Abstract
The ability of a protein to fold into unique functional state or to stay intrinsically disordered is encoded in its amino acid sequence. Both ordered and intrinsically disordered proteins (IDPs) are natural polypeptides that use the same arsenal of 20 proteinogenic amino acid residues as their major building blocks. The exceptional structural plasticity of IDPs, their capability to exist as heterogeneous structural ensembles and their wide array of important disorder-based biological functions that complements functional repertoire of ordered proteins are all rooted within the peculiar differential usage of these building blocks by ordered proteins and IDPs. In fact, some residues (so-called disorder-promoting residues) are noticeably more common in IDPs than in sequences of ordered proteins, which, in their turn, are enriched in several order-promoting residues. Furthermore, residues can be arranged according to their “disorder promoting potencies,” which are evaluated based on the relative abundances of various amino acids in ordered and disordered proteins. This review continues a series of publications on the roles of different amino acids in defining the phenomenon of protein intrinsic disorder and concerns glutamic acid, which is the second most disorder-promoting residue.
Keywords: glutamic acid, intrinsically disordered protein, protein function, protein structure, protein-protein interaction
Introduction
Intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs) are new exciting members of the protein kingdom.1,2 They are highly abundant in nature,3-7 possess numerous intriguing properties,8 are intimately involved in various cellular processes9-23 and are commonly found to be related to the pathogenesis of various diseases.13,24-29 The common theme of protein disorder-based functionality is recognition, and IDPs/IDPRs are frequently involved in complex protein-protein, protein-nucleic acid and protein-small molecule interactions. Some of these interactions can induce a disorder–order transition in the entire IDP or in its part.5,9-12,15,23,30-36 Furthermore, intrinsic disorder opens a unique capability for one protein to be involved in interaction with several unrelated binding partners and to gain different bound structures.22,37 Some IDPs can form highly stable complexes; others are involved in signaling interactions where they undergo constant “bound–unbound” transitions, thus acting as dynamic and sensitive “on-off” switches. These proteins typically return to their intrinsically disordered state after the completion of a particular function. Many of the IDPs/IDPRs can gain different conformations depending on the environmental peculiarities.30,37 All this constitutes an important arsenal of the unique physiological properties of IDPs/IDPRs that determines their ability to exert different functions in different cellular contests according to a specific conformational state.8 The folding-at-binding principle is believed to help IDPs or IDPRs to obtain maximal specificity in a protein–protein interaction without very high affinity.20 This combination of high specificity with low affinity defines the broad utilization of intrinsic disorder in regulatory interactions where turning a signal off is as important as turning it on.10 Although some partial folding during the IDP/IDPR-based interactions is a widespread phenomenon, with significant fraction (~1/3) of the interacting residues in IDPs/IDPRs adopting α-helix, β-strand and irregular structures,31,32 there are still many other IDPs/IDPRs that are involved in the formation of “fuzzy complexes,” where an IDP/IDPR keeps a certain amount of disorder in its bound conformation.35,38-40
Often, the interacting regions in IDPs are observed as loosely structured fragments in their unbound forms. These disorder-based binding sites are known as molecular recognition elements or features (MoREs or MoRFs),30,31 preformed structural elements41 or pre-structured motifs (PreSMos).42 Although the existence of such loosely structured regions suggests that IDPs can adopt their bound structure(s) at a free-energy cost that is not too high, it is important to remember that increasing the stability of the bound conformation does not necessarily enhance the binding affinity.23 Another important feature of the disorder-based interactions is their increased speed due to the greater capture radius and the ability to spatially search through interaction space (the so-called “fly-casting” mechanism)43 and to the fact that fewer encounter events are required for the binding because of lack of orientational restrains.44 Linking all these considerations with the recent report showing that IDP affinities are tuned mostly by association rates45 suggests that the degree of pre-adoption of binding conformations in IDPs has to be limited, but not unfavorable.
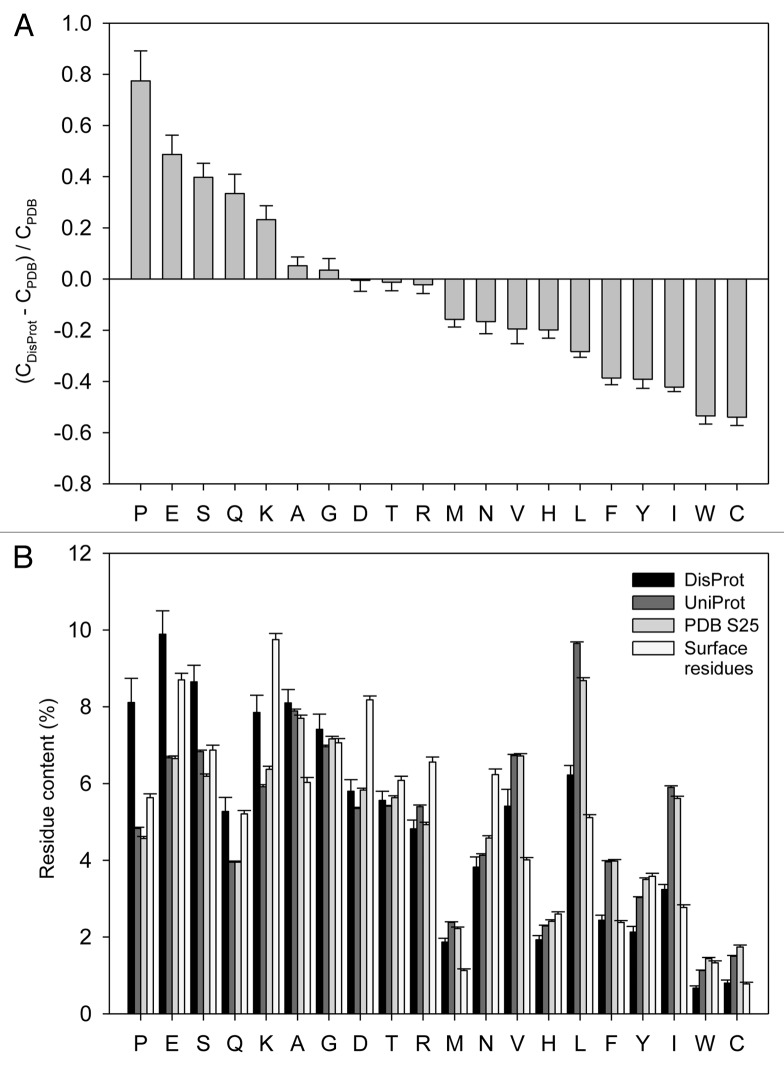
All the functional and structural peculiarities of IDPs/IDPRs are encoded in their amino acid sequences. It was recognized long ago that there are significant differences between ordered proteins/domains and IDPs/IDPRs at the level of their amino acid sequences.5,10,46 In fact, in comparison with ordered proteins, IDPs/IDPRs are characterized by noticeable biases in their amino acid compositions,5,8,10,46-48 containing less of so-called “order-promoting” residues (cysteine, tryptophan, isoleucine, tyrosine, phenylalanine, leucine, histidine, valine, asparagines and methionine, which are mostly hydrophobic residues which are commonly found within the hydrophobic cores of foldable proteins) and more of “disorder-promoting” residues (lysine, glutamine, serine, glutamic acid and proline, which are mostly polar and charged residues, which are typically located at the surface of foldable proteins) (Fig. 1A).
Figure 1. Amino acid determinants defining structural and functional differences between the ordered and intrinsically disordered proteins. (A) Fractional difference in the amino acid composition (compositional profile) between the typical IDPs from the DisProt database49 and a set of completely ordered proteins50 calculated for each amino acid residue. The fractional difference was evaluated as (CDisProt-CPDB)/CPDB, where CDisProt is the content of a given amino acid in a DisProt databse, and CPDB is the corresponding content in the data set of fully ordered proteins. Positive bars correspond to residues found more abundantly in IDPs, whereas negative bars show residues, in which IDPs are depleted. Amino acid types were ranked according to their decreasing disorder-promoting potential.47 (B). Amino acid compositions of several data sets discussed in the text (DisProt,49 UniProt,51 PDB Select 2550 and surface residues48).
Glutamic acid is second of the most common disorder-promoting residues. Figure 1B and Table 1 represent the result of a statistical analysis of the amino acid compositions of proteins in four standard data sets (DisProt,49 UniProt,51 PDB Select 2550 and surface residues48) and shows that the glutamic acid content in these data sets is 9.89 ± 0.61%, 6.67 ± 0.04%, 6.65 ± 0.07% and 8.70 ± 0.17%, respectively (cprofiler.org/help.html).48 In other words, IDPs/IDPRs contain 1.48- and 1.49-times more glutamic acid residues than the average natural proteins from UniProt or ordered proteins from PDB, respectively. Furthermore, the glutamic acid content in IDPs/IDPRs is 1.14-times higher than that on the surfaces of ordered proteins.
Table 1. Amino acid compositions of the standard data sets (modified from ref. 48).
| Residuea | Disorder propensityb | SwissProtc | PDB S25d | Surface residuese | DisProtf |
|---|---|---|---|---|---|
| Pro (P) | 1.000 | 4.83 ± 0.03 | 4.57 ± 0.05 | 5.63 ± 0.10 | 8.11 ± 0.63 |
| Glu (E) | 0.781 | 6.67 ± 0.04 | 6.65 ± 0.07 | 8.70 ± 0.17 | 9.89 ± 0.61 |
| Ser (S) | 0.713 | 6.83 ± 0.04 | 6.19 ± 0.06 | 6.87 ± 0.13 | 8.65 ± 0.43 |
| Gln (Q) | 0.665 | 3.95 ± 0.03 | 3.95 ± 0.05 | 5.21 ± 0.09 | 5.27 ± 0.37 |
| Lys (K) | 0.588 | 5.92 ± 0.05 | 6.37 ± 0.08 | 9.75 ± 0.16 | 7.85 ± 0.45 |
| Ala (A) | 0.450 | 7.89 ± 0.05 | 7.70 ± 0.08 | 6.03 ± 0.13 | 8.10 ± 0.35 |
| Gly (G) | 0.437 | 6.96 ± 0.04 | 7.16 ± 0.07 | 7.06 ± 0.11 | 7.41 ± 0.40 |
| Asp (D) | 0.407 | 5.35 ± 0.03 | 5.83 ± 0.05 | 8.18 ± 0.10 | 5.80 ± 0.30 |
| Thr (T) | 0.401 | 5.41 ± 0.02 | 5.63 ± 0.05 | 6.08 ± 0.11 | 5.56 ± 0.24 |
| Arg (R) | 0.394 | 5.40 ± 0.04 | 4.93 ± 0.06 | 6.56 ± 0.13 | 4.82 ± 0.23 |
| Met (M) | 0.291 | 2.38 ± 0.02 | 2.22 ± 0.04 | 1.13 ± 0.04 | 1.87 ± 0.10 |
| Asn (N) | 0.285 | 4.13 ± 0.04 | 4.58 ± 0.06 | 6.23 ± 0.15 | 3.82 ± 0.27 |
| Val (V) | 0.263 | 6.73 ± 0.03 | 6.72 ± 0.06 | 4.01 ± 0.06 | 5.41 ± 0.44 |
| His (H) | 0.259 | 2.29 ± 0.02 | 2.41 ± 0.04 | 2.60 ± 0.06 | 1.93 ± 0.11 |
| Leu (L) | 0.195 | 9.65 ± 0.04 | 8.68 ± 0.08 | 5.11 ± 0.08 | 6.22 ± 0.25 |
| Phe (F) | 0.117 | 3.96 ± 0.03 | 3.98 ± 0.04 | 2.38 ± 0.05 | 2.44 ± 0.13 |
| Tyr (Y) | 0.113 | 3.03 ± 0.02 | 3.50 ± 0.04 | 3.58 ± 0.08 | 2.13 ± 0.15 |
| Ile (I) | 0.090 | 5.90 ± 0.04 | 5.61 ± 0.06 | 2.77 ± 0.07 | 3.24 ± 0.13 |
| Trp (W) | 0.004 | 1.13 ± 0.01 | 1.44 ± 0.03 | 1.33 ± 0.05 | 0.67 ± 0.06 |
| Cys (C) | 0.000 | 1.50 ± 0.02 | 1.74 ± 0.05 | 0.78 ± 0.04 | 0.80 ± 0.08 |
a Residues are arranged according to their decreasing intrinsic disorder propensity; bDisorder propensity is calculated based on the fractional difference in the amino acid compositions between the disordered and ordered proteins; cSwissProt 51 is the set closest to the distribution of amino acids in nature among the four data sets;51dPDB Select 25 is a subset of proteins from the Protein Data Bank with less than 25% sequence identity, biased toward the composition of proteins amenable to crystallization studies;50eSurface residues determined by the Molecular Surface Package over a sample of PDB structures of monomeric proteins suitable for protein surface analysis; fDisProt 3.4 comprised of a set of experimentally determined disordered regions.49
This article continues a series of publications on the intrinsic disorder alphabet dedicated to the exploration of the amino acid determinants of protein intrinsic disorder. I overview below some functions of glutamic acid in IDPs/IDPRs (as well as in ordered proteins and domains) and show that there is a variety of glutamic acid-specific functions in disordered proteins and regions.
Structural Properties of Glutamic Acid
Chemical structure of glutamic acid
Glutamic acid (glutamate, Glu, E, see Fig. 2A) is one of the 20 proteinogenic amino acids encoded by the standard genetic code and its codons are GAA and GAG. Glutamic acid is a dibasic nonessential amino acid that has a molecular mass of 147.13 Da (molecular mass of Glu residue is 129.12 Da), surface of 190 Å2, volume of 138.4 Å3, pKa of side chain of 4.6 and pI 3.08 at 25 °C. Intriguingly, free glutamic acid is not very soluble, possessing solubility of 0.864 g/100 g at 25 °C, which is significantly lower than the solubility of free prolines (162.3 g/100 g at 25 °C), and the solubility of the vast majority of free amino acids (www.fli-leibniz.de/IMAGE_AA.html).
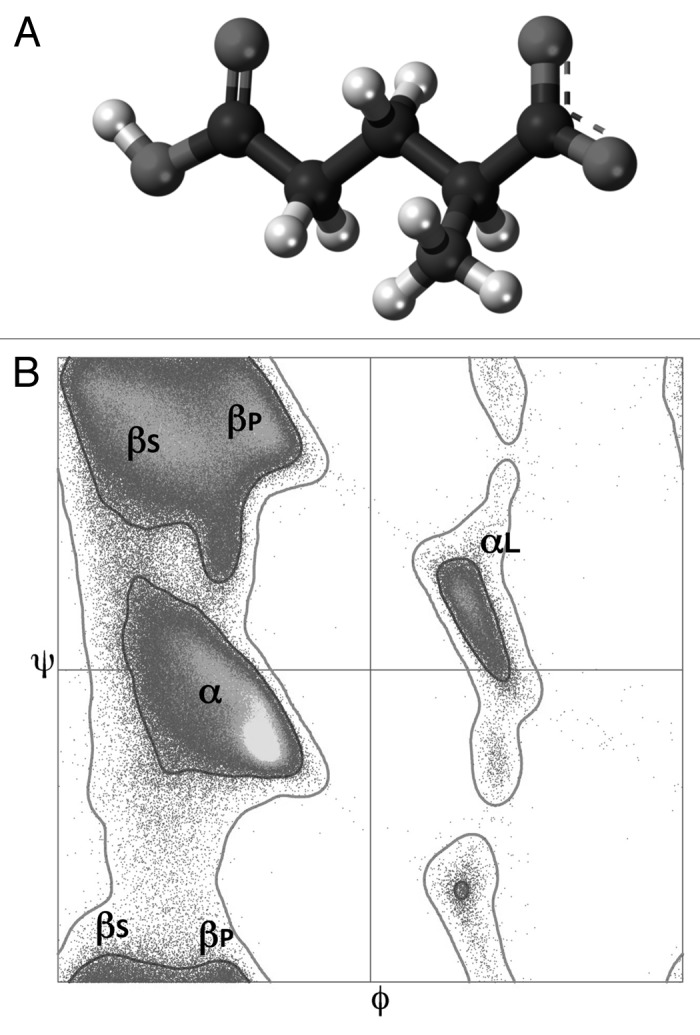
Figure 2. Structural properties of glutamic acid. (A) Chemical structure of the glutamic acid residue. (B) Ramachandran plots for backbone conformations of the 18 non-glycine and non-proline amino acids. Marked regions of density correspond to the right-handed α-helix region (α), mirror image of α (αL), region largely involved in β-sheet formation (βS), and region associated with extended polyproline-like helices, but also observed in β-sheet (βP).
The side chain of glutamic acid contains two methylene group and the carboxylic acid functional group (see Fig. 2A) that exists in a negatively charged deprotonated carboxylate form at pHs greater than its pKa 4.6 (and thus Glu is negatively charged at the physiological pH ranging from 7.35–7.45). Therefore, glutamic acid is one of two acidic amino acids found in proteins that play important roles as general acids in enzyme active centers, as well as in maintaining the solubility and ionic character of proteins. In fact, glutamic acid residue has a non-polar surface of 69 Å2, and the estimated hydrophobic effect associated with the burial of this residue is 1.74 kcal/mol.52 In ordered proteins, glutamic acids are predominantly located on protein surface so that they have access to the solvent. In fact, 93% of glutamic acids in known structures of folded proteins are classified as exposed since they have solvent exposed areas of >30 Å2, and only 4% of glutamic acids in folded proteins possess solvent exposed areas of <10 Å2 and therefore are buried.53 The carboxylate anions and salts of glutamic acid are known as glutamates.
Biological Significance of Free Glutamate
Glutamic acid is one of the most common natural amino acids and the most abundant amino acid in the diet. Besides being an important component of proteins and polypeptides (see below), being a substrate for the production of the Krebs-cycle-related α-ketoglutarate intermediate, glutamine and proline, and being the precursor for the synthesis of the inhibitory γ-aminobutyric acid (GABA) in GABA-ergic neurons, glutamate is the principal excitatory neurotransmitter within the vertebrate nervous system.54 In fact, glutamate is known to act on several different types of receptors and has excitatory effects at ionotropic receptors [such as N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), and kainite, which all incorporate ion channels that are permeable to cations] and modulatory effects at metabotropic receptors [which are G protein–coupled glutamate receptors (mGluR) that modify neuronal and glial excitability through G protein subunits acting on membrane ion channels and second messengers such as diacylglycerol and cAMP].54
At chemical synapses of the glutamatergic neurons, glutamate is stored in vesicles and is released from the pre-synaptic cell by nerve impulses. In the opposing post-synaptic cell, binding of glutamate lead to activation of specific glutamate receptors such as NMDA or AMPA. Glutamate plays an important role in synaptic plasticity in the brain and is involved in various cognitive functions, such as learning and memory.55 In fact, long-term potentiation (one of the plasticity forms) takes place at glutamatergic synapses in the neocortex, hippocampus and other parts of the brain.55 Another important role of glutamate is its ability to generate volume transmission, where extrasynaptic signaling is created via the summation of glutamate released from a neighboring synapse.56
In addition to glutamate receptors, neuronal and glial membranes contain glutamate transporters that are responsible for rapid remove of glutamate from extracellular space.57 Under stress conditions (such as brain injury or disease), glutamate transporters work in reverse leading to the accumulation of the excess glutamate in the extracellular space and promoting entrance of calcium to the cell via the NMDA receptor channels. This process is known as excitotoxicity, and it results in neuronal damage and eventual cell death. The excitotoxicity might occur as part of the ischemic cascade that is associated with stroke, autism, amyotrophic lateral sclerosis, lathyrism, some forms of mental retardation and Alzheimer’s disease.58 The decreased glutamate release is associated with phenylketonuria leading to the developmental disruption of glutamate receptor expression.59,60
Glutamic Acid in Structure of the Ordered Proteins
Glutamic acid in the Ramachandran plot
The structure of a protein can be described using torsion angles—ϕ and ψ—of its backbone that provides a simple view of the conformation of a protein. In sequence order, ϕ is the Ni-1-Ci-Cαi-Ni torsion angle, and ψ is the Ci-Cαi-Ni-Ci+1 torsion angle. Since most combinations of ϕ and ψ are sterically forbidden, the 2D plot of the torsion angles of the protein backbone, known as the Ramachandran plot,61 provides a simple view of the conformation of a protein, since the ϕ-ψ angles cluster into distinct regions in the Ramachandran plot, where each region corresponds to a particular secondary structure. In the generic Ramachandran plot (see Fig. 2B) that refers to the 18 non-glycine and non-proline amino acids, there are four distinct regions of density (the α (right-handed α-helix region), αL (mirror image of α), βS (region largely involved in β-sheet formation) and βP (region associated with extended polyproline-like helices but also observed in β-sheet). The shape of the generic Ramachandran plot is determined mainly by the presence of specific steric clashes61 and backbone dipole–dipole interactions.62-64
Glutamic acid in electrostatic interactions and hydrogen bonds
Glutamic acid participates in electrostatic interactions, which are also known as ionic bonds, or salt bridges, or salt linkages, or ion pairs. An electrostatic interaction is a non-covalent bond that is based on the attraction of two oppositely charged groups. It can easily be broken and reformed and is characterized by the optimal distance of 2.8 Å between the interacting groups. The strength of these interactions depends on the distance of the two charges and the properties of the medium between them. In proteins, electrostatic interactions typically occur between COO- in the side chain of glutamic and aspartic acids and NH3+ in the side chains of lysines and arginines.
Hydrogen bond (H-bond) is another non-covalent bond. This interaction depends on the sharing of one hydrogen atom (H-atom) between two other atoms, where the H-atom has a covalent bond to one of them (which therefore serves as the H-bond donor), and where the other atom, to which the H-atom has a weaker bond, serves as the acceptor, A. Hydrogen bond is weaker than a covalent bond but stronger than a van der Waals bond. Similar to electrostatic interactions, H-bonds can easily be broken and reformed. Among established geometrical criteria for H-bond are a set of optimal distances between the non-H atom of donor and acceptor (Dono–Acceptor <3.9 Å) and between the H atom of donor and acceptor (H–Acceptor <2.5 Å).65 Being negatively charged at physiological pH, glutamic acid can serve as a hydrogen bond acceptor, whereas at acidic pH, it also can be a hydrogen bond donor.
Glutamic acid and protein secondary structure
Although protein secondary structure is determined by hydrogen bonds between donor and acceptor groups in the protein backbone, different amino acids are known to favor the formation of different secondary structure elements, such as α-helices, β-pleated sheets or loops. The α-helix-formers include alanine, cysteine, leucine, methionine, glutamic acid, glutamine, histidine and lysine, whereas valine, isoleucine, phenylalanine, tyrosine, tryptophan and threonine favor β-structure formation, and serine, glycine, uncharged aspartic acid, asparagine and proline are found most often in β-turns. It was pointed out that there is no apparent relationship between the chemical nature of the amino acid side chain and its secondary structure preferences. For example, although glutamic and aspartic acids are closely related chemically, glutamic acid is more likely to be found in helices and aspartic acid is predominantly located in β-turns. In fact, the helical propensity of glutamic acid is 0.40, whereas aspartic acid has an helical propensity of 0.69, the third largest value after proline and glycine.66 Note that the helical propensity is defined as the difference in free energy Δ(ΔG) estimated in kcal/mol per residue in an α-helical configuration relative to alanine, which has been set to zero because it is usually the amino acid with the most favorable helix propensity.66 Here, the higher helical propensity values correspond to more positive free energies and therefore are related to residues which are less favored in α-helix.
Glutamic acid in α-helix caps
Since α-helices in peptides and proteins have an overall dipole moments caused by the cumulative effects of all the individual dipoles from the carbonyl groups of the peptide bond pointing along the helix axis, the overall helical structure is destabilized due to the noticeable entropic effects. The effect of this helical dipole moment can be approximated by placing 0.5–0.7 positive unit charge near the N-terminus and 0.5–0.7 negative unit charge near the C-terminus of the helix.67,68 One of the Nature’s strategies to neutralize this helix dipole is the specific capping of the N-terminal ends of α-helices by negatively charged residues, such as glutamic acids.67,68
Furthermore, careful analysis of α-helices revealed that their first and last four residues differ from the remaining residues by being unable to make intrα-helical hydrogen bonds. Instead, these first four (> N-H) groups and last four (> C = O) groups in an α-helix are often capped by alternative hydrogen bond partners.69-71 Physico-chemical and statistical analysis suggested that certain residues are more preferable at the C- and N-termini of an α-helix (the helical C- and N-caps).70 For example, based on the analysis of series of mutations in the two N-caps of barnase, it was concluded that a single N-cap can stabilize the protein by up to ~2.5 kcal/mol.70 Importantly, the presence of a negative charge of the N-cap was shown to add ~1.6 kcal/mol of stabilization energy mostly due to the compensation effects for the macroscopic electrostatic dipole of the helix.70
From a global survey among proteins of known structure, seven distinct capping motifs are identified—three at the helix N-terminus and four at the C-terminus.71 One of these motifs is the helix-capping motif Ser-X-X-Glu, a sequence that occurs frequently at the N-termini of α-helices in proteins.71-73 Thermodynamic analysis of this Ser-X-X-Glu motif from the GCN4 leucine zipper dimer revealed that the free energy of helix stabilization associated with the hydrogen-bonding and hydrophobic interactions in this capping structure is −1.2 kcal/mol, illustrating that helix capping might play a significant role in protein folding.72 Based on the analysis of 431 α-helices the normalized frequencies for finding particular residues at the Ccap position, the average fraction of buried surface area and the hydrogen bonding patterns of the Ccap residue side-chain were calculated.74 This analysis revealed that the residue found in the Ccap position is on average 70% buried and that there is a noticeable correlation between the relative burial of this residue and its hydrophobicity.74 Furthermore, Ccap residues with polar side-chains were shown to be involved in hydrogen bonding, where the longer side-chains of glutamic acid, glutamin, arginine, lysine and histidine form hydrogen bonds with residues located more than four residues apart, whereas the shorter side-chains of aspartic acid, asparagine, serine and threonine form hydrogen bonds with residues located close in sequence.74 Finally, based on the analysis of α-helical propensity of a series of dodecapeptides containing alanine, asparagine, aspartate, glutamine, glutamate and serine at the N-terminus and arginine, lysine and alanine at the C-terminus, it was concluded that the α-helix-stabilizing abilities of these residues can be ranged as follows: aspartate > asparagine > serine > glutamate > glutamine > alanine at the N-terminus and arginine > lysine > alanine at the C-terminus.75
Glutamic acid and protein solubility
Based on the analysis of solubility-changing substitutions in proteins it has been pointed out that together with two other hydrophilic residues (aspartic acid and serine) glutamic acid contributes significantly more favorably to protein solubility than other hydrophilic residues (asparagine, glutamine, threonine, lysine and arginine).76 Based on this observation, an important strategy for solubility enhancement was proposed, were the hydrophilic residues that do not contribute favorably to protein solubility can be replaced with the hydrophilic residues that contribute more favorably.76
Glutamic Acid and Functions of Ordered Proteins
Glutamic acids inside the pores of ion channels
Being negatively charged at physiological pH, glutamic acid is perfectly suited for binding metal ions. This property is used in specific regulation of a variety of ion channels. For example, in cyclic nucleotide-gated (CNG) channels (which are found in vertebrate photoreceptors and olfactory epithelium,77 elsewhere in the nervous system78-80 and in a variety of other cell types including kidney, testis and heart,81 and whose activation represents the final step in the transduction pathways in both vision and olfaction82-84), a single glutamic acid strategically located in the pore represents the binding site for multiple monovalent cations, the blocking site for external divalent cations and the site for the effect of protons on permeation.82 This is not too surprising since the pore region of the channel controls both the single-channel conductance and the pore diameter of the channel.85 Importantly, CNG channels are permeable to Ca2+, which is an important element in the activation of intracellular targets, and which in addition to permeating CNG channels can profoundly block the current flow carried by monovalent cations through the CNG channels.83 This capability of Ca2+ to block the monovalent cation flow is determined by the high-affinity binding of Ca2+ to a single acidic amino acid residue located in the pore of the channel, which is Glu363 for the rod CNG channel and Glu333 for the catfish olfactory CNG channel.86 This same glutamic acid residue is also responsible for the external rapid proton block of CNG channels, another characteristic that the CNG channels share with Ca2+ channels.86
Glutamic acid also plays an important regulatory role in the voltage-dependent calcium channels that are located in the plasma membrane and form a highly selective conduit by which Ca2+ ions enter all excitable cells and some nonexcitable cells.87 For these channels to operate, Ca2+ ions must enter selectively through the pore, bypassing competition with other extracellular ions. The high selectivity of a unique Ca2+ filter is determined by the four glutamic acid residues located at homologous positions within each of the four pore-forming segments and which form a single or multiple Ca2+-binding site(s) that entrap calcium ions, thus giving them a possibility to be electrostatically repulsed through the intracellular opening of the pore.87
In the bacterial KcsA and inwardly rectifying K+ (Kir) channels, glutamic acid is also involved in the action of the selectivity filter.88 Here, the network of residues stabilizing the pore of KcsA involves a Glu71-Asp80 carboxyl-carboxylate interaction behind the selectivity filter, whereas the structure of the pore in Kir channels is stabilized by a Glu-Arg salt bridge.88 Therefore, although Glu is quite conserved among both types of channels, the network of interactions is not translatable from one channel to the other. This clearly shows that different potassium channels are characterized by diverse gating patterns.88
The presence of a highly conserved glutamic acid residue in the middle of a transmembrane domain is a characteristic feature of a family of transmembrane glycoproteins with two immunoglobulin-like domains, such as basigin (Bsg, also known as CD147 or EMMPRIN), embigin and neuroplastin.89
Finally, a critical glutamic acid residue was recently identified in CLC proteins, which constitute a large structurally defined family of Cl− ion channels and H+/Cl− antiporters which are found in prokaryotes and eukaryotes,90 and which perform their functions in the plasma membrane or in various intracellular organelles such as vesicles of the endosomal/lysosomal pathway or in synaptic vesicles.91 Mutations in human CLC channels are known to cause a set of very diverse diseases such as myotonia (muscle stiffness), Bartter syndrome (renal salt loss) with or without deafness, Dent's disease (proteinuria and kidney stones), osteopetrosis and neurodegeneration, and possibly epilepsy.91 The side chain of the aforementioned critical glutamic acid occupies a third Cl− ion binding site in the closed state of the channel and moves away to allow Cl− binding.90
Glutamic acid valve
Glutamic acid is known to play a unique role in regulation of the cytochrome-c oxidase (CcO) activity. CcO is the last enzyme of the respiratory electron transport chain in mitochondria (or bacteria) located in the inner mitochondrial (or bacterial) membrane, and it is responsible for reducing ~90% of the oxygen taken up in aerobic life. This protein powers the production of ATP by generating an electrochemical proton gradient across the membrane via the catalysis of the oxygen reduction to water that takes place in the binuclear center (BNC) of the enzyme. CcO uses four electrons taken up from the cytochrome c located at the positively charged P-side (outside) of the membrane and four “chemical” protons taken from the negatively charged N-side (inside) to reduce the dioxygen to two water molecules. In addition to this oxygen reduction reaction, four “pump” protons are translocated from the N-side to the P-side across the membrane against the opposing membrane potential, doubling the total amount of charge separated by the enzyme.92-95 Therefore, the main role of CcO is to serve as a proton pump and a generator of the electrochemical proton gradient or charge separation across the membrane, which is achieved via two separate processes. First, the reduction of oxygen to water by electrons and protons taken up from opposite sides of the membrane leads to the net translocation of one electrical charge across the membrane per electron consumed. Second, an additional proton is translocated vectorially across the membrane for each electron consumed, resulting in a net transport of two electrical charges per electron.96 The protons for the chemical reaction are extracted from the N-side of the membrane via two proton pathways, the D- and K-channels. The D-channel starts at a highly conserved residue, Asp 91 (bovine numbering; subunit I) near the N side, and continues to another highly conserved residue Glu242 that donates protons to the BNC, whereas the key residue in the K-channel is a highly conserved lysine (K319).95 The D-channel is responsible for the delivery of four “pump” protons that are first transferred from Glu242 to a “loading” site above the BNC and then delivered to the P side via a proton-exit channel. The mystery of this mechanism is in the ability of Glu242 located at the end of the D-channel to somehow sort “pump” protons from “chemical” protons.95 To explain this behavior, the glutamate valve model has been proposed according to which the side chain of Glu242 shuttles between a state protonically connected to the D channel, and a state connected to the BNC and the pump site.97 In this proton valve model, the Glu242 motion depends on its protonation state, where the unprotonated residue remains predominantly in a “down” conformation, pointing toward the N side, and therefore facilitating the uptake of a proton, whereas protonation shifts the Glu242 to the “up” conformation, where the side chain of this important residue is swung toward the P side by ~4 Å.97
Glutamic acid in the active sites of enzymes
In addition to serve multiple structural roles and being involved in regulation of various channels, glutamic acid residues, being positioned within or in the close proximity to the active sites, might have roles in the catalytic activities of various enzymes. One of the illustrative examples of the functional roles of glutamic acid can be found in bacterial nitric oxide reductase (NOR), which is a membrane-integrated enzyme that catalyzes the reduction of nitric oxide NO to nitrous oxide N2O using a type of anaerobic respiration where cytotoxic NO is immediately decomposed after its production from nitrite NO2− via the nitrite reductase-catalyzed reaction.98-100 Three different NOR types are found in bacteria, with the cytochrome c dependent NOR (cNOR) that consists of two subunits, NorB and NorC, being the most extensively studied enzyme. Precise description of the complex catalytic mechanism of this important enzyme is outside the scopes of this review, and therefore only a small piece of the entire picture, where the roles of glutamic acid are emphasized, is briefly described below. The characteristic feature of cNORs is the presence of five conserved glutamic acid residues (Glu135, Glu138, Glu211, Glu215 and Glu280 in P. aeruginosa cNOR) within the NorB subunit consisting of 12 trans-membrane helices and containing the heme b and the binuclear center (heme b3/FeB) buried in the hydrophobic interior of its trans-membrane region.100 Here, Glu211 is involved in the coordination of FeB and its carboxylate functions as the shuttle for catalytic protons from Glu280 to the bound-NO; Glu280, which interacts with Glu211 but is not involved in direct interaction with FeB, is an important player of the Thr330–Ser277–Glu280–Glu211 network that acts as a delivery pathway for protons utilized in the catalytic NO reduction; the carboxylate group of Glu215, which is located at the backside of Glu211, contributes to the electro-negative environment of the binuclear center of cNOR, and to the low redox potential of heme b3 iron; finally Glu135 and Glu138 are positioned in the loop connecting the transmembrane helices III and IV, with Glu135 serving as one of the Ca2+ ligands (which is crucial for maintaining the configuration of heme b and b3) and assisting in the water-mediated proton transfer through interactions with a number of water molecules, and with Glu138 serving as a key residue for maintaining the unique conformation of the long loop through interactions with the residues in transmembrane helix II, which would stabilize the coordination of Glu135 to Ca2+.100
Mono-ADP-ribosyltransferase, which is responsible for the mono-ADP-ribosylation of proteins, possesses a critical glutamic acid at the catalytic cleft which functions to position NAD for nucleophilic attack at the N-glycosidic linkage for either ADP-ribose transfer or NAD hydrolysis.101 The pronounced Na+/K+ selectivity of Na,K-ATPase relies on the strategic positioning of glutamic acid residues.102 Here, intramembrane Glu327 in transmembrane segment M4, Glu779 in M5, Asp804 and Asp808 in M6 are essential for tight binding of K+ and Na+, whereas Asn324 and Glu327 in M4, together with Thr774, Asn776 and Glu779 in the 771-YTLTSNIPEITP motif of M5 contribute to the Na+/K+ selectivity.102 In the family of thiamin diphosphate enzymes, a highly conserved glutamate is known to promote the C2-H ionization and the thiamin diphosphate activation.103
The direct catalytic role of glutamic acid can be seen in matrix metalloproteinases, which are ubiquitous endopeptidases characterized by an active site where a Zn2+ atom, coordinated by three histidines, plays the catalytic role, assisted by a glutamic acid that acts as a general base.104 For example, one of the well-known zinc-binding metalloproteases that uses a glutamic acid residue as the fourth ligand to coordinate the zinc ion is thermolysin. In thermolysin, glutamic acid is 20 amino acids downstream from the second histidine in the first motif and present in a small conserved motif (NEXXSD).105 In the zincin and PDF groups of metalloproteases, the catalytic zinc-binding site contains the HEXXHXXG motif.105 Also, a glutamic acid residue may be catalytically active in the substrate-binding cleft of plant lysozymes.106 Each enzyme in the α-amylase family of multidomain hydrolases and transferases has one glutamic acid and two aspartic acid residues necessary for activity.107 The irreversible dealkylation reaction catalyzed by the O6-alkylguanine-DNA alkyltransferase (AGT) that directly repairs alkylation damage at the O6-position of guanine is accomplished by an active-site cysteine that participates in a hydrogen bond network with invariant histidine and glutamic acid residues, reminiscent of the serine protease catalytic triad.108 The spore germination protease (GPR) that degrades small, acid soluble proteins (SASP) protecting spore's DNA against damage, is a structurally and functionally unique protease that utilizes glutamic acid residue to catalyze SASP degradation.109 In the hydrolytic aldehyde dehydrogenases (ALDHs), catalytic but flexible glutamic acid residues located within the active site serve as the general base that activates the hydrolytic water molecule in the deacylation step.110
In nudix hydrolases (which is a family of Mg2+-requiring enzymes that catalyze the hydrolysis of nucleoside diphosphates linked to other moieties) there is a specific motif, Nudix box (GX5EX7REUXEEXGU, where U is a bulky hydrophobic residue), that forms a loop–α helix–loop structural motif that functions as a common Mg2+-binding and catalytic site.111 It was emphasized that the overall catalytic powers of Nudix hydrolases consists in accelerating the reaction rate by 109 to 1012 times. The reactions are accelerated 103-105-times by general base catalysis by a glutamate residue within, or beyond the Nudix box, or by a histidine beyond the Nudix box. The additional 103-105-fold rate acceleration is due to the Lewis acid catalysis provided by one, two, or three divalent cations. One divalent cation is coordinated by two or three conserved residues of the Nudix box, the initial glycine and one or two glutamate residues, together with a remote glutamate or glutamine ligand located outside the Nudix box.111
Glutamic acids at various binding sites
Hemopexin is an important multifunctional plasma protein involved in the sequestering of heme released into the plasma from hemoglobin and myoglobin as the result of intravascular or extravascular hemolysis and due to skeletal muscle trauma or neuromuscular disease. It also possesses hyaluronidase activity, serine protease activity, pro-inflammatory and anti-inflammatory activity and is involved in the suppression of lymphocyte necrosis, inhibition of cellular adhesion, and binding of divalent metal ions. Finally, hemopexin possesses two highly exposed Arg–Gly–Glu sequences that may promote interaction with cell surfaces.112
Glutamic acid plays an important role in defining the retinal binding site geometry of rhodopsin, which is the photoreceptor in vertebrate rod cells responsible for vision at low light intensities. 11-cis-retinal is the photoreactive chromophore located in the interior of the protein where it is covalently attached to a lysine side chain through a protonated Schiff base (PSB) linkage.113 Based on the 13C-NMR chemical shift data, it was concluded that Glu113 of rhodopsin is involved in charge interactions with the retinal PSB, which are crucial for maintaining rhodopsin in the inactive state in the dark and whose breaking leads to the protein activation.113
A centrally located glutamic acid residue in position 6 of transmembrane segment VII of the main ligand-binding crevice of the chemokine 7TM receptors (GluVII:06) is crucial for recognition and binding of small molecule non-peptide ligands that contain one or two centrally located, positively charged nitrogen atoms and are characterized by relatively similar elongated overall structure with terminal aromatic moieties.114 Furthermore, since this GluVII:06 is crucial for the binding and hence the function of a number of non-peptide ligands in several chemokine receptors, such as the CCR1, CCR2 and CCR5 receptors, it serves as a selective anchor point for the centrally located, positively charged nitrogen of the small molecule ligands.114
Glutamic acid and metal binding
The role of glutamic acid residues in coordination of various metal ions was already emphasized in sections discussing ion channels. A few other illustrative examples are listed below. Based on the analysis of the complexes formed between integrins (which are central molecules in the adhesion processes that mediate cell–cell and cell–extracellular matrix communication) and their ligands, it has been concluded that divalent cations are critical for integrin interactions with almost all ligands. Importantly, although divalent cations are bound to integrins, their coordination sphere is not completed and the interactions between integrin and its ligands typically involve completing the metal ion coordination with an acidic ligand residue.115 For example, complexes between the human intercellular adhesion molecule-1 (ICAM-1) and the I domain of its integrin receptor αLβ2 are stabilized by a critical glutamate residue that completes the magnesium coordination in integrin.116 Similarly, in the crystal structure of a complex between the I domain of a2b1 integrin and a triple-helical collagen peptide containing a critical GFOGER motif, glutamate residue from the collagen peptide completes the coordination sphere of the I domain metal ion.117 Based on these observations it has been concluded that a metal-glutamate handshake represents a basic mechanism of integrin I domain interaction with its binding partners.115 Furthermore, it is believed now that the general mechanism by which integrins, these αβ-heterodimeric cell-surface receptors that are vital to the survival and function of nucleated cells, recognize their structurally diverse ligands relies on specific glutamic-acid- or aspartic-acid-based sequence motifs that function in a divalent cation-dependent and conformationally sensitive manner.118
The levels of intracellular zinc in living cells are crucial for managing various cellular processes, such as growth, development and differentiation. Zinc is involved in protein, nucleic acid, carbohydrate and lipid metabolism and also plays a role in the control of gene transcription and the coordination of other biological processes controlled by proteins containing DNA-binding zinc finger motifs, RING fingers and LIM domains.119 The physiologically relevant intracellular levels of zinc are controlled by specific zinc transporters which mostly transport zinc into cells from outside.105 Members of one of the subfamilies of these transporters, LIV-1 subfamily of ZIP zinc Transporters (LZT), being similar to other ZIP transporters in secondary structure and ability to transport metal ions across the plasma membrane or intracellular membranes, possess a unique HEXPHEXGD motif containing conserved proline and glutamic acid residues, that fits the consensus sequence for the catalytic zinc-biding site of matrix metalloproteinases (HEXXHXXGXXH), and which is unprecedented in other zinc transporters.105
In addition to this set of specific examples, one should keep in mind that all structures of the Ca2+-binding domains have in common a high negative surface potential usually associated with Asp or Glu residues.120 Therefore, important glutamic acid residues responsible for calcium coordination can be found in various members of the major Ca2+-binding proteins, such as EF-hand domains, EGF-like domains, γ-carboxyl glutamic acid (GLA)-rich domains, cadherin domains, Ca2+-dependent (C)-type lectin-like domains and Ca2+-binding pockets of family C G-protein-coupled receptors.120
A particularly intriguing role was described for the N-terminal glutamic acid residues in the canonical Ca2+-protein, α-lactabumin,121 which is frequently used as a model protein in folding studies and in studies on the effect of calcium binding on protein structure, stability and folding. For example, α-lactabumin was shown to possess significantly different thermal and structural stability in its calcium-bound and calcium-free apo-forms,122 with the apo-protein possessing molten globule-like properties at slightly elevated temperatures.123,124 This strong dependence of the α-lactabumin structural properties on metal-binding is determined by the simple fact that in the apo-form, many acidic side chains have unfavorable charge–charge interactions, with 11 residues (Glu1, Glu7, Glu11, Asp63, Asp64, Asp78, Asp82, Asp83, Asp84, Asp87 and Asp88) possessing significantly unfavorable charge–charge repultion.125 Although calcium binding has the most pronounced effect on residues directly involved in cation coordination (Asp82, Asp87 and Asp88) and strongly affects the other two residues in the Ca2+-binding loop, Asp83 and Asp84, Ca2+ binding has relatively minor effects on residues more distant from the Ca2+-binding site (Glu1, Glu7, Glu11, Asp63 and Asp64), which mostly preserve unfavorable electrostatic interactions seen in the apo-form.125 It was also shown that the mutation-induced neutralization of unfavorable charge–charge interactions in the N-terminus (residues 1–11 of which are characterized by a high proportion of negatively charged residues that cluster on the surface of the native protein) results in stabilization of both the apo- and Ca2+-bound protein.125 Unexpectedly, the ΔGlu1 mutant, where the Glu1 residue was removed, leaving an N-terminal methionine in its place, possessed almost one order of magnitude higher affinity for calcium and higher thermostability (both in the absence and presence of calcium) than the native protein isolated from milk.121 This unique tuning of the α-lactabumin structure and calcium binding suggested that the N-terminal region of this protein might have a direct effect on the calcium-binding loop (and perhaps other regions of the structure).121
Glutamic Acid-Based Posttranslational Modifications of Proteins
The side chains of glutamic acid residues are subjected to several PTMs. Some cytoplasmic and nuclear proteins are known to be methylated, i.e., enzymatically modified by the addition of methyl groups from S-adenosylmethionine. Methylation reactions typically occur on carboxyl groups (such as the side chain of glutamic acid) and modulate the activity of the target protein. Glutamate methyl ester formation plays a major role in chemotactic signal transduction in prokaryotes. For example, methyl-accepting chemotaxis proteins are a family of chemotactic-signal transducers that respond to changes in the concentration of attractants and repellents in the environment, transduce a signal from the outside to the inside of the cell, and facilitate sensory adaptation through the variation of the level of methylation.126,127 In some proteins and peptides, glutamic acids can be amidated. Also, some glutamine residues in proteins undergo spontaneous (nonenzymatic) deamidation to glutamate with rates that depend upon the sequence and higher-order structure of the protein. Functional groups within the protein can catalyze this reaction, acting as general acids, bases, or stabilizers of the transition state.128 In rare cases, glutamate residues can be modified by cyclization via condensation of the α-amino group with the side-chain carboxyl group giving rise to the pyrrolidone carboxylic acid (pyro-Glu). However, it was emphasized that pyro-Glu is exclusively found at the N-terminal end of the thermal polymers when glutamic acid is a predominant amino acid in a mixture of amino acids subjected to thermal polymerization.129
Another important glutamic acid-based PTM is gamma-carboxylation catalyzed by the vitamin K-dependent carboxylase that transforms specific glutamate residues in proteins to gamma-carboxy glutamic acid (Gla) in the presence of reduced vitamin K, molecular oxygen and carbon dioxide.130 This modification is widely distributed in the animal kingdom and has a wide range of physiological implications, such as hemostasis, bone calcification and signal transduction.130
In addition to be a target for various PTMs, glutamic acid itself can be used as an important protein modifier, giving raise to polyglutamylation, which is a specific PTM where polyglutamate chains of variable lengths are added to the modified protein.131 Polyglutamylation is evolutionarily conserved and is commonly found in the microtubule (MT) building block, tubulin. This PTM, being primarily found within the tubulin C-terminal tail that participates in binding of many structural and motor MT-associated proteins, is believed to be crucial for the functional adaptation of MTs. Polyglutamylation is catalyzed by a family of specific enzymes and in addition to tubulin can be found in some other proteins.131
Glutamic Acid in Thermophilic and Hyperthermophilic Organisms
High content of charged residues is one of the tricks used by Nature to make stable proteins in thermophilic and hyperthermophilic organisms.132 In fact, based on the correspondence analysis of the 56 completely sequenced genomes available from the three domains of life (seven eukaryotes, 14 archaeal and 35 bacterial species) it has been concluded and the amino acid composition permits discrimination between the three known lifestyles (mesophily, thermophily or hyperthermophily).132 The most specific amino acid compositional biases that represent specific signatures of thermophilic and hyperthermophilic proteomes are a relative abundance in glutamic acid, concomitantly with a depletion in glutamine and a significant correlation between the relative abundance in glutamic acid (negative charge) and the increase in the lumped “pool” lysine + arginine (positive charges). Being absent in mesophiles, these correlations could represent a physico-chemical basis of protein thermostability. Curiously, the distribution of the remaining charged amino acid, i.e., aspartic acid, appears to be quite homogeneous throughout all the species suggesting that this residue does not participate significantly in the aforementioned compensatory negative/positive (charged) correlation in thermophiles and hyperthermophiles.132 On average, thermophilic and hyperthermophilic proteomes were shown to contain 1.9%, 7.8%, 4.8% and 12.6% of glutamine, glutamic acid, aspartic acid and lysine + arginine residues, respectively. Importantly, some of these numbers are rather different from those found in IDPs/IDPRs, as shown in Table 1.
Glutamic Acid and Structure of IDPs/IDPRs
Although some amount of glutamic acid residues is crucial for the structure and function of ordered proteins/domains, when a protein or a peptide contains a large number of glutamic acid residues and, as a consequence, possesses a small number of hydrophobic residues, it is likely to be disordered at physiological pH due to strong charge-charge repulsion and weak hydrophobic attraction. An illustrative example of such charge-infused proteins is Glu-rich human prothymosin α, in which 64 out of 111 residues are charged (there are 19 Asp, 35 Glu, 2 Arg and 8 Lys residues), the overall content of hydrophobic residues (Leu, Ile and Val) is very low, and aromatic residues (Trp, Tyr, Phe and His) and cystein are absent.133 Based on this amino acid composition, it was not a big surprise to find that prothymosin α behaved as a highly disordered coil-like chain, since one cannot expect that a highly charged polypeptide (that contains 60% of Glu+Asp residues) will have a strong tendency to fold under physiological conditions.133,134 The lack of stable structure also explains the extreme thermal and acid stability of prothymosin α, since one cannot break what is non-existent.133 The peculiar amino acid composition of prothymosin α, this biologically active random coil, was one of the defining factors behind the charge-hydropathy plot (CH-plot) development.5 In fact, based on the analysis of prothymosin α and of 90 other non-globular proteins that lacked almost any ordered secondary structure under physiological conditions in vitro, it was concluded that a combination of high net charge and low hydropathy represents the necessary and sufficient factor for a polypeptide to behave as a natively unfolded protein.5 Strategically positioned glutamic acid residues can modulate conformational stability and function of ordered proteins too. In fact, the role of a glutamic/aspartic acid cluster located outside the Ca2+-binding site, and of the N-terminal Glu1 residue in destabilizing the structure and weakening the calcium-binding capabilities of α-lactabumin has been already discussed (see above).121,125 Therefore based on these observations, protein regions and whole proteins enriched in glutamic acids are expected to be substantially disordered.
Poly-γ-Glutamate, a Natural Wonder and a Biopolymer of Commercial Interest
Poly-γ-glutamate (PGA) is a natural homopolymer synthesized by several bacteria, one archaea (Natrialba aegyptiaca) and one eukaryote (Cnidaria).135 One of the most known sources of PGA is the Japanese specialty natto, a fermentation product made by Bacillus subtilis grown on soybean.135 PGA is a highly soluble polyanionic polymer that sequesters water molecules and can be found in surface-bound and released forms. In structural studies, polyglutamic acid is traditionally used as a biopolymer with a well-characterized secondary structure response to changes in the environmental pH, where PGA is in a random coil-like conformation at neutral pH, but gains monomeric α-helical structure at acidic pH and is transformed into a β-sheet structure at alkaline pH.136-138 Curiously, the addition of polylysine to an aqueous solution of polyglutamic acid homopolypeptide at neutral pH was shown to be accompanied by the instantaneous formation of a gel-like precipitate with intermolecular antiparallel β-structure.139
In bacteria, PGA may be composed of only D-, only L- or both D- and L-glutamate enantiomers, and PGA filaments may be poly-γ-L-glutamate filaments (PLGA), PDGA filaments or poly-γ-L-D-glutamate (PLDGA) filaments.135 The production and maintenance of sufficient D-glutamate pool levels required for the normal bacterial growth is controlled by the glutamate racemase, which is a member of the cofactor-independent, two-thiol-based family of amino acid racemases.140 This enzyme is conserved and essential for growth across the bacterial kingdom and has a conserved overall topology and active site architecture. Therefore, it represents an attractive target for the development of specific inhibitors that could act as possible therapeutic agents.140
In Gram-negative bacteria, the complex responsible for the polyglutamate synthesis is encoded in specific loci. If the PGA is associated with the bacterial surface and forms a capsule, then the corresponding genes are named cap (for “capsule”); however, if the PGA is released, then the corresponding genes are named pgs (for polyglutamate synthase).135 The minimal gene sets contain four genes termed cap or pgs B, C, A and E, with all cap genes and the four pgs genes (pgsB, pgsC, pgsAA, pgsE) being organized into operons.141
Since PGA is an IDP, whose biochemical and biophysical properties are environment-dependent, and since PGA can be found in an anchored to the bacterial surface form or in a released form, this biopolymer can play different roles in different organisms and in different environments.135 For example, when anchored to the bacterial surface, PGA forms a capsule and act as a virulence factor.135,142 In fact, the virulence of Bacillus anthracis (a Gram-positive sporulating bacterium, which is the causal agent of anthrax) was found to be determined by its capsule composed solely of PGA.143 Similarly, the virulence of Staphylococcus epidermidis (another Gram-positive bacterium that causes severe infection after penetrating the protective epidermal barriers of the human body) is dependent on the PGA-based capsule.144 Furthermore, PGA in capsules of these bacteria consists of either a mixture of L- and D-enantiomers (S. epidermidis)144 or solely D-enantiomer (B. anthracis),145 which makes them particularly non-immunogenic.135
The released form of PGA is used by the producing organism for rather different purposes, starting from the sequestration of toxic metal ions that increases the resistance of some soil bacteria to harsh conditions,146 to serving as a source of glutamate for bacteria in a starvation state during late stationary phase,147 to playing a role in decrease of the high local salt concentrations that helps extremophilic bacteria and archaea to survive in a hostile environment,148,149 and in Hydra, to control explosion of the special stringing cells, nematocysts, that are used to capture prey, for locomotion and for defense.150
In addition to have multiple functional roles, bacterially produced PGA has found its way to serve as an important biodegradable component151 with multifarious potential applications in foods, pharmaceuticals, healthcare, water treatment and other fields.152,153 A large commercial advantage of PGA is that this natural biopolymer is nontoxic, biocompatible and nonimmunogenic. It can be produced by various bacterial strains in a controllable way.152 As a result, PGA is commonly used in cosmetics/skin care, bone care, nanoparticle for drug delivery system, hydrogel, etc.154 For example, the PGA-based Medusa system has been recently developed for slow release of therapeutic proteins and peptides.155 Here, a poly L-glutamate backbone is grafted with hydrophobic α-tocopherol molecules, creating a colloidal suspension of nanoparticles in water that contain hydrophobic nanodomains suitable for the reversible binding of various drug molecules.155 The potential multifarious applications of PGA in the areas of biomedical materials, drug delivery carriers, and biological adhesives have been studied extensively.156 In general, γ-PGA is recognized now as an important biomaterial in drug delivery applications, with γ-PGA-based nanoparticles being considered as promising delivery carriers for anticancer therapeutics.157 Recently, a high molecular weight γ-PGA was shown to be used as an immune-stimulating agent.154
Finally, conjugation of paclitaxel, a widely used chemotherapeutic agent whose therapeutic index is limited by low tumor exposure and high systemic exposure, with biodegradable poly-l-glutamic acid generates paclitaxel poliglumex (PPX, CT-2103).158 This macromolecular drug conjugate enhances tumor exposure to the drug, since the release of paclitaxel from the polymeric backbone was shown to be dependent on the PPX degradation by the lysosomal protease cathepsin B, which is upregulated in many tumor types.158
Glutamic Acid and Functions of IDPs/IDPRs
Glutamic acid as a part of the protein degradation targeting signals, PEST motifs
PEST sequences (i.e., sequences enriched in proline (P), glutamic acid (E), serine (S) and threonine (T)) are known to serve as specific degradation signals.159-162 These degradation signals define cellular instability of many proteins and direct them either to the ubiquitin-proteasome degradation or to the calpain cleavage.161,162 This controlled protein degradation is important for activation and deactivation of regulatory proteins involved in signaling pathways that control cell growth, differentiation, stress responses and physiological cell death.159-162 PEST-containing sequences were shown to be solvent exposed and conformationally flexible, which preclude them from been resolved in X-ray structures.159 Based on the comprehensive bioinformatics analysis of experimentally characterized disordered and globular regions and of PDB chains containing PEST regions, it has been concluded that the PEST motif is most frequently located within IDPRs.161 Furthermore, analysis of the proline-rich motif Pro-X-Pro-X-Pro in PEST sequences revealed that these sequences contain glutamic acids much more often than aspartic acids.161 In addition to this Pro-X-Pro-X-Pro motif, many PEST sequences are highly enriched in negatively charged residues and are characterized by a very specific distribution of negative charged patterns.161
Glutamic acids in entropic bristle domains
The entropic bristle domain (EBD) concept was proposed to describe a characteristic behavior of some highly mobile protein regions. The EBD is not a structurally stable entity in the conventional sense, since for this protein region there are no folded states that exist for any appreciable amount of time. Instead, the EBD represents a time-average 3D region of a protein derived from the thermally driven motion of certain polypeptide chains, including those that are part of an otherwise stable folded protein.163 Therefore, the EBD which is defined by the time-averaged occupancy of space by a polypeptide chain, can exclude lager molecules while allowing small molecules and water to move freely through it. It was proposed that since functions of EBD depend on the intrinsically rapid thermal motion of the polypeptide, and the free energy changes that result when that motion is confined, this domain can be used to control binding events, confer mechanical properties, and sterically control molecular interactions.163
Obviously, to be able to serve as an EBD, a given fragment of a protein has to possess specific amino acid composition that would preclude it from folding. Therefore, EBDs are expected to possess low hydropathy and high net charge; i.e., in the CH-plot, they can be found well above the boundary separating compact and extended disordered proteins. One of the illustrative examples of biologically active EBDs (which are not tightly folded, but expected to have a very extended conformation) is given by side-arms of neurofilament (NF) proteins.164 The side-arms of the NF heavy polypeptide, NF-H (which are ~600 amino acids long), were shown by rotary shadow electron microscopy to be ~85 nm long. Since there was not enough mass to form a stiff folded structure to occupy such a volume, it was proposed that the side-arms were not folded but were in constant thermal motion.164 Analysis of the amino acid sequence of the porcine NF medium polypeptide (NF-M, which has an apparent molecular mass of 160 kDa and is one of the two high molecular mass components of mammalian neurofilaments) revealed that this protein has several peculiar features.165 The N-terminal 436 residues contain a non-α-helical arginine-rich headpiece (residues 1–98) with multiple β-turns followed by a highly α-helical rod domain that forms double-stranded coiled-coils (residues 99–412), followed by a C-terminal tailpiece extension (approximately 500 residues) that represents an autonomous domain of unique amino acid composition, being characterized by a high content of lysines and particularly glutamic acids.165 In human NF-M, there are 185 glutamic acids (20.2%), most of which are concentrated within the C-terminal tail, where glutamate accounts for 26.4% (133 out of 504 residues). Similarly, human NF-H (a polypeptide comprising 1,026 residues) has 189 glutamic acids, 143 of which are found in the 613 residues-long C-terminal tail of this protein, whereas in the human NF-L (NF light polypeptide which has 543 residues), there are 99 glutamic acids, with almost half of which (46) being located within the acidic C-terminal subdomain (the last 100 residues of the protein). In addition to neurofilament polypeptides, EBDs were found in microtubule-associated protein 2 (MAP2)166 and NuMa.167 Analysis of the amino acid compositions of these proteins revealed that they follow the trend established by NFs and contain significant amount of glutamic acid residues (220 out of 1,827 residues in human MAP2 are glutamates and there are 291 glutamic acids in the 2,115 residues-long human NuMa).
Recently, we proposed that EBDs can be used as protein solubility enhancers.168 In fact, we showed that highly charged protein sequences (both natural and artificial) can act as EBDs, and that translational fusion of such sequences to target proteins can serve as an effective solubilizing means by creating both large favorable surface area for water interactions and large excluded volumes around the partner.168 This suggests that intrinsically disordered EBDs (which extend away from the partner and sweep out large molecules) can enable the target protein to fold free from interference.168 All artificial fusions used in our study had low sequence complexity and high net charge, but were diversified using distinctive amino acid compositions and lengths.168 Among successful solubilizers were artificial EBDs containing the most disorder-promoting residues (Glu, Pro, Gln and Ser) in the proportion Glu:Pro:Gln:Ser = 2:2:1:1; i.e., sequences containing >33% glutamic acids.168 Therefore, it seems that glutamic acid is crucial for the successful function of EBD-containing proteins.
Glutamic acids in intrinsically disordered chaperones
The high content of glutamic acids in artificial EBDs designed as solubilization means was chosen because of the earlier observation that proteins with high net charge densities can function as effective intra- and intermolecular chaperones.169-172 For example, polyglutamate among other polyanions was shown to act as a chaperone and to accelerate the in vitro refolding of the Arc repressor protein.173 Small heat shock proteins (HSPs) have flexible C-terminal extensions that, although variable in length and sequence, are rich in acidic amino acids.169 The sHSP α-crystallin can act as a chaperone on the fibroblast growth factor 1 (FGF-1), and this chaperone action is mediated by electrostatic interactions between the basic regions of the growth factor and acidic regions of α-crystallin.174 Nucleolar chaperone B23 (294 residues, 31 of which are glutamic acids) has two acidic regions (residues 120–132 and 161–188) that contain 8 glutamic residues each and that are necessary for the B23 chaperone-like activity.175 Tubulin has chaperone-like activity being able to suppress the aggregation of soluble lens proteins, equine liver alcohol dehydrogenase, malic dehydrogenase and insulin, but only if its acidic C-terminus (that contains 39% and 33.3% of glutamic acid residuess in the porcine α- and β-tubulins, respectively) was intact.176-178 Many polyanionic propeptides were shown to serve as intramolecular chaperones to aid folding of the respective proteins.179-182 For example, propeptides of human neutrophil defensins contain up to 15.8% glutamic acids. Also, the C-terminal solubilizing domain of human α-synuclein (residues 100–140) contains 24.4% glutamates, whereas ERD10 (260 residues) and ERD14 dehydrins (185 residues) from Arabidopsis thaliana contain 19.6% and 21.1% glutamic acids respectively.
Some functions of glutamate-rich peptides
This section presents several illustrative examples of important biological functions attributed to glutamate-rich peptides.
Phytochelatins
Heavy metal detoxification in higher plants is dependent on a set of heavy-metal-complexing peptides, phytochelatins, with structure of (γ-glutamic acid-cysteine)n-glycine (n = 2–11) [(γ-Glu-Cys)n-Gly].183 The longest of these peptides possesses a molecular mass of 2.6 kDa, a pI 3.26 and a net charge of −11. These peptides are induced by the exposure of plants to several metals of the transition and main groups (Ib-Va, Z = 29−83) of the periodic table of elements. Phytochelatins are synthesized by a constitutive enzyme, γ-glutamylcysteine dipeptidyl transpeptidase, that uses glutathione (GSH) as a substrate and catalyzes the following reaction: γ-Glu-Cys-Gly + (γ-Glu-Cys)n-Gly→(γ-Glu-Cys)n+1-Gly + Gly.183
Fertilization promoting peptide
Another important glutamate-rich peptide is fertilization promoting peptide (FPP; pGlu-Glu-ProNH2), which is produced by the prostate gland and secreted into seminal plasma.184 FPP was shown to stimulate capacitation, which is the penultimate step in the maturation of mammalian spermatozoa required to render them competent to fertilize an oocyte. Furthermore, although FPP inhibits spontaneous loss of acrosome (an organelle that develops over the anterior half of the head in the spermatozoa), cells retain high fertility in vitro.184
GALA peptide
Recently, a synthetic 30 amino acid-long GALA peptide with a glutamic acid-alanine-leucine-alanine (EALA) repeat was designed to analyze how viral fusion protein sequences interact with membranes.185 This GALA peptide was long enough to span a bilayer when in the α-helical state, and the EALA repeat was adjusted so that the peptide would have a hydrophobic face of sufficient hydrophobicity to interact with the bilayer when the peptide was in an α-helix. Glu residues were used in GALA as a pH-responsive elements.185 When the pH is reduced from 7.0 to 5.0, GALA converts from a water soluble random coil conformation to an amphipathic α-helix that binds to bilayer membranes. Functional analysis revealed that GALA promoted fusion between small unilamellar vesicles and was able to form a transmembrane pore comprised of ~10 GALA α-helical monomers that were oriented perpendicularly to the plane of the membrane.185 Based on these observations, it has been proposed that pH-controlled membrane permealization induced by GALA can serve as a model for the design of environmentally responsive peptidic vehicles for drugs and genes delivery.185
Other type of PESTs: PTP-PESTs
Protein tyrosine phosphatases (PTP) with proline-, glutamate-, serine- and threonine-rich sequence, PTPs-PEST, are a ubiquitously expressed critical regulators of cell adhesion and migration.186,187 This family of PTPs includes three intracellular phosphatases known as proline-enriched phosphatase (PEP) in mice or lymphoid tyrosine phosphatase (LYP) in humans (also known as PTPN22 and PTPN8), PTP-PEST (also referred to as PTPN12) and PTP-hematopoietic stem cell fraction (PTP-HSCF, which is also known by several other names, such as also termed brain-derived phosphatase 1 (BDP1), PTP20, PTP-K1, fetal liver phosphatase 1 (FLP1) and PTPN18.186 All these phosphatases possess a common structural organization that includes an N-terminally located phosphatase domain, followed by a highly divergent central region that contains various motifs for interactions with other proteins, and a conserved C-terminal domain known as carboxyl-terminal homology (CTH) domain.186 Human PTP-LYP (PTPN22/PTPN8) is a 807 residues-long protein that contains 59 and 40 glutamic and aspartic acids and 45, 83 and 32 prolines, serines and threonines, respectively. Human PTP-PEST (PTPN12) consists of 780 residues and has 67, 49, 66, 72 and 54 glutamates, aspartates, prolines, serines and threonines, respectively, most of which are located outside the catalytic domain, with respectively 44, 32, 53, 59 and 39 glutamates, aspartates, prolines, serines and threonines being found in the non-catalytic region (residues 294–780). Finally, among the 460 residues of the human PTP-HSCF (BDP1/PTP20/ PTP-K1/FLP1/PTPN18), there are 27 glutamic acids, 21 aspartic acids, 32 prolines, 29 serines and 25 threonines. Importantly, glutamate-rich, non-catalytic regions of all these PTPs are known to be involved in interactions with multiple binding partners. For example, PTP-LYP is involved in interaction with Grb2, c-Cbl, and the C-terminal Src kinase (Csk), which is the inhibitory protein tyrosine kinase (PTK). The interaction between the PTP-LYP and Csk is mediated by the proline-rich motif in PEP and by the Src homology 3 (SH3) domain of Csk.186 PTP-PEST promiscuously associates with various proteins involved in the organization of the cytoskeleton, such as Cas (and Cas-related proteins Sin and CasL), paxillin (and paxillin-related polypeptides Hic-5 and leupaxin) and the PTKs FAK and Pyk2. This protein also associates with Shc, Grb2 and Csk.186 Finally, PTP-HSCF is involved in association with Csk and Tec.186
Multifarious functions of glutamic acid-rich proteins
Delta factor
In addition to γ-PGA, Bacillus subtilis produces another important polyanion, delta factor, which is an important component of the bacterial RNA polymerase.188 This delta factor is a 20.4 kDa highly acidic (pI = 3.6) protein that contains two distinct regions, a 13 kDa N-terminal domain with uniform charge distribution and a Glu-Asp-rich C-terminal region. The overall contents of glutamic and aspartic acids in delta factor are 20.8% and 17.9% respectively, whereas these numbers increase to 34.3% and 37.3% in the Glu-Asp-rich C-terminal domain. The ordered N-terminal domain contains 32% α-helix and 16% β-sheet, whereas the C-terminal 8.5 kDa domain is highly charged (net charge of −47) and therefore is largely unstructured.188 Importantly, the C-terminal intrinsically disordered domain has an important biological function, since the ability of delta factor to displace RNA from RNA polymerase requires the activities of both the N-terminal core-binding domain and the polyanionic C-terminal region.188
MARCKS
Myristoylated alanine-rich C kinase substrate (MARCKS) is an abundant 32 kDa protein which is unusually rich in alanine and glutamic acid, with glutamic acid and alanine in this proteins accounting for 16.0% and 30.7% residues, respectively. MARCKS is a very prominent cellular substrate for protein kinase C (PKC), and its 22 serine residues and 2 threonines are phosphorylated. Human MARCKS is an acidic protein with a pI of 4.46 which in addition to Ala-Glu enriched N- and C-terminal domains possesses a compact “effector domain” (ED), which is responsible for interaction with calmodulin, is located near the middle of the sequence and is enriched in lysines, serines and phenylalanines.189 MARCKS is a typical IDP with a labile conformation and little ordered structure. In addition to calmodulin this protein can interact with synapsin and actin, and can serve as filamentous actin (F-actin) cross-linking protein. Furthermore, being myristoylated, MARCKS is able to interact with membrane and serves as a cytoskeleton-membrane linkage crucial for controlling cell shape changes.189
ARGLU1
Transcriptional activators and RNA polymerase II are bridged via the central transcriptional coactivator complex, the Mediator complex. It has been recently shown that the arginine and glutamate rich 1 protein (ARGLU1) colocalizes with the Mediator subunit 1 (MED1) in the nucleus, being in contact with the far C-terminal region of MED1.190 This ARGLU1-MED1 interaction is crucial for the estrogen-dependent gene transcription and breast cancer cell growth.190 Human ARGLU1 is a 270 residues-long protein that contains 53 arginines and 54 glutamates. There are two regions with significant composition biases in this protein, an arginine-rich region (residues 3–74) that contains 25 arginines and a glutamic acid-rich region (residues 27–251) containing 49 glutamic acids.
PELP1
Proline-, glutamic acid- and leucine-rich protein-1 (PELP1) plays an important role in mediation of genomic and nongenomic signaling of β-estradiol.191 This potential proto-oncogene functions as a co-regulator of estrogen receptor, and expression of PELP1 is deregulated during breast cancer progression.192 PELP1 contains ten nuclear receptor-interacting boxes (LXXLL motifs), which allow it to interact with estrogen receptor and other nuclear hormone receptors, a zinc finger, a glutamic acid-rich domain and two proline-rich domains.191 There are several consensus PXXP motifs within the proline-rich regions, via which PELP1 couples the estrogen receptor (ER) with SH3 domain-containing kinase signaling proteins, such as Src and PI3K P85 regulatory subunit.191 There are 148 glutamic acids in PELP1 (which is 1,130 residues long), and the majority of them (99) are concentrated within the glutamic acid-rich domain (residues 888–1101).
eIF5
Eukaryotic translation initiation factor 5 (eIF5) is a monomeric protein of about 49 kDa that functions as a GTPase-activating protein (GAP) in translation initiation. eIF5 is involved in initiation of protein synthesis in eukaryotic cells, where, after binding to the 40S initiation complex (40S–eIF3–mRNA–Met-tRNAf–eIF2–GTP) at the AUG codon of an mRNA, it promotes GTP hydrolysis. This initiates a cascade of events that starts from the release of bound initiation factors from the 40S subunit and ends with the joining of the 60S ribosomal subunit to the 40S complex to form the functional 80S initiation complex (80S–mRNA–Met-tRNAf).193 Although eIF5 binds GTP and is able to promote GTP hydrolysis reaction, it does not hydrolyze GTP by itself acting as a typical GTPase-activating protein (GAP). In fact, eIF5 forms a complex with eIF2 via its glutamic acid-rich C-terminal region that binds to the lysine-rich N-terminal region of the β-subunit of eIF2 thus activating the GTPase activity of eIF2.193 In human eIF5, the 3D structure is known for the N-terminal nucleotide binding domain (residues 1–150, PDB ID: 2E9H) and for the W2 domain (residues 232–431, PDB ID: 2IU1). The linker region connecting these two domains is highly disordered and contains one of the functionally important glutamic acid-rich regions (residues 196–202). Overall, there are 11.4% glutamic acid residues in the 431 residues-long amino acid sequence of human eIF5.
Histone-interacting proteins
Since histones are polycations, they are known to be involved in interactions with several polyanionic proteins, particularly with proteins containing glutamic acid-rich domains or regions. For example, the non-epithelial intermediate filament (IF) subunit protein (e.g., human vimentin, which is attached to the nucleus, endoplasmic reticulum and mitochondria, either laterally or terminally and that contains 11.8% glutamic acids) can specifically bind core histones with a stoichiometry of 8 core histones per a nonneuronal IF protein dimer.194 Glutamic acids clearly play a crucial role in this interaction since the 68 kD neurofilament protein, which was already discussed in the EBD section and contains a glutamic acid-rich C-terminal extension, can bind more core histones per dimer (24 molecules of core histones) than the dimer of the non-neuronal IF proteins.194 In the nuclei of Physarum polycephalum, there is an alanine, lysine and glutamic acid-rich nuclear protein (P2) with a molecular mass of ~19.5 kDa that can specifically interact with histones and therefore is co-extracted with histones.195 Based on amino acid sequence analysis, it has been concluded that P2 is a HMG-like protein, which, according to CD measurements, contains only 5% secondary structure and is, therefore, essentially unstructured under in vivo conditions.195
Titin
The gigantic protein titin (there are 34,350 residues in the human protein) is a key component in the assembly and functioning of vertebrate striated muscles. Among numerous cellular functions of titin (also known as connectin) are contribution to the fine balance of forces between the two halves of the sarcomere which is crucial for the elasticity of muscle cells, as well as participation in chromosome condensation and chromosome segregation during mitosis of non-muscle cells. The ability of titin to reversibly extend relies on a set of PEVK segments, rich in proline (P), glutamate (E), valine (V) and lysine (K) residues. The single molecule analysis of the recombinant titin fragment, containing approximately 28-residue PEVK repeats and glutamic acid-rich motifs, revealed that the bending rigidity of the PEVK fragments can be reduced due to calcium-induced conformational changes.196 Furthermore, the glutamic acid-rich motif was shown to be critical for this process. Based on these observations, it has been concluded that the glutamic acid-rich motifs embedded into the PEVK segments make titin a calcium-dependent molecular spring that can adapt to the physiological state of the cell.196 Curiously, titin has 3,193 glutamic acids, 449 of which are found in the glutamic acid-rich region (residues 9974–11917) that contains 31 PEVK motifs. Glutamates are not evenly distributed within the glutamic acid-rich region; e.g., 42 glutamic acids are concentrated within the first 116 residues of this region (residues 9974–10,089). In other words, although the glutamic acid-rich region comprises just 5.6% of the whole titin, it has 14.1% of all the titin’s glutamates.
Bone phosphoproteins
Bone sialoprotein II (BSP II) is an important component of the bone mineralized matrix. This bone-specific glycoprotein contains phosphoserine and sulphotyrosine residues and two regions of contiguous glutamic acid residues (residues 77–84 and 156–169). In one of the first studies dedicated to the analysis of bone phosphoprotein it has been shown that this glycoprotein can be purified from the mixture of proteins extracted by demineralization of rat bone with 0.5 M EDTA in 4 M guanidinium chloride.197 It was also emphasized that this protein possessed an abnormal electrophoretic mobility since although the molecular mass of the phosphoprotein was shown to be about 44 kDa by sedimentation equilibrium analysis, it runs on 5–15% SDS-PAGE (SDS-PAGE) as a protein with a molecular mass of 75 kDa.197 Later studies revealed that BSP is capable of nucleating the bone mineral hydroxyapatite and that this nucleation involves one or both of the glutamic acid-rich sequences suggesting that polycarboxylate sequences might represent a specific site for growth-modulating interactions between proteins and biological hydroxyapatite crystals.198 Similarly, the ability of another acidic, non-collagenous protein of bone and dentin, osteonectin (also known as secreted protein, acidic, rich in cysteine), to bind to hydroxyapatite crystals is determined by its N-terminal region containing glutamic acid-rich sequences.199 SPARC is a highly conserved acidic calcium-binding extracellular-matrix protein.200 This matricellular glycoprotein is composed of three functional domains that are evolutionarily conserved in organisms ranging from nematodes to mammals.201 Starting from the N-terminus, these functional domains are: a Ca2+-binding glutamic acid-rich acidic domain (domain I), a follistatin-like module (domain II), and an extracellular Ca2+-binding (EC) module that contains two EF-hands and two collagen-binding epitopes (domain III). Since domain I was not found in SPARC isolated from the starlet anemone Nematostella vectensis, it has been proposed that SPARC first evolved as a collagen-binding matricellular glycoprotein.201 Human SPARC is a 303 residues-long protein that contains 34 glutamic acids, 15 of which are located within the N-terminal calcium binding region (residues 22–69). Although Xenopus laevis SPARC has a molecular mass of 32.6 kDa, based on SDS-PAGE analysis this protein has a molecular mass of 43 kDa.200
NBP-45
In nuclei of mice cells, there is a nuclear protein NBP-45 related to the nuclear proteins HMG-14/-17. NBP-45 can function as a transcriptional activator, binds specifically to nucleosome core particles,202 preferentially binds to euchromatin and modulates cellular transcription by counteracting linker histone-mediated chromatin compaction.203 NBP-45 is composed of 406 amino acids and has several functional regions and domains: the N-terminal region (residues 1–85) contains three segments that are highly homologous to functionally important domains in the HMG-14/-17 protein family, namely a nuclear localization signal, a nucleosome binding domain and a chromatin unfolding domain, whereas the C-terminal region (residues 86–406) has 43.7% of negatively charged residues.202 In fact, of the 110 glutamic acids and 44 aspartic acids found in NBP-45, 100 glutamic and 40 aspartic acids are located in this highly acidic region.
GARPs in rod photoreceptors
Glutamic acid-rich proteins (GARPs) are common in different organisms and have numerous biological functions. For example, rod photoreceptors contain three different glutamic acid-rich proteins (GARPs), two soluble forms, GARP1 and GARP2, and the N-terminal cytoplasmic domain (GARP part) of the B1 subunit of the cyclic GMP-gated channel (also known as cyclic nucleotide-gated cation channel β-1, CNGB1), that are involved in the control of the Ca2+ propagation from the site of its entry at the cyclic nucleotide-gated channel to the cytosol of the outer segment.204 The cyclic nucleotide-gated (CNG) cation channel of rod photoreceptors is a heterotetramer consisting of homologous subunits, α and β (also known as CNGA1 and CNGB1a). CNGA1 is known to be indispensable for channel activation, whereas CNGB1a plays mostly regulatory structural roles.205 In fact, the N-terminal glutamic acid-rich protein (GARP) domain of CNGB1a and the soluble GARP2 were shown to decrease the opening probability of the CNG channel and therefore these GARPs serve as important autoinhibitors or molecular gate keepers that control the activation of heteromeric rod CNG channels.205 Furthermore, CNGB1 and GARP2, in concert with a retinal tetraspanin (peripherin-2 or peripherinRDS), were shown to contribute to the organization of the specific organelle, outer segment (OS), which possesses a characteristic membranous “stacked pancake” architecture that has to be partially renewed daily to maintain cell function and viability.206 In fact, a mouse knockout of CNGB1 and GARP2 attenuated rod function and caused structural alterations and slowly progressive retinal degeneration.207 Bovine GARP (or CNGB1) is a 1,394 residues-long transmembrane protein which plays important roles in both visual and olfactory signal transduction. CNGB1 has 209 glutamic acids. GARP1 is a 590-residues-long CNGB1 splice variant that possesses 141 glutamic acids. GARP2 is another CNGB1 splice variant that has 299 residues 38 of which are glutamic acids. Native GARP1 and GARP2 purified from bovine rod photoreceptors were shown to be typical IDPs.208
MGARP
Mitochondria-localized glutamic acid-rich protein [MGARP, which is also known as ovary-specific acidic protein (OSAP), corneal endothelium-specific protein 1 (CESP-1) and hypoxia upregulated mitochondrial movement regulator protein (HUMMR)] is one of the highly expressed proteins in retina.209 MGARP is highly enriched in steroidogenic tissues and the visual system, and early in development, this protein is mainly detected in the retina and adrenal gland.210 During the estrous cycle, MGARP levels correlate with estrogen levels in the ovaries. Furthermore, the expression of MGARP is regulated by estrogen in a tissue-specific manner and through a feedback regulatory mechanism.210 As it follows from a long list of names, this protein has numerous important functions. In fact, among functions listed for this protein in the UniProt are (1) plays a role in the trafficking of mitochondria along microtubules, (2) regulates the kinesin-mediated axonal transport of mitochondria to nerve terminals along microtubules during hypoxia, (3) participates in the translocation of TRAK2/GRIF1 from the cytoplasm to the mitochondrion and (4) plays a role in steroidogenesis through maintenance of mitochondrial abundance and morphology.211,212 There are 283 residues in mouse MGARP, 49 of which are glutamic acids exclusively located in the Glu-rich region (residues 79–277). Based on the spectroscopic analysis of this protein, it has been concluded that mouse GARP is an IDP.209
Some other GARPs
The life cycle of the phytopathogenic fungus Verticillium dahliae Kleb causing wilt disease in a wide range of crops, including cotton, includes three vegetative phases: parasitic, saprophytic and dormant.213 One of the genes tagged in a pathogenicity encoded a glutamic acid-rich protein (VdGARP1), which shared no significant similarity to any known proteins.213 This protein was shown to be involved in sensing infertile nutrient conditions in infected cells to promote a transfer from saprophytic to dormant microsclerotia for long-term survival.213 VdGARP1 is a short (91 residues) extremely acidic protein with a pI of 3.3 that contains 52.8% negatively charged residues (31 glutamic acids and 17 aspartic acids).
There are also several other GARPs in various organisms, the functions of which are not known as of yet. Small GARP (a 112-amino acid protein, with a molecular mass of 13.1 kDa and an isoelectric point of 3.94, 29 residues of which are glutamic acids) was found in Euplotes octocarinatus.214 Plasmodium falciparum GARP consists of 679 residues, 169 of which are glutamic acids.215
Rhox8/Tox
Reproductive homeobox 8 protein (Rhox8 or Tox) is a homeodomain protein which is distantly related to the members of the Paired/Pax family belonging to the PEPP subfamily of Paired-like homeobox proteins.216 In mice, Tox is predominately transcribed in the testis and ovary and potentially plays an important role during gametogenesis.216 This 320 residues-long protein contains 113 glutamic acids organized in two poly-glutamic acid stretches (residues 111–139 and 177–201) and several Glu-rich regions, which together with 11 aspartic acids makes Rhox8 highly acidic (pI 3.95).
KIBRA
Kidney and brain protein (KIBRA) is a large (1,113 residues) protein that serves as a potential regulator of the Hippo/SWH (Sav/Wts/Hpo or Salvador/Warts/Hippo) signaling pathway that restricts proliferation and promotes apoptosis therefore being crucial for tumor suppression.217,218 KIBRA has 111 glutamic acids and possesses two N-terminal WW domains, an internal C2-like domain and a C-terminal Glu-rich stretch (residues 819–873).219 Cellular functions of KIBRA are modulated via phosphorylation by protein kinase Czeta (PRKCZ).220 Some cellular activities of KIBRA may be associated with memory performance.220,221 Furthermore, in mammalian cells, this protein co-activates functions of the dynein light chain 1,222 is involved in regulation of the collagen-stimulated activation of the ERK/MAPK cascade223 and modulates directional migration of podocytes.224 KIBRA interacts with histone H3 via its Glu-rich region, and this interaction might play an important role in conferring an optimal transactivation function to the estrogen receptor-α (ER) and also may be involved in the proliferation of ligand-stimulated breast cancer cells.222
SH3BGR
SH3 domain-binding glutamic acid-rich protein (SH3BGR) is a highly acidic (pI 4.09) 239 residues-long protein that possesses 44 glutamates and 15 aspartates and that is expressed in heart and skeletal muscles.225 The majority of glutamates are located within the C-terminal Glu-rich region (residues 170–239), ~43% of which are glutamic acid residues. In addition to SH3BGR, several other members of the SH3BGR family were found in humans. These are the so-called SH3BGR-like proteins, such as SH3BGRL (114 residues, 12 glutamic acids), SH3BGRL2 (107 residues, 10 glutamic acids) and SH3BGRL3 (93 residues, 7 glutamic acids) encoded by chromosomes Xq13.3 6q13–15, and 1p34.3–35, respectively.226 It was shown that the SH3 domain-binding glutamic acid-rich-like protein 3 is upregulated in glioblastoma.227 Also, this protein was noticeably downregulated in the hippocampus and cerebral cortex of APP(E693Δ)-transgenic mice that are used as a model to study the pathological effects of Aβ oligomers in Alzheimer’s disease.228
ABRA
The acidic-basic repeat antigen (ABRA) is a 743-residues-long protein found in the vacuolar space surrounding merozoites in Plasmodium falciparum-infected erythrocytes, being localized in the parasitophorous vacuole and associated with the merozoite surface at the time of schizont rupture.229 Due to its surface location, ABRA is one of the potential vaccine candidates against erythrocytic stages of malaria.230 This protein is one of the antigens enriched in the clusters of merozoites formed with growth inhibitory immune serum and possesses chymotrypsin-like activity,231 which can be inhibited with serine protease inhibitors such as chymostatin and phenyl methyl sulfonyl fluoride (PMSF).232 It was shown that the N-terminal half of the protein is responsible for the protease activity, whereas the highly charged C-terminal part of the protein was not required for this activity.232 Furthermore, the N-terminus contains an erythrocyte-binding domain located within the cysteine-rich N-proximal region of ABRA.229 There are 111 glutamic acids and 108 lysines in ABRA, and in agreement with its name, the amino acid sequence of this protein is characterized by the presence of eight tandem repeats of [VT]-N-D-[ED]-[ED]-D (residues 226–273) and by a lysine-rich C-terminal region (residues 672–721).
KERP1
The parasite Entamoeba histolytica that colonizes the large bowel and provokes an asymptomatic luminal gut infection contains a peculiar lysine and glutamic acid-rich protein 1 (KERP1), which is associated to parasite surface, involved in the parasite adherence to host cells and plays a role in the Entamoeba histolytica liver abscess pathogenesis.233 An interesting feature of KERP1 (184 residues) is a very high content of lysines (25%) and glutamic acids (19%).
Proteins with long simple repeat elements from herpesviruses
One of the mechanisms employed by herpesviruses to evade the immune response, allowing them to persist life-long in their hosts, relies on the use of specific proteins that function as cis-acting inhibitors of antigen presentation.234 Among these inhibitors are the nuclear antigen 1 (EBNA1) and pGZr in the Epstein–Barr virus (EBV) and the latency-associated nuclear antigen 1 (LANA1) of the Kaposi sarcoma herpesvirus.234 The common feature of all these proteins is the presence of long simple repeat elements in their amino acid sequences. For example, pGZr is a 230 amino-acids long glycine, glutamine, and glutamic acid-rich repeat (“GZ” repeat) protein that which is encoded by a large nested open reading frame located in the EBNA1 mRNA and is highly similar (65% amino-acid identity) to the acidic repeat of LANA1.234 Latent nuclear antigen of human herpesvirus 8 (HHV-8) (Kaposi's sarcoma-associated herpesvirus) is a large (1,036 residues) highly acidic protein (pI 3.81) that contains 237 glutamic acids, 179 glutamines, 114 prolines and 90 aspartic acids. In Herpesvirus saimiri (HVS) that infects squirrel monkeys, the functional homolog of Epstein–Barr virus EBNA1 and Kaposi's sarcoma-associated herpesvirus LANA1 proteins is the 501 residues-long product of the open reading frame 73 known as ORF73 or latency associated nuclear antigen.235 ORF73 contains a repeat domain composed of a glutamic acid and glycine repeat linked to a glutamic acid and alanine repeat (EG-EA repeat).235 There are 171, 83 and 43 glutamic acids, glycines and alanines in this latency associated nuclear antigen. Although there is low sequence identity between LANA1, EBNA1 and ORF73, all three proteins determine the poor recognition of viruses by CD8+ cytotoxic T lymphocytes (CTL). However, the mechanisms of their action are rather different. In the Epstein–Barr virus and Kaposi's sarcoma-associated herpesvirus the repeat domains were shown to enhance the stability of EBNA1 and LANA1 and decrease their translation rates, whereas the EG-EA repeat has no effect on the stability of HVS ORF73 or its rate of translation, but results in decreased steady-state levels of ORF73 mRNA.235 Intriguingly, the motif EEAEEAEEE of HVS ORF73 was sufficient to cause a reduction in recognition of ORF73 by CD8+ CTL, suggesting that the EG-EA repeat of HVS ORF73 is crucial for the immune evasion.235
Nsp3a
The N-terminal domain of the severe acute respiratory syndrome coronavirus (SARS-CoV) nonstructural protein 3 (nsp3a) is a typical IDP of 183 residues characterized by the presence of an ubiquitin-like globular domain (residues 1–112) and a flexible, highly extended Glu-rich domain (residues 113–183).236 Nsp3a is a highly acidic protein (pH 3.72) that contains 40 glutamic acids, 28 of which are located within the C-terminal Glu-rich domain.
PPE antigens
Proline and glutamic acid rich proteins (or PPE-repeat containing proteins, or PPE proteins) are important T-cell antigens produced by Mycobacterium avium subsp Paratuberculosis (Map).237 One of the PPEs is a 34.9 kDa protein (359 residues, pI 4.31) which following recombinant expression in E. coli was shown to elicit significant delayed type hypersensitivity skin reaction in mice sensitized with Map, suggesting that this recombinant PPE protein of Map was definitely associated with cellular immune response.237 Curiously, this PPE contains 73 alanines, 44 glycines, 37 prolines, 20 aspartic acids but just 10 glutamic acids.
Pt2L4
Cassava storage roots differentially produce an interesting Pt2L4 protein238 with low sequence complexity characterized by a reduced amino acid alphabet (just 13 amino acids). This 107 residue-long protein contains 56 glutamic acids, 30 alanines, 24 valines, 20 prolines, 18 serines and 15 lysines, but does not have any arginines, asparagines, cysteins, histidines, phenylalanines, tyrosines and tryptophanes.
Glutamic acid-rich protein from cassava roots
Based on the analysis of changes in the cassava root proteome during physiological deterioration of cassava root after harvesting, it has been concluded that the glutamic acid-rich protein was one of the proteins that were upregulated after harvesting.239
Cp190
Eukaryotic genomes contain a set of specific functional elements, chromatin insulators or boundary elements that regulate gene transcription by interfering with promoter-enhancer communication.240 In Drosophila melanogaster, the centrosome-associated zinc finger protein Cp190 protein (Cp190) is a component of the gypsy chromatin insulator complex, which is composed of Cp190, mod(mdg4) and su(Hw) and is required for the function of the gypsy chromatin insulator and other endogenous chromatin insulators organized by Su(Hw), CTCF and BEAF32.240 Although Cp190 is a large protein (1,096 residues) that possesses a complex multidomain structure, only three domains were shown to be essential for the insulator function and for the viability of flies: the BTB/POZ domain, an aspartic acid-rich (D-rich) region and a C-terminal glutamic acid-rich (E-rich) region.240 Here, the N-terminal Cp190 fragment containing the BTB/POZ domain and the D-rich region was shown to be involved in regulation of the Cp190 interaction with insulator complexes, whereas the C-terminally located E-rich region was necessary for the Cp190 dissociation from chromosomes during heat-shock.240
Importantly, the 131 glutamic acids are not equally distributed within the protein, with the N-terminal half containing just 26 glutamic acids and with the remaining 105 glutamates being concentrated within the C-terminal half of Cp190. Therefore, although the overall glutamic acid content of this protein is 12%, its C-terminal half is especially enriched in these residues (19.2%). Also, this uneven distribution is seen not only for Glu, but for all the charged residues. In fact, the N-terminal fragment (residues 1–548) has a net charge of +18 (Asp + Glu = 25 + 26 = 51; Arg + Lys = 31 + 38 = 69), whereas the C-terminal half of Cp190 (residues 549–1096) has a net charge of −120 (Asp + Glu = 62+105 = 167; Arg + Lys = 8 + 39 = 47).
Pcp4l1
Purkinje cell produces two closely related proteins containing IQ motifs, Purkinje cell protein 4-like 1 (Pcp4l1) and Pcp4/PEP-19. Although Pcp4/PEP-19 is able to interact with calmodulin and inhibit calmodulin-dependent enzymes, and although the synthetic peptide constituting only the IQ motif of Pcp4l1 binds calmodulin and inhibits calmodulin-dependent kinase II, the full-length Pcp4l1 does not interact with calmodulin.241 The lack of ability of the full length Pcp4l1 to interact with calmodulin was ascribed to its nine-residue glutamic acid-rich sequence that lies outside the IQ motif in Pcp4l1. Mutational analysis showed that calmodulin binding can be restored not only by the deletion of this inhibitory motif, but also by exchanging it with the homologous region of PEP-19 and by simple point mutation converting a single isoleucine (Ile36) within this motif to phenylalanine or to other aromatic residues.241 Therefore, although PEP-19 and Pcp4l1 possess noticeable sequence similarities, their functional properties are very different due to the presence of the Glu-rich element in Pcp4l1 that can functionally suppress an IQ motif.241
Glutamic acid mutations and human diseases
Chronic beryllium disease and Lys96Glu mutation in HLA-DPB1
Chronic beryllium disease (CBD) is a hypersensitivity disorder that affects 2–16% of workers professionally exposed to berillium in the workplace. CBD is characterized by a granulomatous inflammation and accumulation of beryllium-specific CD4+ T cells in the lung.242 The susceptibility to this disease depends on both genetic factors (genetic susceptibility) and the nature of the exposure. Genetic analysis revealed that a single point mutation at the 69th position of the human leukocyte antigen (HLA) class II histocompatibility antigen DP β 1 chain (HLA-DPB1), where lysine is substituted by a glutamic acid, makes the carriers more susceptible to CBD. It has been proposed that the K→E point mutation affects the ability of HLA-DPB1 to present beryllium to pathogenic CD4+ T cells.242
Sickle cell anemia and Glu6Val mutation in hemoglobin
Sickle-cell (SCA) or drepanocytosis is an autosomal recessive genetic blood disease with over-dominance, characterized by red blood cells that assume an abnormal, rigid, sickle shape. The disease is caused by a single point mutation in the β-globin chain of hemoglobin where the hydrophilic and negatively charged amino acid glutamic acid is replaced by the hydrophobic amino acid valine at the sixth position. As a result of this substitution, sickle hemoglobin polymerizes inside the affected erythrocytes. It was pointed out that such sickle hemoglobin polymerization occurs by homogeneous and heterogeneous nucleation mechanisms, which are both highly sensitive to macromolecular crowding.243 In fact, the rates of homogeneous nucleation were shown to be enhanced by 1010 when the initial concentration was augmented by 50% non-polymerizing hemoglobin.243
Retinitis pigmentosa and mutations in a Glu-rich domain of RPGR
Retinitis pigmentosa (RP) is an inherited, degenerative eye disease associated with the progressive loss of photoreceptor genes that causes severe vision impairment and often blindness.244 Among other factors, RP is caused by mutations in the retinitis pigmentosa GTPase regulator (RPGR) gene which accounts for 15–20% of RP cases in Caucasians.245 Genetic analysis revealed that of 240 RPGR mutations 95% are associated with X-linked retinitis pigmentosa (XLRP), 3% are found in cone, cone-rod dystrophy or atrophic macular atrophy, and 2% are related to syndromal retinal dystrophies with ciliary dyskinesia and hearing loss.245 Importantly, all disease-causing mutations occur in one or more RPGR isoforms containing the C-terminal exon open reading frame 15 (ORF15), and 55% occur in a Glu-rich domain within exon ORF15, which accounts for only 31% of the protein.245 RPGR (1,020 residues) contains 123 glutamic acids, more than half of which (70) are located within the C-terminal Glu-rich domain (residues 530–903).
Pyoderma gangrenosum and Glu250Gln mutation in PSTPIP1
Pyoderma gangrenosum is a condition that causes tissue to become necrotic, causing deep ulcers that usually occur on legs. Pyoderma gangrenosum is one of the most common extra-intestinal manifestations of chronic inflammatory bowel disease.246 The disease is caused by the alterations in the pathway that links the members of the proline-rich, glutamic acid-rich, serine-rich and threonine-rich (PEST) family of protein tyrosine phosphatases (which are critical regulators of adhesion and migration) to their substrates. A major player in this pathway is a cytoskeleton-associated adaptor protein, namely proline-serine-threonine phosphatase-interacting protein 1 (PSTPIP1, also known as CD2-binding protein 1, CD2BP1).246 Defects in PSTPIP1 are the cause of PAPA syndrome (PAPAS), also known as pyogenic sterile arthritis, pyoderma gangrenosum and acne or familial recurrent arthritis (FRA).247 PAPAS is characterized by an autosomal dominant inheritance of early onset, primarily affecting skin and joint tissues. Missense mutations Glu250-Gln and Ala230-Thr in PSTPIP1/CD2BP1 were identified in two families.247 These mutations were shown to affect the ability of PSTPIP1to interact with its natural partners.247,248
Concluding Remarks
This review illustrates that glutamic acid is differently used in ordered proteins/domains and in IDPs/IDPRs. In ordered proteins, glutamic acid residues are crucial for protein solubility and, being strategically placed within protein structure, play several structure-forming and structure-stabilizing roles. Here, glutamic acid is involved in electrostatic interactions and hydrogen bond formation, serves as an important α-helix former, and participates in the α-helix cap formation. Glutamic acid is an important functional residue of ordered proteins, where it can be involved in the formation of specific electrostatic valves inside the pores of ion channels, or can play unique catalytic roles in the active sites of enzymes, or be related to metal binding. In IDPs/IDPRs, overabundance of glutamic acids defines the extended conformation of native coils and native pre-molten globules. Glutamic acid is an important part of the PEST motif related to protein degradation. It is crucial for function of entropic bristle domains and several chaperones. Stretches of glutamic acid residues have a lot of specific functions that range from unique metal binding properties of phytochelatins and bone phosphoproteins, to regulation of cell adhesion and migration, to defining specific immunochemical reactivity of several antigens.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Uversky VN. . Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 2011; 43:1090 - 103; http://dx.doi.org/ 10.1016/j.biocel.2011.04.001; PMID: 21501695 [DOI] [PubMed] [Google Scholar]
- 2.Tompa P. . Unstructural biology coming of age. Curr Opin Struct Biol 2011; 21:419 - 25; http://dx.doi.org/ 10.1016/j.sbi.2011.03.012; PMID: 21514142 [DOI] [PubMed] [Google Scholar]
- 3.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, et al. . Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput 1998; •••:437 - 48; PMID: 9697202 [PubMed] [Google Scholar]
- 4.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. . Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform 2000; 11:161 - 71; PMID: 11700597 [PubMed] [Google Scholar]
- 5.Uversky VN, Gillespie JR, Fink AL. . Why are “natively unfolded” proteins unstructured under physiologic conditions?. Proteins 2000; 41:415 - 27; http://dx.doi.org/; PMID: 11025552 [DOI] [PubMed] [Google Scholar]
- 6.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. . Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004; 337:635 - 45; http://dx.doi.org/ 10.1016/j.jmb.2004.02.002; PMID: 15019783 [DOI] [PubMed] [Google Scholar]
- 7.Xue B, Dunker AK, Uversky VN. . Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn 2012; 30:137 - 49; http://dx.doi.org/ 10.1080/07391102.2012.675145; PMID: 22702725 [DOI] [PubMed] [Google Scholar]
- 8.Uversky VN, Dunker AK. . Understanding protein non-folding. Biochim Biophys Acta 2010; 1804:1231 - 64; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.017; PMID: 20117254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ. . Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293:321 - 31; http://dx.doi.org/ 10.1006/jmbi.1999.3110; PMID: 10550212 [DOI] [PubMed] [Google Scholar]
- 10.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. . Intrinsically disordered protein. J Mol Graph Model 2001; 19:26 - 59; http://dx.doi.org/ 10.1016/S1093-3263(00)00138-8; PMID: 11381529 [DOI] [PubMed] [Google Scholar]
- 11.Tompa P. . Intrinsically unstructured proteins. Trends Biochem Sci 2002; 27:527 - 33; http://dx.doi.org/ 10.1016/S0968-0004(02)02169-2; PMID: 12368089 [DOI] [PubMed] [Google Scholar]
- 12.Dyson HJ, Wright PE. . Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197 - 208; http://dx.doi.org/ 10.1038/nrm1589; PMID: 15738986 [DOI] [PubMed] [Google Scholar]
- 13.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. . Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002; 323:573 - 84; http://dx.doi.org/ 10.1016/S0022-2836(02)00969-5; PMID: 12381310 [DOI] [PubMed] [Google Scholar]
- 14.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. . Intrinsic disorder and protein function. Biochemistry 2002; 41:6573 - 82; http://dx.doi.org/ 10.1021/bi012159+; PMID: 12022860 [DOI] [PubMed] [Google Scholar]
- 15.Uversky VN. . Natively unfolded proteins: a point where biology waits for physics. Protein Sci 2002; 11:739 - 56; http://dx.doi.org/ 10.1110/ps.4210102; PMID: 11910019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uversky VN, Oldfield CJ, Dunker AK. . Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 2005; 18:343 - 84; http://dx.doi.org/ 10.1002/jmr.747; PMID: 16094605 [DOI] [PubMed] [Google Scholar]
- 17.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. . Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005; 272:5129 - 48; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04948.x; PMID: 16218947 [DOI] [PubMed] [Google Scholar]
- 18.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, et al. . Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 2007; 6:1882 - 98; http://dx.doi.org/ 10.1021/pr060392u; PMID: 17391014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. . Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res 2007; 6:1899 - 916; http://dx.doi.org/ 10.1021/pr060393m; PMID: 17391015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Faeder JR, Camacho CJ. . Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A 2009; 106:19819 - 23; PMID: 19903882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim PM, Sboner A, Xia Y, Gerstein M. . The role of disorder in interaction networks: a structural analysis. Mol Syst Biol 2008; 4:179; http://dx.doi.org/ 10.1038/msb.2008.16; PMID: 18364713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. . Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008; 9:Suppl 1 S1; http://dx.doi.org/ 10.1186/1471-2164-9-S1-S1; PMID: 18366598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright PE, Dyson HJ. . Linking folding and binding. Curr Opin Struct Biol 2009; 19:31 - 8; http://dx.doi.org/ 10.1016/j.sbi.2008.12.003; PMID: 19157855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. . Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry 2006; 45:10448 - 60; http://dx.doi.org/ 10.1021/bi060981d; PMID: 16939197 [DOI] [PubMed] [Google Scholar]
- 25.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. . Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 2007; 6:1917 - 32; http://dx.doi.org/ 10.1021/pr060394e; PMID: 17391016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uversky VN, Oldfield CJ, Dunker AK. . Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008; 37:215 - 46; http://dx.doi.org/ 10.1146/annurev.biophys.37.032807.125924; PMID: 18573080 [DOI] [PubMed] [Google Scholar]
- 27.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. . Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept Lett 2009; 16:1533 - 47; http://dx.doi.org/ 10.2174/092986609789839377; PMID: 20001916 [DOI] [PubMed] [Google Scholar]
- 28.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, et al. . Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics 2009; 10:Suppl 1 S7; http://dx.doi.org/ 10.1186/1471-2164-10-S1-S7; PMID: 19594884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. . Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics 2009; 10:Suppl 1 S12; http://dx.doi.org/ 10.1186/1471-2164-10-S1-S12; PMID: 19594871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. . Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005; 44:12454 - 70; http://dx.doi.org/ 10.1021/bi050736e; PMID: 16156658 [DOI] [PubMed] [Google Scholar]
- 31.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, et al. . Analysis of molecular recognition features (MoRFs). J Mol Biol 2006; 362:1043 - 59; http://dx.doi.org/ 10.1016/j.jmb.2006.07.087; PMID: 16935303 [DOI] [PubMed] [Google Scholar]
- 32.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, et al. . Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 2007; 6:2351 - 66; http://dx.doi.org/ 10.1021/pr0701411; PMID: 17488107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mészáros B, Simon I, Dosztányi Z. . Prediction of protein binding regions in disordered proteins. PLoS Comput Biol 2009; 5:e1000376; http://dx.doi.org/ 10.1371/journal.pcbi.1000376; PMID: 19412530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosztányi Z, Mészáros B, Simon I. . ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009; 25:2745 - 6; http://dx.doi.org/ 10.1093/bioinformatics/btp518; PMID: 19717576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uversky VN. . Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev 2011; 40:1623 - 34; http://dx.doi.org/ 10.1039/c0cs00057d; PMID: 21049125 [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN. . Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des 2012; In press PMID: 23170892 [DOI] [PubMed] [Google Scholar]
- 37.Hsu WL, Oldfield C, Meng J, Huang F, Xue B, Uversky VN, et al. . Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput 2012; •••:116 - 27; PMID: 22174268 [PubMed] [Google Scholar]
- 38.Mészáros B, Simon I, Dosztányi Z. . The expanding view of protein-protein interactions: complexes involving intrinsically disordered proteins. Phys Biol 2011; 8:035003; http://dx.doi.org/ 10.1088/1478-3975/8/3/035003; PMID: 21572179 [DOI] [PubMed] [Google Scholar]
- 39.Hazy E, Tompa P. . Limitations of induced folding in molecular recognition by intrinsically disordered proteins. Chemphyschem 2009; 10:1415 - 9; http://dx.doi.org/ 10.1002/cphc.200900205; PMID: 19462392 [DOI] [PubMed] [Google Scholar]
- 40.Tompa P, Fuxreiter M. . Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 2008; 33:2 - 8; http://dx.doi.org/ 10.1016/j.tibs.2007.10.003; PMID: 18054235 [DOI] [PubMed] [Google Scholar]
- 41.Fuxreiter M, Simon I, Friedrich P, Tompa P. . Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol 2004; 338:1015 - 26; http://dx.doi.org/ 10.1016/j.jmb.2004.03.017; PMID: 15111064 [DOI] [PubMed] [Google Scholar]
- 42.Chi SW, Kim DH, Lee SH, Chang I, Han KH. . Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus. Protein Sci 2007; 16:2108 - 17; http://dx.doi.org/ 10.1110/ps.072983507; PMID: 17766372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoemaker BA, Portman JJ, Wolynes PG. . Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A 2000; 97:8868 - 73; http://dx.doi.org/ 10.1073/pnas.160259697; PMID: 10908673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Liu Z. . Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: a critical assessment of the “fly-casting” mechanism. J Mol Biol 2009; 393:1143 - 59; http://dx.doi.org/ 10.1016/j.jmb.2009.09.010; PMID: 19747922 [DOI] [PubMed] [Google Scholar]
- 45.Prakash MK. . Insights on the role of (dis)order from protein-protein interaction linear free-energy relationships. J Am Chem Soc 2011; 133:9976 - 9; http://dx.doi.org/ 10.1021/ja201500z; PMID: 21648484 [DOI] [PubMed] [Google Scholar]
- 46.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, et al. . Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput 1998; •••:473 - 84; PMID: 9697205 [PubMed] [Google Scholar]
- 47.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. . Intrinsic disorder and functional proteomics. Biophys J 2007; 92:1439 - 56; http://dx.doi.org/ 10.1529/biophysj.106.094045; PMID: 17158572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vacic V, Uversky VN, Dunker AK, Lonardi S. . Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 2007; 8:211; http://dx.doi.org/ 10.1186/1471-2105-8-211; PMID: 17578581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. . DisProt: the Database of Disordered Proteins. Nucleic Acids Res 2007; 35:Database issue D786 - 93; http://dx.doi.org/ 10.1093/nar/gkl893; PMID: 17145717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. . The Protein Data Bank. Nucleic Acids Res 2000; 28:235 - 42; http://dx.doi.org/ 10.1093/nar/28.1.235; PMID: 10592235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. . The Universal Protein Resource (UniProt). Nucleic Acids Res 2005; 33:Database issue D154 - 9; http://dx.doi.org/ 10.1093/nar/gki070; PMID: 15608167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karplus PA. . Hydrophobicity regained. Protein Sci 1997; 6:1302 - 7; http://dx.doi.org/ 10.1002/pro.5560060618; PMID: 9194190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bordo D, Argos P. . Suggestions for “safe” residue substitutions in site-directed mutagenesis. J Mol Biol 1991; 217:721 - 9; http://dx.doi.org/ 10.1016/0022-2836(91)90528-E; PMID: 2005621 [DOI] [PubMed] [Google Scholar]
- 54.Meldrum BS. . Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr 2000; 130:Suppl 1007S - 15S; PMID: 10736372 [DOI] [PubMed] [Google Scholar]
- 55.McEntee WJ, Crook TH. . Glutamate: its role in learning, memory, and the aging brain. Psychopharmacology (Berl) 1993; 111:391 - 401; http://dx.doi.org/ 10.1007/BF02253527; PMID: 7870979 [DOI] [PubMed] [Google Scholar]
- 56.Okubo Y, Sekiya H, Namiki S, Sakamoto H, Iinuma S, Yamasaki M, et al. . Imaging extrasynaptic glutamate dynamics in the brain. Proc Natl Acad Sci U S A 2010; 107:6526 - 31; http://dx.doi.org/ 10.1073/pnas.0913154107; PMID: 20308566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shigeri Y, Seal RP, Shimamoto K. . Molecular pharmacology of glutamate transporters, EAATs and VGLUTs. Brain Res Brain Res Rev 2004; 45:250 - 65; http://dx.doi.org/ 10.1016/j.brainresrev.2004.04.004; PMID: 15210307 [DOI] [PubMed] [Google Scholar]
- 58.Hynd MR, Scott HL, Dodd PR. . Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer’s disease. Neurochem Int 2004; 45:583 - 95; http://dx.doi.org/ 10.1016/j.neuint.2004.03.007; PMID: 15234100 [DOI] [PubMed] [Google Scholar]
- 59.Glushakov AV, Dennis DM, Sumners C, Seubert CN, Martynyuk AE. . L-phenylalanine selectively depresses currents at glutamatergic excitatory synapses. J Neurosci Res 2003; 72:116 - 24; http://dx.doi.org/ 10.1002/jnr.10569; PMID: 12645085 [DOI] [PubMed] [Google Scholar]
- 60.Glushakov AV, Glushakova O, Varshney M, Bajpai LK, Sumners C, Laipis PJ, et al. . Long-term changes in glutamatergic synaptic transmission in phenylketonuria. Brain 2005; 128:300 - 7; http://dx.doi.org/ 10.1093/brain/awh354; PMID: 15634735 [DOI] [PubMed] [Google Scholar]
- 61.Ramachandran GN, Ramakrishnan C, Sasisekharan V. . Stereochemistry of polypeptide chain configurations. J Mol Biol 1963; 7:95 - 9; http://dx.doi.org/ 10.1016/S0022-2836(63)80023-6; PMID: 13990617 [DOI] [PubMed] [Google Scholar]
- 62.Maccallum PH, Poet R, Milner-White EJ. . Coulombic interactions between partially charged main-chain atoms not hydrogen-bonded to each other influence the conformations of alpha-helices and antiparallel beta-sheet. A new method for analysing the forces between hydrogen bonding groups in proteins includes all the Coulombic interactions. J Mol Biol 1995; 248:361 - 73; http://dx.doi.org/ 10.1016/S0022-2836(95)80056-5; PMID: 7739046 [DOI] [PubMed] [Google Scholar]
- 63.Ho BK, Thomas A, Brasseur R. . Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the alpha-helix. Protein Sci 2003; 12:2508 - 22; http://dx.doi.org/ 10.1110/ps.03235203; PMID: 14573863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho BK, Brasseur R. . The Ramachandran plots of glycine and pre-proline. BMC Struct Biol 2005; 5:14; http://dx.doi.org/ 10.1186/1472-6807-5-14; PMID: 16105172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McDonald IK, Thornton JM. . Satisfying hydrogen bonding potential in proteins. J Mol Biol 1994; 238:777 - 93; http://dx.doi.org/ 10.1006/jmbi.1994.1334; PMID: 8182748 [DOI] [PubMed] [Google Scholar]
- 66.Pace CN, Scholtz JM. . A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 1998; 75:422 - 7; http://dx.doi.org/ 10.1016/S0006-3495(98)77529-0; PMID: 9649402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hol WG. . The role of the alpha-helix dipole in protein function and structure. Prog Biophys Mol Biol 1985; 45:149 - 95; http://dx.doi.org/ 10.1016/0079-6107(85)90001-X; PMID: 3892583 [DOI] [PubMed] [Google Scholar]
- 68.Hol WG. . Effects of the alpha-helix dipole upon the functioning and structure of proteins and peptides. Adv Biophys 1985; 19:133 - 65; http://dx.doi.org/ 10.1016/0065-227X(85)90053-X; PMID: 2424281 [DOI] [PubMed] [Google Scholar]
- 69.Presta LG, Rose GD. . Helix signals in proteins. Science 1988; 240:1632 - 41; http://dx.doi.org/ 10.1126/science.2837824; PMID: 2837824 [DOI] [PubMed] [Google Scholar]
- 70.Serrano L, Fersht AR. . Capping and alpha-helix stability. Nature 1989; 342:296 - 9; http://dx.doi.org/ 10.1038/342296a0; PMID: 2812029 [DOI] [PubMed] [Google Scholar]
- 71.Aurora R, Rose GD. . Helix capping. Protein Sci 1998; 7:21 - 38; PMID: 9514257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu M, Shu W, Ji H, Spek E, Wang L, Kallenbach NR. . Helix capping in the GCN4 leucine zipper. J Mol Biol 1999; 288:743 - 52; http://dx.doi.org/ 10.1006/jmbi.1999.2707; PMID: 10329176 [DOI] [PubMed] [Google Scholar]
- 73.Zhou HX, Lyu P, Wemmer DE, Kallenbach NR. . Alpha helix capping in synthetic model peptides by reciprocal side chain-main chain interactions: evidence for an N terminal “capping box”. Proteins 1994; 18:1 - 7; http://dx.doi.org/ 10.1002/prot.340180103; PMID: 8146119 [DOI] [PubMed] [Google Scholar]
- 74.Ermolenko DN, Thomas ST, Aurora R, Gronenborn AM, Makhatadze GI. . Hydrophobic interactions at the Ccap position of the C-capping motif of alpha-helices. J Mol Biol 2002; 322:123 - 35; http://dx.doi.org/ 10.1016/S0022-2836(02)00734-9; PMID: 12215419 [DOI] [PubMed] [Google Scholar]
- 75.Forood B, Feliciano EJ, Nambiar KP. . Stabilization of α-helical structures in short peptides via end capping. Proc Natl Acad Sci U S A 1993; 90:838 - 42; http://dx.doi.org/ 10.1073/pnas.90.3.838; PMID: 8430094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trevino SR, Scholtz JM, Pace CN. . Measuring and increasing protein solubility. J Pharm Sci 2008; 97:4155 - 66; http://dx.doi.org/ 10.1002/jps.21327; PMID: 18240286 [DOI] [PubMed] [Google Scholar]
- 77.Kaupp UB. . The cyclic nucleotide-gated channels of vertebrate photoreceptors and olfactory epithelium. Trends Neurosci 1991; 14:150 - 7; http://dx.doi.org/ 10.1016/0166-2236(91)90087-B; PMID: 1710853 [DOI] [PubMed] [Google Scholar]
- 78.Kingston PA, Zufall F, Barnstable CJ. . Rat hippocampal neurons express genes for both rod retinal and olfactory cyclic nucleotide-gated channels: novel targets for cAMP/cGMP function. Proc Natl Acad Sci U S A 1996; 93:10440 - 5; http://dx.doi.org/ 10.1073/pnas.93.19.10440; PMID: 8816819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bradley J, Zhang Y, Bakin R, Lester HA, Ronnett GV, Zinn K. . Functional expression of the heteromeric “olfactory” cyclic nucleotide-gated channel in the hippocampus: a potential effector of synaptic plasticity in brain neurons. J Neurosci 1997; 17:1993 - 2005; PMID: 9045728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei JY, Roy DS, Leconte L, Barnstable CJ. . Molecular and pharmacological analysis of cyclic nucleotide-gated channel function in the central nervous system. Prog Neurobiol 1998; 56:37 - 64; http://dx.doi.org/ 10.1016/S0301-0082(98)00029-X; PMID: 9723130 [DOI] [PubMed] [Google Scholar]
- 81.Yau KW. . Cyclic nucleotide-gated channels: an expanding new family of ion channels. Proc Natl Acad Sci U S A 1994; 91:3481 - 3; http://dx.doi.org/ 10.1073/pnas.91.9.3481; PMID: 7513422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zagotta WN. . Molecular mechanisms of cyclic nucleotide-gated channels. J Bioenerg Biomembr 1996; 28:269 - 78; http://dx.doi.org/ 10.1007/BF02110700; PMID: 8807401 [DOI] [PubMed] [Google Scholar]
- 83.Zagotta WN, Siegelbaum SA. . Structure and function of cyclic nucleotide-gated channels. Annu Rev Neurosci 1996; 19:235 - 63; http://dx.doi.org/ 10.1146/annurev.ne.19.030196.001315; PMID: 8833443 [DOI] [PubMed] [Google Scholar]
- 84.Broillet MC, Firestein S. . Cyclic nucleotide-gated channels. Molecular mechanisms of activation. Ann N Y Acad Sci 1999; 868:730 - 40; http://dx.doi.org/ 10.1111/j.1749-6632.1999.tb11352.x; PMID: 10414360 [DOI] [PubMed] [Google Scholar]
- 85.Goulding EH, Tibbs GR, Liu D, Siegelbaum SA. . Role of H5 domain in determining pore diameter and ion permeation through cyclic nucleotide-gated channels. Nature 1993; 364:61 - 4; http://dx.doi.org/ 10.1038/364061a0; PMID: 7686276 [DOI] [PubMed] [Google Scholar]
- 86.Root MJ, MacKinnon R. . Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron 1993; 11:459 - 66; http://dx.doi.org/ 10.1016/0896-6273(93)90150-P; PMID: 7691102 [DOI] [PubMed] [Google Scholar]
- 87.Varadi G, Strobeck M, Koch S, Caglioti L, Zucchi C, Palyi G. . Molecular elements of ion permeation and selectivity within calcium channels. Crit Rev Biochem Mol Biol 1999; 34:181 - 214; http://dx.doi.org/ 10.1080/10409239991209264; PMID: 10473347 [DOI] [PubMed] [Google Scholar]
- 88.Raja M. . Diverse gating in K+ channels: differential role of the pore-helix glutamate in stabilizing the channel pore. Biochem Biophys Res Commun 2011; 413:1 - 4; http://dx.doi.org/ 10.1016/j.bbrc.2011.08.062; PMID: 21872570 [DOI] [PubMed] [Google Scholar]
- 89.Muramatsu T, Miyauchi T. . Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol Histopathol 2003; 18:981 - 7; PMID: 12792908 [DOI] [PubMed] [Google Scholar]
- 90.Pusch M, Jentsch TJ. . Unique structure and function of chloride transporting CLC proteins. IEEE Trans Nanobioscience 2005; 4:49 - 57; http://dx.doi.org/ 10.1109/TNB.2004.842503; PMID: 15816171 [DOI] [PubMed] [Google Scholar]
- 91.Jentsch TJ, Poët M, Fuhrmann JC, Zdebik AA. . Physiological functions of CLC Cl- channels gleaned from human genetic disease and mouse models. Annu Rev Physiol 2005; 67:779 - 807; http://dx.doi.org/ 10.1146/annurev.physiol.67.032003.153245; PMID: 15709978 [DOI] [PubMed] [Google Scholar]
- 92.Wikstrom MK. . Proton pump coupled to cytochrome c oxidase in mitochondria. Nature 1977; 266:271 - 3; http://dx.doi.org/ 10.1038/266271a0; PMID: 15223 [DOI] [PubMed] [Google Scholar]
- 93.Babcock GT, Wikström M. . Oxygen activation and the conservation of energy in cell respiration. Nature 1992; 356:301 - 9; http://dx.doi.org/ 10.1038/356301a0; PMID: 1312679 [DOI] [PubMed] [Google Scholar]
- 94.Brzezinski P, Gennis RB. . Cytochrome c oxidase: exciting progress and remaining mysteries. J Bioenerg Biomembr 2008; 40:521 - 31; http://dx.doi.org/ 10.1007/s10863-008-9181-7; PMID: 18975062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim YC, Hummer G. . Proton-pumping mechanism of cytochrome c oxidase: a kinetic master-equation approach. Biochim Biophys Acta 2012; 1817:526 - 36; http://dx.doi.org/ 10.1016/j.bbabio.2011.09.004; PMID: 21946020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitchell P. . Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961; 191:144 - 8; http://dx.doi.org/ 10.1038/191144a0; PMID: 13771349 [DOI] [PubMed] [Google Scholar]
- 97.Kaila VR, Verkhovsky MI, Hummer G, Wikström M. . Glutamic acid 242 is a valve in the proton pump of cytochrome c oxidase. Proc Natl Acad Sci U S A 2008; 105:6255 - 9; http://dx.doi.org/ 10.1073/pnas.0800770105; PMID: 18430799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zumft WG. . Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 1997; 61:533 - 616; PMID: 9409151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richardson DJ, Watmough NJ. . Inorganic nitrogen metabolism in bacteria. Curr Opin Chem Biol 1999; 3:207 - 19; http://dx.doi.org/ 10.1016/S1367-5931(99)80034-9; PMID: 10348621 [DOI] [PubMed] [Google Scholar]
- 100.Hino T, Nagano S, Sugimoto H, Tosha T, Shiro Y. . Molecular structure and function of bacterial nitric oxide reductase. Biochim Biophys Acta 2012; 1817:680 - 7; http://dx.doi.org/ 10.1016/j.bbabio.2011.09.021; PMID: 22001779 [DOI] [PubMed] [Google Scholar]
- 101.Okazaki IJ, Moss J. . Mono-ADP-ribosylation: a reversible posttranslational modification of proteins. Adv Pharmacol 1996; 35:247 - 80; http://dx.doi.org/ 10.1016/S1054-3589(08)60277-X; PMID: 8920207 [DOI] [PubMed] [Google Scholar]
- 102.Jorgensen PL, Pedersen PA. . Structure-function relationships of Na(+), K(+), ATP, or Mg(2+) binding and energy transduction in Na,K-ATPase. Biochim Biophys Acta 2001; 1505:57 - 74; http://dx.doi.org/ 10.1016/S0005-2728(00)00277-2; PMID: 11248189 [DOI] [PubMed] [Google Scholar]
- 103.Shaanan B, Chipman DM. . Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. FEBS J 2009; 276:2447 - 53; http://dx.doi.org/ 10.1111/j.1742-4658.2009.06965.x; PMID: 19476486 [DOI] [PubMed] [Google Scholar]
- 104.Sbardella D, Fasciglione GF, Gioia M, Ciaccio C, Tundo GR, Marini S, et al. . Human matrix metalloproteinases: an ubiquitarian class of enzymes involved in several pathological processes. Mol Aspects Med 2012; 33:119 - 208; http://dx.doi.org/ 10.1016/j.mam.2011.10.015; PMID: 22100792 [DOI] [PubMed] [Google Scholar]
- 105.Taylor KM, Nicholson RI. . The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta 2003; 1611:16 - 30; http://dx.doi.org/ 10.1016/S0005-2736(03)00048-8; PMID: 12659941 [DOI] [PubMed] [Google Scholar]
- 106.Beintema JJ, Terwisscha van Scheltinga AC. . Plant lysozymes. EXS 1996; 75:75 - 86; http://dx.doi.org/ 10.1007/978-3-0348-9225-4_5; PMID: 8765295 [DOI] [PubMed] [Google Scholar]
- 107.MacGregor EA, Janecek S, Svensson B. . Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochim Biophys Acta 2001; 1546:1 - 20; http://dx.doi.org/ 10.1016/S0167-4838(00)00302-2; PMID: 11257505 [DOI] [PubMed] [Google Scholar]
- 108.Daniels DS, Tainer JA. . Conserved structural motifs governing the stoichiometric repair of alkylated DNA by O(6)-alkylguanine-DNA alkyltransferase. Mutat Res 2000; 460:151 - 63; http://dx.doi.org/ 10.1016/S0921-8777(00)00024-0; PMID: 10946226 [DOI] [PubMed] [Google Scholar]
- 109.Jedrzejas MJ. . Three-dimensional structure and molecular mechanism of novel enzymes of spore-forming bacteria. Med Sci Monit 2002; 8:RA183 - 90; PMID: 12165756 [PubMed] [Google Scholar]
- 110.Muñoz-Clares RA, González-Segura L, Díaz-Sánchez AG. . Crystallographic evidence for active-site dynamics in the hydrolytic aldehyde dehydrogenases. Implications for the deacylation step of the catalyzed reaction. Chem Biol Interact 2011; 191:137 - 46; http://dx.doi.org/ 10.1016/j.cbi.2010.12.024; PMID: 21195066 [DOI] [PubMed] [Google Scholar]
- 111.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, et al. . Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys 2005; 433:129 - 43; http://dx.doi.org/ 10.1016/j.abb.2004.08.017; PMID: 15581572 [DOI] [PubMed] [Google Scholar]
- 112.Mauk MR, Smith A, Mauk AG. . An alternative view of the proposed alternative activities of hemopexin. Protein Sci 2011; 20:791 - 805; http://dx.doi.org/ 10.1002/pro.616; PMID: 21404362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Han M, Smith SO. . High-resolution structural studies of the retinal--Glu113 interaction in rhodopsin. Biophys Chem 1995; 56:23 - 9; http://dx.doi.org/ 10.1016/0301-4622(95)00011-L; PMID: 7662866 [DOI] [PubMed] [Google Scholar]
- 114.Rosenkilde MM, Schwartz TW. . GluVII:06--a highly conserved and selective anchor point for non-peptide ligands in chemokine receptors. Curr Top Med Chem 2006; 6:1319 - 33; http://dx.doi.org/ 10.2174/15680266106061319; PMID: 16918451 [DOI] [PubMed] [Google Scholar]
- 115.Bella J, Berman HM. . Integrin-collagen complex: a metal-glutamate handshake. Structure 2000; 8:R121 - 6; http://dx.doi.org/ 10.1016/S0969-2126(00)00153-2; PMID: 10873865 [DOI] [PubMed] [Google Scholar]
- 116.Bella J, Kolatkar PR, Marlor CW, Greve JM, Rossmann MG. . The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc Natl Acad Sci U S A 1998; 95:4140 - 5; http://dx.doi.org/ 10.1073/pnas.95.8.4140; PMID: 9539703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. . Structural basis of collagen recognition by integrin alpha2beta1. Cell 2000; 101:47 - 56; http://dx.doi.org/ 10.1016/S0092-8674(00)80622-4; PMID: 10778855 [DOI] [PubMed] [Google Scholar]
- 118.Arnaout MA, Goodman SL, Xiong JP. . Coming to grips with integrin binding to ligands. Curr Opin Cell Biol 2002; 14:641 - 51; http://dx.doi.org/ 10.1016/S0955-0674(02)00371-X; PMID: 12231361 [DOI] [PubMed] [Google Scholar]
- 119.Vallee BL, Falchuk KH. . The biochemical basis of zinc physiology. Physiol Rev 1993; 73:79 - 118; PMID: 8419966 [DOI] [PubMed] [Google Scholar]
- 120.Yáñez M, Gil-Longo J, Campos-Toimil M. . Calcium binding proteins. Adv Exp Med Biol 2012; 740:461 - 82; http://dx.doi.org/ 10.1007/978-94-007-2888-2_19; PMID: 22453954 [DOI] [PubMed] [Google Scholar]
- 121.Veprintsev DB, Narayan M, Permyakov SE, Uversky VN, Brooks CL, Cherskaya AM, et al. . Fine tuning the N-terminus of a calcium binding protein: alpha-lactalbumin. Proteins 1999; 37:65 - 72; http://dx.doi.org/; PMID: 10451551 [DOI] [PubMed] [Google Scholar]
- 122.Permyakov EA, Morozova LA, Burstein EA. . Cation binding effects on the pH, thermal and urea denaturation transitions in alpha-lactalbumin. Biophys Chem 1985; 21:21 - 31; http://dx.doi.org/ 10.1016/0301-4622(85)85003-1; PMID: 3971025 [DOI] [PubMed] [Google Scholar]
- 123.Kuwajima K. . The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 1989; 6:87 - 103; http://dx.doi.org/ 10.1002/prot.340060202; PMID: 2695928 [DOI] [PubMed] [Google Scholar]
- 124.Permyakov EA, Berliner LJ. . alpha-Lactalbumin: structure and function. FEBS Lett 2000; 473:269 - 74; http://dx.doi.org/ 10.1016/S0014-5793(00)01546-5; PMID: 10818224 [DOI] [PubMed] [Google Scholar]
- 125.Permyakov SE, Makhatadze GI, Owenius R, Uversky VN, Brooks CL, Permyakov EA, et al. . How to improve nature: study of the electrostatic properties of the surface of alpha-lactalbumin. Protein Eng Des Sel 2005; 18:425 - 33; http://dx.doi.org/ 10.1093/protein/gzi051; PMID: 16093284 [DOI] [PubMed] [Google Scholar]
- 126.Rao CV, Ordal GW. . The molecular basis of excitation and adaptation during chemotactic sensory transduction in bacteria. Contrib Microbiol 2009; 16:33 - 64; http://dx.doi.org/ 10.1159/000219372; PMID: 19494578 [DOI] [PubMed] [Google Scholar]
- 127.Roberts MA, Papachristodoulou A, Armitage JP. . Adaptation and control circuits in bacterial chemotaxis. Biochem Soc Trans 2010; 38:1265 - 9; http://dx.doi.org/ 10.1042/BST0381265; PMID: 20863296 [DOI] [PubMed] [Google Scholar]
- 128.Wright HT. . Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol 1991; 26:1 - 52; http://dx.doi.org/ 10.3109/10409239109081719; PMID: 1678690 [DOI] [PubMed] [Google Scholar]
- 129.Melius P. . Structure of thermal polymers of amino acids. Biosystems 1982; 15:275 - 80; http://dx.doi.org/ 10.1016/0303-2647(82)90042-9; PMID: 6819870 [DOI] [PubMed] [Google Scholar]
- 130.Bandyopadhyay PK. . Vitamin K-dependent gamma-glutamylcarboxylation: an ancient posttranslational modification. Vitam Horm 2008; 78:157 - 84; http://dx.doi.org/ 10.1016/S0083-6729(07)00008-8; PMID: 18374194 [DOI] [PubMed] [Google Scholar]
- 131.Janke C, Rogowski K, van Dijk J. . Polyglutamylation: a fine-regulator of protein function? ‘Protein Modifications: beyond the usual suspects’ review series. EMBO Rep 2008; 9:636 - 41; http://dx.doi.org/ 10.1038/embor.2008.114; PMID: 18566597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tekaia F, Yeramian E, Dujon B. . Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene 2002; 297:51 - 60; http://dx.doi.org/ 10.1016/S0378-1119(02)00871-5; PMID: 12384285 [DOI] [PubMed] [Google Scholar]
- 133.Uversky VN, Gillespie JR, Millett IS, Khodyakova AV, Vasiliev AM, Chernovskaya TV, et al. . Natively unfolded human prothymosin alpha adopts partially folded collapsed conformation at acidic pH. Biochemistry 1999; 38:15009 - 16; http://dx.doi.org/ 10.1021/bi990752+; PMID: 10555983 [DOI] [PubMed] [Google Scholar]
- 134.Gast K, Damaschun H, Eckert K, Schulze-Forster K, Maurer HR, Müller-Frohne M, et al. . Prothymosin alpha: a biologically active protein with random coil conformation. Biochemistry 1995; 34:13211 - 8; http://dx.doi.org/ 10.1021/bi00040a037; PMID: 7548085 [DOI] [PubMed] [Google Scholar]
- 135.Candela T, Fouet A. . Poly-gamma-glutamate in bacteria. Mol Microbiol 2006; 60:1091 - 8; http://dx.doi.org/ 10.1111/j.1365-2958.2006.05179.x; PMID: 16689787 [DOI] [PubMed] [Google Scholar]
- 136.Fasman GD, Lindblow C, Bondenheimer E. . Conformational Studies on Synthetic Poly-Alpha-Amino Acids: Factors Influencing the Stability of the Helical Conformation of Poly-L-Glutamic Acid and Copolymers of L-Glutamic Acid and L-Leucine. Biochemistry 1964; 3:155 - 66; http://dx.doi.org/ 10.1021/bi00890a004; PMID: 14165017 [DOI] [PubMed] [Google Scholar]
- 137.Sage HJ, Fasman GD. . Conformational studies on poly-L-glutamic acid and copolymers of L-glutamic acid and L-phenylalanine. Biochemistry 1966; 5:286 - 96; http://dx.doi.org/ 10.1021/bi00865a037; PMID: 5938944 [DOI] [PubMed] [Google Scholar]
- 138.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas’ AF, Gilmanshin RI. . Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 1991; 31:119 - 28; http://dx.doi.org/ 10.1002/bip.360310111; PMID: 2025683 [DOI] [PubMed] [Google Scholar]
- 139.Ismail AA, Mantsch HH. . Salt bridge induced changes in the secondary structure of ionic polypeptides. Biopolymers 1992; 32:1181 - 6; http://dx.doi.org/ 10.1002/bip.360320907; PMID: 1384749 [DOI] [PubMed] [Google Scholar]
- 140.Fisher SL. . Glutamate racemase as a target for drug discovery. Microb Biotechnol 2008; 1:345 - 60; http://dx.doi.org/ 10.1111/j.1751-7915.2008.00031.x; PMID: 21261855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Candela T, Mock M, Fouet A. . CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol 2005; 187:7765 - 72; http://dx.doi.org/ 10.1128/JB.187.22.7765-7772.2005; PMID: 16267300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Candela T, Moya M, Haustant M, Fouet A. . Fusobacterium nucleatum, the first Gram-negative bacterium demonstrated to produce polyglutamate. Can J Microbiol 2009; 55:627 - 32; http://dx.doi.org/ 10.1139/W09-003; PMID: 19483793 [DOI] [PubMed] [Google Scholar]
- 143.Tomcsik J, Szongott H. . Ueber ein spezifisches Protein der Kapsel des Milzbrandbazillus. Zeitschr Immunität-Forsch 1933; 78:86 - 99 [Google Scholar]
- 144.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, et al. . Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest 2005; 115:688 - 94; PMID: 15696197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zwartouw HT, Smith H. . Polyglutamic acid from Bacillus anthracis grown in vivo; structure and aggressin activity. Biochem J 1956; 63:437 - 42; PMID: 13341899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McLean RJ, Beauchemin D, Clapham L, Beveridge TJ. . Metal-binding characteristics of the gamma-glutamyl capsular polymer of Bacillus licheniformis ATCC 9945. Appl Environ Microbiol 1990; 56:3671 - 7; PMID: 16348371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kimura K, Tran LS, Uchida I, Itoh Y. . Characterization of Bacillus subtilis gamma-glutamyltransferase and its involvement in the degradation of capsule poly-gamma-glutamate. Microbiology 2004; 150:4115 - 23; http://dx.doi.org/ 10.1099/mic.0.27467-0; PMID: 15583164 [DOI] [PubMed] [Google Scholar]
- 148.Kandler O, König H, Wiegel J, Claus D. . Occurrence of Poly-γ-D-Glutamic Acid and Poly-α-L-Glutamine in the Genera Xanthobacter, Flexithrix, Sporosarcina and Planococcus. Syst Appl Microbiol 1983; 4:34 - 41; http://dx.doi.org/ 10.1016/S0723-2020(83)80032-0; PMID: 23196298 [DOI] [PubMed] [Google Scholar]
- 149.Hezayen FF, Rehm BH, Tindall BJ, Steinbüchel A. . Transfer of Natrialba asiatica B1T to Natrialba taiwanensis sp. nov. and description of Natrialba aegyptiaca sp. nov., a novel extremely halophilic, aerobic, non-pigmented member of the Archaea from Egypt that produces extracellular poly(glutamic acid). Int J Syst Evol Microbiol 2001; 51:1133 - 42; http://dx.doi.org/ 10.1099/00207713-51-3-1133; PMID: 11411682 [DOI] [PubMed] [Google Scholar]
- 150.Weber J. . Poly(gamma-glutamic acid)s are the major constituents of nematocysts in Hydra (Hydrozoa, Cnidaria). J Biol Chem 1990; 265:9664 - 9; PMID: 1693614 [PubMed] [Google Scholar]
- 151.Obst M, Steinbüchel A. . Microbial degradation of poly(amino acid)s. Biomacromolecules 2004; 5:1166 - 76; http://dx.doi.org/ 10.1021/bm049949u; PMID: 15244426 [DOI] [PubMed] [Google Scholar]
- 152.Bajaj I, Singhal R. . Poly (glutamic acid)--an emerging biopolymer of commercial interest. Bioresour Technol 2011; 102:5551 - 61; http://dx.doi.org/ 10.1016/j.biortech.2011.02.047; PMID: 21377358 [DOI] [PubMed] [Google Scholar]
- 153.Buescher JM, Margaritis A. . Microbial biosynthesis of polyglutamic acid biopolymer and applications in the biopharmaceutical, biomedical and food industries. Crit Rev Biotechnol 2007; 27:1 - 19; http://dx.doi.org/ 10.1080/07388550601166458; PMID: 17364686 [DOI] [PubMed] [Google Scholar]
- 154.Poo H, Park C, Kwak MS, Choi DY, Hong SP, Lee IH, et al. . New biological functions and applications of high-molecular-mass poly-gamma-glutamic acid. Chem Biodivers 2010; 7:1555 - 62; http://dx.doi.org/ 10.1002/cbdv.200900283; PMID: 20564573 [DOI] [PubMed] [Google Scholar]
- 155.Chan YP, Meyrueix R, Kravtzoff R, Nicolas F, Lundstrom K. . Review on Medusa:a polymer-based sustained release technology for protein and peptide drugs. Expert Opin Drug Deliv 2007; 4:441 - 51; http://dx.doi.org/ 10.1517/17425247.4.4.441; PMID: 17683256 [DOI] [PubMed] [Google Scholar]
- 156.Shih IL, Van YT, Shen MH. . Biomedical applications of chemically and microbiologically synthesized poly(glutamic acid) and poly(lysine). Mini Rev Med Chem 2004; 4:179 - 88; http://dx.doi.org/ 10.2174/1389557043487420; PMID: 14965290 [DOI] [PubMed] [Google Scholar]
- 157.Manocha B, Margaritis A. . Production and characterization of gamma-polyglutamic acid nanoparticles for controlled anticancer drug release. Crit Rev Biotechnol 2008; 28:83 - 99; http://dx.doi.org/ 10.1080/07388550802107483; PMID: 18568849 [DOI] [PubMed] [Google Scholar]
- 158.Chipman SD, Oldham FB, Pezzoni G, Singer JW. . Biological and clinical characterization of paclitaxel poliglumex (PPX, CT-2103), a macromolecular polymer-drug conjugate. Int J Nanomedicine 2006; 1:375 - 83; http://dx.doi.org/ 10.2147/nano.2006.1.4.375; PMID: 17722272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rechsteiner M, Rogers SW. . PEST sequences and regulation by proteolysis. Trends Biochem Sci 1996; 21:267 - 71; PMID: 8755249 [PubMed] [Google Scholar]
- 160.Rogers S, Wells R, Rechsteiner M. . Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 1986; 234:364 - 8; http://dx.doi.org/ 10.1126/science.2876518; PMID: 2876518 [DOI] [PubMed] [Google Scholar]
- 161.Singh GP, Ganapathi M, Sandhu KS, Dash D. . Intrinsic unstructuredness and abundance of PEST motifs in eukaryotic proteomes. Proteins 2006; 62:309 - 15; http://dx.doi.org/ 10.1002/prot.20746; PMID: 16299712 [DOI] [PubMed] [Google Scholar]
- 162.Belizario JE, Alves J, Garay-Malpartida M, Occhiucci JM. . Coupling caspase cleavage and proteasomal degradation of proteins carrying PEST motif. Curr Protein Pept Sci 2008; 9:210 - 20; http://dx.doi.org/ 10.2174/138920308784534023; PMID: 18537676 [DOI] [PubMed] [Google Scholar]
- 163.Hoh JH. . Functional protein domains from the thermally driven motion of polypeptide chains: a proposal. Proteins 1998; 32:223 - 8; http://dx.doi.org/; PMID: 9714161 [DOI] [PubMed] [Google Scholar]
- 164.Brown HG, Hoh JH. . Entropic exclusion by neurofilament sidearms: a mechanism for maintaining interfilament spacing. Biochemistry 1997; 36:15035 - 40; http://dx.doi.org/ 10.1021/bi9721748; PMID: 9424114 [DOI] [PubMed] [Google Scholar]
- 165.Geisler N, Fischer S, Vandekerckhove J, Plessmann U, Weber K. . Hybrid character of a large neurofilament protein (NF-M): intermediate filament type sequence followed by a long and acidic carboxy-terminal extension. EMBO J 1984; 3:2701 - 6; PMID: 6439558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Voter WA, Erickson HP. . Electron microscopy of MAP 2 (microtubule-associated protein 2). J Ultrastruct Res 1982; 80:374 - 82; http://dx.doi.org/ 10.1016/S0022-5320(82)80051-8; PMID: 7131650 [DOI] [PubMed] [Google Scholar]
- 167.Merdes A, Ramyar K, Vechio JD, Cleveland DW. . A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 1996; 87:447 - 58; http://dx.doi.org/ 10.1016/S0092-8674(00)81365-3; PMID: 8898198 [DOI] [PubMed] [Google Scholar]
- 168.Santner AA, Croy CH, Vasanwala FH, Uversky VN, Van YY, Dunker AK. . Sweeping away protein aggregation with entropic bristles: intrinsically disordered protein fusions enhance soluble expression. Biochemistry 2012; 51:7250 - 62; http://dx.doi.org/ 10.1021/bi300653m; PMID: 22924672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Narberhaus F. . Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. [table of contents.] Microbiol Mol Biol Rev 2002; 66:64 - 93; http://dx.doi.org/ 10.1128/MMBR.66.1.64-93.2002; PMID: 11875128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Park SM, Jung HY, Kim TD, Park JH, Yang CH, Kim J. . Distinct roles of the N-terminal-binding domain and the C-terminal-solubilizing domain of alpha-synuclein, a molecular chaperone. J Biol Chem 2002; 277:28512 - 20; http://dx.doi.org/ 10.1074/jbc.M111971200; PMID: 12032141 [DOI] [PubMed] [Google Scholar]
- 171.Jones LS, Yazzie B, Middaugh CR. . Polyanions and the proteome. Mol Cell Proteomics 2004; 3:746 - 69; http://dx.doi.org/ 10.1074/mcp.R400008-MCP200; PMID: 15143156 [DOI] [PubMed] [Google Scholar]
- 172.Volkin DB, Tsai PK, Dabora JM, Gress JO, Burke CJ, Linhardt RJ, et al. . Physical stabilization of acidic fibroblast growth factor by polyanions. Arch Biochem Biophys 1993; 300:30 - 41; http://dx.doi.org/ 10.1006/abbi.1993.1005; PMID: 7678726 [DOI] [PubMed] [Google Scholar]
- 173.Rentzeperis D, Jonsson T, Sauer RT. . Acceleration of the refolding of Arc repressor by nucleic acids and other polyanions. Nat Struct Biol 1999; 6:569 - 73; http://dx.doi.org/ 10.1038/9353; PMID: 10360363 [DOI] [PubMed] [Google Scholar]
- 174.Edwards KL, Kueltzo LA, Fisher MT, Middaugh CR. . Complex effects of molecular chaperones on the aggregation and refolding of fibroblast growth factor-1. Arch Biochem Biophys 2001; 393:14 - 21; http://dx.doi.org/ 10.1006/abbi.2001.2472; PMID: 11516157 [DOI] [PubMed] [Google Scholar]
- 175.Hingorani K, Szebeni A, Olson MO. . Mapping the functional domains of nucleolar protein B23. J Biol Chem 2000; 275:24451 - 7; http://dx.doi.org/ 10.1074/jbc.M003278200; PMID: 10829026 [DOI] [PubMed] [Google Scholar]
- 176.Manna T, Sarkar T, Poddar A, Roychowdhury M, Das KP, Bhattacharyya B. . Chaperone-like activity of tubulin. binding and reactivation of unfolded substrate enzymes. J Biol Chem 2001; 276:39742 - 7; http://dx.doi.org/ 10.1074/jbc.M104061200; PMID: 11509563 [DOI] [PubMed] [Google Scholar]
- 177.Sarkar T, Manna T, Bhattacharyya S, Mahapatra P, Poddar A, Roy S, et al. . Role of the carboxy-termini of tubulin on its chaperone-like activity. Proteins 2001; 44:262 - 9; http://dx.doi.org/ 10.1002/prot.1091; PMID: 11455599 [DOI] [PubMed] [Google Scholar]
- 178.Guha S, Manna TK, Das KP, Bhattacharyya B. . Chaperone-like activity of tubulin. J Biol Chem 1998; 273:30077 - 80; http://dx.doi.org/ 10.1074/jbc.273.46.30077; PMID: 9804759 [DOI] [PubMed] [Google Scholar]
- 179.Wu Z, Powell R, Lu W. . Productive folding of human neutrophil alpha-defensins in vitro without the pro-peptide. J Am Chem Soc 2003; 125:2402 - 3; http://dx.doi.org/ 10.1021/ja0294257; PMID: 12603122 [DOI] [PubMed] [Google Scholar]
- 180.Zhu XL, Ohta Y, Jordan F, Inouye M. . Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature 1989; 339:483 - 4; http://dx.doi.org/ 10.1038/339483a0; PMID: 2657436 [DOI] [PubMed] [Google Scholar]
- 181.Silen JL, Agard DA. . The alpha-lytic protease pro-region does not require a physical linkage to activate the protease domain in vivo. Nature 1989; 341:462 - 4; http://dx.doi.org/ 10.1038/341462a0; PMID: 2507926 [DOI] [PubMed] [Google Scholar]
- 182.Silen JL, Frank D, Fujishige A, Bone R, Agard DA. . Analysis of prepro-alpha-lytic protease expression in Escherichia coli reveals that the pro region is required for activity. J Bacteriol 1989; 171:1320 - 5; PMID: 2646278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zenk MH. . Heavy metal detoxification in higher plants--a review. Gene 1996; 179:21 - 30; http://dx.doi.org/ 10.1016/S0378-1119(96)00422-2; PMID: 8955625 [DOI] [PubMed] [Google Scholar]
- 184.Fraser LR. . Modulation of mammalian sperm function by fertilization promoting peptide (FPP). Andrologia 1998; 30:241 - 7; http://dx.doi.org/ 10.1111/j.1439-0272.1998.tb01166.x; PMID: 9739421 [DOI] [PubMed] [Google Scholar]
- 185.Li W, Nicol F, Szoka FC Jr.. . GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev 2004; 56:967 - 85; http://dx.doi.org/ 10.1016/j.addr.2003.10.041; PMID: 15066755 [DOI] [PubMed] [Google Scholar]
- 186.Veillette A, Rhee I, Souza CM, Davidson D. . PEST family phosphatases in immunity, autoimmunity, and autoinflammatory disorders. Immunol Rev 2009; 228:312 - 24; http://dx.doi.org/ 10.1111/j.1600-065X.2008.00747.x; PMID: 19290936 [DOI] [PubMed] [Google Scholar]
- 187.Zheng Y, Lu Z. . Regulation of tumor cell migration by protein tyrosine phosphatase (PTP)-proline-, glutamate-, serine-, and threonine-rich sequence (PEST). Chin J Cancer 2013; 32:75 - 83; PMID: 23237212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.López de Saro FJ, Woody AY, Helmann JD. . Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J Mol Biol 1995; 252:189 - 202; http://dx.doi.org/ 10.1006/jmbi.1995.0487; PMID: 7545758 [DOI] [PubMed] [Google Scholar]
- 189.Ramsden JJ. . MARCKS: a case of molecular exaptation?. Int J Biochem Cell Biol 2000; 32:475 - 9; http://dx.doi.org/ 10.1016/S1357-2725(99)00152-1; PMID: 10736562 [DOI] [PubMed] [Google Scholar]
- 190.Zhang D, Jiang P, Xu Q, Zhang X. . Arginine and glutamate-rich 1 (ARGLU1) interacts with mediator subunit 1 (MED1) and is required for estrogen receptor-mediated gene transcription and breast cancer cell growth. J Biol Chem 2011; 286:17746 - 54; http://dx.doi.org/ 10.1074/jbc.M110.206029; PMID: 21454576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Brann DW, Zhang QG, Wang RM, Mahesh VB, Vadlamudi RK. . PELP1--a novel estrogen receptor-interacting protein. Mol Cell Endocrinol 2008; 290:2 - 7; http://dx.doi.org/ 10.1016/j.mce.2008.04.019; PMID: 18571832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nagpal JK, Nair S, Chakravarty D, Rajhans R, Pothana S, Brann DW, et al. . Growth factor regulation of estrogen receptor coregulator PELP1 functions via Protein Kinase A pathway. Mol Cancer Res 2008; 6:851 - 61; http://dx.doi.org/ 10.1158/1541-7786.MCR-07-2030; PMID: 18505929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Das S, Maitra U. . Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol 2001; 70:207 - 31; http://dx.doi.org/ 10.1016/S0079-6603(01)70018-9; PMID: 11642363 [DOI] [PubMed] [Google Scholar]
- 194.Traub P, Perides G, Kühn S, Scherbarth A. . Interaction in vitro of non-epithelial intermediate filament proteins with histones. Z Naturforsch C 1987; 42:47 - 63; PMID: 2953133 [DOI] [PubMed] [Google Scholar]
- 195.Heads RJ, Carpenter BG. . Isolation, characterisation and growth-related changes of an HMG-like protein from microplasmodia of Physarum polycephalum. Biochim Biophys Acta 1991; 1079:15 - 22; http://dx.doi.org/ 10.1016/0167-4838(91)90018-U; PMID: 1888760 [DOI] [PubMed] [Google Scholar]
- 196.Labeit D, Watanabe K, Witt C, Fujita H, Wu Y, Lahmers S, et al. . Calcium-dependent molecular spring elements in the giant protein titin. Proc Natl Acad Sci U S A 2003; 100:13716 - 21; http://dx.doi.org/ 10.1073/pnas.2235652100; PMID: 14593205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Prince CW, Oosawa T, Butler WT, Tomana M, Bhown AS, Bhown M, et al. . Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J Biol Chem 1987; 262:2900 - 7; PMID: 3469201 [PubMed] [Google Scholar]
- 198.Hunter GK, Goldberg HA. . Modulation of crystal formation by bone phosphoproteins: role of glutamic acid-rich sequences in the nucleation of hydroxyapatite by bone sialoprotein. Biochem J 1994; 302:175 - 9; PMID: 7915111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fujisawa R, Wada Y, Nodasaka Y, Kuboki Y. . Acidic amino acid-rich sequences as binding sites of osteonectin to hydroxyapatite crystals. Biochim Biophys Acta 1996; 1292:53 - 60; http://dx.doi.org/ 10.1016/0167-4838(95)00190-5; PMID: 8547349 [DOI] [PubMed] [Google Scholar]
- 200.Damjanovski S, Liu F, Ringuette M. . Molecular analysis of Xenopus laevis SPARC (Secreted Protein, Acidic, Rich in Cysteine). A highly conserved acidic calcium-binding extracellular-matrix protein. Biochem J 1992; 281:513 - 7; PMID: 1736898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Koehler A, Desser S, Chang B, MacDonald J, Tepass U, Ringuette M. . Molecular evolution of SPARC: absence of the acidic module and expression in the endoderm of the starlet sea anemone, Nematostella vectensis. Dev Genes Evol 2009; 219:509 - 21; http://dx.doi.org/ 10.1007/s00427-009-0313-9; PMID: 20043230 [DOI] [PubMed] [Google Scholar]
- 202.Shirakawa H, Landsman D, Postnikov YV, Bustin M. . NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J Biol Chem 2000; 275:6368 - 74; http://dx.doi.org/ 10.1074/jbc.275.9.6368; PMID: 10692437 [DOI] [PubMed] [Google Scholar]
- 203.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, et al. . The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol Cell 2009; 35:642 - 56; http://dx.doi.org/ 10.1016/j.molcel.2009.07.002; PMID: 19748358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Haber-Pohlmeier S, Abarca-Heidemann K, Körschen HG, Dhiman HK, Heberle J, Schwalbe H, et al. . Binding of Ca2+ to glutamic acid-rich polypeptides from the rod outer segment. Biophys J 2007; 92:3207 - 14; http://dx.doi.org/ 10.1529/biophysj.106.094847; PMID: 17218469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Michalakis S, Zong X, Becirovic E, Hammelmann V, Wein T, Wanner KT, et al. . The glutamic acid-rich protein is a gating inhibitor of cyclic nucleotide-gated channels. J Neurosci 2011; 31:133 - 41; http://dx.doi.org/ 10.1523/JNEUROSCI.4735-10.2011; PMID: 21209198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ritter LM, Khattree N, Tam B, Moritz OL, Schmitz F, Goldberg AF. . In situ visualization of protein interactions in sensory neurons: glutamic acid-rich proteins (GARPs) play differential roles for photoreceptor outer segment scaffolding. J Neurosci 2011; 31:11231 - 43; http://dx.doi.org/ 10.1523/JNEUROSCI.2875-11.2011; PMID: 21813684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang Y, Rubin GR, Fineberg N, Huisingh C, McGwin G, Pittler SJ, et al. . Age-related changes in Cngb1-X1 knockout mice: prolonged cone survival. Doc Ophthalmol 2012; 124:163 - 75; http://dx.doi.org/ 10.1007/s10633-012-9317-2; PMID: 22367173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Batra-Safferling R, Abarca-Heidemann K, Körschen HG, Tziatzios C, Stoldt M, Budyak I, et al. . Glutamic acid-rich proteins of rod photoreceptors are natively unfolded. J Biol Chem 2006; 281:1449 - 60; http://dx.doi.org/ 10.1074/jbc.M505012200; PMID: 16280326 [DOI] [PubMed] [Google Scholar]
- 209.Qi S, Wang Y, Zhou M, Ge Y, Yan Y, Wang J, et al. . A mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is highly expressed in retina that exhibits a large area of intrinsic disorder. Mol Biol Rep 2011; 38:2869 - 77; http://dx.doi.org/ 10.1007/s11033-010-9948-x; PMID: 20107910 [DOI] [PubMed] [Google Scholar]
- 210.Zhou M, Wang Y, Qi S, Wang J, Zhang S. . The expression of a mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is under the regulation of the HPG axis. Endocrinology 2011; 152:2311 - 20; http://dx.doi.org/ 10.1210/en.2011-0050; PMID: 21447634 [DOI] [PubMed] [Google Scholar]
- 211.Matsumoto T, Minegishi K, Ishimoto H, Tanaka M, Hennebold JD, Teranishi T, et al. . Expression of ovary-specific acidic protein in steroidogenic tissues: a possible role in steroidogenesis. Endocrinology 2009; 150:3353 - 9; http://dx.doi.org/ 10.1210/en.2008-1584; PMID: 19325000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li Y, Lim S, Hoffman D, Aspenstrom P, Federoff HJ, Rempe DA. . HUMMR, a hypoxia- and HIF-1alpha-inducible protein, alters mitochondrial distribution and transport. J Cell Biol 2009; 185:1065 - 81; http://dx.doi.org/ 10.1083/jcb.200811033; PMID: 19528298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gao F, Zhou BJ, Li GY, Jia PS, Li H, Zhao YL, et al. . A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS One 2010; 5:e15319; http://dx.doi.org/ 10.1371/journal.pone.0015319; PMID: 21151869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Xu J, Wang W, Chai BF, Liang AH. . [Cloning and characterization of a novel trinucleotide repeat-containing gene GARP from Euplotes octocarinatus]. Yi Chuan 2007; 29:87 - 91; http://dx.doi.org/ 10.1360/yc-007-0087; PMID: 17284430 [DOI] [PubMed] [Google Scholar]
- 215.Triglia T, Stahl HD, Crewther PE, Silva A, Anders RF, Kemp DJ. . Structure of a Plasmodium falciparum gene that encodes a glutamic acid-rich protein (GARP). Mol Biochem Parasitol 1988; 31:199 - 201; http://dx.doi.org/ 10.1016/0166-6851(88)90170-3; PMID: 2903445 [DOI] [PubMed] [Google Scholar]
- 216.Kang YL, Li H, Chen WH, Tzeng YS, Lai YL, Hsieh-Li HM. . A novel PEPP homeobox gene, TOX, is highly glutamic acid rich and specifically expressed in murine testis and ovary. Biol Reprod 2004; 70:828 - 36; http://dx.doi.org/ 10.1095/biolreprod.103.021048; PMID: 14627546 [DOI] [PubMed] [Google Scholar]
- 217.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. . Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 2010; 18:288 - 99; http://dx.doi.org/ 10.1016/j.devcel.2009.12.012; PMID: 20159598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. . Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 2010; 18:300 - 8; http://dx.doi.org/ 10.1016/j.devcel.2009.12.011; PMID: 20159599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, et al. . Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun 2003; 300:862 - 7; http://dx.doi.org/ 10.1016/S0006-291X(02)02945-5; PMID: 12559952 [DOI] [PubMed] [Google Scholar]
- 220.Büther K, Plaas C, Barnekow A, Kremerskothen J. . KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun 2004; 317:703 - 7; http://dx.doi.org/ 10.1016/j.bbrc.2004.03.107; PMID: 15081397 [DOI] [PubMed] [Google Scholar]
- 221.Johannsen S, Duning K, Pavenstädt H, Kremerskothen J, Boeckers TM. . Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience 2008; 155:1165 - 73; http://dx.doi.org/ 10.1016/j.neuroscience.2008.06.054; PMID: 18672031 [DOI] [PubMed] [Google Scholar]
- 222.Rayala SK, den Hollander P, Manavathi B, Talukder AH, Song C, Peng S, et al. . Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J Biol Chem 2006; 281:19092 - 9; http://dx.doi.org/ 10.1074/jbc.M600021200; PMID: 16684779 [DOI] [PubMed] [Google Scholar]
- 223.Hilton HN, Stanford PM, Harris J, Oakes SR, Kaplan W, Daly RJ, et al. . KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim Biophys Acta 2008; 1783:383 - 93; http://dx.doi.org/ 10.1016/j.bbamcr.2007.12.007; PMID: 18190796 [DOI] [PubMed] [Google Scholar]
- 224.Duning K, Schurek EM, Schlüter M, Bayer M, Reinhardt HC, Schwab A, et al. . KIBRA modulates directional migration of podocytes. J Am Soc Nephrol 2008; 19:1891 - 903; http://dx.doi.org/ 10.1681/ASN.2007080916; PMID: 18596123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Scartezzini P, Egeo A, Colella S, Fumagalli P, Arrigo P, Nizetic D, et al. . Cloning a new human gene from chromosome 21q22.3 encoding a glutamic acid-rich protein expressed in heart and skeletal muscle. Hum Genet 1997; 99:387 - 92; http://dx.doi.org/ 10.1007/s004390050377; PMID: 9050928 [DOI] [PubMed] [Google Scholar]
- 226.Mazzocco M, Maffei M, Egeo A, Vergano A, Arrigo P, Di Lisi R, et al. . The identification of a novel human homologue of the SH3 binding glutamic acid-rich (SH3BGR) gene establishes a new family of highly conserved small proteins related to Thioredoxin Superfamily. Gene 2002; 291:233 - 9; http://dx.doi.org/ 10.1016/S0378-1119(02)00602-9; PMID: 12095696 [DOI] [PubMed] [Google Scholar]
- 227.Khalil AA. . Biomarker discovery: a proteomic approach for brain cancer profiling. Cancer Sci 2007; 98:201 - 13; http://dx.doi.org/ 10.1111/j.1349-7006.2007.00374.x; PMID: 17233837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Takano M, Maekura K, Otani M, Sano K, Nakamura-Hirota T, Tokuyama S, et al. . Proteomic analysis of the brain tissues from a transgenic mouse model of amyloid β oligomers. Neurochem Int 2012; 61:347 - 55; http://dx.doi.org/ 10.1016/j.neuint.2012.05.018; PMID: 22634250 [DOI] [PubMed] [Google Scholar]
- 229.Kushwaha A, Perween A, Mukund S, Majumdar S, Bhardwaj D, Chowdhury NR, et al. . Amino terminus of Plasmodium falciparum acidic basic repeat antigen interacts with the erythrocyte membrane through band 3 protein. Mol Biochem Parasitol 2002; 122:45 - 54; http://dx.doi.org/ 10.1016/S0166-6851(02)00077-4; PMID: 12076769 [DOI] [PubMed] [Google Scholar]
- 230.Kushwaha A, Rao PP, Suresh RP, Chauhan VS. . Immunogenicity of recombinant fragments of Plasmodium falciparum acidic basic repeat antigen produced in Escherichia coli. Parasite Immunol 2001; 23:435 - 44; http://dx.doi.org/ 10.1046/j.1365-3024.2001.00390.x; PMID: 11489167 [DOI] [PubMed] [Google Scholar]
- 231.Nwagwu M, Haynes JD, Orlandi PA, Chulay JD. . Plasmodium falciparum: chymotryptic-like proteolysis associated with a 101-kDa acidic-basic repeat antigen. Exp Parasitol 1992; 75:399 - 414; http://dx.doi.org/ 10.1016/0014-4894(92)90253-7; PMID: 1493872 [DOI] [PubMed] [Google Scholar]
- 232.Kushwaha A, Rao PP, Duttu VS, Malhotra P, Chauhan VS. . Expression and characterisation of Plasmodium falciparum acidic basic repeat antigen expressed in Escherichia coli. Mol Biochem Parasitol 2000; 106:213 - 24; http://dx.doi.org/ 10.1016/S0166-6851(99)00212-1; PMID: 10699251 [DOI] [PubMed] [Google Scholar]
- 233.Santi-Rocca J, Weber C, Guigon G, Sismeiro O, Coppée JY, Guillén N. . The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell Microbiol 2008; 10:202 - 17; PMID: 17711481 [DOI] [PubMed] [Google Scholar]
- 234.Ossevoort M, Zaldumbide A, te Velthuis AJ, Melchers M, Ressing ME, Wiertz EJ, et al. . The nested open reading frame in the Epstein-Barr virus nuclear antigen-1 mRNA encodes a protein capable of inhibiting antigen presentation in cis. Mol Immunol 2007; 44:3588 - 96; http://dx.doi.org/ 10.1016/j.molimm.2007.03.005; PMID: 17449101 [DOI] [PubMed] [Google Scholar]
- 235.Gao J, Coulson JM, Whitehouse A, Blake N. . Reduction in RNA levels rather than retardation of translation is responsible for the inhibition of major histocompatibility complex class I antigen presentation by the glutamic acid-rich repeat of herpesvirus saimiri open reading frame 73. J Virol 2009; 83:273 - 82; http://dx.doi.org/ 10.1128/JVI.01532-08; PMID: 18945762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Serrano P, Johnson MA, Almeida MS, Horst R, Herrmann T, Joseph JS, et al. . Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J Virol 2007; 81:12049 - 60; http://dx.doi.org/ 10.1128/JVI.00969-07; PMID: 17728234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Deb R, Goswami PP. . Expression of a Gene Encoding 34.9 kDa PPE Antigen of Mycobacterium avium subsp. paratuberculosis in E. coli. Mol Biol Int 2010; 2010:628153; http://dx.doi.org/ 10.4061/2010/628153; PMID: 22110958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.de Souza CR, Aragão FJ, Moreira EC, Costa CN, Nascimento SB, Carvalho LJ. . Isolation and characterization of the promoter sequence of a cassava gene coding for Pt2L4, a glutamic acid-rich protein differentially expressed in storage roots. Genet Mol Res 2009; 8:334 - 44; http://dx.doi.org/ 10.4238/vol8-1gmr560; PMID: 19440969 [DOI] [PubMed] [Google Scholar]
- 239.Owiti J, Grossmann J, Gehrig P, Dessimoz C, Laloi C, Hansen MB, et al. . iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J 2011; 67:145 - 56; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04582.x; PMID: 21435052 [DOI] [PubMed] [Google Scholar]
- 240.Oliver D, Sheehan B, South H, Akbari O, Pai CY. . The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol 2010; 11:101; http://dx.doi.org/ 10.1186/1471-2121-11-101; PMID: 21194420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Morgan MA, Morgan JI. . Pcp4l1 contains an auto-inhibitory element that prevents its IQ motif from binding to calmodulin. J Neurochem 2012; 121:843 - 51; http://dx.doi.org/ 10.1111/j.1471-4159.2012.07745.x; PMID: 22458599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fontenot AP, Maier LA. . Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol 2005; 26:543 - 9; http://dx.doi.org/ 10.1016/j.it.2005.08.004; PMID: 16099719 [DOI] [PubMed] [Google Scholar]
- 243.Ferrone FA, Rotter MA. . Crowding and the polymerization of sickle hemoglobin. J Mol Recognit 2004; 17:497 - 504; http://dx.doi.org/ 10.1002/jmr.698; PMID: 15362110 [DOI] [PubMed] [Google Scholar]
- 244.Hartong DT, Berson EL, Dryja TP. . Retinitis pigmentosa. Lancet 2006; 368:1795 - 809; http://dx.doi.org/ 10.1016/S0140-6736(06)69740-7; PMID: 17113430 [DOI] [PubMed] [Google Scholar]
- 245.Shu X, Black GC, Rice JM, Hart-Holden N, Jones A, O’Grady A, et al. . RPGR mutation analysis and disease: an update. Hum Mutat 2007; 28:322 - 8; http://dx.doi.org/ 10.1002/humu.20461; PMID: 17195164 [DOI] [PubMed] [Google Scholar]
- 246.Wollina U, Haroske G. . Pyoderma gangraenosum. Curr Opin Rheumatol 2011; 23:50 - 6; http://dx.doi.org/ 10.1097/BOR.0b013e328341152f; PMID: 21037478 [DOI] [PubMed] [Google Scholar]
- 247.Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S, et al. . Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum Mol Genet 2002; 11:961 - 9; http://dx.doi.org/ 10.1093/hmg/11.8.961; PMID: 11971877 [DOI] [PubMed] [Google Scholar]
- 248.Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, et al. . Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A 2003; 100:13501 - 6; http://dx.doi.org/ 10.1073/pnas.2135380100; PMID: 14595024 [DOI] [PMC free article] [PubMed] [Google Scholar]