Abstract
The evolution of CRISPR–cas loci, which encode adaptive immune systems in archaea and bacteria, involves rapid changes, in particular numerous rearrangements of the locus architecture and horizontal transfer of complete loci or individual modules. These dynamics complicate straightforward phylogenetic classification, but here we present an approach combining the analysis of signature protein families and features of the architecture of cas loci that unambiguously partitions most CRISPR–cas loci into distinct classes, types and subtypes. The new classification retains the overall structure of the previous version but is expanded to now encompass two classes, five types and 16 subtypes. The relative stability of the classification suggests that the most prevalent variants of CRISPR–Cas systems are already known. However, the existence of rare, currently unclassifiable variants implies that additional types and subtypes remain to be characterized.
The CRISPR–Cas modules are adaptive immune systems that are present in most archaea and many bacteria1–5 and provide sequence-specific protection against foreign DNA or, in some cases, RNA6. A CRISPR locus consists of a CRISPR array, comprising short direct repeats separated by short variable DNA sequences (called ‘spacers’), which is flanked by diverse cas genes. CRISPR–Cas immunity involves three distinct mechanistic stages: adaptation, expression and interference7–11. The adaptation stage involves the incorporation of fragments of foreign DNA (known as ‘protospacers’) from invading viruses and plasmids into the CRISPR array as new spacers. These spacers provide the sequence memory for a targeted defence against subsequent invasions by the corresponding virus or plasmid. During the expression stage, the CRISPR array is transcribed as a precursor transcript (pre-crRNA), which is processed and matured to produce CRISPR RNAs (crRNAs). During the interference stage, crRNAs, aided by Cas proteins, function as guides to specifically target and cleave the nucleic acids of cognate viruses or plasmids7,9,12,13. Recent studies suggest that CRISPR–Cas systems can also be used for non-defence roles, such as the regulation of collective behaviour and pathogenicity14–16.
Numerous, highly diverse Cas proteins are involved in the different stages of CRISPR activity (Box 1; see Supplementary information S1 (table)). Briefly, Cas1 and Cas2, which are present in most known CRISPR–Cas systems, form a complex that represents the adaptation module and is required for the insertion of spacers into CRISPR arrays17,18. Protospacer acquisition in many CRISPR–Cas systems requires recognition of a short protospacer adjacent motif (PAM) in the target DNA19–22. During the expression stage, the pre-crRNA molecule is bound to either Cas9 (which is a single, multidomain protein) or to a multisubunit complex, forming the crRNA–effector complex. The pre-crRNA is processed into crRNAs by an endonuclease subunit of the multisubunit effector complex23 or via an alternative mechanism that involves bacterial RNase III and an additional RNA species, the tracrRNA (transactivating CRISPR RNA)24. Finally, at the interference stage, the mature crRNA remains bound to Cas9 or to the multi-subunit crRNA–effector complex, which recognizes and cleaves the cognate DNA10,11,25,26 or RNA26–31.
Box 1. Cas protein families and functional modules.
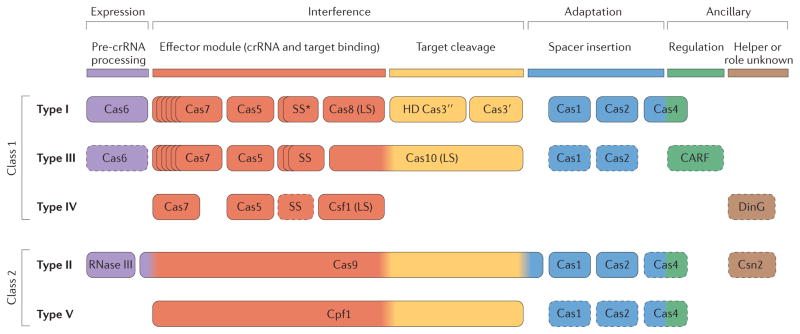
The Cas proteins can be divided into four distinct functional modules: adaptation (spacer acquisition); expression (crRNA processing and target binding); interference (target cleavage); and ancillary (regulatory and other CRISPR-associated functions) (FIG. 1). In recent years, a wealth of structural and functional information has accumulated for the core Cas proteins (Cas1 Cas10) (see Supplementary information S1 (table)), which allows them to be classified into these modules.
The adaptation module is largely uniform across CRISPR Cas systems and consists of the Cas1 and Cas2 proteins, with possible additional involvement of the restriction endonuclease superfamily enzyme Cas4 (REF. 91) and, in type II systems, Cas9 (REFS 63,64). Cas1, which adopts a unique α-helical fold, is an integrase that mediates the insertion of new spacers into CRISPR arrays by cleaving specific sites within the repeats17,89,92. The role of Cas2, which is a homologue of the mRNA interferase toxins of numerous toxin antitoxin systems, is less well understood3,72,93,94. Cas2 has been shown to form a complex with Cas1 in the Escherichia coli type I CRISPR Cas system and is required for adaptation. However, although Cas2 has RNase95 and DNase activities96, its catalytic residues are dispensable for adaptation17, indicating that these activities are not directly involved in this process, at least in this species.
The expression and interference modules are represented by multisubunit CRISPR RNA (crRNA) effector complexes36,38,39,43,46,97,98 (BOX 2) or, in type II systems, by a single large protein, Cas9(REFS 24,25,99). In the expression stage, pre-crRNA is bound to the multisubunit crRNA effector complex, or to Cas9, and processed into a mature crRNA in a step catalysed by an RNA endonuclease23 (typically Cas6; in type I and type III systems) or an alternative mechanism that involves RNase III and a transactivating CRISPR RNA (tracrRNA)24 (in type II systems). However, in at least one type II CRISPR Cas system, that of Neisseria meningitidis, crRNAs with mature 5′ ends are directly transcribed from internal promoters, and crRNA processing does not occur69.
In the interference module, the crRNA effector complex (in type I and type III systems) or Cas9 (in type II systems) combines nuclease activity with dedicated RNA-binding domains. Target binding relies on base pair formation with the spacer region of the crRNA. Cleavage of the target is catalysed by the HD family nuclease (Cas3″ or a domain in Cas3) in type I systems52,100, by the combined action of the Cas7 and Cas10 proteins in type III systems26,39,46,101,104 or by Cas9 in type II systems25. In type I systems, the HD nuclease domain is either fused to the superfamily 2 helicase Cas3′ (REFS 50–52) or is encoded by a separate gene, cas3″, whereas in type III systems a distinct HD nuclease domain is fused to Cas10 and is thought to cleave single-stranded DNA during interference105. In type II systems, the RuvC-like nuclease (RNase H fold) domain and the HNH (McrA-like) nuclease domain of Cas9 each cleave one of the strands of the target DNA 25,106. Remarkably, the large (~950 1,400 amino acids) multidomain Cas9 protein is required for all three of the functional steps of CRISPR-based immunity (adaptation, expression and interference) in type II systems and thus concentrates much of the CRISPR Cas system’s function in a single protein.
The ancillary module is a combination of various proteins and domains that, with the exception of Cas4, are much less common than the core Cas proteins in CRISPR Cas systems. Aside from its putative role in adaptation, Cas4 is thought to contribute to CRISPR Cas-coupled programmed cell death3,94. Other notable components of the ancillary module include: a diverse set of proteins containing the CRISPR-associated Rossmann fold (CARF) domain35,107, which have been hypothesized to regulate CRISPR Cas activity107 (in many type I and type III systems); and the inactivated P-loop ATPase Csn2, which forms a homotetrameric ring that accommodates linear double-stranded DNA in the central hole (in type II systems)108,111. Csn2 is not required for interference but apparently has a role in spacer integration, possibly preventing damage from the double-strand break in the chromosomal DNA6,110. Ancillary module genes are often found outside of CRISPR cas loci, but the functions of these stand-alone genes have not been characterized in depth72,94.
The rapid evolution of most cas genes32–34 and the remarkable variability in the genomic architecture of CRISPR–cas loci poses a major challenge for the consistent annotation of Cas proteins and for the classification of CRISPR–Cas systems13,35. Nevertheless, a consistent classification scheme is essential for expedient and robust characterization of CRISPR–cas loci in new genomes, and thus important for further progress in CRISPR research. Owing to the complexity of the gene composition and genomic architecture of the CRISPR–Cas systems, any single, all-encompassing classification criterion is rendered impractical, and thus a ‘polythetic’ approach based on combined evidence from phylogenetic, comparative genomic and structural analysis was developed13. At the top of the classification hierarchy are the three main types of CRISPR–Cas systems (type I–type III). These three types are readily distinguishable by virtue of the presence of unique signature proteins: Cas3 for type I, Cas9 for type II and Cas10 for type III13. Within each type of CRISPR–Cas system, several subtypes have been delineated based on additional signature genes and characteristic gene arrangements13,35. Recently, in-depth sequence and structural analysis of the effector complexes from different variants of CRISPR–Cas systems has uncovered common principles of their organization and function4,30,31,36–46. In parallel, the biotechnological development of molecular components of type II CRISPR–Cas systems into a powerful new generation of genome editing and engineering tools has triggered intensive research into the functions and mechanisms of these systems, thereby advancing our understanding of the Cas proteins and associated RNAs47,48.
In this Analysis article, we refine and extend the classification of CRISPR–cas loci based on a comprehensive analysis of the available genomic data. As a result of this analysis, we introduce two classes of CRISPR–Cas systems as a new, top level of classification and define two putative new types and five new subtypes within these classes, resulting in a total of five types and 16 subtypes. We employ this classification to analyse the evolutionary relationships between CRISPR–cas loci using several measures. The results of this analysis highlight pronounced modularity as an emerging trend in the evolution of CRISPR–Cas systems. Finally, we demonstrate the potential for automated annotation of CRISPR–cas loci by developing a computational approach that uses the new classification to assign CRISPR–Cas system subtype with high precision.
Classification of CRISPR–cas loci
The classification of CRISPR–Cas systems should ideally represent the evolutionary relationships between CRISPR–cas loci. However, the pervasive exchange and divergence of cas genes and gene modules has resulted in a complex network of evolutionary relationships that cannot be readily (and cleanly) partitioned into a small number of distinct groupings (although such partitioning might be achievable for individual modules, see below). Therefore, we adopted a two-step classification approach that first identified all cas genes in each CRISPR–cas locus and then determined the signature genes and distinctive gene architectures that would allow the assignment of these loci to types and subtypes.
To robustly identify cas genes, which is a non-trivial task owing to high sequence variability, we developed a library of 394 position-specific scoring matrices (PSSM)49 for all 93 known protein families associated with CRISPR–Cas systems (see Supplementary information S2 (table)). Importantly, this set included 229 PSSMs for recently characterized families that were not part of the previous CRISPR–Cas classification13. The PSSMs were used to search the protein sequences annotated in 2,751 complete archaeal and bacterial genomes that were available at the National Center for Biotechnology Information (NCBI) as of 1 February 2014 (see Supplementary information S3 (box) for a detailed description of the methods). A highly significant similarity threshold was used to identify bona fide cas genes. Genes that were located in the same genomic neighbourhood as bona fide cas genes (irrespectively of their proximity to a CRISPR array) and that encoded proteins with moderate similarity to Cas PSSMs were then identified as putative cas genes. This two-step procedure was devised to minimize the false-positive rate, while allowing the detection of diverged variants of Cas proteins.
Gene neighbourhoods around the identified cas genes were merged into 1,949 distinct cas loci from 1,302 of the 2,751 analysed genomes, including 1,694 complete loci. A cas locus was annotated as ‘complete’ if it encompassed at least the full complement of genes for the main components of the interference module (the multisubunit crRNA–effector complex or Cas9). This criterion was adopted because, although the adaptation module genes cas1 and cas2 are the most common cas genes, many otherwise complete (and hence thought to be functionally active) CRISPR–Cas systems lack cas1 and cas2 and seem to instead depend on adaptation modules from other loci in the same genome. Within the set of complete loci, 111 composite loci that contained two or more adjacent CRISPR–Cas units (each consisting of at least a full complement of essential effector complex components) were identified and split into distinct units. Each locus or unit was classified by scoring type-specific and subtype-specific PSSMs that were constructed from multiple sequence alignments of the respective signature Cas proteins (see Supplementary information S2,S4 (tables)). For some of the more diverged signature proteins, multiple PSSMs were required for a single protein to capture the entire diversity of the cognate CRISPR–Cas subtype.
Of the single-unit complete loci, 1,574 (93%) were assigned to a specific subtype or the newly defined putative types IV and V, which are not split into subtypes, eight were identified up to the type only and one remained unclassified by our procedure (a subtype I-D system operon that is adjacent to the remnants of a subtype III-B system operon disrupted by recombination).
Our analysis suggests that the CRISPR–Cas systems can be divided on the basis of the genes encoding the effector modules; that is, whether the systems have several variants of a multisubunit complex (the CRISPR-associated complex for antiviral defence (Cascade) complex, the Csm complex or the Cmr complex) or Cas9. Thus, we introduce a new, broadest level of classification of CRISPR–Cas systems, which divides them into ‘class 1’ and ‘class 2’. Class 1 systems possess multisubunit crRNA–effector complexes, whereas in class 2 systems all functions of the effector complex are carried out by a single protein, such as Cas9. We also find evidence for two putative new types, type IV and type V, which belong to class 1 and class 2, respectively. These observations result in a new classification system in which CRISPR–Cas systems are clustered into five types, each with a distinctive composition of expression, interference and adaptation modules (FIG. 1). These five types are divided into 16 subtypes, including five new subtypes (II-C, III-C and III-D, together with the single subtypes of type IV and type V systems), as detailed below.
Figure 1. Functional classification of Cas proteins.
Protein names follow the current nomenclature and classification13. An asterisk indicates that the putative small subunit (SS) protein is instead fused to Cas8 (the type I system large subunit (LS)) in several type I subtypes33. The type III system LS and type IV system LS are Cas10 and Csf1 (a Cas8 family protein), respectively. Dispensable components are indicated by dashed outlines. Cas6 is shown with a solid outline for type I because it is dispensable in some but not most systems and by a dashed line for type III because most systems lack this gene and use the Cas6 provided in trans by other CRISPR–cas loci. The two colours for Cas4 and three colours for Cas9 reflect that these proteins contribute to different stages of the CRISPR Cas response. The functions shown for type IV and type V system components are proposed based on homology to the cognate components of other systems, and have not yet been experimentally verified. The functional assignments for Cpf1 are tentatively inferred by analogy with Cas9 (only the RuvC (and TnpB)-like domains of the two proteins are homologous). CARF, CRISPR-associated Rossmann fold; pre-crRNA, pre-CRISPR RNA. This research was originally published in Biochem. Soc. Trans. Makarova K. S., Wolf Y. I., & Koonin E. V. The basic building blocks and evolution of CRISPR–Cas systems. Biochem. Soc. Trans. 2013; 41: 1392–1400 © The Biochemical Society.
Class 1 CRISPR Cas systems
Class 1 CRISPR–Cas systems are defined by the presence of a multisubunit crRNA–effector complex. The class includes type I and type III CRISPR–Cas systems, as well as the putative new type IV.
Type I CRISPR–Cas systems
All type I loci contain the signature gene cas3 (or its variant cas3′), which encodes a single-stranded DNA (ssDNA)-stimulated superfamily 2 helicase with a demonstrated capacity to unwind double-stranded DNA (dsDNA) and RNA–DNA duplexes50–52. Often, the helicase domain is fused to a HD family endonuclease domain that is involved in the cleavage of the target DNA50,53. The HD domain is typically located at the amino terminus of Cas3 proteins (with the exception of subtype I-U and several subtype I-A systems, in which the HD domain is at the carboxyl terminus of Cas3) or is encoded by a separate gene (cas3″) that is usually adjacent to cas3′ (FIG. 1).
Type I systems are currently divided into seven subtypes, I-A to I-F and I-U, all of which have been defined previously13. In the case of subtype I-U, U stands for uncharacterized because the mechanism of pre-crRNA cleavage and the architecture of the effector complex for this system remain unknown33. The type I-C, I-D, I-E and I-F CRISPR–Cas systems are typically encoded by a single (predicted) operon that encompasses the cas1, cas2 and cas3 genes together with the genes for the subunits of the Cascade complex (BOX 2). By contrast, many type I-A and I-B loci seem to have a different organization in which the cas genes are clustered in two or more (predicted) operons35. In most type I loci, each of the cas gene families is represented by a single gene.
Box 2. Structural composition of multiprotein crRNA–effector complexes.
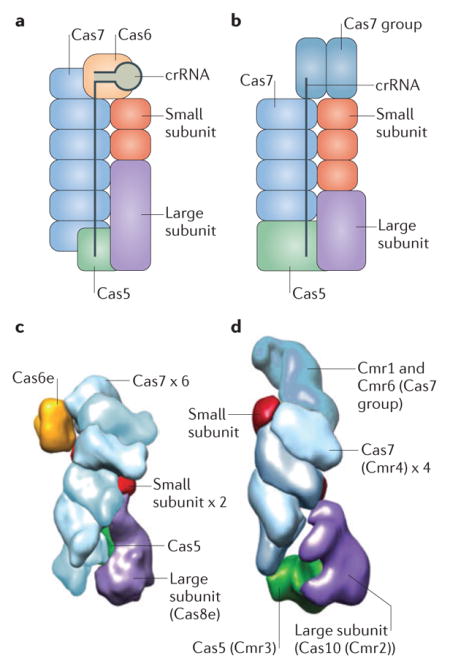
In type I and type III CRISPR Cas systems, multiprotein CRISPR RNA (crRNA) effector complexes mediate the processing and interference stages of the CRISPR defence system. In type I systems, this complex is known as the CRISPR-associated complex for antiviral defence (Cascade; see the figure, part a) complex, whereas in type III-A and type III-B systems the complexes are respectively known as Csm and Cmr (see the figure, part b) complexes. A common structural feature among the Cas proteins found in crRNA effector complexes is the RNA recognition motif (RRM), a nucleic acid-binding domain that is the core fold of the extremely diverse RAMP protein superfamily4,32,34. The RAMPs Cas5 and Cas7 comprise the skeleton of the crRNA effector complexes. In type I systems, Cas6 is typically the active endonuclease that is responsible for crRNA processing, and Cas5 and Cas7 are non-catalytic RNA-binding proteins; however, in type I-C systems, crRNA processing is catalysed by Cas5 (REF. 55). In type III systems, the enzyme that is responsible for processing has not been directly identified but is generally assumed to be Cas6 (REFS 38–40; however, Cas6 is not a subunit of the effector complex in these systems, and in some cases is provided in trans by other CRISPR Cas loci), whereas Cas7 is involved in co-transcriptional RNA degradation during the interference stage26.
In addition to Cas5, Cas6 and Cas7, crRNA effector complexes typically contain two proteins that are designated, according to their size, the large subunit and the small subunit. The large subunit is present in all known type I and type III crRNA effector complexes, whereas the small subunit is missing in some type I loci; a carboxy-terminal domain of the large subunit is predicted to functionally replace the small subunit in complexes where the small subunit is absent33. In type III systems, the large subunit is the putative cyclase-related enzyme encoded bycas10, whereas in type I systems the large subunit is encoded by diversecas8 genes that adopt a complex structure and show no readily detectable similarity to other proteins. Cas10 contains two cyclase-like Palm domains (a form of the RRM domain)112,113, and the conservation of catalytic amino acid residues implies that one of these domains is active whereas the other is inactivated; the catalytic site of the active domain is required for cleavage of double-stranded DNA during interference26, but its activity remains to be characterized in detail. Although it has been speculated that Cas8 is a highly derived homologue of Cas10 (REFS 4,33), and the similarity between the organizations of the types I and III crRNA effector complexes is consistent with this possibility, sequence and structural comparisons fail to provide clear evidence. Some Cas8 proteins of subtype I-B have been shown to possess the single-stranded DNA-specific nuclease activity114 required for interference115. However, whether such activity is a universal feature of the large subunit remains to be determined.
The small subunit proteins are encoded by csm2 (subtypes III-A and III-D), cmr5 (subtypes III-B and III-C), cse2 (subtype I-E) or csa5 (subtype I-A). They are α-helical proteins that have no detectable homologues, although a structural comparison suggests that the small subunit proteins of type I and III systems are homologous to one another116. Despite differences in structural details, the overall shapes and architectures of the Cascade43,45,97, Cmr and Csm complexes36,38,41,98,117 are remarkably similar, as can be seen from electron microscopy images of Escherichia coli Cascade complexes31 (comprising Cas5, Cas6e and six Cas7 proteins, together with Cas8e as the large subunit and two Cse2 proteins as the small subunits; see the figure, partc) and Thermus thermophilus Cmr complexes36 (comprising a Cas5 group protein known as Cmr3 and six Cas7 group proteins, namely Cmr1, Cmr6 and 4 copies of Cmr4, together with a Cas10 group protein known as Cmr2 as the large subunit and Cmr5 as the small subunit; see the figure, partd). This suggests that the ancestral multisubunit effector complex evolved before the divergence of type I and type III CRISPR Cas systems. Figure part c from REF. 31, Nature Publishing Group. Figure part d adapted with permission from REF. 36, Cell Press.
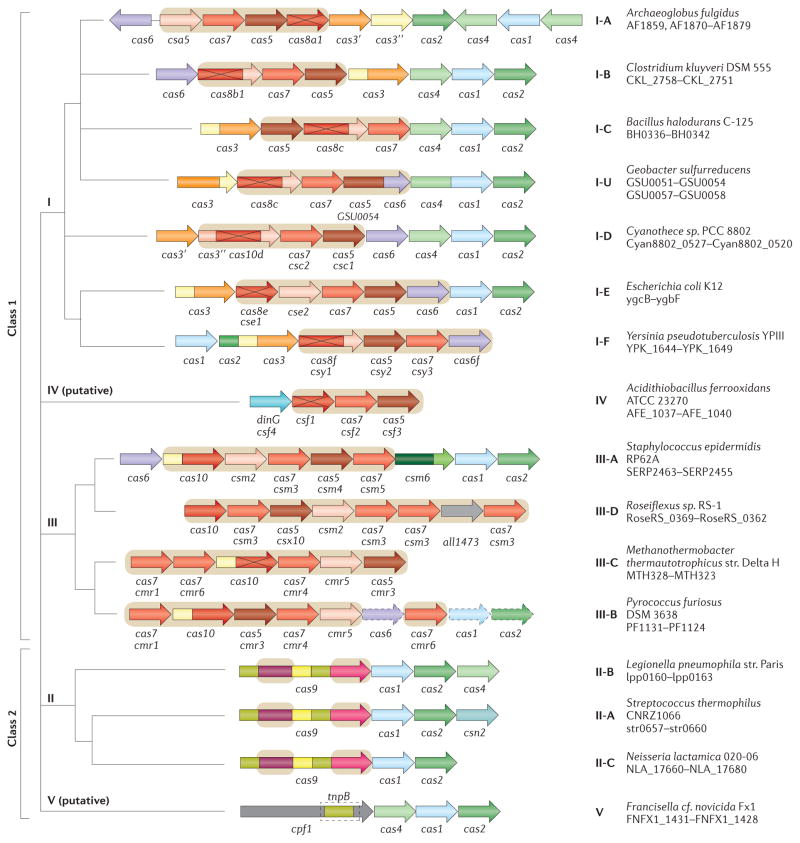
Each type I subtype has a defined combination of signature genes and distinct features of operon organization (FIG. 2; see Supplementary information S4 (table)). Notably, cas4 is absent in I-E and I-F systems, and cas3 is fused to cas2 in I-F systems. Subtypes I-E and I-F are monophyletic (that is, all systems of the respective subtype are descended from a single ancestor) in phylogenetic trees of Cas1 and Cas3, and each has one or more distinct signature genes (see Supplementary information S4,S5,S6 (table, box, box)).
Figure 2. Architectures of the genomic loci for the subtypes of CRISPR–Cas systems.
Typical operon organization is shown for each CRISPR–Cas system subtype. For each representative genome, the respective gene locus tag names are indicated for each subunit. Homologous genes are colour-coded and identified by a family name. The gene names follow the classification from REF. 13. Where both a systematic name and a legacy name are commonly used, the legacy name is given under the systematic name. The small subunit is encoded by either csm2, cmr5, cse2 or csa5; no all-encompassing name has been proposed to collectively describe this gene family to date. Crosses through genes encoding the large subunit (Cas8 or Cas10 family members) indicate inactivation of the respective catalytic sites. Genes and gene regions encoding components of the interference module (CRISPR RNA (crRNA)–effector complexes or Cas9 proteins) are highlighted with a beige background. The adaptation module (cas1 and cas2) and cas6 are dispensable in subtypes III-A and III-B; in particular, they are rarely present in subtype III-B (dashed lines). Dark green denotes the CARF domain. Gene regions coloured cream represent the HD nuclease domain; the HD domain in Cas10 is distinct from that of Cas3 and Cas3″. Also coloured are the regions of cas9 that roughly correspond to the RuvC-like nuclease (lime green), HNH nuclease (yellow), recognition lobe (purple) and protospacer adjacent motif (PAM)-interacting domains (pink). The regions of cpf1 aside from the RuvC-like domain are functionally uncharacterized and are shown in grey, as is the functionally uncharacterized all1473 gene in subtype III-D.
Subtypes I-A, I-B and I-C seem to be descendants of the ancestral type I gene arrangement (cas1–cas2–cas3–cas4–cas5–cas6–cas7–cas8)4,54. This arrangement is preserved in subtype I-B, whereas subtypes I-A and I-C are diverged derivatives of I-B with differential gene loss and rearranged gene orders. A single signature gene for each of these subtypes could not be defined. The only protein that shows no significant sequence similarity between the subtypes is Cas8. However, the Cas8 sequence is highly diverged even within subtypes, so that consistent application of the signature gene approach would result in numerous new subtypes. For example, there are at least 10 distinct Cas8b families within subtype I-B and at least 8 Cas8a families within subtype I-A (see Supplementary information S2 (table)). Thus, notwithstanding its complex evolution, we retain subtype I-B, which is best defined by the ancestral type I gene composition. The three main subdivisions within subtype I-B roughly correspond to the previously described subtypes Hmari, Tneap and Myxan32 (see also TIGRFAM directory), and now could be defined through specific Cas8b families, Cas8b1 (for Hmari), Cas8b2 (for Tneap) and Cas8b3 (for Myxan), with a few exceptions.
A subset of subtype I-B systems defined by the presence of the cas8b1 gene has been described as subtype I-G in the recent classification of archaeal CRISPR–Cas systems35. However, inclusion of bacterial CRISPR–Cas leads to increased diversity within subtype I-B so that if subtype I-G is recognized, consistency would require splitting I-B into several subtypes. Therefore, at present, we classify these variants within subtype I-B.
Subtype I-C seems to be a derivative of subtype I-B that lacks Cas6, which seems to be functionally replaced by Cas5 (REF. 55). Subtype I-A is another derivative of subtype I-B and is typically characterized by the fission of cas8 into two genes that encode degraded large and small subunits, respectively, as well as fission of cas3 into cas3′ and cas3″.
Subtype I-D also has several unique features, including Cas10d (instead of a Cas8 family protein) and a distinct variant of Cas3 (REF. 13) (FIG. 2; see Supplementary information S2,S4 (tables)). Subtype I-U is typified by the presence of an uncharacterized signature gene (GSU0054; TIGRFAM reference TIGR02165) and several other distinctive features that have been analysed in detail previously33 (see Supplementary information S4 (table)). This group is monophyletic in the Cas3 tree and mostly monophyletic in the Cas1 tree (see Supplementary information S5,S6 (boxes)).
The phylogenetic tree of the type I signature protein Cas3′ (and the homologous region of Cas3) has been reported to accurately reflect the subtype classification43, which is suggestive of a degree of evolutionary coherence between the phylogenies of the different genes in the operons of each subtype. However, re-analysis of the Cas3 phylogeny using a larger, more diverse sequence set (see Supplementary information S6 (box)) reveals a complex picture in which subtypes I-A, I-B and I-C are polyphyletic (that is, not descended from a common ancestor). Conceivably, this discrepancy results from a combination of accelerated evolution of many Cas3 variants and horizontal gene transfer.
In addition to the complete type I CRISPR–cas loci, analysis of sequenced genomes has revealed a variety of putative type I-related operons that encode effector complexes but are not associated with cas1, cas2 or cas3 genes and are only in some cases adjacent to CRISPR arrays (see Supplementary information S4 (table)). These solo effector complexes are often encoded on plasmids and/or associated with transposon-related genes. Many of these operons are derivatives of subtype I-F, whereas others are derivatives of subtype I-B (see Supplementary information S4,S7 (tables)). Some of the genomes that have these incomplete type I systems encode Cas1–Cas2 as parts of other CRISPR–cas loci but others lack these genes altogether (see Supplementary information S7 (table)). The functionality of solo effector complexes has not been investigated.
Type III CRISPR–Cas systems
All type III systems possess the signature gene cas10, which encodes a multi-domain protein containing a Palm domain (a variant of the RNA recognition motif (RRM)) that is homologous to the core domain of numerous nucleic acid polymerases and cyclases and that is the largest subunit of type III crRNA–effector complexes (BOX 2). Cas10 proteins show extensive sequence variation among the diverse type III CRISPR–Cas systems, which means that several PSSMs are required to identify these loci. All type III loci also encode the small subunit protein (see below), one Cas5 protein and typically several paralogous Cas7 proteins (FIG. 1). Often, Cas10 is fused to an HD family nuclease domain that is distinct from the HD domains of type I CRISPR–Cas systems and, unlike the latter, contains a circular permutation of the conserved motifs of the domain34,56.
Type III systems have been previously classified into two subtypes, III-A (previously known as Mtube subtype or Csm module) and III-B (previously known as Cmr module or RAMP module), that can be distinguished by the presence of distinct genes encoding small subunits, csm2 (in the case of subtype III-A) and cmr5 (in the case of subtype III-B) (FIG. 2; see Supplementary information S4 (table)). Subtype III-A loci usually contain cas1, cas2 and cas6 genes, whereas most of the III-B loci lack these genes and therefore depend on other CRISPR–Cas systems present in the same genome4, providing strong evidence for the modularity of CRISPR–Cas systems35 (FIG. 2). Both subtype III-A and subtype III-B CRISPR–Cas systems have been shown to co-transcriptionally target RNA26,27,37–39,57 and DNA26,58–61.
The composition and organization of type III CRISPR–cas loci are more diverse than those of type I systems — although there are fewer type III subtypes, each of these is more polymorphic than type I subtypes. This diversity is due to gene duplications and deletions, domain insertions and fusions, and the presence of additional, poorly characterized domains that could be involved either in crRNA–effector complex functions or in associated immunity. At least two type III variants (one from subtype III-A and one from subtype III-B) are common and are here upgraded to subtypes III-D and III-C, respectively, as proposed earlier for archaea35 (FIG. 3; see Supplementary information S8 (table)). The distinctive feature of subtype III-C (previously known as MTH326-like33) is the apparent inactivation of the cyclase-like domain of Cas10 accompanied by extreme divergence of the sequence of this protein. Subtype III-D loci typically encode a Cas10 protein that lacks the HD domain. They also contain a distinct cas5-like gene known as csx10 and often an uncharacterized gene that is homologous to all1473 from Nostoc sp. PCC 7120 (REF. 33). Both of these new subtypes lack cas1 and cas2 genes (FIG. 2) and accordingly are predicted to recruit adaptation modules in trans. The phylogeny of Cas10, the signature gene of type III CRISPR–Cas, is consistent with the subtype classification, with each subtype representing a distinct clade (see Supplementary information S9 (box)).
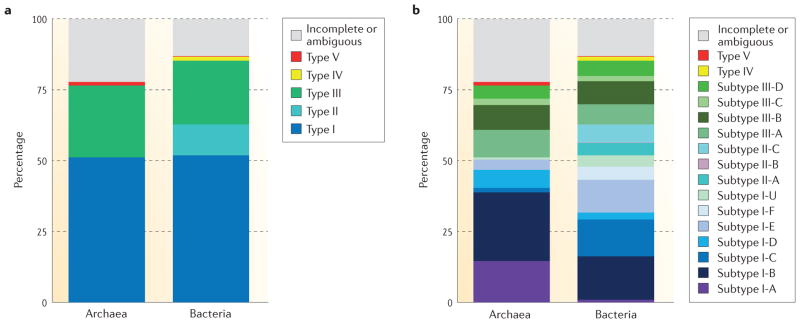
Figure 3. Distribution of CRISPR–Cas systems in sequenced archaeal and bacterial genomes.
a | Distribution by types. Chart showing the proportions of identified CRISPR–cas loci in bacterial or archaeal genomes that encode type I, type II, type III, type IV or type V CRISPR Cas systems. The proportion of loci that encode incomplete systems or that we could not classify unambiguously is also shown. b | Distribution by subtypes. Chart showing the proportions of identified CRISPR–cas loci in bacterial or archaeal genomes that encode each of the subtypes of CRISPR–Cas systems included in the new classification described in this article. Note that type IV and V loci each encompass a single subtype. The proportion of loci that encode incomplete systems or that we could not classify unambiguously is also shown.
Putative type IV CRISPR–Cas systems
Several bacterial genomes contain putative, functionally uncharacterized type IV systems, often on plasmids, as can be typified by the AFE 1037-AFE 1040 operon in Acidithiobacillus ferrooxidans ATCC 23270. Similar to most subtype III-B loci, this system lacks cas1 and cas2 genes and is often not in proximity to a CRISPR array or, in many cases, is encoded in a genome that has no detectable CRISPR arrays (it might be more appropriate to denote the respective loci Cas systems rather than CRISPR–Cas). Type IV systems encode a predicted minimal multisubunit crRNA–effector complex that consists of a partially degraded large subunit, Csf1, Cas5 and — as a single copy — Cas7, and in some cases, a putative small subunit33 (FIG. 1); csf1 can serve as a signature gene for this system. The minimalist architecture of type IV loci is distinct from those of all type I and type III subtypes (FIG. 2; see Supplementary information S4 (table)), which together with the unique large subunit (Csf1) justifies their status as a new type.
There are two distinct variants of type IV CRISPR–Cas systems, one of which contains a DinG family helicase (REF. 62), and a second one that lacks DinG but typically contains a gene encoding a small α-helical protein, which is a putative small subunit 33. Type IV systems could be mobile modules that, similar to subtype III-B systems, use crRNAs from different CRISPR arrays once these become available. This possibility is consistent with the occasional localization of type IV loci adjacent to CRISPR arrays, cas6 genes and (less often) adaptation genes35.
Class 2 CRISPR Cas systems
Class 2 CRISPR–Cas systems are defined by the presence of a single subunit crRNA–effector module. This class includes type II CRISPR–Cas systems, as well as a putative new classification, type V.
Type II CRISPR–Cas systems
Type II CRISPR–Cas systems dramatically differ from types I and III, and are by far the simplest in terms of the number of genes. The signature gene for type II is cas9, which encodes a multidomain protein that combines the functions of the crRNA–effector complex with target DNA cleavage25, and also contributes to adaptation63,64. In addition to cas9, all identified type II CRISPR–cas loci contain cas1 and cas2 (see REF. 65 for a detailed comparative analysis of type II systems) (FIG. 1) and most type II loci also encode a tracrRNA, which is partially complementary to the repeats within the respective CRISPR array65–67.
The core of Cas9, which includes both nuclease domains and a characteristic Arg-rich cluster, most likely evolved from genes of transposable elements that are not associated with CRISPR65. Thus, owing to the significant sequence similarity between Cas9 and its homologues that are unrelated to CRISPR–Cas, Cas9 cannot be used as the only signature for identification of type II systems. Nevertheless, the presence of cas9 in the vicinity of cas1 and cas2 genes is a hallmark of type II loci.
Type II CRISPR–Cas systems are currently classified into three subtypes, which were introduced in the previous classification (II-A and II-B)13 or subsequently proposed on the basis of a distinct locus organization (II-C)65,66,68 (FIG. 2; see Supplementary information S4 (table)). Subtype II-A systems include an additional gene, csn2 (FIG. 2), which is considered a signature gene for this subtype. The long and short variants of Csn2 form compact clusters when superimposed over the Cas9 phylogeny and seem to correspond to two distinct variants of subtype II-A65. However, as with subtype I-B, we chose to keep these two variants within subtype II-A. It was recently shown that all four subtype II-A Cas proteins are involved in spacer acquisition63.
Subtype II-B lacks csn2 but includes cas4, which is otherwise typical of type I systems (FIG. 2). Moreover, subtype II-B cas1 and cas2 are more closely related to type I homologues than to subtype II-A, which is suggestive of a recombinant origin of subtype II-B65. Subtype II-C loci only have three protein-coding genes (cas1, cas2 and cas9) and are the most common type II CRISPR–Cas system in bacteria3,65,66. A notable example of a subtype II-C system is the crRNA-processing-independent system found in Neisseria meningitidis69 (BOX 1).
In the Cas9 phylogeny, subtypes II-A and II-B are monophyletic whereas subtype II-C is paraphyletic with respect to II-A (that is, subtype II-A originates from within II-C)65. Nevertheless, II-C was retained as a single subtype given the minimalist architecture of the effector modules shared by all II-C loci.
Putative type V CRISPR–Cas systems
A gene denoted cpf1 (TIGRFAM reference TIGR04330) is present in several bacterial genomes and one archaeal genome, adjacent to cas1, cas2 and a CRISPR array (for example, in the FNFX1 1431–FNFX1 1428 locus of Francisella cf. novicida Fx1)70 (FIG. 2). These observations led us to putatively define a fifth type of CRISPR–Cas system, type V, which combines Cpf1 (the interference module) with an adaptor module (FIG. 1; see Supplementary information S4 (table)). Cpf1 is a large protein (about 1,300 amino acids) that contains a RuvC-like nuclease domain homologous to the respective domain of Cas9 and the TnpB protein of IS605 family transposons, along with putative counterparts to the characteristic Arg-rich region of Cas9 and the Zn finger of TnpB. However, Cpf1 lacks the HNH nuclease domain that is present in all Cas9 proteins54,65. Given the presence of a predicted single-subunit crRNA–effector complex, the putative type V systems are assigned to class 2 CRISPR–Cas. Some of the putative type V loci also encode Cas4 and accordingly resemble subtype II-B loci, whereas others lack Cas4 and are more similar in architecture to subtype II-C. Unlike Cas9, Cpf1 is encoded outside the CRISPR–Cas context in several genomes, and its high similarity with TnpB suggests that cpf1 is a recent recruitment from transposable elements.
If future experiments were to show that these loci encode bona fide CRISPR–Cas systems and that Cpf1 is a functional analogue of Cas9, then these systems would arguably qualify as a novel type of CRISPR–Cas. Despite the overall similarity to type II CRISPR–Cas systems, the putative type V loci clearly differ from the established type II subtypes more than type II subtypes differ from each other, most notably in the distinct domain architectures of Cpf1 and Cas9. Furthermore, whereas type II systems are specific to bacteria, a putative type V system is present in at least one archaeon, Candidatus Methanomethylophilus alvus35.
Rare, unclassifiable CRISPR Cas systems
The classification of CRISPR–Cas systems outlined above covers nearly all of the CRISPR–cas loci identified in the currently sequenced archaeal and bacterial genomes (FIG. 3). Nonetheless, owing to the rapid evolution of CRISPR–cas loci, which involves extensive recombination, it was not possible to account for all variants.
As a case in point, a putative CRISPR–Cas system was recently identified in Thermococcus onnurineus71. Based on some marginal similarities to protein components of crRNA–effector complexes, this locus was previously described as a Csf module35, which here is classified as type IV. However, only the putative Cas7 protein from this locus (TON 0323) is most similar to the variant characteristic of type IV systems (Csf2), whereas Cas2 and Cas4 are uncharacteristic of type IV loci, and an uncharacterized large protein containing an HD domain is present instead of Csf1. These features suggest classification of the T. onnurineus as a derived type I system (notwithstanding the absence of the signature gene cas3 or its variant cas3′), although it could not be assigned to any known subtype.
Several unusual variants of type III systems also posed a challenge for our classification. For example, the 15-gene locus in Ignisphaera aggregans has previously been classified as subtype III-D35. However, the III-D signature gene csx10, which encodes Cas5, is missing, and the other Cas proteins encoded by this locus show limited similarities to different type III subtypes71. Therefore, the I. aggregans locus seems to encode a type III system but cannot be unequivocally assigned to any subtype. Another distinct type III variant has been identified in several Crenarchaeota, primarily from the order Sulfolobales35. These loci lack detectable small subunits encoded by csm2 or cmr5 but contain a unique cas gene provisionally denoted csx26. Another variant is typified by the CRISPR–cas locus from Thermotoga lettingae35, which is the only known type III system to encode a single Cas7 protein, a feature of type IV systems. These two type III variants share more similarity with subtype III-A than with other subtypes and are currently assigned to this subtype (see Supplementary information S9 (box)); however, subsequent analysis of new genomes along with experimental study might prompt their reclassification into separate subtypes.
This accumulation of unclassifiable variants suggests that the current approaches to CRISPR–Cas system classification will need to be further refined to cope with the challenge of ever increasing diversity.
Distribution in archaea and bacteria
Approximately 47% of analysed bacterial and archaeal genomes encode CRISPR–cas loci. As reported previously13,72, CRISPR–Cas systems are much more prevalent in archaea (87% of genomes) than they are in bacteria (50% of genomes). For those genomes encoding CRISPR–cas loci, the rate of incomplete loci is similar for archaeal and bacterial genomes (17% and 12%, respectively). Complete single-unit loci are most commonly type I systems in both archaeal and bacterial genomes (64% and 60% of the loci, respectively), whereas putative type IV and type V systems are rare (<2% overall). Archaea possess significantly more type III systems than bacteria (34% versus 25% of the complete single-unit CRISPR–cas loci) but lack type II systems (13% in bacteria) (FIG. 3a). Thus, class 2 CRISPR–Cas systems are represented in archaea only by a single instance of the putative type V.
Overall, the most abundant CRISPR–Cas system is subtype I-B (20% of complete single-unit loci), followed by subtypes I-C and I-E (13% and 12%, respectively). In archaea, subtype I-A is the second most abundant after subtype I-B (18% and 30%, respectively), followed by subtypes III-A and III-B; subtype I-F is missing35 (FIG. 3b). Among the three type II subtypes, subtypes II-C and II-A are the most abundant, comprising 7% and 5% of bacterial single-unit cas loci, respectively; subtype II-B is a minority, with only six loci that are restricted to Proteobacteria (0.3%). Finally, archaea encompass a significantly greater fraction of multi-unit loci than bacteria (14% versus 6%). Of the 13% of all CRISPR–cas loci that are incomplete or unclassified, 48% are partial type I loci and 25% are partial type III loci.
Different archaeal and bacterial phyla show distinct trends in the distribution of CRISPR–Cas systems (see Supplementary information S8 (table)). Notably, the Crenarchaeota lack subtypes I-B and I-C systems, which are abundant in other archaea and bacteria, whereas the Euryarchaeota are enriched in subtype I-B loci35. The Actinobacteria show a strong preference for subtype I-E systems, and the Cyanobacteria for subtype III-B systems, whereas the Firmicutes account for most of the subtype II-A systems. Finally, the Proteobacteria lack subtype I-A systems but are strongly enriched in subtype I-F loci. Considering the extraordinary importance of type II CRISPR–Cas systems in biotechnology, it is worth emphasizing that these systems represent a minority of CRISPR–cas loci. They also seem to be specific to bacteria and are significantly over-represented in the Proteobacteria and the Firmicutes.
We expect that the bias of available sequence data towards cultivable microorganisms, especially those of medical or biotechnological importance, affects the currently observed distribution of CRISPR–Cas systems. Nevertheless, the remarkable stability of the overall fraction of CRISPR-possessing microorganisms over several years of observation seems to imply that at least the main trends are captured by the present analysis.
Modular organization and evolution
Similarly to other defence systems, CRISPR–cas loci evolve under strong selection pressure exerted by changing pathogens, resulting in rapid evolution that is largely uncoupled from the evolution of the rest of the respective genomes. Here we examine the evolutionary relationships between different components of the CRISPR–Cas systems and put forward the concept of modular organization, with semi-independent evolution of each module.
cas loci and CRISPR arrays
For the purpose of comparative analysis of CRISPR–Cas systems, CRISPR arrays were predicted in all genomes using CRISPRfinder73,74 following the procedure described in CRISPRmap75 and CRISPRstrand76. For each of the 1,949 cas loci, the nearest CRISPR array was identified, which showed a natural cut-off of 530 base pairs for the distance between cas loci and proximal CRISPR arrays (Supplementary information S8 (table)). Using this cut-off, 1,484 cas loci (75%) were classified as adjacent to a CRISPR array, 383 loci (22%) were present in CRISPR-positive genomes but far from any array, and 82 loci (54 complete and 28 incomplete, 3% total) were present in CRISPR-negative genomes. Although, as expected, the fraction of cas loci in CRISPR-negative genomes was significantly higher for incomplete (6.5%) than complete (2.3%) cas loci (χ2 test P value of 7 × 10−5), the existence of complete cas loci that were not accompanied by a recognizable CRISPR array anywhere in the genome was notable, as it defies the principle that crRNA–effector complexes are universally associated with CRISPR immunity. These CRISPR-less loci could be remnants of recently inactivated CRISPR–Cas systems or might function in a different way to the characterized CRISPR–Cas systems.
Conversely, of the 4,210 detected CRISPR arrays, 1,382 (33%) are adjacent (within 530 base pairs) to a cas locus, 2,365 arrays (56%) are located outside of cas loci in cas-positive genomes, and the remaining 463 arrays (11%) are orphans, present in genomes without detected cas loci. The orphan CRISPR arrays are probably remnants of formerly functional CRISPR–Cas systems.
CRISPR arrays are themselves classified into 18 structural families and 24 sequence families (only 23 were used here because one family could not be associated with any cas loci in our dataset), including unclassified repeats75–77. Both structural and sequence families of CRISPR show significant preferential association with particular types and subtypes of cas loci, although in most cases associations with other types or subtypes can also occur (FIG. 4; see Supplementary information S3,S10 (boxes)).
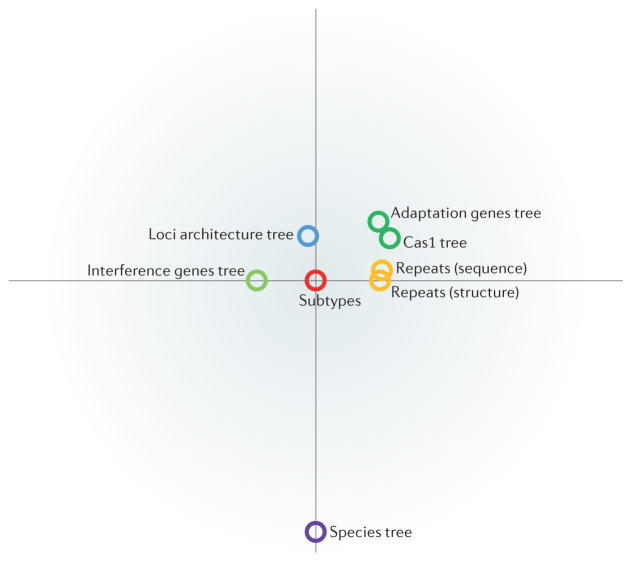
Figure 4. Comparison of different classifications of CRISPR–Cas systems.
This graph shows the strength of correlation between the new classification of CRISPR–Cas systems described here (‘subtypes’; in the centre of the graph) and other classification measures. ‘Interference genes tree’ represents a phylogeny of interference module genes, which encode multisubunit CRISPR RNA (crRNA)–effector complexes or Cas9 proteins. This tree was created using a simple clustering approach based on aggregate protein sequence similarity. ‘Adaptation genes tree’ represents clustering produced by the same method but based on both components of the adaptation module, Cas1 and Cas2. ‘Cas1 phylogeny’ is the phylogenetic tree of Cas1 proteins shown in FIG. 5. ‘Loci architecture tree’ represents clustering based on a quantitative measure we developed to compare the architectures of CRISPR–cas loci. The measure is based on a weighted similarity index of the order of cas genes. ‘Repeats (sequence)’ denotes the classification of CRISPR sequences into 24 families on the basis of sequence similarity. ‘Repeats (structure)’ denotes the classification of CRISPR sequences into 18 families on the basis of structural similarity. The species tree represents the phylogeny of bacterial and archaeal translation systems. The distances depicted are inversely proportional to the degree of similarity. The full similarity matrix is shown in Supplementary information S11 (table).
CRISPR–Cas systems and the species tree
Defence systems of bacteria and archaea evolve under extreme selection pressure from pathogens, particularly viruses, often using non-classic evolutionary processes, such as the seemingly Lamarckian adaptations represented by spacer integrations in CRISPR arrays78, the partially selfish mode of reproduction in which toxin–antitoxin systems are maintained in the genome through their addictive properties79, and pervasive horizontal gene transfer72,80. In line with these trends, evidence of extensive horizontal transfer of CRISPR–cas loci has been reported8,13,34,81–83.
To quantify the propensity of CRISPR–Cas systems to evolve via horizontal — as opposed to vertical — transmission, we compared various system features with a provisional species tree of bacteria and archaea that was reconstructed from concatenated ribosomal protein alignments84. As expected, the classification of the cas loci showed only weak consistency with the species tree (FIG. 4). The association between the species tree and CRISPR repeat types was also weak for both structure-based and sequence-based repeat classification (FIG. 4; see Supplementary information S11 (table)). These observations quantitatively show that horizontal transfer dominates the evolution of CRISPR–cas loci.
Cas1 phylogeny, CRISPR–Cas classification and architecture of cas loci
We examined the key evolutionary trends of the CRISPR–Cas systems in connection with the classification outlined above. Cas1 is the most conserved Cas protein, in terms of both representation in CRISPR–cas loci and amino acid sequence conservation85, and the Cas1 phylogeny generally correlates with the organization of CRISPR–cas loci13. Thus, until recently, Cas1 has been considered to be the signature of the presence of CRISPR–Cas systems in a genome13,32,34. However, in this analysis we identified 86 genomes containing complete (and by inference, functional) effector modules but that lacked cas1. These include genomes encoding the putative type IV systems, most subtype III-B, III-C and III-D systems and rare variants of subtypes I-C and I-F; 14 of these genomes also lack readily identifiable CRISPR arrays (FIG. 2; see Supplementary information S7 (table)).
Conversely, in some archaea and bacteria cas1 genes are located outside CRISPR–cas loci4, often within predicted self-synthesizing transposable elements dubbed casposons86. Casposon-encoded Cas1 proteins probably function as integrases that mediate the mobility of these transposons. The discovery of casposons suggests that the CRISPR–Cas adaptive immunity system arose from the insertion of a casposon near an innate immunity locus that encoded an effector complex87.
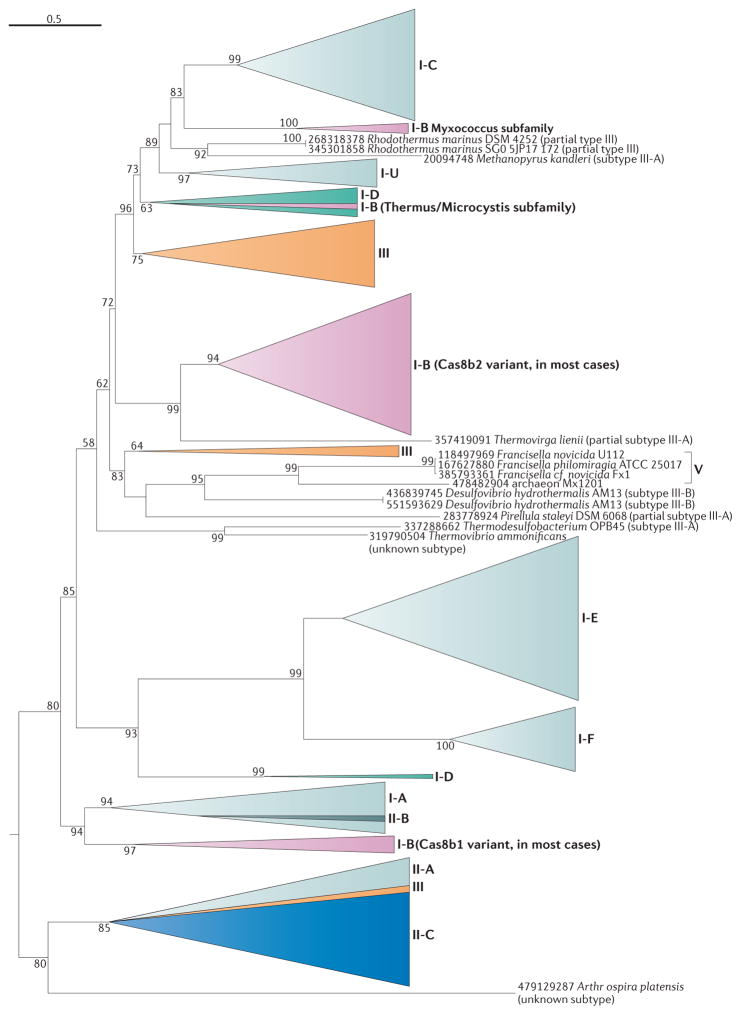
Of the 1,949 CRISPR–cas loci analysed, 1,404 encompass at least one cas1 gene. We constructed a phylogenetic tree of all 1,418 Cas1 sequences (some composite loci contain at least two cas1 genes) and rooted the tree using the modified midpoint procedure (FIG. 5; see Supplementary information S5 (box)). Mapping CRISPR–cas loci onto the Cas1 tree (FIG. 5) demonstrates a considerable agreement between the phylogeny of Cas1 and locus types and subtypes, consistent with previous observations. Thus, cas1 genes of subtypes I-E, I-F, II-B and putative type V are strictly monophyletic, and cas1 genes of subtypes I-C, I-U and II-A are largely monophyletic, with a few exceptions. In addition, cas1 genes of subtypes II-A and II-C form a mostly homogeneous clade, in agreement with a previous analysis65. By contrast, cas1 genes from the other type I subtypes and type III loci are scattered across the tree, suggestive of primarily horizontal evolution13,34,35,88. Thus, although substantial recombination occurs between the adaptation module and the other modules of the cas loci, the combination of the adaptation module with other modules is far from random.
Figure 5. Mapping of the CRISPR–Cas classification onto the phylogenetic tree of Cas1.
Subtypes from the new classification of CRISPR–Cas systems described here were mapped onto a sequence-based phylogenetic reconstruction of 1,418 proteins from the Cas1 family, which is the most conserved Cas protein family. The phylogeny shows a close agreement with the subtype classification, as subtypes I-A, I-C, I-E, I-F, I-U, II-A, II-B, and putative type V are mostly or strictly monophyletic and are shown in gradients of light grey, except for II-B, which is shown in dark grey to indicate its origin from within I-A. The more discordant distribution of Cas1 for other subtypes probably results from horizontal transfer. None of the type III subtypes is monophyletic (in contrast to the Cas10 tree shown in Supplementary information S9 (box)), and so type III subtypes are not indicated. Note that Cas1 is absent in type IV loci and so these putative CRISPR Cas systems are not shown. Triangles denote multiple collapsed branches. Individual genes are labelled with species names and gene identification numbers. Bootstrap values are indicated as percentage points; values below 50% are not shown.
As expected, the phylogeny of Cas1 is a poor match to the species tree of archaea and bacteria. The correlation of the distances between species with those between the corresponding cas1 genes in the tree is much weaker than the correlation between the Cas1 phylogeny and CRISPR–cas locus classification (FIG. 4; see Supplementary information S11 (table)). These observations imply an extensive history of horizontal transfers, many of which involved complete CRISPR–cas loci, whereas a smaller number included the adaptation module alone.
Cas1 is crucial to the adaptation stage of the CRISPR-mediated immune response17,89 and thus could be expected to co-evolve with CRISPR arrays83,88. We mapped structure-based and sequence-based repeat classification of CRISPR arrays adjacent to cas loci to the Cas1 tree. When only fully classified CRISPR repeats are considered, a high degree of consistency is observed between the Cas1 tree topology and repeat classification (FIG. 4; see Supplementary information S11 (table)), which probably reflects the direct recognition of repeats by Cas1 and its mechanistic involvement in the formation of the CRISPR arrays89.
We also developed a quantitative measure to compare the architectures of the cas loci to one another and to generate a similarity dendrogram (see Supplementary information S12 (box)). Overall, the topology of the dendrogram is consistent with the subtype classification of CRISPR–Cas systems (FIG. 4; see Supplementary information S11 (table)). However, the clusters obtained by this method are much narrower than the respective subtypes, which is consistent with a frequent rearrangement of CRISPR–Cas loci. By contrast, clusters obtained from protein similarity searches, using proteins from the interference module, are broader and often directly correspond to individual subtypes (see Supplementary information S12,S13 (boxes)). As expected, the clustering of CRISPR–Cas systems by locus architecture is substantially more compatible with the Cas1 phylogeny than with the species tree (FIG. 4), in agreement with the considerable evolutionary coherence of the CRISPR–Cas systems despite frequent horizontal gene transfer of CRISPR–cas loci and of individual modules.
Automated annotation of CRISPR–cas loci
Given the rapid pace of microbial genome sequencing, tools for the automated annotation of CRISPR–cas locus subtypes in newly sequenced genomes would be highly valuable. Although a careful inspection of combined features is required for accurate subtype annotation, we investigated whether an automated annotation method based on the similarity of the protein sequences of interference modules can faithfully reproduce the existing locus annotation.
To assess the value of the interference module as a proxy for the distribution of CRISPR–cas loci in our classification, we adopted a simple clustering approach based on aggregate sequence protein similarity35. This approach was chosen because of the lack of a universal marker suitable for phylogenetic analysis, as there is great variability in gene composition and module architecture between subtypes. The resulting cluster den-drogram (see Supplementary information S13 (box)) showed a high correlation with the subtype classification (FIG. 4; see Supplementary information S10 (box)). A similar cluster dendrogram constructed for Cas1 and Cas2 (see Supplementary information S14 (text)) showed a strong correlation with the Cas1 phylogeny but a considerably weaker correlation with the classification and architecture of CRISPR–cas loci than observed for the crRNA–effector complex dendrogram (FIG. 4; see Supplementary information S11 (table)). This difference supports our rationale in classifying CRISPR–cas loci on the basis of the interference module rather than Cas1 and demonstrates the ability of interference module protein clustering to closely reflect the new classification.
Having established the strong agreement between the clustering of interference module proteins and our classification, we constructed an automated classifier using prior information on the association between sequence PSSMs and CRISPR–cas loci and the corresponding classification of the effector modules. The classifier achieved 0.998 accuracy, which means that only 4 of 1,942 subtypes were incorrectly assigned (see Supplementary information S4,S15 (table, figure)). However, the accuracy of the method depends on the level of sequence similarity of the analysed Cas proteins to those available in the modelling phase, and predictably drops when the variants are only distantly related to the existing subtypes. Thus, the automated classifier described here has only limited applicability when annotating divergent variants of CRISPR–Cas subtypes.
Conclusions
The principal conclusion from the comparative analysis of the CRISPR–cas loci described here is the dynamic character and pronounced modularity of the evolution of this adaptive immunity system, which is conceivably driven by a perpetual arms race between the host genome and invading plasmids and viruses (dynamic evolution is a general theme in the evolution of defence systems72,80). In particular, the Cas1–Cas2 adaptation module evolved, to a large extent, independently of the operational modules (in particular, crRNA–effector complexes) of CRISPR–Cas systems, in agreement with the probable origin of the system as the result of the integration of a casposon-like mobile element next to an operon encoding a stand-alone effector complex87. The dynamic, modular evolution of CRISPR–Cas is also manifested at the level of the architecture of cas loci and the combination of different families of CRISPR arrays with different cas loci. However, a complementary trend is the frequent horizontal transfer of complete CRISPR–cas loci, which confers a degree of coherence to these systems and ensures that there is almost no congruence between the evolution of CRISPR–Cas and the species phylogeny as represented by the translation system90.
The dynamic and modular character of CRISPR–Cas evolution hampers a straightforward classification based on evolutionary relationships. However, the classification approach we propose here, which combines signature genes with elements of the architecture of cas loci, assigned nearly all of the detected CRISPR–cas loci to specific subtypes. Furthermore, the resulting classification is largely compatible with the results of sequence-based clustering of crRNA–effector complexes, which can be adopted for automated classification of CRISPR–Cas systems from new genomes. The refinement of automated classification using more sophisticated machine learning and other computational techniques could lead to the development of fully automated classification of CRISPR–Cas systems.
In many respects, the new classification closely resembles the 2011 version13, suggesting that the most common variants of CRISPR–Cas systems have already been discovered. However, we introduced a new top level, class, to account for the key differences between multisubunit and single-subunit crRNA–effector modules, as well as two new putative types (type IV and type V) and five new subtypes (II-C, III-C and III-D, together with the single subtypes of type IV and type V systems). Furthermore, the existence of currently unclassifiable variants implies that rare types and subtypes remain to be discovered and characterized, and the number of these is expected to substantially increase with the sequencing of new bacterial and archaeal genomes and metagenomes. In particular, the similarity between Cpf1 of the putative type V system and TnpB, which is usually found in transposons, suggests that multiple variants of single-subunit effector modules, and thus class 2 systems, might have evolved on independent occasions.
The classification of CRISPR–Cas systems and the principles of CRISPR–Cas evolution outlined here are expected to help the identification and focused discovery of new variants, some of which could become novel tools for genome engineering.
Supplementary Material
Acknowledgments
K.S.M., Y.I.W., D.H. and E.V.K. are supported by the National Institutes of Health (NIH) Intramural Research Program at the National Library of Medicine, US Department of Health and Human Services. R.M.T. and M.P.T. are supported by NIH grants RO1 GM54682 and RO1 GM99876. J.v.d.O. was partly supported by SIAM Gravitation Grant 024.002.002 from the Netherlands Organization for Scientific Research (N.W.O.). S.J.J.B. was financially supported by an NWO Vidi grant (864.11.005) and European Research Council (ERC) Stg (639707). A.F.Y. is supported by the Natural Sciences and Engineering Research Council (NSERC) Strategic Network Grant IBN and NSERC Discovery grant. S.M. acknowledges funding from Natural Sciences and Engineering Research Council of Canada (Discovery program) and holds a Tier 1 Canada Research Chair in Bacteriophages. F.J.M.M. is supported by the Ministerio de Economía y Competitividad (BIO2014-53029). R.B. is supported by the Deutsche Forschungsgemeinschaft (DFG) grant (BA 2168/5-2). S.A.S. and R.A.G. were funded primarily by the Danish Natural Science Research Council. O.S.A., F.C., S.J.S., R.B., S.A.S and R.A.G. are grateful to all members of the FOR1680 for helpful discussions.
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
TIGRFAM file directory: ftp://ftp.jcvi.org/pub/data/TIGRFAMs TIGR02165 | TIGR04330
See online article: S1 (table) | S2 (table) | S3 (box) | S4 (table) | S5 (box) | S6 (box) | S7 (table) | S8 (table) | S9 (box) | S10 (box) | S11 (table) | S12 (box) | S13 (box) | S14 (box) | S15 (figure)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Makarova KS. CRISPR–Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013;10:679–686. doi: 10.4161/rna.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR–Cas systems. Biochem Soc Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R. CRISPR–Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 8.Westra ER, et al. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 9.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 10.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 11.Magadán AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3–Cas system. PLoS ONE. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Makarova KS, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westra ER, Buckling A, Fineran PC. CRISPR–Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 15.Sampson TR, Weiss DS. CRISPR–Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol. 2014;4:37. doi: 10.3389/fcimb.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louwen R, Staals RH, Endtz HP, van Baarlen P, van der Oost J. The role of CRISPR–Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev. 2014;78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunez JK, et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 22.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samai P, et al. Co-transcriptional DNA and RNA cleavage during type III CRISPR–Cas immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Duijn E, et al. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteom. 2012;11:1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of Archaea. RNA Biol. 2014;11:156–167. doi: 10.4161/rna.27990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staals RH, et al. Structure and activity of the RNA-targeting Type III-B CRISPR–Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spilman M, et al. Structure of an RNA silencing complex of the CRISPR–Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staals RH, et al. RNA targeting by the type III-A CRISPR–Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamulaitis G, et al. Programmable RNA shredding by the type III-A CRISPR–Cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 40.Benda C, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56:43–54. doi: 10.1016/j.molcel.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Hale CR, Cocozaki A, Li H, Terns RM, Terns MP. Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes Dev. 2014;28:2432–2443. doi: 10.1101/gad.250712.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the helicase family tree. Curr Opin Struct Biol. 2014;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 46.Taylor DW, et al. Structures of the CRISPR–Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sander JD, Joung JK. CRISPR–Cas systems for editing, regulating and targeting genomes. Nat Biotech. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Koonin EV. PSI-BLAST — a tool for making discoveries in sequence databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 50.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong B, et al. Molecular insights into DNA interference by CRISPR-associated nuclease-helicase Cas3. Proc Natl Acad Sci USA. 2014;111:16359–16364. doi: 10.1073/pnas.1410806111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huo Y, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014;21:771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makarova KS, Koonin EV. Annotation and classification of CRISPR–Cas systems. Methods Mol Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam KH, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR–Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR–Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng L, Garrett RA, Shah SA, Peng X, She Q. A novel interference mechanism by a type IIIB CRISPR–Cmr module in Sulfolobus. Mol Microbiol. 2013;87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 61.Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White MF. Structure, function and evolution of the XPD family of iron-sulfur-containing 5′→3′ DNA helicases. Biochem Soc Trans. 2009;37:547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 63.Heler R, et al. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR–Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR–Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briner AE, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 68.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR–Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 71.Makarova KS, et al. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles. 2014;18:877–893. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bland C, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lange SJ, Alkhnbashi OS, Rose D, Will S, Backofen R. CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 2013;41:8034–8044. doi: 10.1093/nar/gkt606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alkhnbashi OS, et al. CRISPRstrand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics. 2014;30:i489–i496. doi: 10.1093/bioinformatics/btu459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leplae R, et al. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koonin EV, Wolf YI. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front Cell Infect Microbiol. 2012;2:119. doi: 10.3389/fcimb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 82.Almendros C, Mojica FJ, Díez-Villaseñor C, Guzmán NM, García-Martínez J. CRISPR–Cas functional module exchange in Escherichia coli. mBio. 2014;5:e00767–e00713. doi: 10.1128/mBio.00767-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 84.Yutin N, Puigbo P, Koonin EV, Wolf YI. Phylogenomics of prokaryotic ribosomal proteins. PLoS ONE. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR–Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koonin EV, Krupovic M. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 2015;16:184–192. doi: 10.1038/nrg3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–556. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Puigbo P, Wolf YI, Koonin EV. Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hooton SP, Connerton IF. Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front Microbiol. 2014;5:744. doi: 10.3389/fmicb.2014.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wiedenheft B, et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 93.Kwon AR, et al. Structural and biochemical characterization of HP0315 from Helicobacter pylori as a VapD protein with an endoribonuclease activity. Nucleic Acids Res. 2012;40:4216–4228. doi: 10.1093/nar/gkr1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beloglazova N, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nam KH, et al. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beloglazova N, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramia NF, et al. Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPR–Cas complex. Cell Rep. 2014;9:1610–1617. doi: 10.1016/j.celrep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu X, Ye K. Cmr4 is the slicer in the RNA-targeting Cmr CRISPR complex. Nucleic Acids Res. 2015;43:1257–1267. doi: 10.1093/nar/gku1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brendel J, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (crispr)-derived rnas (crrnas) in Haloferax volcanii. J Biol Chem. 2014;289:7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Osawa T, Inanaga H, Sato C, Numata T. Crystal structure of the CRISPR–Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 105.Jung TY, et al. Crystal structure of the Csm1 subunit of the Csm complex and its single-stranded DNA-specific nuclease activity. Structure. 2015;23:782–790. doi: 10.1016/j.str.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 106.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Makarova KS, Anantharaman V, Grishin NV, Koonin EV, Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nam KH, Kurinov I, Ke A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J Biol Chem. 2011;286:30759–30768. doi: 10.1074/jbc.M111.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Koo Y, Jung DK, Bae E. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PLoS ONE. 2012;7:e33401. doi: 10.1371/journal.pone.0033401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arslan Z, et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 2013;41:6347–6359. doi: 10.1093/nar/gkt315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee KH, et al. Identification, structural, and biochemical characterization of a group of large Csn2 proteins involved in CRISPR-mediated bacterial immunity. Proteins. 2012;80:2573–2582. doi: 10.1002/prot.24138. [DOI] [PubMed] [Google Scholar]
- 112.Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR–Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 113.Shao Y, et al. Structure of the Cmr2–Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guy CP, Majernik AI, Chong JP, Bolt EL. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186. doi: 10.1093/nar/gkh960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cass SD, et al. The role of Cas8 in type I CRISPR interference. Biosci Rep. 2015;35:e00197. doi: 10.1042/BSR20150043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reeks J, et al. Structure of the archaeal Cascade subunit Csa5: relating the small subunits of CRISPR effector complexes. RNA Biol. 2013;10:762–769. doi: 10.4161/rna.23854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jackson RN, Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.