Abstract
Profilins are small actin-binding proteins found in eukaryotes and certain viruses that are involved in cell development, cytokinesis, membrane trafficking, and cell motility. Originally identified as an actin sequestering/binding protein, profilin has been involved in actin polymerization dynamics. It catalyzes the exchange of ADP/ATP in actin and increases the rate of polymerization. Profilins also interact with polyphosphoinositides (PPI) and proline-rich domains containing proteins. Through its interaction with PPIs, profilin has been linked to signaling pathways between the cell membrane and the cytoskeleton, while its role in membrane trafficking has been associated with its interaction with proline-rich domain-containing proteins. Depending on the organism, profilin is present in a various number of isoforms. Four isoforms of profilin have been reported in higher organisms, while only one or two isoforms are expressed in single-cell organisms. The affinity of these isoforms for their ligands varies between isoforms and should therefore modulate their functions. However, the significance and the functions of the different isoforms are not yet fully understood. The structures of many profilin isoforms have been solved both in the presence and the absence of actin and poly-L-proline. These structural studies will greatly improve our understanding of the differences and similarities between the different profilins. Structural stability studies of different profilins are also shedding some light on our understanding of the profilin/ligand interactions. Profilin is a multifaceted protein for which a dramatic increase in potential functions has been found in recent years; as such, it has been implicated in a variety of physiological and pathological processes.
Keywords: Actin polymerization, Phosphoinositides, Poly-L-proline, Profilin
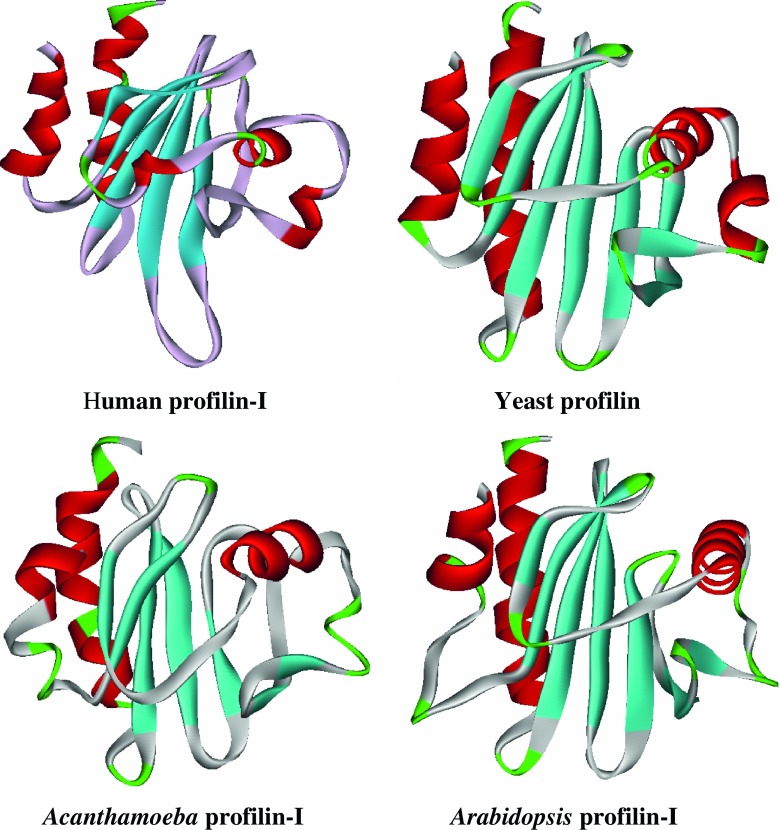
Profilins are small, 14- to 17-kDa proteins expressed in eukaryotes (Tseng et al. 1984; Sonobe et al. 1986; Kwiatkowski and Bruns 1988; Magdolen et al. 1988; Widada et al. 1989; Valenta et al. 1991a, b) and certain viruses (Machesky et al. 1994). Its isoforms comprise 100–131 amino acids. The structure of profilin and some isoforms have been solved both with and without ligands. Although the amino acid sequence varies substantially between profilin isoforms or between profilin from different organisms, structural studies have shown that they assume similar three-dimensional (3D) structures (Fig. 1). Indeed, most profilins consist of seven beta sheets and four helices.
Fig. 1.
Profilin-I isoforms from different organisms showing a similar helix (red) and strand (cyan) structure (PDB database: 1PFL, 1KOK, 2PRF, and 3NUL) with the loops highlighted in green
Profilin was originally identified as being involved in actin polymerization (Carlsson et al. 1977), and its role as a key regulator of F-actin dynamics has been well established. Other profilin ligands have also been identified, including proline-rich domain-containing proteins and phosphatidylinositol polyphosphates. The difference in sequences between profilin isoforms results in profilin having different affinities for its ligands.
The importance of profilins for normal cell proliferation and differentiation, growth, motility, and cytokinesis is well documented. Gene disruption and antisense studies in Dictyostelium revealed that profilin isoforms compensate for each other’s function (Haugwitz et al. 1994). Mutants expressing only one isoform did not show any changes in phenotype. However, mutants lacking both isoforms showed various defects, such as affected motility, increased cell size (×10), increased accumulation of actin, impaired cytokinesis, and developmental arrest before fruiting body formation. These phenotypic defects were rescued by either of the profilin isoforms. The chickadee gene in Drosophilae, which codes for profilin, is essential for female fertility and affects bristle development (Verheyen and Cooley 1994). Gene disruption studies have suggested that an allelic variant of chickadee (Chk bin) in the same locus is responsible for both male and female fertility and that chickadee does not rescue this phenotype (Akiyama and Okada, 1993).
Profilin-I gene knockout studies have suggested that in the mouse, unlike Dictyostelium, profilin isoforms do not compensate for each other’s function (Witke et al. 2001). The profilin-I null mutant was found to be embryonic lethal, while heterozygous (ko/wt) embryos showed reduced survival. These heterozygous (ko/wt) mouse for profilin-I had a 50% reduction in expression level but survived. These results suggest that profilin-I could be affecting mouse development in a dose-dependent manner. (Witke et al. 2001).
Immunogold labeling in fibroblast cells showed that profilin was markedly enriched in lamellipodia or pseudopodial lobes, and a constituent of the microfilament structures was observed to be associated with cellular membranes (Buss et al. 1992). Immunofluorescence staining showed that profilin is expressed in ruffling areas of the peripheral lamellae and nascent stress fibers of spreading cells and that profilin is absent in peripheral belts of stationary cells growing in epithelioid sheets (Mayboroda et al; 1997). Similarly, localization studies using profilin–green fluorescent protein (GFP) fusion molecules with mammalian cells showed profilin localization in areas of high actin dynamics, such as leading lamellae and ruffles induced by epidermal growth factor (Wittenmayer et al. 2000), and that it is a component of intracellular vesicles (Dong et al. 2000). Profilin has also been localized in intranuclear bodies, such as speckles, Cajal bodies, and gems (Giesemann et al. 1999; Skare et al. 2003). The toxicity produced by the overexpression of actin is also alleviated by profilin when it is co-expressed (Magdolen et al. 1988). Profilin is involved in transporting actin into the nucleus via exportin 6 and is associated with the snRNP complex and the regulation of transcription factor expression (Skare et al. 2003) This finding adds another dimension to this small actin-binding protein. Profilin was one of the proteins found to be expressed when living cells are subjected to hyperoxia (Vorum et al. 2004), but the function of the overexpression or regulation during stress is not yet understood (Gareus et al. 2006).
Profilin isoforms
Four profilin genes have been reported in the mouse and humans to date. The isoforms are generally expressed by different genes; however, differentially spliced isoforms are also known to exist. Human, bovine, mouse, and rat, profilin-II has been shown to be alternatively spliced into profilin-IIA and -IIB (Di Nardo et al. 2000; Lambrechts et al. 2000b). In humans, profilin-I is expressed in all cells, whereas other isoforms are tissue-specific. Profilin IIA and IIB are brain specific and shown to be required for neuronal development (Witke et al. 2001). Profilin-II forms complexes with proteins known to be involved in membrane trafficking, such as synapsin and dynamin-I. Profilin-III in humans and mouse are expressed in the testis and kidney and exclusively in developing spermatids (Braun et al. 2002). Profilin-III and -IV show only 30% amino acid identity among themselves and with other mammalian profilins (Obermann et al. 2005). Profilin-IV plays a major role in acrosome formation and sperm morphogenesis. Results of Northern blot and in situ transcript hybridization analyses suggest that profilin-III and -IV are transcribed in the germ cells. However, the timing of their expression during post-natal development of rat testis and in the rat spermatogenetic cycle was distinct. In the human testis, profilin-IV mRNA expression correlates with the presence of germ cells. Profilin-III and -IV may regulate testicular actin cytoskeleton dynamics and play a role in acrosome generation and spermatid nuclear shaping. (Obermann et al. 2005)
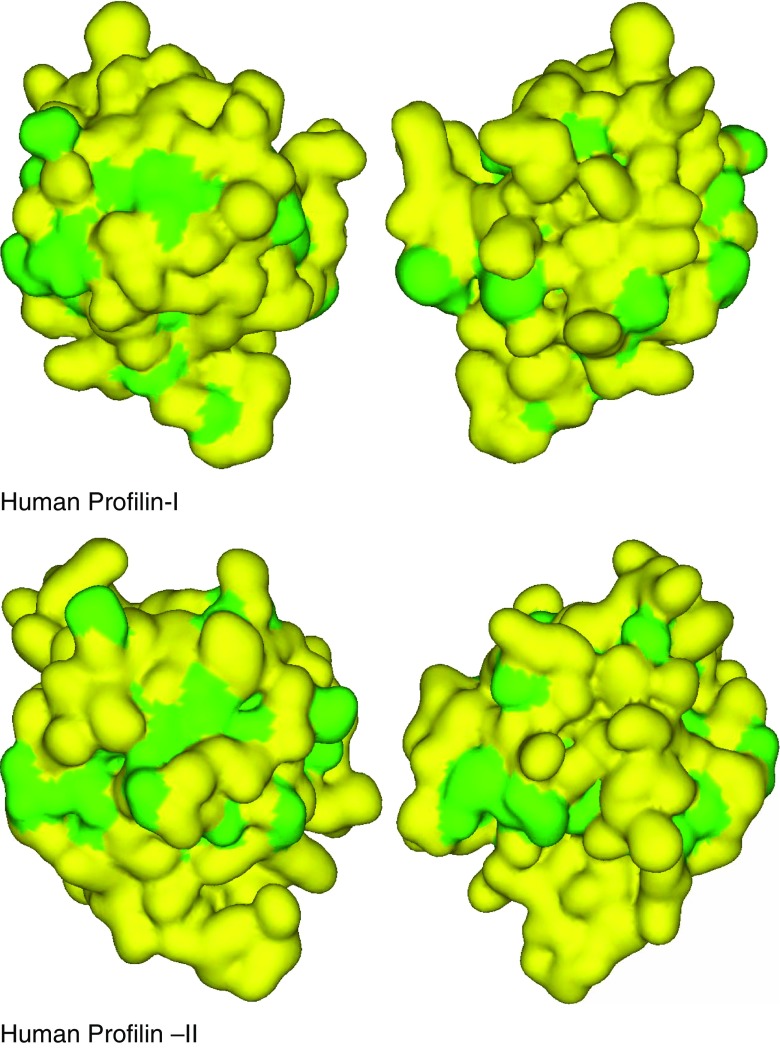
The biochemical and structural characteristics of profilin-I and -II from different organisms have been compared. Gieselmann et al. (1995) reported that human profilin-I shows approximately a fivefold higher affinity for actin than profilin-II. In the mouse, profilin-II has been shown to have a higher affinity for dynamin-I than profilin-I (Witke et al. 1998). Structural analyses of human profilin isoforms by radiography suggest that the substitution of profilin-I S29 by Y29 in profilin-II contributes to the higher affinity of profilin-II for proline-rich sequences (Nodelman et al. 1999). Although human profilin-I and -II fold into similar 3D structures, the surface properties, such as exposure of hydrophobic patches (Fig. 2), and biochemical properties of each isoform are distinct. However, further detailed structural and functional studies are required to reveal the significance of the presence of profilin isoforms in eukaryotes.
Fig. 2.
Two 180° views of human profilin-I and II isoforms showing hydrophobic surfaces (green) and hydrophilic surfaces (yellow). PDB database: 1PFL and 1D1J-chain A
Three profilin isoforms, PFN-1, PFN-2, and PFN-3, have been identified in Caenorhabditis elegans, of which PFN-1 is essential and PFN-2 and PFN-3 are not (Polet et al. 2006). Immunostaining has revealed different expression patterns for the profilin isoforms. In embryos, PFN-1 localizes to the cytoplasm and to the cell–cell contacts during the early stages of embryogenesis, and to the nerve ring during later stages of embryogenesis. During late embryogenesis, PFN-3 expression has been specifically detected in the walls of muscle cells. In adult worms, PFN-1 is expressed in the neurons, the vulva, and the somatic gonad, PFN-2 in the intestinal wall, the spermatheca, and the pharynx, and PFN-3, in a dot-like fashion, in the body wall muscle cells (Polet et al. 2006). Dictyostelium amoebae has two profilin isoforms (I and II); profilin-I is essential for growth and development, whereas profilin-II is not. Saccharomyces cerevisiae and S. pombe are known to each have only a single profilin isoform (Magdolen et al. 1988; Ezezika et al. 2009).
Profilin and actin polymerization dynamics
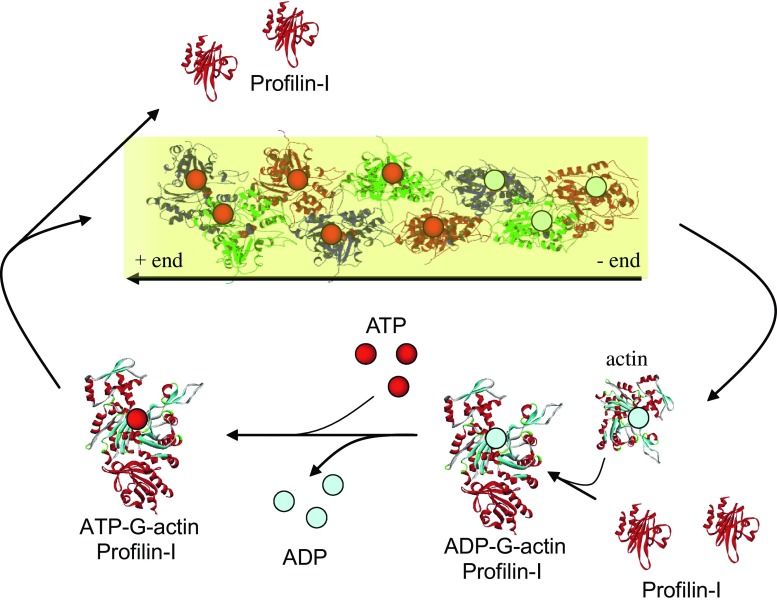
Profilins can bind to and sequester actin monomers, thereby decreasing the concentration of free actin monomers that are available for filament elongation (Carlsson et al. 1977). They replenish the pool of ATP-actin monomers by increasing the rate of nucleotide exchange by 1000-fold compared with the rate of nucleotide exchange based on simple diffusion (Goldschmidt-Clermont et al. 1992). The profilin–ATP–actin complex can interact with the fast-growing, barbed, or plus end of the actin filament and release the ATP–actin monomer, which is then added to the filament (Fig. 3). Consequently, the elongating filament consists of ATP-actin. Along the filament, the ATP is slowly hydrolyzed by the intrinsic ATPase activity of actin, which generates ADP–actin in the older part of the filament. ADP–actin can be released slowly from the pointed or minus end of the filament by depolymerization or at an accelerated rate by actin-depolymerizing proteins (Witke 2004).
Fig. 3.
Schematic representation of actin polymerization and ADP/ATP exchange by profilin. Red, solid ribbon Profilin, ribbon with helices in red and sheets in cyan actin, grey spheres ADP, red spheres ATP
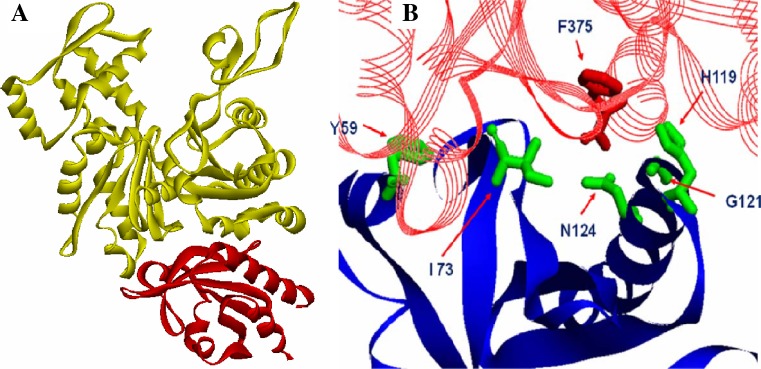
The crystal structure of the bovine beta–actin–profilin complex has been solved (Fig. 4a; Chik et al. 1996). Mutation studies have confirmed that the C-terminal residue of actin, F375, plays a major role in profilin binding (Lambrechts et al. 2002). In addition, five other residues in profilin, namely Y59, I73, H119, G121, and N124, are also important for binding actin, and any mutation of any of these (Fig. 4b) impairs the actin binding ability of profilin (Eads et al. 1998; Nodelman et al. 1999; Ezezika et al. 2009).
Fig. 4.
a A ribbon diagram of the atomic structure of profilin (red) co-crystallized with monomeric actin (yellow). For clarity, the ATP in the actin cleft is not represented. b Ribbon and stick model showing the actin and profilin contacts, with the essential amino acids highlighted as green sticks (profilin) and red sticks (actin residues). PDB database: 1HLU
Intracellular pathogens, such as Listeria monocytogenes and Shigella flexneri, use actin polymerization as a mechanism to propel themselves through the cytoplasm and to spread to neighboring cells without entering the extracellular space. These pathogens can induce polarized polymerization of actin on their surface, which pushes them forward, leaving an actin comet tail behind. In L. monocytogenes, the bacterially encoded membrane protein ActA is essential for this type of motility (Kocks 1994). Based on profilin-depletion experiments, the recruitment of profilin to ActA seems to be an important step in bacterial motility (Theriot et al. 1994). However, the results of an in vitro motility assay using L. monocytogenes and purified components, such as actin, capping protein, cofilin, and the actin-related protein (Arp)2/3, suggested that profilin was not essential for bacterial movement but that it could increase its efficiency (Loisel et al. 1999).
Profilin and phosphatidylinositol 4, 5-bisphosphate [PI(4, 5)P2]
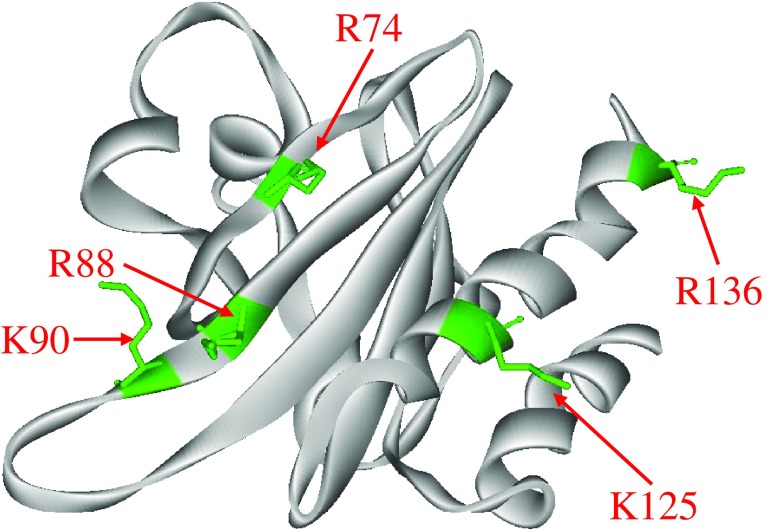
The interaction of profilin with PI(4,5)P2 is based on the binding of the negatively charged headgroup of the phosphoinositide to basic amino acids (Fig. 5). The more positively charged Acanthamoeba profilin-II isoform has the highest affinity for PI(4, 5)P2. Similarly, the more basic human profilin-I isoform interacts better with PI(4, 5)P2 than profilin-IIA (Lambrechts et al. 1997). Yu et al. (1992) compared the PI(4, 5)P2 binding sites of profilin and gelsolin and postulated that residues 126–136 (KCYEMSHLRR) of human profilin-I are a modified version of the PI(4, 5)P2-binding motif in gelsolin (KSGLKYKK). Using photoactivatable homologs of PI (4, 5)P2, Chaudhary et al. (1998) subsequently hypothesized that the carboxy terminal basic residues in human profilin-I is involved in contacting the negative head groups of PI(4, 5)P2. A competition between poly-L-proline and PI(4, 5)P2 for binding to profilin (Lambrechts et al. 1997) has been observed, which is consistent with the proposal that the carboxy terminus of profilin is involved in PI(4, 5)P2 binding (Cedergren-Zeppezauer et al. 1994). Based on a comparison of the crystal structure of the two Acanthamoeba profilin isoforms, it has been proposed that a surface with a positive electrostatic potential, formed by residues 71, 80, 81, and 115, corresponding to human profilin residues 74, 88, 90, and 125, were the main PI(4, 5)P2 binding sites in Acanthamoeba profilin (Fedorov et al. 1994). This surface largely overlaps with the actin binding surface. Hence, this model explains the observed competition between actin and PI(4, 5)P2 for binding to profilin (Fig. 5; Lassing and Lindberg 1985). Mutagenesis of the yeast homolog partially confirmed this model with residue 71, but not residue 80, being implicated in phosphoinositide binding (Haarer et al. 1993).
Fig. 5.
Profilin structure highlighting the binding sites for phosphatidylinositol 4, 5-bisphosphate [PI(4,5)P2]. Green sticks Residues involved in PI(4,5)P2 binding, solid ribbon backbone of profilin. PDB database: 1PFL
Based on the in silico model of profilin-II derived from profilin-I, Lambrechts et al. (1997) suggested that Glu56 in mammalian profilin-IIA was responsible for the weaker interaction of this isoform because the negative charge of the Glu56 residue reduces the large, positively charged surface around the hypothetical PI(4, 5)P2 binding site. In human profilin, however, only Arg88—and not Arg74—are argued to be involved in PI(4, 5)P2-binding since only the mutant in Arg88 showed decreased inhibition of PI(4, 5)P2 hydrolysis by PLCγ (Sohn et al. 1995)
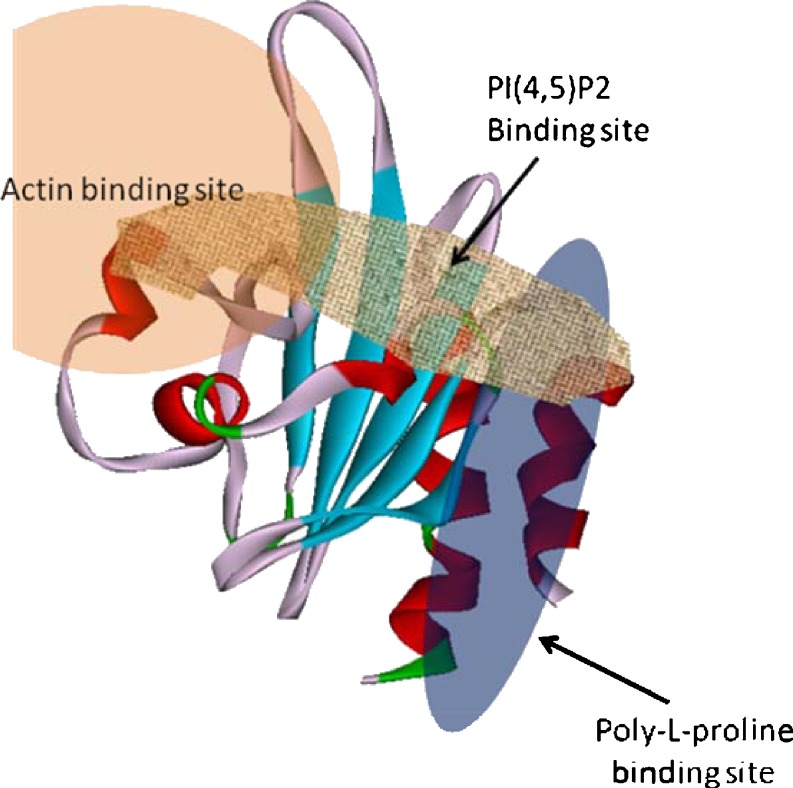
In addition to Arg88, Arg136 in the carboxy terminal helix makes a major contribution to PI(4, 5)P2-binding. Given that the R136D mutant, but not R88A, displays wild-type actin binding activity, it has been proposed that the PI(4, 5)P2 and actin binding sites only partly overlap (Fig. 6). There is also an association between PI(4, 5)P2-binding and the binding of proline-rich ligands. Indeed, the profilin-IIA mutant W3A, which is defective in poly-L-proline binding, shows increased PI(4, 5)P2-binding. Based on observed conformational changes following poly-L-proline and PI(4, 5)P2-binding, Lambrechts et al. (2002) proposed that the correct orientation of the terminal α-helices is important for ligand binding. This is strengthened by the fact that the addition of a myc-tag to the carboxy terminal helix of profilin-IIA abolishes poly-L-proline binding completely (Lambrechts et al. 2002).
Fig. 6.
Human profilin-1 showing actin and poly-L-proline binding sites overlapping with PI(4,5)P2 binding sites
In vitro studies have revealed that PI(4, 5)P2 dissociates actin:profilin complexes (Lassing and Lindberg 1985) and demonstrated the interaction between profilin-I and PI(4, 5)P2 in both micelles and vesicles (Lassing and Lindberg 1988; Goldschmidt-Clermont et al. 1990). PI(4,5)P2 was shown to disrupt the ‘profilactin’ complex, suggesting a mechanism by which ATP–actin monomers are released; this release, in turn, could promote local actin polymerization. More recently, Lu et al. (1996) showed that phosphatidylinositol (3,4)-bisphosphate and phosphatidylinositol (3,4,5)-triphosphate bind to profilin with even higher affinity than PI(4, 5)P2 and that phosphatidylinositol (3,4,5)-triphosphate inhibits profilin sequestering activity much better than PI(4, 5)P2.
In addition, PI(4, 5)P2, bound to profilin, can only be hydrolyzed by phospholipase Cγ1 (PLCγ1) when this lipase is phosphorylated and activated, which occurs in response to transmembrane signaling (Goldschmidt-Clermont et al. 1990; 1991). This leads to two, not mutually exclusive scenarios: (1) profilins are involved in phosphoinositide metabolism and (2) PI(4, 5)P2 hydrolysis causes translocation of profilin from the membrane to the cytosol where it can interact with actin or other ligands. These scenarios suggest an important role for profilin–phosphoinositide interaction in vivo (Janmey et al. 1995; Ostrander et al. 1995). The structural basis for this interaction has, however, only been partly resolved.
Profilin and poly-L-proline
Profilins bind poly-L-proline stretches, and many proteins containing proline-rich domains have been identified as ligands. However, the Vaccinia virus (Machesky et al. 1994) and mouse profilin-IIB spliced form do not show any binding affinity to poly-L-proline (Di Nardo et al. 2000). The enabled/vasodilator stimulated phosphoprotein (Ena/VASP) family is a well-established ligand for profilin (Reinhard et al. 1995; Gertler et al. 1996; Lambrechts et al. 2000a). In neuronal cells, profilin-II has been shown to interact with ligands involved in signal transduction, membrane trafficking, and the recycling of vesicles, such as ROCK2, synapsins, POP-130/CyFIP1, and dynamin-1 (Gareus et al. 2006).
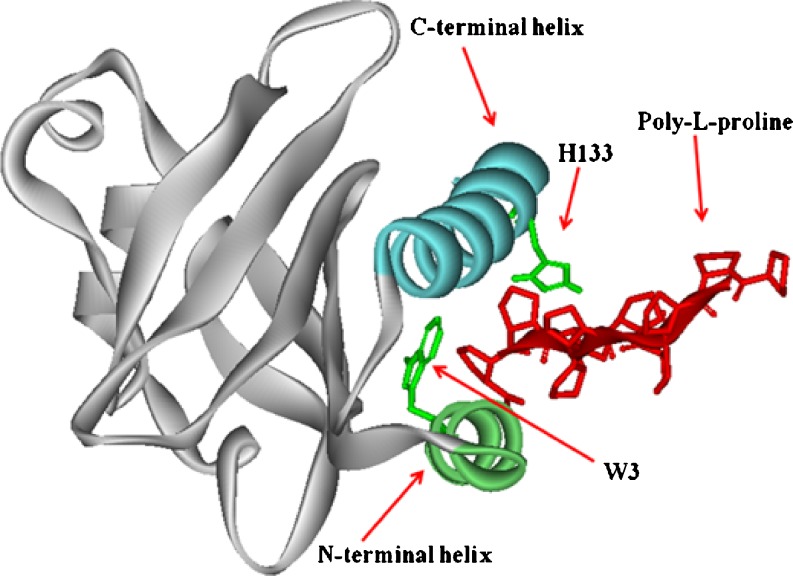
The poly-L-proline binding site has been investigated in crystallographic, mutagenesis, and spectroscopic studies, revealing that a hydrophobic pocket between the amino and carboxy terminal α-helices forms the binding site for poly-L-proline (Fig. 7; Bjorkegren et al. 1993; Haarer et al. 1993; Ostrander et al. 1999).
Fig. 7.
Profilin structure showing the binding of poly-L-proline peptide in the groove between the two alpha helices of the N and C terminus. PDB database: 1AWI-chain A
H133 and W3 are two of the amino acids that have been shown to be involved in binding. Deletion of the last three residues in yeast profilin produces a 100-fold decrease in poly-L-proline affinity. Kaiser and Pollard (1996) reported that the deletion and substitution of as few as three residues at the C-terminus of amoeba profilin reduced poly-L-proline binding.
Profilin mutants unable to bind poly-L-proline but still capable of binding actin and PI(4,5)P2 are lethal, suggesting that poly-L-proline binding is an essential function of profilin. However, the functional mechanisms and cellular pathways regulated by profilin–poly-L-proline interaction remain unclear.
Phosphorylation of profilin
Calf profilin is phosphorylated in the presence of PI(4, 5)P2 (Hansson at al. 1988; Singh et al. 1996) at serine 137 by protein kinase C (PKC); in contrast, plant profilin has been reported to be phosphorylated on multiple tyrosine residues. However, the precise role of such phosphorylations is not yet understood (Guillen et al. 1999).
So far in this article, we have highlighted the importance of poly-L-proline binding and of the carboxy terminal residues involved in this binding. Phosphorylation of the carboxy terminal serine of profilin may therefore be an important regulatory switch controlling profilin interactions with ligands containing proline-rich domains.
Spectrofluorimetric, immunoprecipitation, and Western blot studies have revealed that the phosphorylation of profilin increases its affinity towards actin and poly-L-proline (Sathish et al. 2004). However, the opposite result was obtained by Shao et al. ( 2008) with S137D mutant: the S137D mutant failed to interact with the proline-rich domain-containing huntingtin protein, and there was no profilin-induced huntingtin aggregate formation observed. Further, Sathish et al. (2004) observed increased levels of the actin critical monomer concentration in the presence of phosphoprofilin, indicating that phosphoprofilin may be a better actin sequester. Binding isotherm data and Scatchard plot analysis of the changes in profilin tryptophan fluorescence in the presence of increasing amounts of poly-L-proline revealed that phosphoprofilin possesses a high affinity towards poly-L-proline sequences (Sathish et al. 2004).
Oligomerization of profilins
Several studies (Babich et al. 1996, Mitterman et al. 1998, Wopfner et al. 2002) have reported the spontaneous oligomerization of profilins. In 1996, Babich et al. reported the presence of dimers and tetramers in profilin preparations isolated from human platelets. These authors suggested that the oligomers are formed via sulphydryl and/or ionic bonds and proposed that profilin tetramers are the oligomeric state that binds actin. Oligomerization has also been reported for birch pollen and human and yeast profilins expressed and purified from Escherichia coli (Mitterman et al. 1998). These authors showed that the disulphide bond formation between profilin can be disrupted under reducing conditions but that both reduced and oxidized profilins bind to actin. Wopfner et al. (2002) found that profilin from mugwort pollen formed dimers and tetramers, and they suggested that the oligomeric forms may have a higher allergenic potential than monomers. Although there is evidence that profilin does form oligomers, the presence of these oligomers in cells and their functions remain to be elucidated.
Stabilities of profilin and profilin mutants
The stability of profilins has been investigated by urea denaturation, acid-induced denaturation, and/or thermal denaturation. The mid-point urea denaturation concentration for most of the profilin is approximately 3.5 M. However, some mutants and isoforms show large variations in stability. The denaturation of S. cerevisiae profilin in urea was monitored by fluorescence, and the mid-point urea concentration was found to be 3. 4 M (Eads et al. 1998). These authors also reported a free energy change for unfolding of 5.9 and 1.8 kcal/ mol. Urea-induced unfolding studies were performed on various mutations of S. pombe profilin (Lu and Pollard 2001). The mid-point urea denaturation concentrations for most of the profilin mutants varied from 3.3 to 4.5 M. However, the L121R mutant showed a mid-point of denaturation at much lower urea concentration (1.6 M). The stabilities of some profilin mutants have also been indirectly tested by the different concentrations of urea used to elute them from the ploy-L-proline column. Human profilin-I mutants Y6F and K25Q were eluted with 3.5 M urea, whereas wt profilin was eluted with 7.5 M urea (Sohn et al. 1995).
The mid-point of urea denaturation concentration reported for human profilin-I was 3.5 M (McLachlan et al 2007), while that for the mutants R136D, R88A/R136D, and R88E/R136D were 3.2, 2.8 and 3.5 M, respectively (Lambrechts et al. 2002). Using acid-induced denaturation, an intermediate state was found in profilin-I at pH 4, with a free energy change of 2 kcal/mol (McLachlan et al. 2007). Acanthamoeba profilin-I and -II have a stability similar to that of human profilin-I despite their large difference in sequences (Kaiser and Pollar 1996).
Studies have also been carried out on plant profilin. The thermal unfolding of apple profilin gave a Tm of 65°C (Ma et al. 2006) while oxidized (polymeric) and reduced (monomeric) birch profilin gave a Tm of 42.6°C and 50.5°C, respectively (Mitterman et al. 1998). The mid-point urea denaturation concentration in Z. maize profilin wt was found to be slightly higher than that of human profilin-I with a concentration of 4 M (Karakesisoglou et al. 1996; Kovar et al, 2001), while the concentration for Z. maize profilin mutants Y6Q, K86A (Valenta et al. 1991a) Y6F (Ayscough 1998) and D8A (Ho et al. 1989) were 2.8, 3.7, 3.8, and 3.5 M urea, respectively. These studies clearly demonstrate that mutations may not change the overall folding of the proteins but could severely affect stability and function.
Profilin and cancer
Janke et al. (2000) reported a reduced expression level in tumorigenic breast cancer cells compared to control cells. The identification of the differential expression of profilin genes in fetal rat stomach and normal adult rat stomach has lead to the finding that profilin could be involved in the tumorigenic transformation of mucous cells (Tanaka et al. 1992). Experiments with the temperature-sensitive cell line tsFT101 derived from a mouse mammary carcinoma cell line revealed a wobble mutation in profilin at cysteine 39 complemented by a normal profilin gene, thereby providing evidence that profilin is involved in cancer suppression (Cao et al. 1997). Janke et al. (2000) reported that the level of profilin expressed was low in tumorigenic breast cancer cell lines and that the transfection of profilin-I cDNA into CAL51 cells, a tumorigenic breast cancer cell line, had a prominent effect on cell growth, cytoskeletal organization and spreading, and the suppression of tumorigenicity. Feldner et al. (2002) suggested that the suppressor effect of profilin on tumor cell migration might be through profilin’s function in controlling actin dynamics. Gene silencing studies have revealed that profilin-I is involved in endothelial cell migration and proliferation (Ding et al. 2006) while Zou et al. (2007) and Bae et al. (2009) have shown that profilin-I is also involved in breast cancer cell migration.
Metastasis is one of the major issues in cancer progression. The implied involvement of profilin in cell motility, proliferation, and development has been established unambiguously in lower and higher eukaryotes. When a pancreatic adenocarcinoma cell line (PaCa44) was treated with a chemotherapeutic agent, 5-aza-2′-deoxycytidine (DAC), with the aim of evaluating the effect of this drug on cell growth and protein expression, cell proliferation was strongly inhibited and profilin-I was one of the genes shown to be silenced (Cecconi et al. 2003). Using stable isotope labeling with amino acids in cell culture in pancreatic cancer cells, it has been confirmed that profilin-I is downregulated (Gronborg et al. 2006). The effect of profilin overexpression on the migration of breast cancer cells were studied by Roy and Jacobson (2004), who found that overexpressing profilin in BT474 cells prevented cell migration. They also observed that even a moderate level of profilin overexpression significantly impaired the ability of BT474 cells to spread on a fibronectin-coated substrate and migrate in response to epidermal growth factor (EGF). Furthermore, Wittenmayer et al. (2004) found that the profilin functional actin binding site is required to reduce the tumerogenicity of breast cancer cells.
The results from experiments using membrane-permeable profilin-I (PTD4-PfnI) suggest that profilin-I induces lamellipodia formation independently of the presence of EGF in primary bovine trabecular meshwork cells (Syriani et al. 2008). The effects are time- and dosage-dependent and specific to the profilin-I isoform: the H133S mutation in the poly-L-proline binding domain showed a reduced ability to induce lamellipodia, and the H199E mutation in the actin binding domain failed to induce membrane spreading (Syriani et al. 2008). Profilin-I levels were found to be lower in hepatocarcinoma (SMMC-7721) cells and the all-trans retinoic acid-induced inhibition of proliferation and migration was found to be mediated through profilin-I expression. The overexpression of profilin-I led to the inhibition of cell proliferation and migration, and RNAi-based PFN1 silencing was shown to rescue the inhibitory effect (Wu et al. 2006). The overexpression of wild-type and mutant profilin-I in breast cancer cell lines suggests that intact actin binding site of profilin-I is required to suppress cell motility and that both actin and poly-L-proline binding sites are required for profilin-induced enhancement in cell spreading (Zou et al. 2007).
Das et al. (2009) have recently demonstrated that the overexpression of profilin-I in breast cancer cells dramatically suppresses growth-factor-induced PI(3,4,5)P3 generation. Since membrane phosphoinositides are critical regulators of the actin cytoskeleton, profilin-I could be an important nexus between phosphoinositide-derived signaling and cell motility. Such evidence suggests that profilin-I is a potential cancer suppressor which controls cell migration and proliferation through modulation of actin dynamics.
Conclusions
Profilin was first identified as an actin-binding protein 32 years ago. Since then, its actin binding properties have been extensively studied. New functions have been identified during the last decade, and profilin now appears to occupy a major position at the crossroad of multiple cellular pathways involving cytoskeletal reorganization as well as membrane and nuclear trafficking, vesicle recycling, and signal transduction. However, very little is still known about the molecular mechanisms involving profilin in those many different roles. Deciphering the molecular signals that direct profilin and its isoforms toward specific functions is the task that lies ahead of us.
References
- Akiyama T, Okada M. A mutation in the Drosophila profilin homolog gene affect gametogenesis and bristle development in both sexes. Dev Growth Differ. 1993;35:637–645. doi: 10.1111/j.1440-169X.1993.00637.x. [DOI] [PubMed] [Google Scholar]
- Ayscough KR. In vivo functions of actin-binding proteins. Curr Opin Cell Biol. 1998;10:102–111. doi: 10.1016/S0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Babich M, Foti LR, Sykaluk LL, Clark CR. Profilin forms tetramers that bind to G-actin. Biochem Biophys Res Commun. 1996;218:125–131. doi: 10.1006/bbrc.1996.0022. [DOI] [PubMed] [Google Scholar]
- Babich M, Foti LR, Wong L, Pack GR. In vitro translation and computational analyses of human profilin multimers. Proc West Pharmacol Soc. 2005;48:39–43. [PubMed] [Google Scholar]
- Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219:354–364. doi: 10.1002/jcp.21677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkegren C, Rozycki M, Schutt CE, Lindberg U, Karlsson R. Mutagenesis of human profilin locates its poly(L-proline)-binding site to a hydrophobic patch of aromatic amino acids. FEBS Lett. 1993;333:123–126. doi: 10.1016/0014-5793(93)80388-B. [DOI] [PubMed] [Google Scholar]
- Braun A, Aszódi A, Hellebrand H, Berna A, Fässler R, Brandau O. Genomic organization of profilin-III and evidence for a transcript expressed exclusively in testis. Gene. 2002;283:219–225. doi: 10.1016/S0378-1119(01)00855-1. [DOI] [PubMed] [Google Scholar]
- Buss F, Temm-Grove C, Henning S, Jockusch BM. Distribution of profilin in fibroblasts correlates with the presence of highly dynamic actin filaments. Cell Motil Cytoskeleton. 1992;22:51–61. doi: 10.1002/cm.970220106. [DOI] [PubMed] [Google Scholar]
- Cao Y, Motomura K, Ohtsuru A, Matsumoto T, Yamashita S, Kosaka M. Profilin gene expression and regulation in a temperature-sensitive breast cancer cell line: tsFT101. Pflugers Arch. 1997;434:341–345. doi: 10.1007/s004240050406. [DOI] [PubMed] [Google Scholar]
- Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin: a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- Cecconi D, Astner H, Donadelli M, Palmieri M, Missiaglia E, Hamdan M, Scarpa A, Righetti PG. Proteomic analysis of pancreatic ductal carcinoma cells treated with 5-aza-2′-deoxycytidine. Electrophoresis. 2003;24:4291–4303. doi: 10.1002/elps.200305724. [DOI] [PubMed] [Google Scholar]
- Cedergren-Zeppezauer ES, Goonesekere NC, Rozycki MD, Myslik JC, Dauter Z, Lindberg U, Schutt CE. Crystallization and structure determination of bovine profilin at 2.0 A resolution. J Mol Biol. 1994;240:459–475. doi: 10.1006/jmbi.1994.1461. [DOI] [PubMed] [Google Scholar]
- Chaudhary A, Chen J, Gu QM, Witke W, Kwiatkowski DJ, Prestwich GD. Probing the phosphoinositide 4, 5-bisphosphate binding site of human profilin I. Chem Biol. 1998;5:273–281. doi: 10.1016/S1074-5521(98)90620-2. [DOI] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE. The structure of an open state of beta-actin at 2.65 A resolution. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218:436–443. doi: 10.1002/jcp.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Gareus R, Kwiatkowski D, Witke W. Alternative splicing of the mouse profilin II gene generates functionally different profilin isoforms. J Cell Sci. 2000;113:3795–3803. doi: 10.1242/jcs.113.21.3795. [DOI] [PubMed] [Google Scholar]
- Ding Z, Lambrechts A, Parepally M, Roy P. Silencing profilin-1 inhibits endothelial cell proliferation, migration and cord morphogenesis. J Cell Sci. 2006;119:4127–4137. doi: 10.1242/jcs.03178. [DOI] [PubMed] [Google Scholar]
- Dong J, Radau B, Otto A, Muller E, Lindschau C, Westermann P. Profilin I attached to the Golgi is required for the formation of constitutive transport vesicles at the trans-Golgi network. Biochim Biophys Acta. 2000;1497:253–260. doi: 10.1016/S0167-4889(00)00056-2. [DOI] [PubMed] [Google Scholar]
- Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen K, Rubenstein PA, Haarer BK, Almo SC. Structure Determination and haracterization of Saccharomyces cerevisiae Profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: Insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284:2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov AA, Magnus KA, Graupe MH, Lattman EE, Pollard TD, Almo SC. X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates. Proc Natl Acad Sci USA. 1994;91:8636–8640. doi: 10.1073/pnas.91.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldner JC, Brandt BH. Cancer cell motility-on the road from c-erbB-2 receptor steered signaling to actin reorganization. Exp Cell Res. 2002;272:93–108. doi: 10.1006/excr.2001.5385. [DOI] [PubMed] [Google Scholar]
- Gareus R, Di Nardo A, Rybin V, Witke W. Mouse profilin 2 regulates endocytosis and competes with SH3-ligand binding to dynamin 1. J Biol Chem. 2006;28:2803–2811. doi: 10.1074/jbc.M503528200. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/S0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Gieselmann R, Kwiatkowski DJ, Janmey PA, Witke W. Distinct biochemical characteristics of the two human profilin isoforms. Eur J Biochem. 1995;229:621–628. doi: 10.1111/j.1432-1033.1995.tb20506.x. [DOI] [PubMed] [Google Scholar]
- Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, Jockusch H. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Kim JW, Machesky LM, Rhee SG, Pollard TD. Regulation of phospholipase C-gamma 1 by profilin and tyrosine phosphorylation. Science. 1991;251:1231–1233. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The control of actin nucleotide exchange by thymosin-β4 and profilin: A potential regulatory mechanism for actin polymerization in cells. Mol Biol Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- Guillen G, Valdes-Lopez V, Noguez R, Olivares J, Rodriguez-Zapata LC, Perez H, Vidali L, Villanueva MA, Sanchez F. Profilin in Phaseolus vulgaris is encoded by two genes (only one expressed in root nodules) but multiple isoforms are generated in vivo by phosphorylation on tyrosine residues. Plant J. 1999;19:497–508. doi: 10.1046/j.1365-313X.1999.00542.x. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Petzold AS, Brown SS. Mutational analysis of yeast profilin. Mol Cell Biol. 1993;13:7864–7873. doi: 10.1128/mcb.13.12.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson A, Skoglund G, Lassing I, Lindberg U, Ingelman-Sundberg M. Protein kinase C-dependent phosphorylation of profilin is specifically stimulated by phosphatidylinositol bisphosphate (PIP2) Biochem Biophys Res Commun. 1988;150:526–531. doi: 10.1016/0006-291X(88)90425-1. [DOI] [PubMed] [Google Scholar]
- Haugwitz M, Noegel AA, Karakesisoglou J, Schleicher M. Dictyostelium amoebae that lack G-actin-sequestering profilins show defects in F-actin content, cytokinesis, and development. Cell. 1994;79:303–314. doi: 10.1016/0092-8674(94)90199-6. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Janke J, Schluter K, Jandrig B, Theile M, Kolble K, Arnold W, Grinstein E, Schwartz A, Estevez-Schwarz L, Schlag PM, Jockusch BM, Scherneck S. Suppression of tumorigenicity in breast cancer cells by the microfilament protein profilin-1. J Exp Med. 2000;191:1675–1686. doi: 10.1084/jem.191.10.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Protein regulation by phosphatidylinositol lipids. Chem Biol. 1995;2:61–65. doi: 10.1016/1074-5521(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Pollard TD. Characterization of actin and poly-L-proline binding sites of Acanthamoeba profilin with monoclonal antibodies and by mutagenesis. J Mol Biol. 1996;256:89–107. doi: 10.1006/jmbi.1996.0070. [DOI] [PubMed] [Google Scholar]
- Karakesisoglou I, Schleicher M, Gibbon BC, Staiger CJ. Plant profilins rescue the aberrant phenotype of profilin-deficient Dictyostelium cells. Cell Motil Cytoskeleton. 1996;34:36–47. doi: 10.1002/(SICI)1097-0169(1996)34:1<36::AID-CM4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kocks C. Intracellular motility: Profilin puts pathogens on the actin drive. Curr Biol. 1994;4:465–468. doi: 10.1016/S0960-9822(00)00105-6. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Drobak BK, Collings DA, Staiger CJ. The characterization of ligand-specific maize (Zea mays) profilin mutants. Biochem J. 2001;358:49–57. doi: 10.1042/0264-6021:3580049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Bruns GA. Human profilin: Molecular cloning, sequence comparison and chromosomal analysis. J Biol Chem. 1988;263:5910–5915. [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts AI, Kwiatkowski A, Lanier LM, Bear JE, Vandekerckhove J, Ampe C, Gertler FB. cAMP-dependent protein kinase phosphorylation of EVL, a Mena/VASP relative, regulates its interaction with actin and SH3 domains. J Biol Chem. 2000;275:36143–36151. doi: 10.1074/jbc.M006274200. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Braun A, Jonckheere V, Aszodi A, Lanier LM, Robbens J, Van Colen I, Vandekerckhove J, Fässler R, Ampe C. Profilin II is alternatively spliced, resulting in profilin isoforms that are differentially expressed and have distinct biochemical properties. Mol Cell Biol. 2000;20:8209–8219. doi: 10.1128/MCB.20.21.8209-8219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C. Mutational analysis of human profilin I reveals a second PI(4, 5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem. 2002;3:1–12. doi: 10.1186/1471-2091-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4, 5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specificity of the interaction between phosphatidylinositol 4, 5-bisphosphate and the profilin:actin complex. J Cell Biochem. 1988;37:255–267. doi: 10.1002/jcb.240370302. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Shieh WR, Rhee SG, Yin HL, Chen CS. Lipid products of phosphoinositide 3-kinase bind human profilin with high affinity. Biochemistry. 1996;35:14027–14034. doi: 10.1021/bi961878z. [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zuidmeerw L, Bohle B, Bolhaarz STH, Gadermaier G, Gonzalez-Manceboz E, Fernandez-Rivasz M, Knulstz AC, Himly M, Aserok R, Ebner C, van Reew R, Ferreira F, Breiteneder H, Hoffmann-Sommergruber K. Characterization of recombinant Mal d 4 and its application for componentresolved diagnosis of apple allergy. Clin Exp Allergy. 2006;36:1087–1096. doi: 10.1111/j.1365-2222.2006.02541.x. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Cole NB, Moss B, Pollard TD. Vaccinia virus expresses a novel profilin with a higher affinity for polyphosphoinositides than actin. Biochemistry. 1994;33:10815–10824. doi: 10.1021/bi00201a032. [DOI] [PubMed] [Google Scholar]
- Magdolen V, Oechsner U, Müller G, Bandlow W. The intron-containing gene for yeast profilin (PFY) encodes a vital function. Mol Cell Biol. 1988;8:5108–5115. doi: 10.1128/mcb.8.12.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayboroda O, Schluter K, Jockusch BM. Differential colocalization of profilin with microfilaments in PtK2 cells. Cell Motil Cytoskeleton. 1997;37:166–177. doi: 10.1002/(SICI)1097-0169(1997)37:2<166::AID-CM9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- McLachlan GD, Cahill SM, Girvin ME, Almo SC. Acid-induced equilibrium folding intermediate of human platelet profilin. Biochemistry. 2007;46:6931–6943. doi: 10.1021/bi0602359. [DOI] [PubMed] [Google Scholar]
- Mitterman I, Fetrow JS, Schaak DL, Almo SC, Kraft D, Heberle-Bors E, Valenta R. Oligomerization of profilins from birch, man and yeast: Profilin, a ligand for itself? Sex Plant Reprod. 1998;11:183–191. doi: 10.1007/s004970050140. [DOI] [Google Scholar]
- Nodelman IM, Bowman GD, Lindberg U, Schutt CE. X-ray structure determination of human profilin II: a comparative structural analysis of human profilins. J Mol Biol. 1999;294:1271–1285. doi: 10.1006/jmbi.1999.3318. [DOI] [PubMed] [Google Scholar]
- Obermann H, Raabe I, Balvers M, Brunswig B, Schulze W, Kirchhoff C. Novel testis-expressed profilin IV associated with acrosome biogenesis and spermatid elongation. Mol Hum Reprod. 2005;11:53–64. doi: 10.1093/molehr/gah132. [DOI] [PubMed] [Google Scholar]
- Ostrander DB, Gorman JA, Carman GM. Regulation of profilin localization in Saccharomyces cerevisiae by phosphoinositide metabolism. J Biol Chem. 1995;270:27045–27050. doi: 10.1074/jbc.270.45.27045. [DOI] [PubMed] [Google Scholar]
- Ostrander DB, Ernst EG, Lavoie TB, Gorman JA. Polyproline binding is an essential function of human profilin in yeast. Eur J Biochem. 1999;262:26–35. doi: 10.1046/j.1432-1327.1999.00354.x. [DOI] [PubMed] [Google Scholar]
- Polet D, Lambrechts A, Ono K, Mah A, Peelman F, Vandekerckhove J, Baillie DL, Ampe C, Ono S. Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil Cytoskeleton. 2006;63:14–28. doi: 10.1002/cm.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Jacobson K. Overexpression of profilin reduces the migration of invasive breast cancer cells. Cell Motil Cytoskeleton. 2004;57:84–95. doi: 10.1002/cm.10160. [DOI] [PubMed] [Google Scholar]
- Sathish K, Padmaa B, Veerendra M, Bhargavic V, Radhikaa KVN, Wasiaa R, Sairamd M, Surya SS. Phosphorylation of profilin regulates its interaction with actin and poly (L-proline) Cell Signal. 2004;16:589–596. doi: 10.1016/j.cellsig.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Shao J, Welch WJ, Diprospero NA, Diamond MI. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol Cell Biol. 2008;28:5196–5208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Chauhan A, Murakami N, Styles J, Elzinga M, Chauhan VP. Phosphoinositide-dependent in vitro phosphorylation of profilin by protein kinase C: Phospholipid specificity and localization of the phosphorylation site. Recept Signal Transduct. 1996;6:77–86. [PubMed] [Google Scholar]
- Skare P, Kreivi JP, Bergstrom A, Karlsson R. Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res. 2003;286:12–21. doi: 10.1016/S0014-4827(03)00102-2. [DOI] [PubMed] [Google Scholar]
- Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ. Localization of a binding site for phosphatidylinositol 4, 5-bisphosphate on human profilin. J Biol Chem. 1995;270:21114–21120. doi: 10.1074/jbc.270.36.21114. [DOI] [PubMed] [Google Scholar]
- Sonobe S, Takahashi S, Hatano S, Kuroda K. Phosphorylation of amoeba G-actin and its effect on actin polymerization. J Biol Chem. 1986;261:14837–14843. [PubMed] [Google Scholar]
- Syriani E, Gomez-Cabrero A, Bosch M, Moya A, Abad E, Gual A, Gasull X, Morales M. Profilin induces lamellipodia by growth factor-independent mechanism. FASEB J. 2008;22:1581–1596. doi: 10.1096/fj.06-7654com. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sasaki H, Kino I, Sugimura T, Terada M. Genes preferentially expressed in embryo stomach are predominantly expressed in gastric cancer. Cancer Res. 1992;52:3372–3377. [PubMed] [Google Scholar]
- Theriot JA, Rosenblatt J, Portnoy DA, Goldschmidt-Clermont PJ, Mitchison TJ. Involvement of profilin in the actin-based motility of L. monocytogenes in cells and in cell-free extracts. Cell. 1994;76:505–517. doi: 10.1016/0092-8674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Tseng PC-H, Runge MS, Cooper JA, Williams RC, Pollard TD. Physical, immunochemical, and functional properties of Acanthamoeba proflin. J Cell Biol. 1984;98:214–221. doi: 10.1083/jcb.98.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta R, Breiteneder H, Pettenburger K, Breitenbach M, Rumpold H, Kraft D, Scheiner O. Homology of the major birch-pollen allergen, Bet vI, with the major pollen allergens of alder, hazel, and hornbeam at the nucleic acid level as determined by cross-hybridization. J Allergy Clin Immunol. 1991;87:677–682. doi: 10.1016/0091-6749(91)90388-5. [DOI] [PubMed] [Google Scholar]
- Valenta R, Duchene M, Breitenbach M, Pettenburger K, Koller L, Rumpold H, Scheiner O, Kraft D. A low molecular weight allergen of white birch (Betula verrucosa) is highly homologous to human profilin. Int Arch Allergy Appl Immunol. 1991;94:368–370. doi: 10.1159/000235406. [DOI] [PubMed] [Google Scholar]
- Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- Vorum H, Ostergaard M, Hensechke P, Enghild JJ, Riazati M, Rice GE. Proteomic analysis of hyperoxia-induced responses in the human choriocarcinoma cell line JEG-3. Proteomics. 2004;4:861–867. doi: 10.1002/pmic.200300639. [DOI] [PubMed] [Google Scholar]
- Widada JS, Ferraz C, Liautard JP. Total coding sequence of profilin cDNA from Mus musculus macrophage. Nucleic Acids Res. 1989;17:2855. doi: 10.1093/nar/17.7.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 2004;14:461–469. doi: 10.1016/j.tcb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Witke W, Podtelejnikov AV, Di Nardo A, Sutherland JD, Gurniak CB, Dotti C, Mann M. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I is essential for cell survival and cell division in early mouse development. Proc Natl Acad Sci USA. 2001;98:3832–3836. doi: 10.1073/pnas.051515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenmayer N, Rothkegel M, Jockusch BM, Schluter K. Functional characterization of green fluorescent protein-profilin fusion proteins. Eur J Biochem. 2000;267:5247–5256. doi: 10.1046/j.1432-1327.2000.01600.x. [DOI] [PubMed] [Google Scholar]
- Wittenmayer N, Jandrig B, Rothkegel M, Schluter K, Arnold W, Haensch W, Scherneck S, Jockusch BM. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15:1600–1608. doi: 10.1091/mbc.E03-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopfner N, Willeroidee M, Hebenstreit D, van Ree R, Aalbers M, Briza P, Thalhamer J, Ebner C, Richter K, Ferreira F. Molecular and immunological characterization of profilin from mugwort pollen. Biol Chem. 2002;383:1779–1789. doi: 10.1515/BC.2002.199. [DOI] [PubMed] [Google Scholar]
- Wu N, Zhang W, Yang Y, Liang YL, Wang LY, Jin JW, Cai XM, Zha XL. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics. 2006;6:6095–6106. doi: 10.1002/pmic.200500321. [DOI] [PubMed] [Google Scholar]
- Yu F, Sun HQ, Janmey PA, Yin HL. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J Biol Chem. 1992;267:14616–14621. [PubMed] [Google Scholar]
- Zou L, Jaramillo M, Whaley D, Wells A, Panchapakesa V, Das T, Roy P. Profilin-1 is a negative regulator of mammary carcinoma aggressiveness. Br J Cancer. 2007;97:1361–1371. doi: 10.1038/sj.bjc.6604038. [DOI] [PMC free article] [PubMed] [Google Scholar]