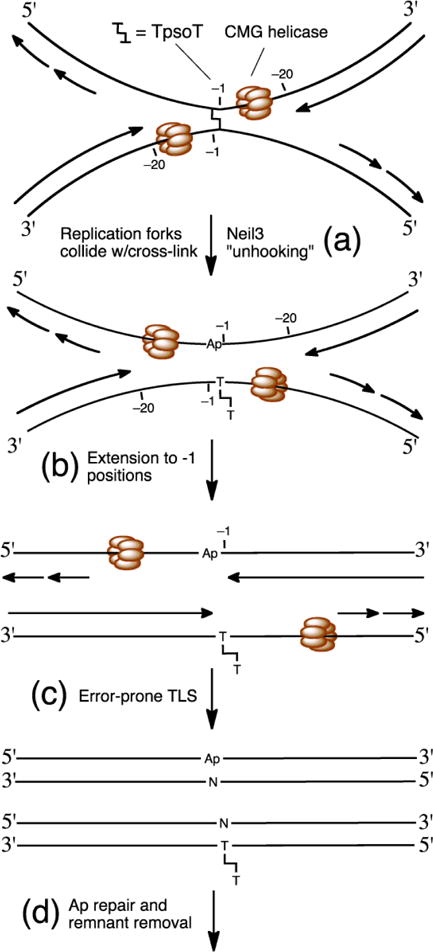
Model for replication-dependent, incision-independent repair of a psoralen-derived interstrand cross-link. (a) CMG helicase stalls at cross-link. (b) NEIL3 unhooks the cross-link. (c) TLS and extension past the Ap site and psoralen adduct remnant. (d) Repair of the Ap site by a pathway involving APE, pol β, and ligase III (upper duplex) and NER or BER to remove the psoralen cross-link remnant (lower duplex).
4. Evidence for a direct role for the catalytic action of a BER glycosylase in cross-link repair
A recent study by Walter and coworkers provides evidence that the catalytic activity of a BER glycosylase enzyme can play a central role in the repair of interstrand DNA cross-links [94]. In this work, the direct catalytic action of the BER glycosylase NEIL3 was implicated in the repair of both psoralen-derived and abasic site-derived interstrand DNA cross-links in Xenopus egg extracts.
Psoralens are a class of plant-derived DNA cross-linking agents [95]. The planar furocoumarin system intercalates between the base pairs of duplex DNA where 300–400 nm light can induce reactions between the pi-bonds of the natural product and the 5,6-double bonds of thymine and cytosine residues to generate both covalent monoadducts and interstrand cross-links (Figure 4A) [96–99]. Interstrand cross-links arise preferentially at 5′-TA sequences[100] via sequential photoinduced [2+2] cycloadditions of thymine residues with the 4′,5′-double in the furan ring and the 3,4-double bond of the pyrone system [95, 98] (Figure 4B). DNA duplexes containing a psoralen cross-link display local unwinding at the cross-link site but, overall, retain a B-form structure [96–102]. However, the cross-link structure in duplex DNA may not be directly relevant to the X-shaped and Y-shaped structures generated at a stalled replication fork, which are the true substrates for replication-coupled repair [48].
Figure 4.
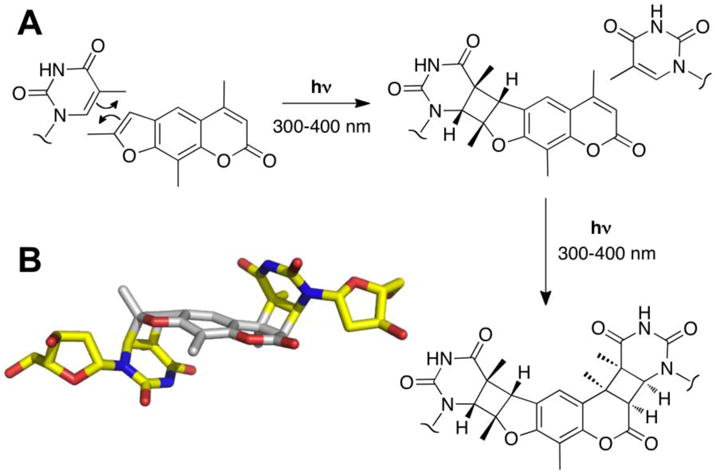
Formation and structure of a psoralen-derived interstrand cross-link. Panel A: Two sequential photoinduced [2+2] cycloaddition reactions generate interstrand cross-links preferentially at 5′-TA sequences in duplex DNA. The reaction is shown for the psoralent derivative trioxsalen. Panel B: The three-dimensional structure of a psoralen-derived cross-link in duplex DNA. Thymine residues are shown with carbons colored yellow and psoralen with carbons colored in gray (image prepared by modification of pdb 204d).
In the Xenopus egg extract system of Semlow et al., the repair of a psoralen cross-link was distinct from that previously observed for a cisplatin interstrand cross-link [94]. Relatively small amounts of homologous recombination intermediates were seen and, while FANCD2 was ubiquitinated, depletion of the FANCD2/FANCI complex had a minimal effect on repair. This led the authors to consider the possibility that the psoralen cross-link might be unhooked by the action of a BER glycosylase. This process would generate an abasic site on one strand and a modified thymine residue on the other (Figure 3). Several lines of evidence supported this supposition. Depletion of the TLS polymerase REV1 caused stalling of replication at or near the −1 position on both strands. Consistent with the proposed glycosylase-mediated generation of an abasic site on one of the strands, the repair intermediates could be digested by the enzyme apurinic endonuclease (APE) [103–105]. Furthermore, introduction of 2′-deoxy-2′-fluoroarabinofuranosyl thymine (FdT) residues at the cross-link site shunted repair of the cross-link to the incision-dependent (Fanconi anemia) pathway shown in Figure 1. This result strongly implicated glycosylase activity in repair of the psoralen cross-link because the electron-withdrawing effect of the 2′-fluoro substituent in FdT (Figure 2C) inhibits the action of BER glycosylases through destabilization of the oxocarbenium ion-like transition state of these enzymatic reactions [81, 82, 106, 107].
Repair of an Ap-derived cross-link was also investigated by Walter and coworkers [94]. Ap sites are ubiquitous endogenous lesions in cellular DNA [19, 103, 108] and recent work has characterized interstrand cross-links arising from the reaction of the Ap aldehyde residue with the exocyclic amino groups of adenine and guanine residues in duplex DNA (Scheme 1) [17, 109, 110]. The Ap-derived cross-links are chemically stable in duplex DNA [110, 111] and the dA-Ap cross-link was previously shown to block DNA replication by the strand-displacing polymerase ϕ29, stalling primer extension at the −1 position immediately preceding the cross-link [112]. The stability of the Ap-derived cross-links may derive from the fact that they exist as cyclic aminoglycosides rather than in the ring-opened imine form (Scheme 1) [111, 113, 114]. The ubiquitous nature of Ap sites in genomic DNA makes endogenous cellular formation of Ap-derived cross-links an intriguing possibility.
Scheme 1.
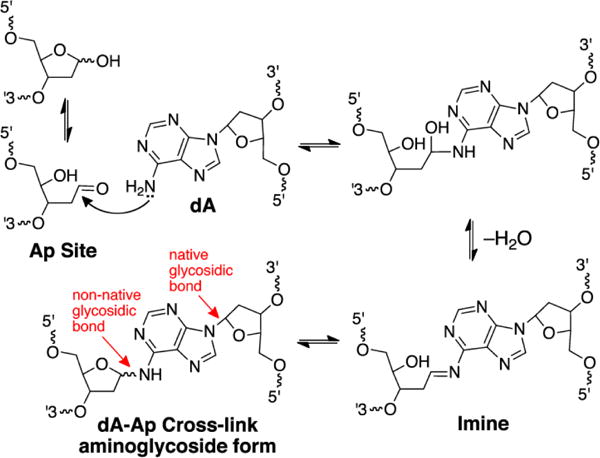
Formation and structure of the dA-Ap interstrand cross-link.
In the Xenopus system, approximately 20% of the dA-Ap cross-link was repaired by the FANCD2/FANCI incision-dependent pathway, while 80% of the repair proceeded via the incision-independent route [94]. As in the case of the psoralen cross-link described above, the repair intermediates could be digested by APE. Depletion of Rev1 prevented approximately half of the glycosylase-dependent repair, consistent with unhooking of the non-native glycosylase linkage in the cross-link to generate an Ap site on one strand and a native adenine residue on the other (Figure 5 and Scheme 1).
Figure 5.
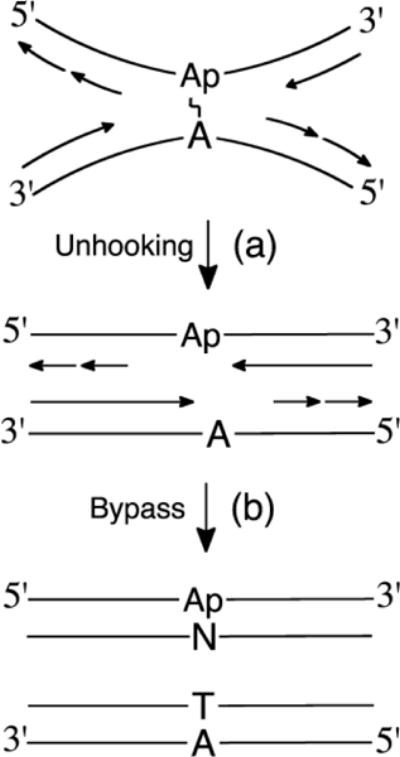
Model for replication-dependent, incision-independent repair of an Ap-derived interstrand cross-link. (a) NEIL3 unhooking of the cross-link by cleavage of the non-native glycosidic bond. (b) TLS and extension past the Ap site and normal extension past the native adenine residue. Repair of the Ap-containing duplex may be completed by the BER proteins APE, pol β, and DNA ligase.
5. The glycosylase NEIL3 is involved in unhooking psoralen and Ap-derived interstrand cross-links
Semlow et al. provided evidence that the glycosylase NEIL3 is responsible for unhooking of the psoralen and Ap-derived cross-links in the Xenopus egg extracts [94]. For example, immunodepletion of NEIL3 led to a decrease in repaired products and addition of exogenous active NEIL3 enzyme to the assay reversed this effect [94].
NEIL3 has a number of properties that are consistent with its newly defined role in replication-dependent cross-link repair. The enzyme is expressed in early S and G2 phases of the cell cycle [115, 116], during DNA replication and unpublished data cited in the Semlow publication [94] notes that NEIL3 is associated with the replisome. As discussed above, BER enzymes typically act on DNA substrate conformations in which the target base is extruded from the double helix (Figure 2B) [81–83]. Clearly, a nucleobase involved in a cross-link cannot be extruded in the typical manner. However, DNA structure at a stalled replication fork is atypical [48]. When NEIL3 unhooks cross-links at a stalled replication fork, the enzyme presumably must act upon a nucleobase that is located near the junction of a strand-separated Y-shaped or X-shaped DNA structure [2, 3, 34–36, 48]. Significantly, NEIL3 displays preferences for the removal of damaged bases from non-duplex substrates including single-stranded DNA, bubble structures, and G-quadruplexes [115, 117, 118]. There are a few examples of glycosylases that operate without extrusion of the damaged base [119–122] and it will be interesting to learn how the catalytic machinery of NEIL3 gains access to the glycosidic bond of cross-linked nucleobases in the non-canonical DNA structures generated at a stalled replication fork.
It is possible to rationalize the observation that NEIL3 has the catalytic power to deglycosylate nucleobases involved in the psoralen and Ap-derived cross-links [94]. To understand these activities it may be useful to compare the cross-link structures to previously characterized NEIL3 substrates such as FapyA, FapyG, thymine glycol, and dihydrothymine [115, 118]. The non-native glycosidic bond in the dA-Ap cross-link resembles the glycosidic bonds in the NEIL3 substrates FapyA and FapyG, while the saturated thymine rings in the psoralen cross-link resemble the known substrates thymine glycol and dihydrothymine (Figure 6). A previous observation that the related BER glycosylase NEIL1 removes psoralen monoadducts from DNA provides further support for the idea that NEIL3 has the catalytic power to deglycosylate a psoralen-thymine adduct [93].
Figure 6.
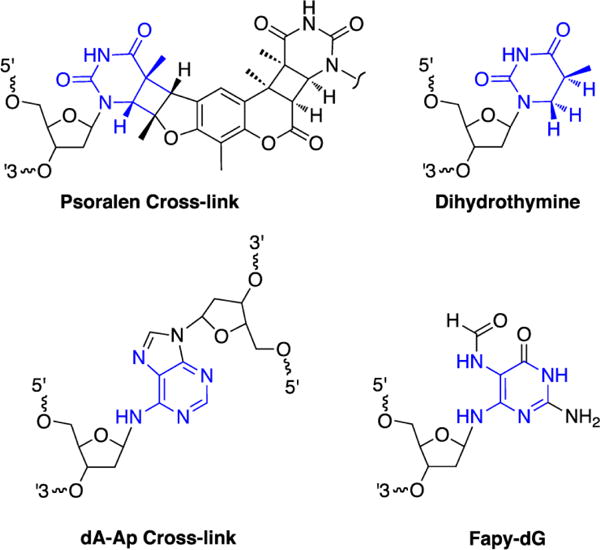
Psoralen- and Ap-derived cross-links (left side) are structurally analogous to known NEIL3 substrates (right side).
6. Comparison of incision-dependent and incision-independent cross-link repair pathways
The evidence presented by Semlow et al. suggests that the incision-independent, glycosylase unhooking pathway involving NEIL3 represents the front-line mechanism for repair of the two cross-links studied in the Xenopus system [94]. If the incision-independent pathway is thwarted, for example when glycosylase-resistant FdT sugars are present at the psoralen cross-link site, the slower incision-dependent (Fanconi) pathway is engaged (in general, these repair processes take place over the course of several hours) [94].
It may be advantageous to unhook cross-links at a stalled replication fork via an incision-independent pathway. In the incision-independent glycosylase pathway, the bifunctional (lyase) property of NEIL3 [115] is evidently suppressed, thus avoiding double-strand break formation (Figures 3 and 5) [94]. In contrast, the actions of structure-specific endonucleases [45, 47] in the incision-dependent Fanconi pathway generate a double-strand break (Figure 1). Avoidance of double-strand break formation in the glycosylase-dependent unhooking mechanism eliminates the need to engage homologous recombination, a process that is quite complex in its own right and can lead to insertions and deletions in the genome [69, 123]. Both the incision-dependent (Fanconi) and incision-independent cross-link repair pathways include error-prone, potentially mutagenic steps [64, 65, 124], involving the bypass of psoralen adduct remnants [62, 63, 125, 126] and/or Ap sites [127, 128].
It may be important to mesh the new evidence implicating NEIL3 in cross-link unhooking with literature supporting the involvement of Fanconi proteins, XPF/ERCC1, and double-strand breaks in the repair of psoralen-derived cross-links [129–131]. Three possible explanations were offered in this regard [94]: i. in some clonogenic survival experiments, the number of cross-links generated by the psoralen-UV treatment may overwhelm the capacity of NEIL3, necessitating engagement of the incision-dependent Fanconi pathway, ii. some cross-links generated in chromatin may not be sterically accessible to NEIL3, thus requiring recognition and repair involving Fanconi proteins, iii. Fanconi proteins such as FANCD2 may have critical functions in cross-link repair other than unhooking. For example, FANCD2 can modulate chromatin dynamics [132] and may interact with FANCM in the reversal of stalled replication forks [133].
It is also interesting to note that Neil3−/− knockout mice have been generated [134, 135]. These mice do not display a profound phenotype, although loss of proliferating neuronal progenitors was noted after hypoxia-ischemia, leading to the suggestion that “NEIL3 exercises a highly specialized function through accurate molecular repair of DNA in rapidly proliferating cells” [134]. The lack of a severe phenotype in Neil3−/− mice, such as would be expected to arise from cross-link repair deficiencies (e.g. Fanconi anemia) [41, 44, 45, 136, 137], could be due to the strongly overlapping functions of the Fanconi anemia and NEIL3-dependent repair pathways [94]. In addition, it is possible that overlapping functions of NEIL3 and the related NEIL1 glycosylase in cross-link unhooking could blunt the effects of NEIL3 deletion. NEIL1 displays substrate specificity similar to NEIL3 in terms of the damaged nucleobases that it excises and, like NEIL3, displays the ability to deglycosylate damaged bases located in non-duplex structures (in bubbles and near nicks) [138]. Interestingly, NEIL1 is associated with the replisome and a “cowcatcher” role has been proposed, in which the enzyme carries out pre-replication removal of oxidatively-induced lesions from single-stranded regions of DNA at the replication fork [139].
7. New possibilities suggested by the role of NEIL3 in cross-link repair: considering other enzyme activities that could catalyze replication-dependent unhooking of interstrand cross-links
The participation of NEIL3 in cross-link unhooking offers the possibility of an entirely new role for BER glycosylases and other DNA-processing enzymes. There are several properties that might favor the participation of any given enzyme in cross-link unhooking. i. Physical association with the replisome or Fanconi anemia proteins such as activated FANCD2/FANCI could direct enzyme activity to a cross-linked nucleotide located at a stalled replication fork. ii. Unhooking enzymes must have catalytic activity on a cross-linked nucleotide located at the junction of Y-shaped, X-shaped, or Holliday junction (chickenfoot) structures at a stalled replication fork [2, 3, 34–36, 48, 140]. In the case of NEIL3, such activity was foreshadowed by studies demonstrating its ability to remove damaged nucleobases from non-canonical substrates such as single-stranded DNA, quadruplex DNA, and bubbles [115, 117, 118]. Borrowing terminology used to describe endonucleases such as XPF-ERCC1 [45, 47], we could say that an enzyme such as NEIL3 has “structure-specific glycosylase” activity. iii. An enzyme involved in incision-independent unhooking of a cross-link must have the catalytic power to cleave one of the covalent bonds involved in the interstrand cross-link. Although the mechanisms by which NEIL3 gains access to the glycosidic bonds in the psoralen-derived and dA-Ap cross-links remain unknown, precedents (Figure 6 and discussion above) indicate that the enzyme possesses the fundamental chemical power to cleave the glycosidic bonds in these cross-links.
Each BER glycosylase has the catalytic power to process a limited structural group of nucleobases [74–80]. There are many structurally diverse cross-links [8, 20] and the reactivity of each particular cross-link will define its susceptibility toward enzymatic unhooking. In this regard, it is important to recognize that interstrand cross-links are not a generic, interchangeable, or homogeneous group. Differences in the chemical structure and reactivity of various cross-links are important. It is almost certain that many cross-links will be chemically resistant to the unhooking action of NEIL3. However, other glycosylases may have the catalytic power to unhook cross-links that are refractory to NEIL3. For example, AAG may act upon cross-links derived from nitrogen mustards (Figure 7A) [141–143]. This is suggested by the ability of AAG to catalyze hydrolysis of the glycosidic bonds in N7-alkyl-dG and N3-alkyl-dA monoadducts [144]. Similarly, AAG has the chemical potential to unhook the N1-dG attachment in a cross-link derived from the anticancer drug 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU, Figure 7B) [145, 146], as suggested by the ability of the enzyme to deglycosylate N1-methyl-dG [144]. AAG typically operates on extruded nucleobases[81, 147] and it is completely unknown whether the active site of this enzyme can accommodate (and act upon) a cross-linked nucleotide at a stalled replication fork. In this context, however, it may be noteworthy that AAG is able to remove lesions located in non-duplex (i.e. single-stranded) substrates [144].
Figure 7.
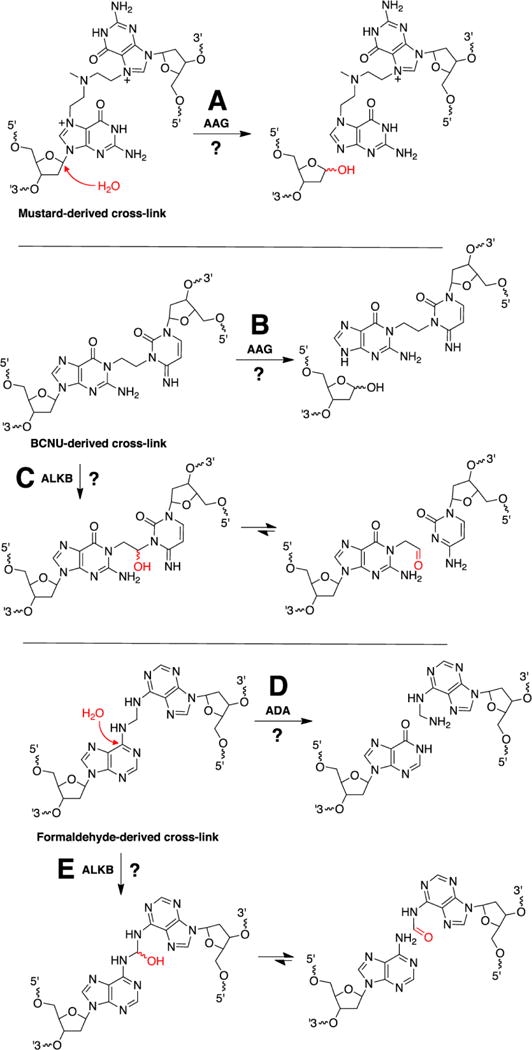
Speculative mechanisms for incision-independent unhooking of structurally diverse interstrand cross-links.
Some cross-links may not be amenable to unhooking by glycosylase enzymes. For example, there are no BER enzymes that deglycosylate platinated nucleobases generated by the antitumor agent cisplatin [74–80]. Similarly, there are no known BER glycosylases that act on N6-alkyladenine, N2-alkylguanine, or N3-cytosine residues [74–80]. However, other classes of DNA-processing enzymes conceivably could unhook cross-links containing these types of chemical attachments. For example, the iron(II) and α-ketoglutarate-dependent dioxygenase activity of ALKB enzymes can carry out the oxidative dealkylation of N3-alkylcytosine residues [148, 149]. This chemistry could enable unhooking of the N1-dG-ethyl-N3-dC cross-links derived from BCNU (Figure 7C). Some isoforms of human ALKB enzymes act on non-duplex substrates [148, 149].
The dioxygenase activity of ALKB enzymes also has the potential to unhook formaldehyde-derived [21] N6-dA-CH2-N6-dA, N2-dG-CH2-N2-dG, N2-dG-CH2-N4-dC, and N6-dA-CH2-N4-dC cross-links (Figure 7E). ALKB enzymes are known to catalyze the oxidative dealkylation of N6-alkyl-dA and N2-alkyl-dG residues [148, 149].
Enzymes with adenosine deaminase activity have the potential to unhook N6-dA-CH2-N6-dA cross-links derived from formaldehyde (Figure 7D) [21]. However, it must be noted that the enzyme adenosine deaminase (ADA) has weak activity on oligomeric DNA substrates [150]. ADAR enzymes are RNA editing enzymes that deaminate adenine residues in oligomeric RNA substrates [151]. Interestingly, Beal’s group showed that ADAR-2 can deaminate a 2′-deoxyadenosine residue located at the 5′-end of an RNA substrate [152].
The deaminase activity associated with activation-induced cytidine deaminases (AID/APOBEC family of proteins) [153–155] has the potential to unhook N4-dC-CH2-N6-dA cross-links derived from formaldehyde [21]. Some RNA-editing enzymes also have the potential to catalyze deamination of cytosine residues in DNA [156] and might participate in the unhooking of N4-dC-CH2-N6-dA cross-links if this activity could be selectively directed at a cross-linked cytosine residue.
These selected examples (perhaps unnecessarily limited to consideration of enzymatic activities associated with known DNA-processing enzymes) serve to illustrate how diverse enzymatic activities could carry out the unhooking of various interstrand cross-links. However, we emphasize that the above discussion is highly speculative because the postulated “structure-specific” activities deviate, substantially in some cases, from the established substrate specificities of these enzymes [157].
8. Summary
Deep appreciation of various DNA repair pathways has been built upon the in vitro characterization of the fundamental chemical, biochemical, and structural properties of individual repair proteins [158–161]. Along these lines, it will be important to elucidate the biochemical activities of NEIL3 (and other enzymes) on cross-linked DNA substrates that resemble a stalled replication fork or transcription bubble. In addition, NEIL3 presents an excellent opportunity to elucidate the structural mechanisms by which a BER enzyme can access the glycosidic bonds in a cross-linked duplex. In addition, chemical biologists can help address the roles of other BER glycosylases and ALKB family enzymes in the replication-independent unhooking of interstrand cross-links. For instance, methods are in place for the preparation of duplex DNA substrates housing a site-specific and structurally defined interstrand cross-link [17, 110, 146, 162–174]. This will enable shuttle vector-based methods, in conjunction with genetic manipulation, for interrogating the roles of BER glycosylases and ALKB family proteins in the removal of interstrand cross-link lesions in cells [164, 175]. Likewise, genetic modulation of these DNA repair enzymes, together with measurements of interstrand cross-links in cellular DNA by liquid chromatography-tandem mass spectrometry (LC-MS/MS) [72, 176–181], also offers the opportunity for exploring the roles in these enzymes in the unhooking and repair of interstrand cross-links. At the same time, it will be important for studies in cellular systems (and cell extracts) to establish how proteins present at a stalled replication fork choreograph the action of NEIL3 and other enzymes involved in cross-link unhooking, and to gather data from living organisms that shed light on the biological roles and potential medicinal relevance of these repair pathways.
Highlights.
Current models for replication-dependent cross-link repair involve activation of the Fanconi anemia pathway is activated and cross-links are “unhooked” by the action of structure-specific endonucleases such as XPF-ERCC1 that make incisions flanking the cross-link.
Recent work provides evidence for a new, incision-independent unhooking mechanism involving the intrusion of a base excision repair enzyme, NEIL3, into the world of cross-link repair.
The evidence suggests that the glycosylase action of NEIL3 unhooks interstrand cross-links derived from an abasic site or the psoralen derivative trioxsalen.
Acknowledgments
We apologize to authors of many important papers in the field that were not cited due to space limitations. KSG and YW are grateful to the National Institutes of Health for support during the writing of this review (ES 021007). We thank Professor Orlando Schärer for insightful comments on the manuscript.
Abbreviations
- BCNU
carmustine or 1,3-bis(2-chloroethyl)-1-nitrosourea
- BER
base excision repair
- CMG helicase
Cdc45-MCM-GINS helicase
- NEIL
Nei-like
- TLS
translesion synthesis
- HR
homologous recombination
- NER
nucleotide excision repair
- FdT
2′-deoxy-2′-fluoroarabinofuranosyl thymine
- Ap
DNA abasic site
- APE
apurinic endonuclease, Fapy, formamidopyrimidine
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest
References
- 1.Watson JD, Crick FHC. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, Li H, O’Donnell ME. The architecture of a eucarytoic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao NY, O’Donnell M. Snapshot: The replisome. Cell. 2010;141:1088. doi: 10.1016/j.cell.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imani-Nejad M, Johnson KM, Price NE, Gates KS. A new cross-link for an old cross-linking drug: the nitrogen mustard anticancer agent mechlorethamine generates cross-links derived from abasic sites in addition to the expected drug-bridged cross-links. Biochemistry. 2016;55:7033–7041. doi: 10.1021/acs.biochem.6b01080. [DOI] [PubMed] [Google Scholar]
- 5.Geiduschek EP. “Reversible” DNA. Proc Nat Acad Sci USA. 1961;47:950–955. doi: 10.1073/pnas.47.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohn KW. Beyond DNA cross-linking: History and prospects of DNA-targeted cancer treatment. Cancer Res. 1996;56:5533–5546. [PubMed] [Google Scholar]
- 7.Brookes P, Lawley PD. The alkylation of guanine and guanylic acid. J Chem Soc. 1961:3923–3928. [Google Scholar]
- 8.Schärer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 9.Osawa T, Davies D, Hartley JA. Mechanism of cell death resulting from DNA interstrand cross-linking in mammalian cells. Cell Death Dis. 2011;2:e187. doi: 10.1038/cddis.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutation Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- 11.Grossman KF, Ward AM, Matkovic ME, Folias AE, Moses RE. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutation Res. 2001;487:73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- 12.McVey M. Strategies for DNA interstrand cross-link repair: Insights from worms, flies, frogs, and slime molds. Env Mol Mutagenesis. 2010;51:646–658. doi: 10.1002/em.20551. [DOI] [PubMed] [Google Scholar]
- 13.Clauson C, Schärer OD, Niedernhofer LJ. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harbor Perspectives in Biology. 2013;5:a012732/012731–a012732/012725. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams HL, Gottesman ME, Gautier J. The differences between ICL repair during and outside of S phase. Trends Biochem Sci. 2013;38:386–393. doi: 10.1016/j.tibs.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand crosslinks in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muniandy PA, Liu J, Majumdar A, Liu S-t, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Interstrand DNA–DNA Cross-Link Formation Between Adenine Residues and Abasic Sites in Duplex DNA. J Am Chem Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gates KS. Covalent Modification of DNA by Natural Products. In: Kool ET, editor. Comprehensive Natural Products Chemistry. Pergamon; New York: 1999. pp. 491–552. [Google Scholar]
- 19.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajski SR, Williams RM. DNA cross-linking agents as antitumor drugs. Chem Rev. 1998;98:2723–2795. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 21.Chaw YFM, Crane LE, Lange P, Shapiro R. isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- 22.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Interstrand cross-links induced by alpha, beta-unsaturated aldehydes derived from lipid peroxidation and environmental sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen HJC, Chen YC. Analysis of glyoxal-induced DNA cross-links by capillary liquid chromatography nanospray ionization tandem mass spectrometry. Chem Res Toxicol. 2009;22:1334–1341. doi: 10.1021/tx900129e. [DOI] [PubMed] [Google Scholar]
- 24.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. Scope and mechanism of interstrand crosslink formation by the C4′-oxidized abasic site. J Am Chem Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, Swenberg JA, Crossan GP, Patel KJ. Endogenous formaldehyde is a hemapoietic stem cell genotoxin and metabolic carcinogen. Mol Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong IS, Ding H, Greenberg MM. Oxygen independent DNA interstrand cross-link formation by a nucleotide radical. J Am Chem Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dextraze ME, Gantchev T, Girouard S, Hunting D. DNA interstrand cross-links induced by ionizing radiation: an unsung lesion. Mutation Res. 2010;704:101–107. doi: 10.1016/j.mrrev.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Panasci L, Xu ZY, Bello V, Aloyz R. The role of DNA repair in nitrogen mustard drug resistance. Anticancer Drugs. 2002;13:211–220. doi: 10.1097/00001813-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Usanova S, Piée-Staffa A, Sied U, Thomale J, Schneider A, Kaina B, Köberle B. Cisplatin sensitivity of testis tumour cells is due to deficiency in interstrand-crosslink repair and low ERCC1-XPF expression. Mol Cancer. 2010;9:248. doi: 10.1186/1476-4598-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman HS, Colvin OM, Kaufmann SH, Ludeman SM, Bullock N, Bigner DD, Griffith OW. Cyclophosphamide resistance in medulloblastoma. Cancer Res. 1992;52:5373–5378. [PubMed] [Google Scholar]
- 31.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wynne P, Newton C, Ledermann JA, Olaitan A, Mould TA, Hartley JA. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br J Cancer. 2007;97:927–933. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torres-Ramos SJ, Cousineau L, Caplan S, Panasci L. Correlation of resistance to nitrogen mustards in chronic lymphocytic leukemia with enhanced removal of melphalan-induced DNA cross-links. Biochem Pharmacol. 1989;15:3122–3123. doi: 10.1016/0006-2952(89)90025-7. [DOI] [PubMed] [Google Scholar]
- 34.Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of replication-coupled DNA interstrand cross-link repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan Z, Bai L, Sun J, Georgescu R, Liu J, O’Donnell ME, Li H. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali FA, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC, Rueda D, Costa A. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knipscheer P, Räschle M, Smogorzewska A, Enolu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi Anemia Pathway Promotes Replication-Dependent DNA Interstrand Cross-Link Repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longerich S, Kwon Y, Tasi MS, Hiaing AS, Kupfer GM, Sung P. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–5670. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang C-C, Li Z, Lopez-Martinez D, Nicholson WV, Vénien-Bryan C, Cohn MA. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun. 2016;7 doi: 10.1038/ncomms12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–349. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 42.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–201. [PubMed] [Google Scholar]
- 43.Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niedernhofer LJ, Lalai AS, Hoeijmakers JHJ. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim BH, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair. 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Douwel DK, Boonen RACM, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. XPF-ERCC1 Acts in Unhooking DNA Interstrand Crosslinks in Cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodskinson MRG, Silhan J, Crossan GP, Garayoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. Mouse SLX4 Is a Tumor Suppressor that Stimulates the Activity of the Nuclease XPF-ERCC1 in DNA Crosslink Repair. Mol Cell. 2014;54:472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sengerová B, Wang AT, McHugh PJ. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011;10:3999–4008. doi: 10.4161/cc.10.23.18385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D’Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 Participates in the Fanconi Anemia Pathway of Cross-Link Repair. Mol Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 54.Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene. 2015;569:153–161. doi: 10.1016/j.gene.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, Pavletich NP. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science. 2014;346:1127–1130. doi: 10.1126/science.1258973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pizzolato J, Mukherjee S, Schärer OD, Jiricny J. FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand cross-links in vitro. J Biol Chem. 2015;290:22602–22611. doi: 10.1074/jbc.M115.663666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 Acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 58.Murina O, von Aesch C, Karakus U, Ferretti LP, Bolck HA, Hänggi K, Sartori AA. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014;7:1030–1038. doi: 10.1016/j.celrep.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 59.Wang AT, Sengerová B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlink JH, Schofield CJ, Gilead O, Hartley JA, McHugh PJ. Human SNM1A and XPF–ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011;25:1859–1870. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho TV, Schärer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Env Mol Mutagenesis. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 61.Ho TV, Guainazzi A, Derkunt SB, Enoiu M, Schärer OD. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 2011;39:7455–7464. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy U, Schärer OD. Involvement of Translesion Synthesis DNA Polymerases in DNA Interstrand Crosslink Repair. DNA Repair (Amst) 2016;44:33–41. doi: 10.1016/j.dnarep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mogi S, Butcher CE, Oh DH. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand cross-links in human cells. Exp Cell Res. 2008;314:887–895. doi: 10.1016/j.yexcr.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Budzowska M, Graham TGW, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34:1971–1985. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma S, Canman CE. REV1 and DNA polymerase zeta in DNA interstrand cross-link repair. Env Mol Mutagenesis. 2010;53:725–740. doi: 10.1002/em.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases η, ζ, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ziv O, Geacintov N, Nakajima S, Yasui A, Livneh Z. DNA polymerase zeta cooperates with polymerases kappa and iota in translesion DNA synthesis across pyrimidine photodimers in cells from XPV patients. Proc Nat Acad Sci USA. 2009;106:11552–11557. doi: 10.1073/pnas.0812548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hinz JM. Role of homologous recombination in DNA interstrand cross-link repair. Env Mol Mutagenesis. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- 69.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13:729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- 71.Long DT, Räschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang JC, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand cross-links. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rohleder F, Kuper J, Kisker C, Huang JC, Seidman M, Xue Y, Wang W, Round A. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 2016;44:3219–3232. doi: 10.1093/nar/gkw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015;43:10083–10101. doi: 10.1093/nar/gkv1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;4:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krokan HE, Bjørås M. Base Excision Repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wallace SS. Base excision repair: a critical player in many games. DNA Repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 79.Schermerhorn KM, Delaney S. A chemical and kinetic perspective on base excision repair of DNA. Acc Chem Res. 2014;47:1238–1246. doi: 10.1021/ar400275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.David SS, O’Shea VL, Kundu S. Base excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stivers JT, Jiang YJ. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 83.Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: Mechanisms of nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 84.Piersen CE, McCullough AK, Lloyd RS. AP lyases and dRPases: commonality of mechanism. Mutation Res. 2000;459:43–53. doi: 10.1016/s0921-8777(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 85.Manoharan M, Ransom SC, Mazumder A, Gerlt JA, Wilde JA, Withka JA, Bolton PH. The characterization of abasic sites in DNA heteroduplexes by site specific labeling with carbon-13. J Am Chem Soc. 1988;110:1620–1622. [Google Scholar]
- 86.McCullough AK, Sanchez A, Dodson ML, Marapaka P, Taylor JS, Lloyd RS. The reaction mechanism of DNA glycosylase/AP lyases at abasic sites. Biochemistry. 2001;40:561–568. doi: 10.1021/bi002404+. [DOI] [PubMed] [Google Scholar]
- 87.Wilson DM, III, Seidman MM. A novel link to base excision repair? Trends Biochem Sci. 2010;35:247–252. doi: 10.1016/j.tibs.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maor-Shoshani A, Meira LB, Yang X, Samson LD. 3-Methyladenine DNA glycosylase is important for cellular resistance to psoralen interstrand cross-links. DNA Repair. 2008;7:1399–1406. doi: 10.1016/j.dnarep.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McNeill DR, Paramasivam M, Baldwin J, Huang JC, Vyjayanti VN, Seidman MM, Wilson DM., III NEIL1 responds and binds to psoralen-induced DNA interstrand crosslinks. J Biol Chem. 2013;288:12426–12436. doi: 10.1074/jbc.M113.456087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kothandapani A, Dangeti VS, Brown AR, Banze LA, Wang XH, Sobol RW, Patrick SM. Novel role of base excision repair in mediating cisplatin cytotoxicity. J Biol Chem. 2011;286:14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kothandapani A, Patrick SM. Evidence for base excision repair processing of DNA interstrand cross-links. Mutation Res. 2013;743–744:44–52. doi: 10.1016/j.mrfmmm.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Couvé S, Macé-Aimé G, Roselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Couvé-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Semlow DR, Zhang J, Budzowska M, Drohat AC, Walter JC. Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell. 2016;167:498–511. doi: 10.1016/j.cell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hearst JE. Photochemistry of the psoralens. Chem Res Toxicol. 1989;2:69–75. doi: 10.1021/tx00008a001. [DOI] [PubMed] [Google Scholar]
- 96.Haran TE, Crothers DM. Phased psoralen cross-links do not bend the DNA double-helix. Biochemistry. 1988;27:6967–6971. doi: 10.1021/bi00418a044. [DOI] [PubMed] [Google Scholar]
- 97.Spielmann HP, Dwyer TJ, Hearst JE, Wemmer DE. Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry. 1995;34:12937–12953. [PubMed] [Google Scholar]
- 98.Spielmann HP, Dwyer TJ, Sastry SS, Hearst JE, Wemmer DE. DNA structural reorganization upon conversion of a psoralen furan-side monoadduct to an interstrand cross-link: implications for DNA repair. Proc Nat Acad Sci USA. 1995;92:2345–2349. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hwang GS, Kim JK, Choi BS. The solution structure of a psoralen cross-linked DNA duplex by NMR and relaxation matrix refinement. Biochem Biophys Res Commun. 1996;219:191–197. doi: 10.1006/bbrc.1996.0204. [DOI] [PubMed] [Google Scholar]
- 100.Sinden RR, Hagerman PJ. Interstrand psoralen cross-links do not introduce appreciable bends in DNA. Biochemistry. 1984;23:6299–6303. doi: 10.1021/bi00321a002. [DOI] [PubMed] [Google Scholar]
- 101.Lukin M, de los Santos C. NMR Structures of Damaged DNA. Chem Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 102.Tomic MT, Wemmer DE, KIm SH. Structure of a psoralen cross-linked DNA in solution by nuclear magnetic resonance. Science. 1987;238:1722–1725. doi: 10.1126/science.3686011. [DOI] [PubMed] [Google Scholar]
- 103.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccaromyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 104.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic sites in DNA. Mutation Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 105.Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutation Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 106.Schärer OD, Kawate T, Gallinari P, Jiricny J, Verdine GL. Investigation of the mechanisms of DNA binding of the human G/T glycosylase using designed inhibitors. Proc Nat Acad Sci USA. 1997;94:4878–4883. doi: 10.1073/pnas.94.10.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Onizuka K, Yeo J, David SS, Beal PA. NEIL1 Binding to DNA Containing 2′-Fluorothymidine Glycol Stereoisomers and the Effect of Editing. ChemBioChem. 2012;13:1338–1348. doi: 10.1002/cbic.201200139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 109.Dutta S, Chowdhury G, Gates KS. Interstrand crosslinks generated by abasic sites in duplex DNA. J Am Chem Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y, Gates KS. On the Formation and Properties of Interstrand DNA-DNA Cross-links Forged by Reaction of an Abasic Site With the Opposing Guanine Residue of 5′-CAp Sequences in Duplex DNA. J Am Chem Soc. 2013;135:1015–1025. doi: 10.1021/ja308119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Price NE, Catalano MJ, Liu S, Wang Y, Gates KS. Chemical and structural characterization of interstrand cross-links formed between abasic sites and adenine residue in duplex DNA. Nucleic Acids Res. 2015;43:3434–3441. doi: 10.1093/nar/gkv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Z, Johnson KM, Price NE, Gates KS. Characterization of interstrand DNA-DNA cross-links derived from abasic sites using bacteriophage ϕ 29 DNA polymerase. Biochemistry. 2015;54:4259–4266. doi: 10.1021/acs.biochem.5b00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Catalano MJ, Liu S, Andersen N, Yang Z, Johnson KM, Price NA, Wang Y, Gates KS. Chemical structure and properties of the interstrand cross-link formed by the reaction of guanine residues with abasic sites in duplex DNA. J Am Chem Soc. 2015;137:3933–3945. doi: 10.1021/jacs.5b00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Catalano MJ, Ruddraraju KV, Gates KS. Crystal structure of a nucleoside model of the interstrand cross-link formed by the reaction of 2′-deoxyguanosine and an abasic site in duplex DNA. Acta Cryst E. 2016;E72:624–627. doi: 10.1107/S205698901600517X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu M, Doublié S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutation Res. 2013;743–744:4–11. doi: 10.1016/j.mrfmmm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Neurauter CG, Luna L, Bjoras M. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst) 2012;11:401–409. doi: 10.1016/j.dnarep.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 117.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao XS, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Nat Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mullins EA, Shi R, Parsons ZD, Yuen PK, David SS, Igarashi Y, Eichman BF. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015;527:254–258. doi: 10.1038/nature15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Parsons ZD, Bland JM, Mullins EA, Eichman BF. A Catalytic Role for C-H/π Interactions in Base Excision Repair by Bacillus cereus DNA Glycosylase AlkD. J Am Chem Soc. 2016;138:11485–11488. doi: 10.1021/jacs.6b07399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang S, Liu K, Xiao L, Yang L, Li H, Zhang F, Lei L, Lo SC, Feng X, Li A, He J. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 2015;44:187–197. doi: 10.1093/nar/gkv949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 123.Jonnalagadda VS, Matsuguchi T, Engelward BP. Interstrand crosslink-induced homologous recombination carries an increased risk of deletions and insertions. DNA Repair. 2005;4:594–605. doi: 10.1016/j.dnarep.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 124.Shen X, Li L. Mutagenic repair of interstrand crosslinks. Env Mol Mutagenesis. 2010;51:493–499. doi: 10.1002/em.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith LA, Makarova AV, Samson LD, Thiesen KE, Char A, Bessho T. Bypass of a psoralen DNA interstrand cross-link by DNA polymerases beta, iota, and kappa in vitro. Biochemistry. 2012;51:8931–8938. doi: 10.1021/bi3008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNa interstrand cross-link repair. Mol Cell Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV 1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: Evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Nat Acad Sci USA. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang N, Lu X, Peterson CA, Legerski RJ. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol Cell Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bessho T. Induction of DNA Replication-mediated Double Strand Breaks by Psoralen DNA Interstrand Cross-links. J Biol Chem. 2003;278:5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- 131.Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, W W, Li L. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sato K, Ishiai M, Toda K, Furukoshi S, Osakabe A, Tachiwana H, Takizawa Y, Kagawa W, Kitao H, Dohmae N, Obuse C, Kimura H, Takata M, Kurumizaka H. Histone chaperone activity of Fanconi anemia proteins, FANCD2 and FANCI, is required for DNA crosslink repair. EMBO J. 2012;31:3524–3636. doi: 10.1038/emboj.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sobeck A, Stone S, Landais I, de Graaf B, Hoatlin ME. The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J Biol Chem. 2009;284:25560–25568. doi: 10.1074/jbc.M109.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sejersteda Y, Hildrestranda GA, Kunkea D, Veslemøy R, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Åsegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjøråsa M. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Nat Acad Sci USA. 2011;108:18802–18807. doi: 10.1073/pnas.1106880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jailand CMO, Scheffler K, Benestad SL, Moldal T, Ersdal C, Gunnes G, Suganthan R, Bjørås M, Tranulisa MA. Neil3 induced neurogenesis protects against prion disease during the clinical phase. Sci Rep. 2016;6:37844. doi: 10.1038/srep37844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Neveling K, Endt D, Hoehn H, Schindler D. Genotype-phenotype correlations in Fanconi anemia. Mutation Res. 2009;668:73–91. doi: 10.1016/j.mrfmmm.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 137.Duxin JP, Walter JC. What is the DNA repair defect underlying Fanconi anemia. Curr Opin Cell Biol. 2015;37:49–60. doi: 10.1016/j.ceb.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 139.Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, Li GM, Boldogh IB, Tomkinson AE, Mitra S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Nat Acad Sci USA. 2013;110:E3090–3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Balcome S, Park S, Quirk Dorr DR, Hafner L, Phillips L, Tretyakova N. Adenine-containing DNA-DNA crosslinks of antitumor nitrogen mustards. Chem Res Toxicol. 2004;17:950–962. doi: 10.1021/tx0499463. [DOI] [PubMed] [Google Scholar]
- 142.Millard JT, Raucher S, Hopkins PB. Mechlorethamine cross-links deoxyguanosine residues at 5′-GNC sequences in duplex DNA fragments. J Am Chem Soc. 1990;112:2459–2460. [Google Scholar]
- 143.Rink SM, Solomon MS, Taylor MJ, Rajur SB, McLaughlin LW, Hopkins PB. Covalent structure of a nitrogen mustard-induced DNA interstrand cross-link: an N7-to-N7 linkage of deoxyguanosine residues at the duplex sequence 5′-d(GNC) J Am Chem Soc. 1993;115:2551–2557. [Google Scholar]
- 144.Lee CYI, Delaney JC, Kartalou M, Lingaraju GM, Maor-Shoshani A, Essigmann JM, Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fischhaber PL, Gall AS, Duncan JA, Hopkins PB. Direct demonstration in synthetic oligonucleotides that N,N′-bis(2-chloroethyl)-nitrosourea cross-links N1 of deoxyguanosine to N3 of deoxycytidine on opposite strands of duplex DNA. Cancer Res. 1999;17:4363–4368. [PubMed] [Google Scholar]
- 146.Hentshel S, Alzeer J, Angelov T, Schärer OD, Luedtke NW. Synthesis of DNA interstrand cross-links using a photocaged nucleobase. Angew Chem Int Ed Eng. 2012;51:3466–3469. doi: 10.1002/anie.201108018. [DOI] [PubMed] [Google Scholar]
- 147.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Nat Acad Sci USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: Repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yi C, He C. DNA repair by reversal of DNA damage. Cold Spring Harb Perspect Biol. 2013;5:a012575. doi: 10.1101/cshperspect.a012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wolfenden R, Sharpless TK, Allan R. Substrate binding by adenosine deaminase. J Biol Chem. 1987;242:977–983. [PubMed] [Google Scholar]
- 151.Goodman RA, Macbeth MR, Beal PA. ADAR proteins: structure and catalytic mechanism. Curr Top Microbiol Immunol. 2012;353:1–33. doi: 10.1007/82_2011_144. [DOI] [PubMed] [Google Scholar]
- 152.Yi-Brunozzi HY, Easterwood LM, Kamilar GM, Beal PA. Synthetic substrate analogs for the RNA-editing adenosine deaminase ADAR-2. Nucleic Acids Res. 1999;27:2912–2917. doi: 10.1093/nar/27.14.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–371. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bransteittter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine in single-stranded DNA but requires the action of RNase. Proc Nat Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rubio MAT, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GAM, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Nat Acad Sci USA. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.O’Brien PJ. Catalytic promiscuity and the divergent function of DNA repair enzymes. Chem Rev. 2006;106:720–752. doi: 10.1021/cr040481v. [DOI] [PubMed] [Google Scholar]
- 158.Lindahl T. DNA glycosylases, endonucleases for Apurinic/apyridinic sites, and base excision repair. Prog Nucleic Acids Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- 159.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Perspect Biol. 2000;65:127–134. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 160.Lindahl T, Ljunquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 161.Sancar A. It Is Chemistry but Not Your Grandfather’s Chemistry. Biochemistry. 2017;56:1–2. doi: 10.1021/acs.biochem.6b01262. [DOI] [PubMed] [Google Scholar]
- 162.Gamboa Varela J, Gates KS. A Simple, High-Yield Synthesis of DNA Duplexes Containing a Covalent, Thermally-Reversible Interstrand Cross-link At a Defined Location Angew. Chem Int Ed Eng. 2015;54:7666–7669. doi: 10.1002/anie.201502566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gamboa Varela J, Gates KS. Simple, high-yield syntheses of DNA duplexes containing interstrand DNA-DNA cross-links between an N4-aminocytidine residue and an abasic site. Curr Protoc Nucleic Acid Chem. 2016;65:5.16.11–15.16.15. doi: 10.1002/cpnc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Enoiu M, Ho TV, Long DT, Walter JC, Scharer OD. Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol Biol. 2012;920:203–219. doi: 10.1007/978-1-61779-998-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Angelov T, Guainazzi A, Schärer OD. Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org Lett. 2009;11:661–664. doi: 10.1021/ol802719a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mukherjee S, Guainazzi A, Schärer OD. Synthesis of struturally diverse major groove DNA interstrand crosslinks using three different aldehyde precursors. Nucleic Acids Res. 2014;42:7429–7435. doi: 10.1093/nar/gku328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stevens K, Madder A. Furan-modified oligonucleotides for fast, high-yielding and site-selective DNA inter-strand cross-linking with non-modified complements. Nucleic Acids Res. 2009;37:1555–1565. doi: 10.1093/nar/gkn1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tomás-Gamasa M, Serdjukow S, Su M, Müller M, Carell T. “Post-it” type connected DNA created with a reversible covalent cross-link. Angew Chem Int Ed Eng. 2014;53:796–800. doi: 10.1002/anie.201407854. [DOI] [PubMed] [Google Scholar]
- 169.Haque MM, Sun H, Liu S, Wang Y, Peng X. Photoswitchable formation of a DNA interstrand cross-link by a coumarin-modified oligonucleotide. Angew Chem Int Ed Eng. 2014;53:7001–7005. doi: 10.1002/anie.201310609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ye M, Guillaume J, Liu Y, Sha R, Wang R, Seeman NC, Canary JW. Site specific inter-strand cross-links of DNA duplexes. Chem Sci. 2013;4:1319–1329. doi: 10.1039/C2SC21775A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Coleman RS, Kesicki EA. Template-directed cross-linking of oligonucleotide site-specific covlaent modification of dG-N7 within duplex DNA. J Org Chem. 1995;60:6252–6253. [Google Scholar]
- 172.McManus FP, Khaira A, Noronha AM, Wilds CJ. Preparation of covalently linked complexes between DNA and O6-alkylguanine-DNA alkyltransferase using interstrand cross-linked DNA. Bioconjugate Chem. 2013;24:224–233. doi: 10.1021/bc300553u. [DOI] [PubMed] [Google Scholar]
- 173.Noll DM, Noronha AM, Miller PS. Synthesis and characterization of DNA duplexes containing an N4-C-ethyl-N4-C interstrand cross-link. J Am Chem Soc. 2001;123:3405–3411. doi: 10.1021/ja003340t. [DOI] [PubMed] [Google Scholar]
- 174.Guainazzi A, Schärer OD. Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cell Mol Life Sci. 2010;67:3683–3697. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.You C, Wang Y. Mass spectrometry-based quantitative strategies for assessing the biological consequences and repair of DNA adducts. Acc Chem Res. 2016;49:205–213. doi: 10.1021/acs.accounts.5b00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cao H, Hearst JE, Corash L, Wang Y. LC-MS/MS for the Detection of DNA Interstrand Cross-Links Formed by 8-Methoxypsoralen and UVA Irradiation in Human Cells. Anal Chem. 2008;80:2932–2938. doi: 10.1021/ac7023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lai C, Cao H, Hearst JE, Corash L, Luo H, Wang Y. Quantitative Analysis of DNA Interstrand Cross-Links and Monoadducts Formed in Human Cells Induced by Psoralens and UVA Irradiation. Anal Chem. 2008;80:8790–8798. doi: 10.1021/ac801520m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu S, Wang Y. A Quantitative Mass Spectrometry-Based Approach for Assessing the Repair of 8-Methoxypsoralen-Induced DNA Interstrand Crosslink and Monoadducts in Mammalian Cells. Anal Chem. 2013;85:6732–6739. doi: 10.1021/ac4012232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tretyakova N, Villalta PW, Kotapati S. Mass spectrometry of structurally modified DNA. Chem Rev. 2013;113:2395–2436. doi: 10.1021/cr300391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Goggin M, Anderson C, Park S, Swenberg JA, Walker V, Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry analysis of the adenine-guanine cross-links of 1,2,3,4-diepoxybutane in tissues of butadiene-exposed B6C3F1 mice. Chem Res Toxicol. 2008;21:1163–1170. doi: 10.1021/tx800051y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Paz MM, Ladwa S, Champeil E, Liu Y, Rockwell S, Boamah EK, Bargonetti J, Callahan JF, Roach J, Tomasz M. Mapping DNA adducts of mitomycin C and decarbamoyl mitomycin C in cell lines using liquid chromatography/electrospray tandem mass spectrometry. Chem Res Toxicol. 2008;21:2370–2378. doi: 10.1021/tx8002615. [DOI] [PMC free article] [PubMed] [Google Scholar]


