Abstract
Bacterial biofilms are structured communities of cells enclosed in a self-produced hydrated polymeric matrix that can adhere to inert or living surfaces. D-Amino acids were previously identified as self-produced compounds that mediate biofilm disassembly by causing the release of the protein component of the polymeric matrix. However, whether exogenous D-amino acids could inhibit initial bacterial adhesion is still unknown. Here, the effect of the exogenous amino acid D-tyrosine on initial bacterial adhesion was determined by combined use of chemical analysis, force spectroscopic measurement, and theoretical predictions. The surface thermodynamic theory demonstrated that the total interaction energy increased with more D-tyrosine, and the contribution of Lewis acid–base interactions relative to the change in the total interaction energy was much greater than the overall nonspecific interactions. Finally, atomic force microscopy analysis implied that the hydrogen bond numbers and adhesion forces decreased with the increase in D-tyrosine concentrations. D-Tyrosine contributed to the repulsive nature of the cell and ultimately led to the inhibition of bacterial adhesion. This study provides a new way to regulate biofilm formation by manipulating the contents of D-amino acids in natural or engineered systems.
Keywords: bacterial adhesion, D-tyrosine, extracellular polymeric substances, thermodynamics, biofilm
Introduction
Bacterial adhesion to solid surfaces is an important issue in a number of disciplines, ranging from biomedical engineering to wastewater treatment technologies (Costerton et al., 1999; Hall-Stoodley et al., 2004; Xu and Liu 2011b). Many microbes naturally grow as biofilms and may have beneficial traits for cleanup of wastewater, off-gas, soils, etc. (Ralebitso-Senior et al., 2012; Xu and Liu 2011b). They are also known for their ability to convert agriculturally derived materials to products of commercial value such as alcohols and organic acids (Rosche et al., 2009). Although certain biofilms can be beneficial, other biofilms pose severe problems, such as the corrosion and obstruction of water pipes or fouling of membranes in filtration systems (Hori and Matsumoto 2010; Xu and Liu 2011a). Thus, understanding the mechanism of biofilm formation and effectively regulating its formation and disassembly for human purposes is important for many biotechnology fields.
Biofilms are aggregates of microbes embedded in a matrix of extracellular polymeric substances (EPS), which consist of different types of biopolymers including polysaccharides, proteins, lipids, and extracellular DNA (Flemming and Wingender 2010; Pope et al., 2011). Found on almost all types of submerged surfaces, biofilms are a majority of the biomass in natural and engineered systems (Lee et al., 2011; Peulen and Wilkinson 2011). During biofilm formation, bacterial adhesion onto abiotic and biotic surfaces is the key determinant in initial cell attachment (Razatos et al., 1998). The physicochemical factors of bacterial cells and the surfaces govern this critical initial step in biofilm formation processes (Herzberg and Elimelech 2007; Lutterodt et al., 2009; Nielsen et al., 2011).
Biofilm formation requires several physiological and physicochemical steps (Bos et al., 1999). Bacterial adhesion has been shown to be a function of the physicochemical and thermodynamic properties of both bacterial surfaces and substrates based on analysis of surface charge and hydrophilicity measurements (i.e., the cell surface free energy); both parameters are significantly influenced by EPS content on bacterial surfaces (Harvey et al., 2011; Sheng and Yu 2006; Sheng et al., 2010; Terada et al., 2012; Wang et al., 2012). Recently, it has been reported that some molecular signals govern quorum-sensing, which serves as a switching mechanism between successive phases of biofilm development (Davies et al., 1998; McDougald et al., 2012). As one of the quorum-sensing molecules, self-produced D-amino acids (D-AAs), can promote biofilm disassembly and reduce microbial attachment (Kolodkin-Gal et al., 2010; Xu and Liu 2011a; Xu and Liu 2011b). In natural environments and engineered systems, various species of bacteria interact with each other, with the majority of excreted amino acids in the L-isomer form, with only some bacteria excreting D-AAs. These excreted D-AAs may affect other bacterial species in the microbial community and cause biofilm disruption. The precise role of exogenous D-AAs in the initial bacterial adhesion to substratum surface is not clear yet, and how D-AAs trigger variations in surface interactions of bacteria still needs to be revealed.
Initial bacterial adhesion is generally determined by the Derjaguin–Landau–Verwey–Overbeek (DLVO) theory, which is expressed as the sum of Lifshitz–van der Waals interactions and electrostatic interactions (Feriancikova et al., 2013). The classical DLVO theory was projected to describe the stability of colloidal suspensions. It involves the estimation of the energy from the electrostatic double-layer force and an attractive Lifshitz–van der Waals force (in the case of two identical surfaces). The Lewis acid– base interaction has been added to the DLVO theory (extended DLVO theory, XDLVO) to reconcile the discrepancy between the theoretical prediction and experimental results (Chen and Strevett 2001; Chia et al., 2011). In addition to theoretical predictions, experimental studies regarding interactions between bacteria and substrates have been conducted. For instance, atomic force microscopy (AFM) has been utilized for surface adhesion force measurement for bacteria attached to solid substrates (Gordesli and Abu-Lail 2012; Volle et al., 2008). However, the information on bacteria interfacial interactions and the assessment of adhesion forces in the presence of D-AAs is scarce.
This study aims to clarify whether exogenous D-AAs could inhibit the initial bacterial adhesion and to elucidate the interaction forces and energy mechanisms of bacterial adhesion by combining the use of XDLVO predictions and AFM. This work here has characterized the discrepancies of bacterial surface characteristics and the thermodynamic parameters over a range of D-tyrosine concentrations. Here, Escherichia coli (E. coli) and D-tyrosine were used as the model microbe and D-AAs, respectively. As a typical bacterium of microbial adhesion experiments, E. coli cannot produce and release D-AAs (Lam et al., 2009; Prigent-Combaret et al., 2000) and D-tyrosine has potent activity toward biofilm disassembly (Kolodkin-Gal et al., 2010). The results obtained from this study provide a better understanding of the role of D-tyrosine on bacterial adhesion from a thermodynamic perspective, which may provide a new way to regulate biofilm formation by manipulating the content of D-AAs in natural or engineered systems.
Materials and Methods
Bacterial Growth and Surface Characteristics
Escherichia coli JM109 was provided by China General Microbiological Culture Collection Center and was used in this study as the test bacterium. Cells were cultured in a standard Luria-Bertani (LB) medium with different concentrations of D-tyrosine (Aladdin; 99%) (0, 10, 25, 50 μM) at 37°C, 200 rpm, and for 18 h until stationary phase was reached, and then were collected by centrifugation at 5000 g for 15 min. L-Tyrosine (Aladdin; 99%) (0, 10, 25, 50 μM) was used as an additional control. The cells were washed twice using 10 mM KCl (the pH was unadjusted, ranging from 5.4 to 5.8) to remove any residual medium from the bacterial surface. Finally, the cells were resuspended in select solutions for cell surface characterization and adhesion experiments. To confirm the effect of exogenous D-tyrosine on bacterial cell adhesion behaviors, control experiments were conducted using the cells harvested from mineral medium (containing per liter 5 g of glucose, 2 g of (NH4)2SO4, 0.2 g of MgSO4·7H2O, 4 g of K2HPO4, 4.6 g of KH2PO4, 1 g of sodium citrate, and pH = 7.2) with different concentrations of L-tyrosine or D-tyrosine (0, 10, 25, 50 μM) respectively. Each data point is composed of three independent samples.
Bacteria mass was described as the dry weight, and the optical density (OD600) of the washed bacterial suspension was measured as absorbance by the spectrophotometer (UV-2000, Unic, Shanghai, China) at 600 nm in all the experiments. All chemical reagents in this study were of analytical grade.
Electrophoretic mobility (EPM) was measured by the ZetaSizer 3000HSA (Malvern, England, UK) and repeated at least five times at 25°C. Zeta potential (ζ potential) was calculated in a 10 mM KCl solution via EPM using the Helmholtz-Smoluchowski equation (Nakatani et al., 1978). EPS was extracted by the cation exchange resin technique (CER) (Sheng et al., 2006). After filtrating through 0.22-mm cellulose acetate membranes, the supernatant was used as the EPS fraction for chemical analyses. Polysaccharides and proteins were determined by the anthrone-sulfuric (DuBois et al., 1956) and Lowery methods (Lowry et al., 1951), respectively. Glucose and bovine serum albumin were used as the standards. These processes were performed in duplicate and each data point is composed of three independent samples.
The hydrophilicity of bacteria was measured as the microbial adhesion to hydrocarbon test (the percent of total cells partitioned into n-dodecane) (Kim et al., 2009; Tran et al., 2013). In brief, 1 mL of n-dodecane (analysis grade) was added to 4 mL of a cell suspension, which was then mixed for 2 min with a vortex. The suspension was then allowed to separate for 15 min at room temperature. The percentage of bacteria partitioning into the hydrocarbon phase was calculated after the optical density of the suspension in the water phase was measured at 600 nm. All experiments were conducted in triplicate.
The contact angle was calculated by a contact angle meter (JC2000C, Shanghai, China), which was used to evaluate interfacial tension and surface energy. Prior to the measurement, the prepared acetate cellulose membrane membranes were air-dried for 45 min. To measure contact angles of E. coli, the bacterial layer was prepared on a 0.22-μm acetate cellulose membrane by adding 10 mL of cell suspension (OD600 = 1.0), and then placed on a 1% agar plate for cell preservation before measurement. The contact angles were measured using the sessile drop technique with a drop of purified water (γ LW = 21.8 and γ + = γ− 25.5 mJ/m2), formamide (γ LW = 39.0, γ+ = 2.3 and γ− = 39.6 mJ/m2), and diiodomethane (γ LW = 50.8 and γ+ = γ− = 0 mJ/m2) (Brant and Childress 2002). Each measurement was made on at least 10 droplets at different locations on the bacterial layer for each of the three liquids, which was used to calculate standard deviations. The other measurements are described in detail in the supplemental material.
Quartz Sand Treatment
Ultrapure quartz sand with a 1 mm diameter (Aladdin; ≥ 99%) was used as the collector surface for bacterial adherence. The sand was thoroughly immersed in 12 N HCl for at least 24 h, cleaned by deionized water, and then baked at 800°C for approximately 6 h to remove the organic substances on the surface. To increase the bacterial cells adhesion onto the quartz sands, the cleaned quartz sand was chemically modified with 4% (v/v) γ-aminopropyltriethoxyasilane (APTES) (Aladdin; ≥99%) in acetone at 45°C for 24 h, then cleaned by soaking first in acetone, followed by a thorough rinsing with deionized water, and finally air-dried. Zeta potential of quartz sand was determined by an electrokinetic analyzer (Anton Paar SurPASS, Austria, Graz).
Cell Adhesion and Desorption Experiments
The influence of D-tyrosine or L-tyrosine on cell adhesion to quartz sands was evaluated by batch adhesion experiments with E. coli harvested from growth media (LB and mineral medium) with different L-isomer or D-isomer tyrosine concentrations (0, 10, 25, 50 μM). The experiments were conducted by adding 10 g of sand into 50 mL glass bottles with 10 mL of cell solution, and then gently vibrating with a shaker (8 rpm) for 3 h to reach equilibrium, based on the result of preliminary experiments.
The bacteria adhered on the collector surface irreversibly, and was washed by a 0.1 mM KCl solution after rinsing with the background solutions (10 mM KCl). The experiments were conducted with the washed quartz sands after the cell adhesion tests, and the sands were gently vibrated with shaker for 3 h to determine the desorption efficiency. The cell adhesion and desorption tests were conducted according to the methods provided in the SI.
AFM Measurements
Silicon wafers were used as the substrates for AFM measurements, and were treated with ultrapure water: hydrogen peroxide: ammonia water (1:3:1) for at least 15 min after cleaning with sonication in ultrapure water and ethanol, and were then immersed in a 25% sodium hydroxide solution for at least 24 h. After cleaning with ethanol and ultrapure water, the wafers were soaked in 1% APTES for 2 h to be silanized. The amino-functionalized wafers were cleaned by ultrapure water to remove the excess saline, and finally the silanized substrates were kept in 120°C for 15 min. Before and after making the AFM measurements on the bacteria, force measurements were made on a bacteria-free area of silicon wafers, to ensure that the tip had not been contaminated by contacting the bacteria layer on the silica wafers. The spring constant (k) for the tips was 0.15 ± 0.02 nN/nm; tips were calibrated before each experiment by the equipment.
The cells used for AFM measurements were stirred in 2.5% v/v glutaraldehyde for 2 h at 4°C. The gluaraldehyde does not affect the physiochemical properties of the E. coli cells, but provides a method for ensuring no movement of the cells during measurements (Razatos et al., 1998). Prior to the AFM experiments, a drop of the bacteria suspension (20 μL) was deposited onto the silanized silicon slides and left to settle at room temperature for 20 min. The bacteria-coated slide was rinsed with sterile water to remove any weakly bounded bacteria and air dried before measuring the same day.
All AFM measurements were performed at room temperature using a scanning probe microscope (Veeco Instruments Inc., NY) with Si3N4 cantilevers (SDL-A, Bruker AXS Inc., Fitchburg). This microscope had a dimension icon controller and extender module. The tip elements are showed in the supplemental material. In this study, five cells were examined from each medium. And on each cell, 15 points were located to perform AFM measurements. AFM images were acquired by using the Scan Asyst Mode, while the force–distance curves were evaluated by tipping mode. The adhesion forces were calculated from the retraction curves which represented the sum of all interaction forces (specific and nonspecific) between the bacterial surface and the silicon nitride tips. The specific and nonspecific forces were measured by the Poisson analysis of adhesion forces (MATLAB, Natick, MA) (Gordesli and Abu-Lail 2012).
XDLVO Profiles
According to the XDLVO approach, the total energy of interaction (WTOT) between spherical particles (bacteria) and an infinite planar surface can be calculated by the sum of electrostatic interactions (WEL), Lifshitz–van der Waals interactions (WLW), and Lewis acid– base interactions (WAB).
| (1) |
| (2) |
| (3) |
| (4) |
where, ζB and ζG are the zeta potential of bacteria surfaces (B) and quartz sands (G), respectively. Due to Born repulsion, y0, the minimum equilibrium cut-off distance between bacterial cells and the collector, equals 0.157 nm in previous work (Feriancikova et al., 2013). R is the cell radius, and h is the separation distance between the cell and the solid surface. κ is defined as the Debye length or the “thickness” of the double layers and can be estimated by ionic strength and the concentration of background solution. λ is typically between 0.2~1 nm, and is used as 0.6 nm in this work, which is a characteristic decay length of Lewis acid–base interactions in solution. The other parameters in these formulas have been described in supplemental material in details.
Statistical Analysis
In the figures and tables, data are presented along with error bars associated with one standard deviation. Statistical differences between mean values were analyzed using a Student’s t-test.
Results
Cell Adhesion and Desorption Tests
To evaluate the effect of D-tyrosine on the growth of E. coli, the bacteria cells cultivated from LB media with different concentrations of D-tyrosine (0, 10, 25, 50 μM) for at least 24 h, and the biomass at different time point were determined by optical density. The growth curves indicated that D-tyrosine did not inhibit bacterial growth (Fig. S1), which is consistent with previous report (Kolodkin-Gal et al., 2010).
The adhesion tests revealed changes in E. coli adhesion behavior over the range of D-tyrosine concentrations (Fig. 1). It is evident that the adhesion rates decreased sharply from 26.3% to 10.0% with an increase in the D-tyrosine concentration from 0 μM to 25 μM. Above 25 μM D-tyrosine, there was no measurable cell adhesion (8.8%) (Fig. 1a). Therefore, the existence of D-tyrosine has an obvious effect on microbial adhesion (P < 0.05). Moreover, the desorption rate increased from 43% to 100% based on the adhered bacteria over the range of D-tyrosine concentrations (Fig. 1a); the amount of irreversibly adhered cells decreased from 14.7% to 0%. Nevertheless, there was no significant difference for adhesion and desorption efficiency among the cells grown in the presence of L-tyrosine with different concentrations (Fig. 1b). This result shows that D-tyrosine has a significant effect on bacterial adhesion (P < 0.05).
Figure 1.
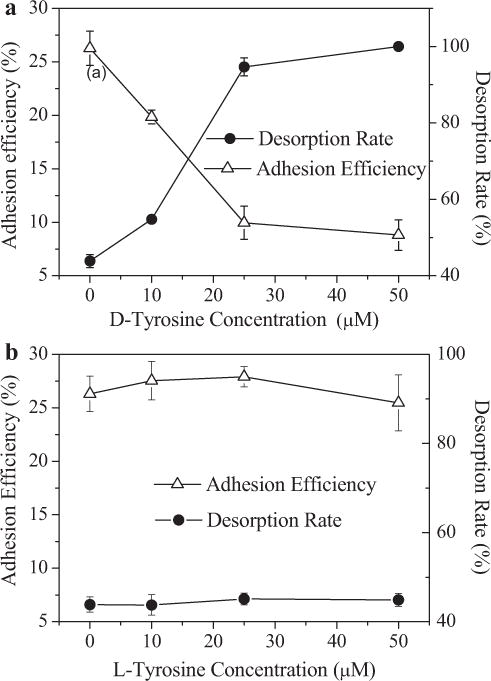
Adhesion efficiency and desorption rate of E. coli onto a quartz collector surface, determined as a function of D-tyrosine (a), and L-tyrosine (b). Experiments were conducted at unadjusted pH (5.4–5.8), room temperature (25°C), and with bacteria cultivated from LB media with different concentrations of D-tyrosine. Error bars represent standard errors of the means.
In order to identify whether there was any effect of exogenous D-AAs on cells in the LB broth, further experiments with E. coli cells harvested from mineral medium with D-tyrosine or L-tyrosine (0, 10, 25,50 μM) were conducted (Fig. S2). The results indicated that D-tyrosine could affect microbial adhesion even in a chemically defined medium.
The above results demonstrate that bacteria exposed to D-tyrosine exhibited poor adhesion ability. The bacterial adhesion is attributed to the concurrent existence of deposition, as well as irreversible and reversible adhesion. In this study, the deposition could be neglected since the desorption rate of bacteria with 50 μM D-tyrosine was nearly 100%. As to the bacteria without D-tyrosine, almost 40% of the adhered bacteria were detached under lower ionic strength solution, indicating that the bulk of the adhered microbes are reversibly bound. When cultivated at the high D-tyrosine concentration, the fraction of the reversibly adhered bacteria increased as the D-tyrosine concentration increased (P < 0.05), showing a higher reversibility for the cells (Fig. 1a).
Influence of D-Tyrosine on Surface Properties of E. coli Cells
To better understand the effect of D-tyrosine on bacterial adhesion behavior, the extracellular polysaccharide and protein contents of bacteria were determined (Fig. 2). Compared with the cells that were not exposed to D-tyrosine, a decrease (P < 0.05) in protein was observed for the bacteria cultured with D-tyrosine. However, there was only slight difference in protein of cells grown with different D-tyrosine concentrations. For polysaccharide it was less sensitive to changes (P > 0.05) with D-tyrosine concentrations. This result indicated that D-tyrosine most likely inhibited protein secretion.
Figure 2.
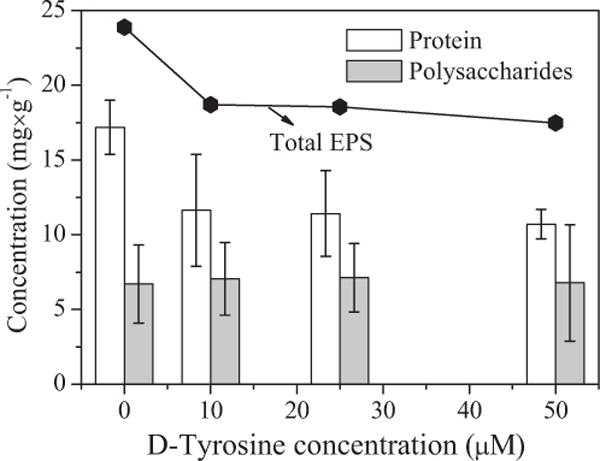
The content of total EPS, polysaccharides, and proteins of the E. coli cells from LB media with different D-tyrosine concentrations (0, 10, 25, 50 μM). Error bars represent standard errors of the means.
As indicated by the measured zeta potential (Fig. 3), all the E. coli cells used in this study were negatively charged, while the quartz sand modified with amino-silane was positively charged under the experimental conditions. The electrostatic interaction between bacteria and sand was attractive, which contributed to bacteria adhesion. Over the range of D-tyrosine concentrations, the absolute magnitude of the cell zeta potential increased (from 30.0 mV to 43.6 mV) with the increase in D-tyrosine concentration.
Figure 3.
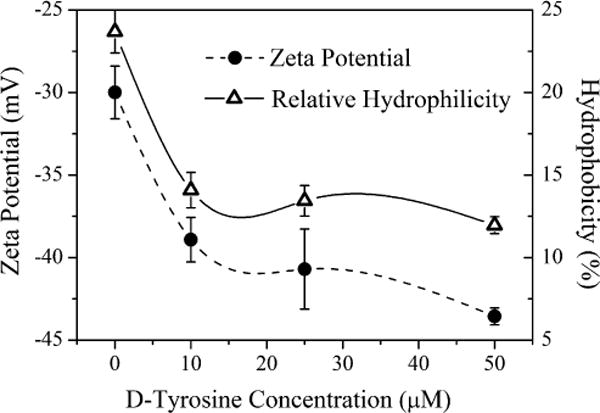
The relative hydrophobicity and zeta potential values of E. coli cells as a function of D-tyrosine concentration (0, 10, 25, 50 μM) in LB media. All experiments were conducted at unadjusted pH and room temperature. Error bars represent standard errors of the means.
The hydrophobicity of E. coli cells from experiments with different concentrations of D-tyrosine (Fig. 3) showed that the surface hydrophobicity decreased from 23.7% to approximately 12.0%, as the D-tyrosine concentration increased, which is consistent with zeta potential results. The result of relative hydrophibicity is also in consistence with that of water contact angles test (Fig. S3). The more negative surface potential denotes that the increased fractions of polar functional groups led to a more hydrophilic bacteria surface (Liu et al., 2007). In addition, the relative low EPS content was responsible for high hydrophilicity at high D-tyrosine concentrations (Chen and Walker 2007; Sheng and Yu 2006).
XDLVO Interaction Energy Profiles between E. coli Cells and Sand
The XDLVO theory prediction was applied to evaluate the interaction energy between cells and the quartz sand (Fig. 4), which were consistent with the bacterial adhesion experimental data for E. coli cells. With an increase in D-tyrosine concentration, the height of the repulsive energy barrier between cells and the quartz sand exhibited a significant change (P < 0.05) (from 24.5 kT to 3328.4 kT). No energy barrier for microbial adhesion (i.e., completely favorable condition) in the absence of D-tyrosine was observed. A higher energy barrier means a more stable suspension. Hence, it was difficult for the bacteria exposed to D-tyrosine to attach to surfaces.
Figure 4.
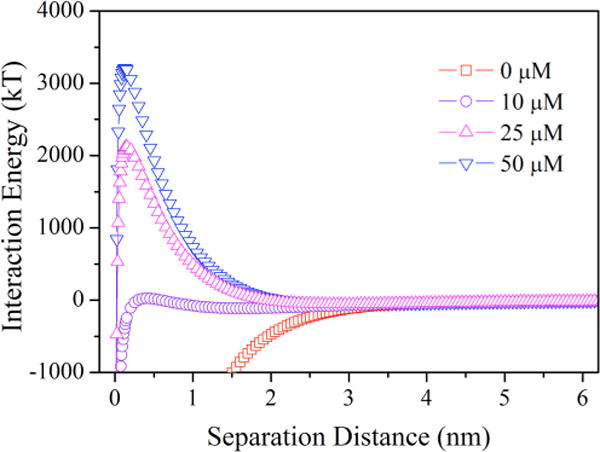
Total interaction energy profiles as a function of separation distance for E. coli cells grown under a range of D-tyrosine concentrations, suspended in 10 mM KCl.
In addition, secondary energy minima in the interaction energy predictions illustrate the desorption ability of microbial cells from collector surfaces (Liu et al., 2010). The values of the secondary energy minima and positions are summarized in Table SI. The percentage of bacteria (cultivated in 10 μM D-tyrosine) reversibly adhering on the collector was 11.5% when the depth of secondary energy minimum was −114.5 kT at the separation distance of 1.6 nm (Table SI). However, when a secondary energy minimum of −54.7 kT existed at 2.9 nm, 9.4% of adhered cells were reversibly attached in 25 μM D-tyrosine. These results indicate that the shallower secondary energy minimum caused a decrease in the reversible attachment onto the surface and promoted the detachment from the surface.
As part of the total interaction energy, WAB, WLW, and WEL values for E. coli as a function of distance between bacteria and sands were determined (Fig. 5). The Lifshitz–van der Waals interactions (WLW) were attractive and nearly identical, while the absolute values of attractive electrostatic interactions (WEL) were variable with an increase in D-tyrosine concentration. However, Lewis acid–base interactions were attractive without D-tyrosine (favorable conditions for adhesion). With increasing D-tyrosine concentrations, Lewis acid–base interactions turned repulsive and the absolute values increased. Therefore, the contribution of WAB to the total energy was very significant.
Figure 5.
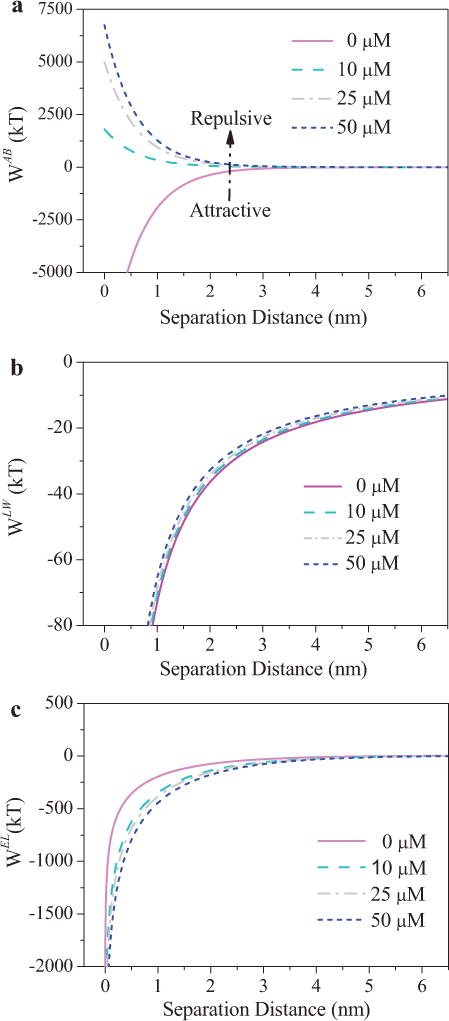
Contribution of WAB (a), WLW (b), and WEL (c) on the total interaction energy between E. coli cells and quartz sand exposed to different D-tyrosine concentrations, which adhered on quartz sands in 10 mM KCl at uncontrolled pH values.
Probing the Adhesion Force with AFM
Since the Lewis acid–base interaction energy plays the most important role on inhibiting bacterial adhesion in the presence of D-tyrosine, it is indicated that the acid-base interaction including hydrophobic interaction and hydration effects, which are mainly electron–donor/electron–accepter interaction (Hori and Matsumoto 2010). The chemical bonds on the AFM tips are mainly O—H and Si—N, while the bonds on modified quartz surface O—H, Si—N, N—H, etc. The tips and modified quartz would form the similar bonds with the bacteria surface. Therefore, here we used AFM to quantitatively probe the adhesion force between bacteria and quartz sands. The shape and size of bacteria were assessed by AFM scans (see Fig. S4 in the supplemental material) and implied that D-tyrosine did not affect their morphology. In addition to topographic imaging, AFM force–distances were used to determine interactions between the silicon nitride probe and bacteria surface in the air. The retraction curves were variable, since the adhesion force or “liftoff” (the point where the tips break free of tip–sample interaction) was observed (Table I). The magnitude of the interaction forces was highly dependent on D-tyrosine concentration, which decreased from 61.8 nN to 28.1 nN. As shown in Table I, the adhesion force decreased sharply (P < 0.05) compared with that without D-tyrosine, while this adhesion force varied slightly (P > 0.1) in the presence of D-tyrosine.
Table I.
Results from the AFM force–distance curve over the range of D-tyrosine concentrations.
| Parameters | 0 μM | 10 μM | 25 μM | 50 μM |
|---|---|---|---|---|
| Adhesion force (nN) | 61.8 | 40.2 | 33.5 | 28.1 |
| Nonspecific forces (nN) | −45.8 | −40.5 | −28.6 | −17.5 |
| Specific forces (nN) | 0.9 | 1.0 | 2.1 | 3.3 |
| Separation energy (×10−18 J) | 8479.2 | 3189.8 | 1452.8 | 1689.3 |
| Hydrogen bands numbers | 847919 | 318985 | 145275 | 168928 |
The total separation energy is estimated as the total area of the attraction-force region in the retraction force by Equation 5 and these parameters obtained from the retraction curves are presented in Table I:
| (5) |
in which, I is the distance (nm) moved by the cantilever in every interval, Fs,i is the separation force, kc is the spring constant (nN/nm) and the distance is dd,i (nm).
By using Poisson statistical analyses, nonspecific (i.e., Lifshitz– van der Waals force and electrostatic forces) and specific forces (i.e., Lewis acids–base interaction force) of the adhesion forces were quantified through linear regression (Table I). The results illustrated that the nonspecific and specific forces decreased over the range of D-tyrosine concentrations. The nonspecific forces were attractive for all conditions investigated, while the specific forces were repulsive for the cells cultured with D-tyrosine acids. On average, the nonspecific forces were stronger than the specific forces; mean-while, 61.8% of nonspecific forces decreased and the specific forces increased from 0.9 nN to 3.3 nN.
The maximum energy of the original microbial cells was about 8500 × 10−18 J and the adhesion energy decreased sharply (P < 0.05) for the bacteria cultured in the presence of D-tyrosine (Table I). Consequently, the hydrogen bond numbers which equaled the adhesion energy divided by the value of Gabbis energy (10–20) were reduced (Wen et al., 2011).
Discussion
The experimental and theoretical results show that the interaction energies of bacterial cells changed significantly (P < 0.05) in the presence of D-tyrosine. These results also demonstrate the crucial role of D-tyrosine in inhibition of the initial bacterial adhesion.
Influence of Total Interaction Energy on Bacterial Adhesion Behavior
It has been recognized that a two-step mechanism of bacterial attachment to surfaces is involved in biofilm formation (Gordesli and Abu-Lail 2012). The first step involves long-range nonspecific interactions such as Lifshitz–van der Waals and electrostatic interactions, as described by XDLVO theory. The second step is described as irreversible bacterial adhesion, due to much stronger specific short-range interactions (hydrogen bonds between cell and surface and/or EPS and surfaces, capillary forces, etc.) which could be interpreted as Lewis acid–base interactions (calculated in XDLVO theory) and polymer interactions (Dunne 2002).
The total interaction energy for E. coli cells (Fig. 4) indicates that D-tyrosine had a negative contribution to bacterial adhesion and inhibited irreversible adhesion. This was confirmed by the adhesion and desorption tests (Fig. 1). Additionally, the depth of the secondary energy minima above the separation distances may determine the adhesion and desorption rates for cells. Reversible adhesion was reduced with the decrease of the depth of secondary energy minimum because of D-tyrosine concentration variations (see Table SI).
The AFM measurement also indicated a decreased adhesion rate in the presence of D-tyrosine, which was consistent with above results. The quantification of specific and nonspecific forces revealed decreasing attractive nonspecific forces performed the reversible adhesion, and the increasing specific forces inhabited the irreversible adhesion. This quantitative analysis enabled us to further understand the micro-scale mechanisms behind D-tyrosine-inhibited bacterial adhesion.
Contribution of WAB, WLW, and WEL to Bacterial Adhesion
The Hamaker constant, proportional to the Lifshitz–van der Waals interactions, is a parameter that describes the strength of the interaction between bacteria and surface. The discrepancy of effective Hamaker constants is nearly non-existent (P > 0.1) (see Table SI), causing a slight decrease of the Lifshitz–van der Waals interaction energy with an increase of D-tyrosine. For the surface covered by layers that contain larger quantities water, the effective Hamaker constant was reduced (Hermansson 1999), which was consistent with the trend of cell surface hydrophobicity. Moreover, the intensity of the attractive Lifshitz–van der Waals interactions decayed slowly with the interaction distance and became zero for distances of about 10 nm (Fig. 5b). Furthermore, the zeta potential of bacterial cells increased (P < 0.05) (i.e., more negative) as D-tyrosine concentration increased, which should enhance the attractive electrostatic interaction force. However, with increasing D-tyrosine concentrations, the electrostatic interactions decay more slowly with the separation distance, due to the more negative zeta potentials (Fig. 5c). The resulting profile showed that the Lifshitz– van der Waals interactions play a more important role than the electrostatic interactions in bacterial reversible adhesion, because the former are longer range.
Although WLW and WEL were identical and not sensitive to D-tyrosine concentration variations, the contribution of WAB to the total interaction energy is flexible (Fig. 5). As the dominant interaction energy (Fig. 5a), WAB interaction energy increased substantially (P < 0.05) with the increasing D-tyrosine concentration (over 0 μM), indicating that D-tyrosine reduced bacterial adhesion by preventing bacteria from approaching the surface at sufficiently close distances. The higher intensity of the Lewis acid– base interactions decays with longer interacting distances with the increasing D-tyrosine concentrations. After comprehensive analysis for those interaction energies, the cell surface hydrophilicity appears to be responsible for the change of energy barriers as well as primary and secondary energy minima observed for all the E. coli cells.
Influence of EPS on Adhesion Behavior
EPS are the bridge between the cell and collector surfaces to overcome the energy barrier, causing the bacteria to adhere to the surface irreversibly (Azeredo et al., 1999; Hori and Matsumoto 2010). In our study, the EPS production decreased in the presence of D-tyrosine. Consequently, the reduction of EPS may directly cause decreased bacterial adhesion. A previous report illustrated that high exopolymer production of Sphingomonas paucimobilis was quite beneficial to bacterial adhesion by overcoming the energy barrier of 300 kT (Hori and Matsumoto 2010). The percentage of irreversibly adhered bacteria was 14.7% in 10 μM D-tyrosine, whereas, for the bacteria in 25 μM and 50 μM D-tyrosine, the energy barriers were much higher, and the irreversibly adhered bacteria were nonexistent (0.5% and 0%). This observed behavior is consistent with those reported previously on EPS (Tsuneda et al., 2003).
Role of Hydrogen Bonds in Adhesion Behavior
The hydrogen bonds are also important for the bacterial adhesion behaviors, and are mainly responsible for the Lewis acids–base interactions (Hermansson 1999). The results of cell surface hydrophobicity showed a strong electron donating potential with high γ– and small electron acceptor ability (the value of γ+ is almost 0) (data not shown), which was the same as quartz sand. The increased hydrophobic interactions (divided into Lifshitz–van der Waals and Lewis acid–base components), would be attributed to the stronger hydrogen bonding between bacterial surfaces and the experimental solutions with increasing D-tyrosine concentrations. This prevented bacterial adhesion. Additionally, the sand surface contains hydroxyl and amine groups that have the ability to donate and accept protons, respectively (Hwang et al., 2010). Thus, a competition between the water and bacterial cells for hydrogen bond sites on the collector surface would occur. The amide carbonyl oxygen found on the surface of the EPS acted as stronger electron donors for the hydroxyl groups on quartz sand, while the hydroxyl groups of the EPS offered sites for forming hydrogen bonds with amine groups on the quartz sand (Chen and Walker 2007). This led to the preferential adhesion of bacteria onto the quartz sand.
With the decreasing amount of EPS, the reduction in hydrogen bonds (Table I) may be responsible for the poor bacterial adhesion. This result was consistent with the AFM analysis suggesting hydrogen bonds between the bacteria and quartz sand. Nearly 80% of the hydrogen bonds decreased with the increase in D-tyrosine concentrations from 0 μM to 50 μM. This provided evidence that hydrogen bonds were the key factors affecting bacterial adhesion.
Furthermore, a decrease in bacterial adhesion was also influenced by the detaching behavior demonstrated by AFM measurements. The forces measured by AFM were comprised of DLVO forces (Lifshitz–van der Waals and electrostatic interaction) and non-DLVO forces (Lewis acid–base interaction, steric interaction, etc.). The decrease in EPS excretion and Lifshitz–van der Waals interactions will reduce the adhesion force (Table I). Consequently, bacteria on the surface tended to detach from quartz sand (Fig. 1). The experimental and theoretical results indicated that significant increases in hydrophilicity and decreases in hydrogen bond numbers (i.e., decreased EPS excretions) resulted in a significant decrease in the bacterial attachment efficiency.
Conclusions
In this study the effect of the exogenous amino acid D-tyrosine on initial bacterial adhesion was systematically investigated from a thermodynamic point of view. The total interaction energy increased with more D-tyrosine, and the contribution of Lewis acid-base interactions relative to the change in the total interaction energy was much greater than the overall nonspecific interactions. The hydrogen bond numbers and adhesion forces decreased with the increase in D-tyrosine concentrations. It was revealed that D-tyrosine contributed to the repulsive nature of the cell and ultimately led to the inhibition of bacterial adhesion.
Supplementary Material
Acknowledgments
The authors wish to thank the National Natural Science Foundation of China (51208283, and 51178254), Specialized Research Fund for the Doctoral Program of Higher Education (20120131120017), Natural Science Foundation of Shandong Province (JQ201116), for the partial support of this study. Alicia A. Taylor was supported by the National Research Service Award Institutional Training Grant (T32 ES018827).
Contract grant sponsor: National Natural Science Foundation of China Grant numbers: 51208283; 51178254
Contract grant sponsor: Specialized Research Fund for the Doctoral Program of Higher Education
Contract grant number: 20120131120017
Contract grant sponsor: Natural Science Foundation of Shandong Province
Contract grant number: JQ201116
Contract grant sponsor: National Research Service Award Institutional Training Grant
Contract grant number: T32 ES018827
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
The authors declare no conflicts of interest.
References
- Azeredo J, Visser J, Oliveira R. Exopolymers in bacterial adhesion: Interpretation in terms of DLVO and XDLVO theories. Colloid Surf B Biointerfaces. 1999;14:141–148. [Google Scholar]
- Bos R, Mei HC, Busscher HJ. Physico-chemistry of initial microbial adhesive interactions–its mechanisms and methods for study. FEMS Microbiol Rev. 1999;23:179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- Brant JA, Childress AE. Assessing short-range membrane-colloid interactions using surface energetics. J Membr Sci. 2002;203:257–273. [Google Scholar]
- Chen G, Strevett KA. Impact of surface thermodynamics on bacterial transport. Environ Microbiol. 2001;3:237–245. doi: 10.1046/j.1462-2920.2001.00181.x. [DOI] [PubMed] [Google Scholar]
- Chen G, Walker SL. Role of solution chemistry and ion valence on the adhesion kinetics of groundwater and marine bacteria. Langmuir. 2007;23:7162–7169. doi: 10.1021/la0632833. [DOI] [PubMed] [Google Scholar]
- Chia TWR, Tuan Vu, McMeekin N, Fegan T, Dykes N. Stochasticity of bacterial attachment and its predictability by the extended Derjaguin–Landau–Verwey– Overbeek theory. Appl Environ Microbiol. 2011;77:3757–3764. doi: 10.1128/AEM.01415-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton J, Greenberg E. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Dunne WM. Bacterial adhesion: seen any good biofilms lately. Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feriancikova L, Bardy SL, Wang L, Li J, Xu S. Effects of outer membrane protein TolC on the transport of Escherichia coli within saturated quartz sands. Environ Sci Technol. 2013;47:5720–5728. doi: 10.1021/es400292x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Gordesli FP, Abu-Lail NI. Combined poisson and soft-particle DLVO analysis of the specific and nonspecific adhesion forces measured between L. monocytogenes grown at various temperatures and silicon nitride. Environ Sci Technol. 2012;46:10089–10098. doi: 10.1021/es300653w. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Harvey RW, Metge DW, Mohanram A, Gao X, Chorover J. Differential effects of dissolved organic carbon upon re-entrainment and surface properties of groundwater bacteria and bacteria-sized microspheres during transport through a contaminated, sandy aquifer. Environ Sci Technol. 2011;45:3252–3259. doi: 10.1021/es102989x. [DOI] [PubMed] [Google Scholar]
- Hermansson M. The DLVO theory in microbial adhesion. Colloid Surf B Biointerfaces. 1999;14:105–119. [Google Scholar]
- Herzberg M, Elimelech M. Physiology and genetic traits of reverse osmosis membrane biofilms: A case study with Pseudomonas aeruginosa. ISME J. 2007;2:180–194. doi: 10.1038/ismej.2007.108. [DOI] [PubMed] [Google Scholar]
- Hori K, Matsumoto S. Bacterial adhesion: From mechanism to control. Biochem Eng J. 2010;48:424–434. [Google Scholar]
- Hwang G, Lee CH, Ahn IS, Mhin BJ. Analysis of the adhesion of Pseudomonas putida NCIB 9816-4 to a silica gel as a model soil using extended DLVO theory. J Hazard Mater. 2010;179:983–988. doi: 10.1016/j.jhazmat.2010.03.101. [DOI] [PubMed] [Google Scholar]
- Kim HN, Hong Y, Lee I, Bradford SA, Walker SL. Surface characteristics and adhesion behavior of Escherichia coli O157:H7: Role of extracellular macromolecules. Biomacromolecules. 2009;10:2556–2564. doi: 10.1021/bm900516y. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Wahman DG, Bishop PL, Pressman JG. Free chlorine and monochloramine application to nitrifying biofilm: Comparison of biofilm penetration, activity, and viability. Environ Sci Technol. 2011;45:1412–1419. doi: 10.1021/es1035305. [DOI] [PubMed] [Google Scholar]
- Liu XM, Sheng GP, Luo HW, Zhang F, Yuan SJ, Xu J, Zeng RJ, Wu JG, Yu HQ. Contribution of extracellular polymeric substances (EPS) to the sludge aggregation. Environ Sci Technol. 2010;44:4355–4360. doi: 10.1021/es9016766. [DOI] [PubMed] [Google Scholar]
- Liu XM, Sheng GP, Yu HQ. DLVO approach to the flocculability of a photosynthetic H-2-producing bacterium, Rhodopseudomonas acidophila. Environ Sci Technol. 2007;41:4620–4625. doi: 10.1021/es070107n. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lutterodt G, Basnet M, Foppen JWA, Uhlenbrook S. The effect of surface characteristics on the transport of multiple Escherichia coli isolates in large scale columns of quartz sand. Water Res. 2009;43:595–604. doi: 10.1016/j.watres.2008.11.001. [DOI] [PubMed] [Google Scholar]
- McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- Nakatani H, Barber J, Forrester J. Surface charges on chloroplast membranes as studied by particle electrophoresis. Biochim Biophys Acta. 1978;504:215–225. doi: 10.1016/0005-2728(78)90019-1. [DOI] [PubMed] [Google Scholar]
- Nielsen L, Li X, Halverson LJ. Cell–cell and cell–surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ Microbiol. 2011;13:1342–1356. doi: 10.1111/j.1462-2920.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Peulen TO, Wilkinson KJ. Diffusion of nanoparticles in a biofilm. Environ Sci Technol. 2011;45:3367–3373. doi: 10.1021/es103450g. [DOI] [PubMed] [Google Scholar]
- Pope PB, Totsika M, de Carcer Aguirre, Schembri D, Morrison MA. Muramidases found in the foregut microbiome of the Tammar wallaby can direct cell aggregation and biofilm formation. ISME J. 2011;5:341–350. doi: 10.1038/ismej.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Ralebitso-Senior TK, Senior E, Di Felice R, Jarvis K. Waste gas biofiltration: Advances and limitations of current approaches in microbiology. Environ Sci Technol. 2012;46:8542–8573. doi: 10.1021/es203906c. [DOI] [PubMed] [Google Scholar]
- Razatos A, Ong YL, Sharma MM, Georgiou G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc Natl Acad Sci U S A. 1998;95:11059–11064. doi: 10.1073/pnas.95.19.11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche B, Li XZ, Hauer B, Schmid A, Buehler K. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 2009;27:636–643. doi: 10.1016/j.tibtech.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Sheng GP, Yu HQ. Relationship between the extracellular polymeric substances and surface characteristics of Rhodopseudomonas acidophila. Appl Microbiol Biotechnol. 2006;72:126–131. doi: 10.1007/s00253-005-0225-1. [DOI] [PubMed] [Google Scholar]
- Sheng G-P, Yu H-Q, Wang C-M. FTIR-spectral analysis of two photosynthetic H-2-producing strains and their extracellular polymeric substances. Appl Microbiol Biotechnol. 2006;73:204–210. doi: 10.1007/s00253-006-0442-2. [DOI] [PubMed] [Google Scholar]
- Sheng GP, Yu HQ, Li XY. Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: A review. Biotechnol Adv. 2010;28:882–894. doi: 10.1016/j.biotechadv.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Terada A, Okuyama K, Nishikawa M, Tsuneda S, Hosomi M. The effect of surface charge property on Escherichia coli initial adhesion and subsequent biofilm formation. Biotechnol Bioeng. 2012;109:1745–1754. doi: 10.1002/bit.24429. [DOI] [PubMed] [Google Scholar]
- Tran CTH, Kondyurin A, Chrzanowski W, Bilek MMM, McKenzie DR. Influence of pH on yeast immobilization on polystyrene surfaces modified by energetic ion bombardment. Colloids Surf B Biointerfaces. 2013;104:145–152. doi: 10.1016/j.colsurfb.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Tsuneda S, Aikawa H, Hayashi H, Yuasa A, Hirata A. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol Lett. 2003;223:287–292. doi: 10.1016/S0378-1097(03)00399-9. [DOI] [PubMed] [Google Scholar]
- Volle CB, Ferguson MA, Aidala KE, Spain EM, Nunez ME. Quantitative changes in the elasticity and adhesive properties of Escherichia coli ZK1056 prey cells during predation by Bdellovibrio bacteriovorus 109. J Langmuir. 2008;24:8102–8110. doi: 10.1021/la8009354. [DOI] [PubMed] [Google Scholar]
- Wang LL, Wang LF, Ren XM, Ye XD, Li WW, Yuan SJ, Sun M, Sheng GP, Yu HQ, Wang XK. PH dependence of structure and surface properties of microbial EPS. Environ Sci Technol. 2012;46:737–744. doi: 10.1021/es203540w. [DOI] [PubMed] [Google Scholar]
- Wen Z, Stack AG, Yongsheng C. Interaction force measurement between E. coli cells and nanoparticles immobilized surfaces by using AFM. Colloids Surf B Biointerfaces. 2011;82:316–324. doi: 10.1016/j.colsurfb.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Xu H, Liu Y. D-amino acid mitigated membrane biofouling and promoted biofilm detachment. J Membr Sci. 2011a;376:266–274. [Google Scholar]
- Xu H, Liu Y. Reduced microbial attachment by D-amino acid-inhibited AI-2 and EPS production. Water Res. 2011b;45:5796–5804. doi: 10.1016/j.watres.2011.08.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


