Abstract
Linear mixed models (LMMs) are widely used for regression analysis of data that are assumed to be clustered or correlated. Assessing model fit is important for valid inference but to date no confirmatory tests are available to assess the adequacy of the fixed effects part of LMMs against general alternatives. We therefore propose a class of goodness-of-fit tests for the mean structure of LMMs. Our test statistic is a quadratic form of the difference between observed values and the values expected under the estimated model in cells defined by a partition of the covariate space. We show that this test statistic has an asymptotic chi-squared distribution when model parameters are estimated by maximum likelihood or by least squares and method of moments, and study its power under local alternatives both analytically and in simulations. Data on repeated measurements of thyroglobulin from individuals exposed to the accident at the Chernobyl power plant in 1986 are used to illustrate the proposed test.
Keywords: asymptotic efficiency, information matrix, maximum likelihood estimators, method of moments, model fit, random effects
1. Introduction
The linear mixed model (LMM) (McCulloch and Searle, 2001) extends the linear model by including random effects in addition to the usual fixed effects in the linear predictors. By incorporating random effects LMMs can accommodate clustered or correlated data. Developments in model fitting algorithms and their implementations in statistical packages (e.g. lme in R; PROC Mixed in SAS 9.2; SAS Institute, Cary, NC) have greatly facilitated the applications of LMMs.
Two important steps in modeling are selecting a model and checking its fit. Often model selection is done by comparing nested models, via likelihood ratio or score tests, as part of model building, and approaches are also available for comparing non-nested models (Cox, 1961; Godfrey, 1988). Variables are often selected for inclusion into a model if their p-value obtained from a Wald test meets some significance criterion. AIC, BIC and other model selection principles (Rao and Wu, 1989; Shao, 1997) also focus on selection of covariates. Once a model is selected, its fit should be assessed. For fixed effects models this is done by checking residuals and formal goodness of fit tests, such as score or Wald tests or likelihood ratio tests based on nested models. Khuri, Mathew and Sinha (1998) discussed likelihood ratio testing for fixed effects within LMMs. The literature for assessing the fit of LMMs against general alternatives is limited, and is mostly concerned with specification of the random effect distributions. Likelihood ratio testing for the presence of random effects in LMMs has been discussed by Self and Liang (1987) and Crainiceanu and Ruppert (2004). Jiang (2001) and Ritz (2004) assessed the distributional assumptions for the random effects in LMMs. Claeskens and Hart (2009) proposed tests for normality of the random effects and/or error terms. Lombardía and Sperlich (2008) introduced a test for the hypothesis of a linear fixed effect part in a generalized linear mixed model against the alternative of a semiparametric fixed effect part. Pan and Lin (2005) propose checking the adequacy of 2-level generalized linear mixed models based on the maximum absolute partial sums of residuals over a scalar projection of covariates. Their approach allows for assessing overall model fit as well as the functional form of individual components of the fixed effects part. However, to date there is no general easily computable test for checking the fit of the fixed-effect part of a model against unspecified alternatives, including omitted covariates or interaction terms or misspecifications of the functional form of covariates. Such a test is needed as a model-building tool.
Examination of the residuals of a model is a standard way to judge the quality of model fit. This can be done in many different ways. One useful way is to classify the response into mutually exclusive events defined in terms of the covariates and then assess for each category the deviation of the observed values and the expected values under the model. For survival data, Schoenfeld (1980) presented a class of omnibus chi-squared goodness of fit tests for the proportional hazards regression model, based on the observed minus the expected values of the covariates at each failure time. In this article, we adopt the idea of Schoenfeld (1980) and develop a goodness of fit test for the mean structure of LMMs by comparing the observed and expected values computed from the model within cells of a partition of the covariate space.
The rest of the paper is organized as follows. In Section 2 we present the linear mixed model, introduce the goodness of fit test statistic, and derive its asymptotic properties, including its theoretical power under local alternatives. We first assume that the random effects components and the error term are normally distributed and parameters are estimated by maximum likelihood (Section 2.2). We then relax the assumption of normality and only require finite higher order moments for the random effect and the error term and estimate parameters using least squares and method of moments (Section 2.3). We study the power of the test in simulations in Section 3, present a data example in Section 4 and close with a discussion in Section 5.
2. Goodness of fit test statistic for linear mixed models
2.1. The linear mixed model
We consider the linear mixed model (LMM) with additive random effects,
| (1) |
where YN×1 is the vector of observations; is the design matrix for the fixed effects part of the model, where xi denotes the p × 1 covariate vector for individual i; β is a p × 1 vector of unknown fixed effects parameters; Zr is the known N × mr design matrix for the random effect αr, an mr × 1 random vector, for r = 1, …, R. The random effects α1, …, αR are i.i.d. and independent of the error term ε. In the next section we assume that the components αkr of αr and ε are normally distributed. Within the LMM with a single random effect, we later require no distributional assumptions on the random effect and the error terms, but only the finiteness of their 4 + δ moments for some δ > 0. We let θ = (β, ψ) be the parameters of model (1), where is the vector of all variance components.
An important special case of model (1) is the 2-level LMM, that includes only a single random effect,
| (2) |
where, using a slightly different notation, the 1 × p vector denotes covariates for the jth observation within the ith cluster. The first entry in xij is set to be 1 to accommodate an intercept term in the model. We let yi = (yi1, …, yini) denote the vector of observations for the ith cluster. The normally distributed cluster specific random effects are assumed to be independent of the error terms . Then under model (2), Y is also normal, Y ~ N(Xβ, V) with a block diagonal covariance matrix V, where each of the m ni × ni blocks Vi, i = 1, …, m, has entries on the diagonal and entries elsewhere. Throughout this paper, we regard models (1) and (2) as conditional specifications of the distribution of Y given X.
2.2. Test statistic and its asymptotic behavior when parameters are estimated by maximum likelihood
2.2.1. LMM with a single random effect
We first discuss the 2-level LMM in (2) when both the random effect and the error term are normally distributed and derive our test statistic for the setting where the model parameters are estimated by maximum likelihood (MLE). Here X is considered to be fixed.
Under Assumptions 1.1-1.6 stated below in Theorem 1, consistency and asymptotic normality of the MLE follow from Miller (1977), i.e. , where J denotes the limiting Fisher information matrix. Wand (2007) showed that under model (2), and are asymptotically uncorrelated and thus
| (3) |
where
| (4) |
We assume that Jββ is positive definite (Assumption 1.5, Theorem 1).
To test the goodness of fit of the mean structure of the LMM (2), we first divide the covariate space into L disjoint regions E1, …, EL. These regions are based on categorizations of single covariates or of composites of multiple covariates that may or may not be included in the current model. For example, for a single continuous covariate with support on the interval (a, b), the cells E could be defined by El = (cl, cl+1], l = 1, …, L − 1 where a = c1 < c2 < … < cL−1 < cL = b. For a categorical (discrete) variable X that takes the values cl for l = 1, …, L, the partition can be defined by El = {X = cl}. We compute the observed and expected sums in each region El as
| (5) |
| (6) |
where I denotes the indicator function. When the cell partition is based on covariates not included in the model (2), then we let xij denote the vector of all available covariates and the covariates used in the model, and use , where β* corresponds to the coefficients of . However, for notational simplicity we employ the expressions (5) and (6) throughout.
Letting f = (f1, …, fL) and e(β) = (e1(β), …, eL(β)), the observed minus the expected vector is
| (7) |
Since the true parameter vector β0 is unknown, to create a test statistic we must replace it by a consistent asymptotically normal estimator, the MLE as in (7) within Section 2.2 and the generalized least squares estimator (15) in Section 2.3. We further make Assumption 1.7 which, together with Assumptions 1.4 and 1.5, ensures the existence of components of the limiting variance covariance matrix for the test statistic.
Theorem 1. We make the following assumptions:
Assumption 1.1. The true parameter point θ0 = (β0, ψ0) is an interior point of .
Assumption 1.2. and .
Assumption 1.3. X is fixed and has full rank.
Assumption 1.4. .
Assumption 1.5. Jββ = limN→∞ XTV−1 X/N exists and is positive definite.
Assumption 1.6. The 2 × 2 matrix M with elements defined below exists and is positive definite;
where G0 = I is the N × N identity matrix and G1 is the block-diagonal matrix with m blocks and each block is an ni × ni matrix of all 1s. After some algebra,
It is easy to see that matrix M is the average of nonnegative definite matrices. Under Assumption 1.4, M is positive definite if and only if . Thus the main restriction in Assumption 1.6 is the requirement that M exists.
Assumption 1.7. For any cell partition E1, … EL of the covariate space, exists for l = 1, …, L.
For model (2), under Assumptions 1.1-1.7, as N → ∞,
where
and
The off-diagonal and diagonal elements of H, Hlk and Hll, are
| (8) |
| (9) |
and
| (10) |
The proof of Theorem 1 is given in Appendix A.
Corollary 1. Consistent estimators for the quantities given in (8), (9), (10) and (4) are
Corollary 2. For model (2), under Assumptions 1.1-1.7 in Theorem 1, as N → ∞, , where is an L × L matrix that is consistently estimated by based on Corollary 1.
The proof of Corollary 2 is given in Appendix B.
Our goodness of fit test statistic is then given by the quadratic form
| (11) |
where denotes the Moore-Penrose generalized-inverse (also called ‘pseudo-inverse’) of a slight modification of the consistent variance estimator . We define in the following paragraph. Under the null hypothesis that model (2) is the true model, T has an asymptotic central distribution, where with probability converging to 1 as N → ∞, . This result applies to the modification of any consistent estimator of the Σ matrix, such as restricted maximum likelihood (REML) estimators.
The modification of variance estimates we describe next applies in several places in this paper. The issue is always to avoid numerical instabilities and asymptotic distributional anomalies due to rank differences between consistent nonnegative-definite variance estimators and the true asymptotic variance Σ. Assume in what follows that there exists a known threshold ξ (say, 10−4) smaller than all non-zero singular values of Σ. Since Σ is nonnegative definite, all singular values are nonnegative. Denoting the spectral decomposition of as , where is an orthonormal basis of eigenvectors of and ckN are the corresponding eigenvalues, the modification of is defined as , and its Moore-Penrose pseudo-inverse is then . We prove in Proposition 1, Appendix D, that is coordinate-free and unique, that rank with probability converging to 1, and that the asymptotic distribution of T in (11) is and would persist if the Moore-Penrose choice of pseudo-inverse were replaced by any other generalized-inverse of as defined in Rao (1973, Sec. 1.b). In addition, the assumption of a fixed known threshold ξ can be replaced by allowing ξ = ξN to depend non-randomly on N and converge to 0 sufficiently slowly. (The rate would depend on the specific consistent estimator .)
2.2.2. LMM with multilevel additive random effects
We now consider the general LMM with multilevel additive random effects given in equation (1) with a fixed covariate matrix X.
Again, the covariate space, comprised of covariates not all of which need be included in the model, is divided into L disjoint regions E1, …, EL, and for l = 1, 2, …, L, we define the observed and expected vectors f = (f1, …, fL) and e(β) = (e1(β), …, eL(β)) as , and . The impact of the choice of L is discussed further in the simulation section (Section 3).
Conditions in Miller (1977) that ensure the consistency and asymptotic normality of the MLE of θ = (β, ψ) are given in the Supplementary Material S1 (assumptions A.1-A.7). We additionally make assumptions A.8-A.9 (Supplementary Material S1) to ensure the existence of large-sample averages involving xk and I[xk∈Ej].
Theorem 2. For model (1), under Assumptions A.1-A.9 given in the Supplementary Material S1,
| (12) |
as N → ∞, where is the MLE of β, is a consistent estimator of , is the modification of as defined in the last paragraph of Section 2.2.1, denotes the Moore-Penrose pseudoinverse of , and for r ≡ rank(Σ), , as N → ∞. Here H = limN→∞ FVFT, with
| (13) |
and the l-th row of Λ given by
| (14) |
The proof of Theorem 2 is similar to that of Theorem 1 and is given in Supplementary Material S2.
2.3. Test statistic and its asymptotic properties for two-level LMM with parameters estimated by least squares and method of moments
We consider the LMM (2), but now only require that E(αi) = E(∊ij) = 0, , , and that there is a δ > 0, such that and , instead of assuming normality of αi and ∊ij. To compensate for the weaker distributional assumptions, we assume for simplicity in probability limit theorems that the covariate vectors xij are random, that {(xi1, …, xini), ni} are i.i.d. and that and , where xi denotes the ni × p matrix of covariates for the ith cluster.
We estimate β by the generalized least squares estimator
| (15) |
where V depends on the variance components . They are estimated by the method of moments by equating the right-hand sides of
| (16) |
| (17) |
respectively with their estimates based on the sum of squares within groups (SSW) and the sum of squares between groups (SSB) in the analysis of variance, given by
The notation stands for and , and similar averages apply to the covariates x. Because different clusters are independent, and , and have finite second moments, SSW/m and SSB/m satisfy the law of large numbers. The estimating equations (15), (16) and (17) can be solved iteratively for and yield consistent estimates (Richardson and Welsh, 1994; Jiang, 1996).
To obtain the test statistic, we again compute the observed and expected values in each of the L cells of the covariate space as in (5) and (6).
Theorem 3. For the LMM (2), under Assumptions 3.1-3.3 in Appendix D, as N → ∞, , where . Thus , where is a consistent estimator of Σ, is the modification of as defined in the last paragraph of the section 2.2.1, denotes the Moore-Penrose pseudoinverse of , and N → ∞.
As X is a matrix of random variables in this section, Assumption 3.3 ensures that Assumptions 1.4-1.7 hold, which are needed when X are assumed to be fixed in Section 2.1. The matrices H, Λ and Jββ are the same as for the two-level LMM (2) when parameters are estimated by maximum likelihood under the assumption of normality of αi and ∊ij and are specified in (8), (9), (10) and (4). The proof of the theorem is given in Appendix E.
Remark 1. Theorem 1 and Theorem 3 are still valid when empirical quantiles instead of fixed cut-o s are used to define cell partition, is we assume that the empirical quantiles of coordinates of xi converge to unique limits or under Assumption 3.3.
2.4. Power of the test
For the multi-level LMM (1), we derive the theoretical power under local, and more specifically under contiguous alternatives for the test in (12) for the situation where some covariates that influence the outcome y are omitted from model (1). This case also covers omitted interactions of covariates or omitted higher order terms and is thus practically relevant.
Let X be the true N × p covariate matrix and X* be a submatrix of X of dimension N × p* used to fit model (1), with p* < p. The null hypothesis is H0: θN = θ0. We assess the power of T under the alternative
| (18) |
with θ0 = (β0, ψ0), where several components of β0 are 0. The vector corresponding to X*. Here is the vector difference between the parameter values under the alternative hypothesis and the parameter values under the null hypothesis.
Based on the derivation for Theorem 2, we have that under H0, . By applying Le Cam’s third lemma (see Appendix F for details), we find that under the alternative hypothesis H1 in (18),
| (19) |
where
| (20) |
with Λ given by expression (14). Λ* is computed using X* in place of X in (14).
Thus under H1, T* has a limiting noncentral χ2 distribution
| (21) |
where and the non centrality parameter is . For a given type I error level α, the power is thus , where is the 1 − α quantile of the central distribution and P denotes the non central distribution. In the computation of the power we use the Moore-Penrose inverse of a modification as in (11) of a consistent estimator in (20).
As an illustration, we show the asymptotic power to detect lack of fit for an omitted covariate for the two-level LMM (2). We assume Y ~ N(XT, V), where X = (1, x1, x2, x3) and V is the block diagonal covariance matrix. The xij = (xij1, xij2, xij3), i = 1, …, m; j = 1, …, ni are i.i.d. and drawn from a multivariate normal distribution
| (22) |
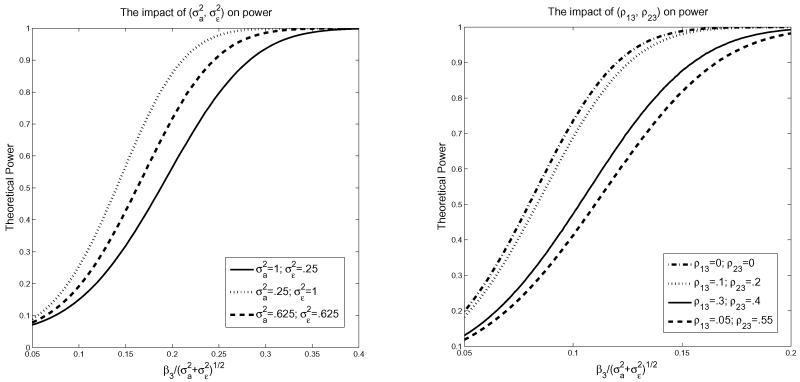
and xij and ni are independent. In fitting the model, we omit x3 leading to X* = (1, x1, x2) and a = (0, 0, 0, β3) in (20). For this setting, τ in (20) and Σ* in (19) can be computed explicitly as a functions of the moments of X and ni (section 2.2.3., Tang, 2010). We study the impact of the magnitude of the variance components and and the correlations ρ13 and ρ23 in (22) between the omitted covariate x3 and the covariates in the model (x1 and x2) on the theoretical power when the cell partition is based on theoretical quantiles of the omitted covariate x3 with L = 8 cells. For ρ13 = 0.5 and ρ23 = 0.6, Figure 1 (left panel) plots the theoretical power against for three choices of all corresponding to the same overall variance and varying β3 on the x-axis. For any fixed pair of , the power of the test, not surprisingly, increases as a function of β3, the coefficient of the omitted covariate x3. This can also be seen by taking a first order Taylor expansion of the theoretical power formula around λ = 0, as the power for λ close to zero depends linearly on λ = τT(Σ*)−1τ, which is a function of . Figure 1 (left panel) shows that for any fixed β3 the power increases when the random effect decreases compared to the error term . Figure 1 (right panel) plots the power for , for different choices of (ρ13, ρ23). The power increases as decreases. For ρ13 = 0 and ρ23 = 0, that is, when x3 is uncorrelated with x1 and x2, the power is not affected by the individual values of and , but only depends on the sum . The theoretical formulas for power under contiguous-alternatives given here will generally be close to the actual power only for very large sample sizes. However, numerical studies presented in the next section show that these formulas often also agree with empirical power in samples of moderate size (m = 50, N = 150 to N = 200).
Figure 1.
Left: Theoretical power as a function of ; Right: Power as a function of (ρ13, ρ23).
3. Simulations to assess power and robustness of the test statistic
For a given number of clusters m, we first generated the cluster sizes ni from a uniform distribution on {2, 3, 4, 5} for i = 1, …, m and then drew independent covariates xij for all simulations presented below. We present scenarios for which we believe out test would be practically most relevant: models with omitted main effects (Scenario I), omitted interaction terms and main effects (Scenarios II and III) and misspecified functional forms of a covariate (Scenario IV). We covered a range of effect sizes to provide a fair assessment of the performance of our test.
3.1. Main effects only (Scenario I)
Here xij = (x1ij, x2ij, x3ij) were drawn from the multivariate normal distribution given in (22). Given X = (1, x1, x2, x3), β, σα = 1 and σ∊ = 0.5, we generated Y from a multivariate normal distribution, Y ~ N(XT β, V).
3.1.1. Size and power of T
To check the size of the test for various choices of cell partition based on X, we let ρ13 = ρ23 = 0 in (22), β = (β0, β1, β2, β3) = (1, 1, 1, 1), m = 500 and fit model (2) with all covariates X in the model. Table 1 gives the number of cells and the covariates that are the basis of the cell partition in the first column. Cell partitions in the computation of the test statistic T were based on empirical quantiles of the respective components of X. For all cell partitions in Table 1 the empirical sizes were close to the nominal α levels of 0.05 and 0.1.
Table 1.
Empirical size of the test under different cell partitions (Scenario I). m = 500, E(N) = 1750, β3 = 1, ρ13 = ρ23 = 0, σα = 1, σε = .5, K = 2000. L denotes the number of cells for the test statistic.
| L | α | Emp. Size | α | Emp. Size |
|---|---|---|---|---|
| 8 (x1) | 0.05 | 0.052 | 0.1 | 0.103 |
| 3×4 (x1, x2) | 0.05 | 0.053 | 0.1 | 0.108 |
| 5×4 (x1, x3) | 0.05 | 0.045 | 0.1 | 0.094 |
| 6×7 (x2, x3) | 0.05 | 0.047 | 0.1 | 0.096 |
To assess the power of the test, we generated data from model (2) that includes all three covariates but then omitted x3 in fitting the model to the data. We set (ρ13, ρ23) = (0.5, 0.6), β = (β0, β1, β2, β3) = (1, 1, 1, 0.25). We then generated K = 2000 datasets for a given X. We repeated this data generation process for D = 1000 independently drawn design matrices X. For a given X, we computed the theoretical power of T* in (21) based on the asymptotic χ2 distribution with the true values of and . The empirical moments for X were used in the calculation of the non-centrality parameter λ. For a given X and each generated Y, we calculated the estimated theoretical power based on the asymptotic χ2 distribution with the variance components estimated based on the given Y and empirical moments of X in (20). We then repeated the calculation of the estimated theoretical power for each of the K = 2000 generated Y and by taking the average, we obtained a mean estimated theoretical power for that given X. For each given design matrix X, we also calculated the empirical power based on K = 1000 iterations on Y. For m = 20 clusters the cell partition was based on the empirical quantiles of the omitted x3 with L = 8 cells to avoid empty cells, but for m = 50 or 500, we used theoretical quantiles of x3 as cell boundaries for computational ease.
The mean theoretical power (“Theo.Pow.”), the mean estimated theoretical power (“Theo.Pow.hat”) and the empirical power (“Empi.Pow.n”) agreed very well, even when m is small (Table 2). However, only for m = 500 was there adequate power to detect lack of fit when β3 = 0.25, which is substantially smaller than the coefficients β1 = β2 = 1 of x1, and x2, the covariates included in the model. When the effect of the omitted covariate was larger, β3 = 0.8, the test statistic had approximately 80% power even for m = 50 clusters.
Table 2.
Power and robustness study (Scenario I) with L = 8,K = 2000, D = 1000, (ρ13 ,ρ23) = (.5,.6),σα = 1, σε = .5. Standard deviation (std.dev.) relates to variation across the randomly generated 500 covariate matrices X
| Power | m = 500, EN = 1750 |
m = 50, EN = 175 |
m = 20, EN = 70 |
|||
|---|---|---|---|---|---|---|
| β3 = .25 | mean | std.dev. | mean | std.dev. | mean | std.dev. |
| Theo.Pow. | 0.800 | 0.040 | 0.120 | 0.023 | 0.086 | 0.018 |
| Theo.Pow.hat | 0.799 | 0.039 | 0.125 | 0.023 | 0.090 | 0.019 |
| Empi.Pow.n | 0.799 | 0.037 | 0.111 | 0.022 | 0.063 | 0.017 |
|
| ||||||
| Misspecification of the error term distribution | ||||||
|
| ||||||
| Empi.Pow.t3 | 0.798 | 0.036 | 0.112 | 0.023 | 0.062 | 0.019 |
| Empi.Pow.t5 | 0.799 | 0.036 | 0.111 | 0.022 | 0.063 | 0.018 |
|
| ||||||
| Misspecification of the random intercept distribution | ||||||
|
| ||||||
| Empi.Pow.t3 | 0.817 | 0.033 | 0.132 | 0.027 | 0.076 | 0.021 |
| Empi.Pow.t5 | 0.799 | 0.036 | 0.116 | 0.023 | 0.067 | 0.018 |
| Power | m = 500, EN = 1750 |
m = 50, EN = 175 |
m = 20, EN = 70 |
|||
|---|---|---|---|---|---|---|
| β3 = .8 | mean | std.dev. | mean | std.dev. | mean | std.dev. |
| Theo.Pow. | 1 | 0 | 0.847 | 0.104 | 0.541 | 0.189 |
| Theo.Pow.hat | 1 | 0 | 0.821 | 0.096 | 0.512 | 0.151 |
| Empi.Pow.n | 1 | 0 | 0.820 | 0.102 | 0.444 | 0.161 |
|
| ||||||
| Misspecification of the error term distribution | ||||||
|
| ||||||
| Empi.Pow.t3 | 1 | 0 | 0.821 | 0.102 | 0.449 | 0.164 |
| Empi.Pow.t5 | 1 | 0 | 0.821 | 0.102 | 0.444 | 0.162 |
|
| ||||||
| Misspecification of the random intercept distribution | ||||||
|
| ||||||
| Empi.Pow.t3 | 1 | 0 | 0.852 | 0.074 | 0.545 | 0.155 |
| Empi.Pow.t5 | 1 | 0 | 0.824 | 0.094 | 0.472 | 0.159 |
3.1.2. Robustness of T with respect to error and random effects distributions
In Table 2 we also assessed the impact of misspecification of the error distribution on the power of the test statistic. Using the same setting as in the power calculations given above, we generated ∊ from a t distribution with k = 3 or 5 degrees of freedom (d.f.) instead of from a . We rescaled the variance of ∊ so that the noise had the same variance as in the normal case. The power of the test under a t-distribution was virtually the same as with a normal error distribution indicating that our test is robust to symmetric violations of normality. For example, for m = 50 with β3 = 0.8, the power was 0.83 for the normal error distribution and for t-distributions with 3 and 5 d.f. (Table 2). We also used the same misspecification for the random effects distribution, and observed very similar results (Table 2). We chose the t distribution because it is symmetric but has heavier tails than the normal distribution and it satisfies the conditions given in Section 2.3 on the existence of moments of the random effects and errors.
3.1.3. Impact of choice of the cell partition on power
As is true for Pearson’s chi-squared test, the choice of cell partition strongly impacts the performance of our goodness of fit test. To illustrate the impact of the cell partition on the power of our test we generated y from a model with E(y) = 1 + x1 + x2 + 0.15x3, with σα = 1 and σ∊ = 0.5, but then omitted x3 in the subsequent model fitting. We studied cell partitions based on only x1, only x2, only x3, both x1 and x2, both x1 and x3, or both x2 and x3, all based on empirical quartiles of the covariates. Table 3 shows that a lack of fit is detectable by our test statistic only when the cell partition involves the omitted covariate x3, and power decreases as correlations (ρ13, ρ23) between the covariates increase.
Table 3.
Impact of cell partition on empirical power when covariate x3 with β3 = .15 is omitted from model fitting (Scenario I) with m = 500, σα = 1, σε = 0.5, K = 2000.
| Cell Variables |
ρ13 = 0, ρ23 = 0 |
ρ13 = 0.2, ρ23 = 0.3 |
ρ13 = 0.4, ρ23 = 0.5 |
|||
|---|---|---|---|---|---|---|
| L=12 | L=42 | L=12 | L=42 | L=12 | L=42 | |
| x 1 | 0.056 | 0.060 | 0.046 | 0.044 | 0.045 | 0.044 |
| x 2 | 0.055 | 0.046 | 0.048 | 0.048 | 0.051 | 0.050 |
| x 3 | 0.985 | 0.871 | 0.936 | 0.748 | 0.630 | 0.367 |
| x1, x2 | 0.054 | 0.050 | 0.052 | 0.046 | 0.051 | 0.048 |
| x1, x3 | 0.968 | 0.821 | 0.896 | 0.732 | 0.578 | 0.382 |
| x2, x3 | 0.962 | 0.843 | 0.913 | 0.752 | 0.642 | 0.435 |
3.2. Normally distributed covariates with an omitted interaction term (Scenario II)
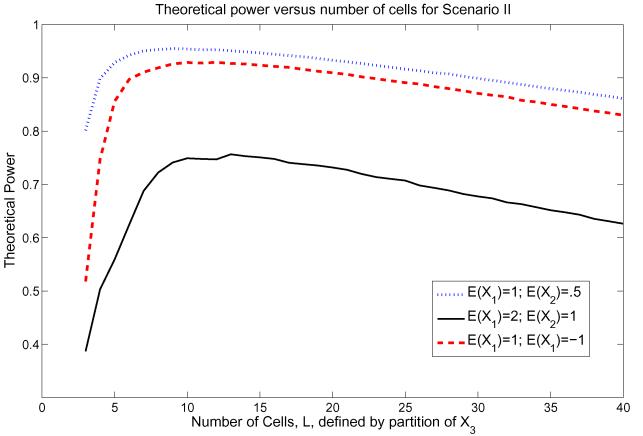
We generated y from a linear model with E(y) = 1 + x1 + x2 + β3x1x2, where x1 and x2 are independent and , i = 1, 2. We first let β3 = 0.2 and set σα = 1, σ∊ = 0.5. We then fit model (2) without x3 = x1x2. The test had adequate power only when the cell partition is based on empirical quantiles of x1 and x2, or on the omitted interaction term x3 = x1x2, but not if the cell partition was based on either x1 or x2 alone, for ρ12 = 0 and ρ12 = .3 (Table 1, Web Supplementary Material). Figure 2 shows the power of the test as a function of the number of cells computed based on quantiles of the omitted covariate x3 for various values of μi = E(Xi), i = 1, 2. The power was higher for smaller absolute values of μ1 and μ2 and was largest for L = 11 cells for μ1 = 2 and μ2 = 1 and L = 7 cells for μ1 = 1 and μ2 = 0.5.
Figure 2.
The impact of number of cells for cell partition on theoretical power
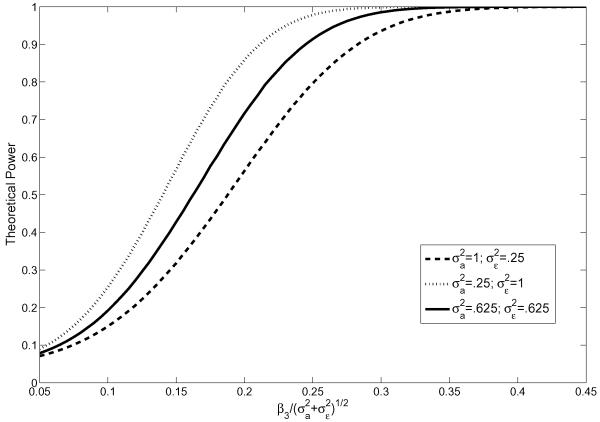
Figure 3 plots the theoretical power against for three choices of corresponding to the same overall variance and varying β3 on the x-axis when the cell partition was based on x3 with L = 8 cells using fixed cell boundaries. Our conclusions are consistent with those in Section 3.1. For any fixed pair of , the power of the test increased as a function of β3, the coefficient of the omitted covariate x3. For any fixed β3, the power increased when the random effect decreases compared to the error term .
Figure 3.
The impact of on theoretical power, ρ12 = 0 (Scenario II)
3.3. Omitted main effect and interaction term (Scenario III)
Here we generated y from a LMM that includes three covariates x1, x2 and x3 through E(y) = 1 + x1 + x2 + 0.05x3 + 0.1x1x3, with σα = 1 and σ∊ = 0.5. We then omitted x3 and any interactions with x3 from the model fitting and investigate the power of our test in working model M1 with covariates x1, x2, and working model M2 with x1, x2 and . The cell partition for M1 and M2 was based on empirical quantiles of x1 with L = 8 cells. We simulated x3 ~ N(0, σ2) with σ = 1.5, and let x1 = ex3. The covariate was generated independently of x1 and x3.
Under this setting, with m = 500 clusters, our test had power 1 to detect lack of fit of the working model M1. The Wald test for inclusion of the quadratic term in model M2 had power 1. As x3 is not available, a Wald test cannot be applied for any term related to x3. We thus would select model M2 (with covariates x1, x2, ) and a Wald-type test is not able to further assist in testing model inadequacy. However, our proposed test had power of 0.938 to detect lack of fit of M2, under the cell partition based on x1, a covariate in M2.
This example, in which the omitted covariate is not available in the dataset but a correlated variable is, shows the usefulness of our test in addition to Wald type tests. However, our test has reasonable power against omitted-covariate alternatives only when the partitions of the covariate space are based on variables that are correlated with the omitted covariates.
3.4. Misspecified functional form of a covariate (Scenario IV)
We generated y from a LMM with with σα = 1 and σ∊ = .5, and studied the power of our test when instead of only x3 is used in the working model. x = (x1, x2, x3) was generated from the multivariate normal distribution given in (22) with (ρ13, ρ23) = (.5, .6). We used empirical quantiles of x3 to define L = 8 cells for m = 500 or 50 clusters.
When m = 500, our test had approximately 87% power in detecting model inadequacy (Table 4). The Wald test to assess the significance of x3 however had of only approximately 6% power. Thus based on Wald test, x3 would not be included in the model and therefore it is unlikely that the higher order term would be considered.
Table 4.
Power and robustness study (Scenario IV). L = 8, K = 2000, D = 1000, (ρ13, ρ23) = (.5, .6), β3 = 0.1, σα = 1, σε = .5. Note: The standard deviation (std.dev.) relates to variation across the randomly generated 1000 covariate matrices X. Each number was obtained as the mean over 1000 simulated covariate matrices X and for each generated X, K = 2000 iterations were used to simulate the response vector Y.
| m= 500, EN = 1750 |
m = 50, EN = 175 |
|||
|---|---|---|---|---|
| Mean Power | std.dev. | Mean Power | std.dev. | |
| Cell Variable x3 | 0.873 | 0.035 | 0.122 | 0.028 |
| Wald Test on x3 | 0.059 | 0.024 | 0.065 | 0.026 |
|
| ||||
| Misspecification of the error term distribution | ||||
|
| ||||
| εij simulated from t3 | ||||
|
| ||||
| Cell Variable x3 | 0.873 | 0.035 | 0.123 | 0.030 |
| Wald Test on x3 | 0.059 | 0.024 | 0.065 | 0.026 |
|
| ||||
| εij simulated from t5 | ||||
|
| ||||
| Cell Variable x3 | 0.873 | 0.035 | 0.122 | 0.029 |
| Wald Test on x3 | 0.060 | 0.025 | 0.066 | 0.030 |
|
| ||||
| Misspecification of the random effect distribution | ||||
|
| ||||
| αi simulated from t3 | ||||
|
| ||||
| Cell Variable x3 | 0.878 | 0.032 | 0.139 | 0.033 |
| Wald Test on x3 | 0.061 | 0.027 | 0.067 | 0.031 |
|
| ||||
| αi simulated from t5 | ||||
|
| ||||
| Cell Variable x3 | 0.872 | 0.035 | 0.126 | 0.030 |
| Wald Test on x3 | 0.061 | 0.026 | 0.067 | 0.031 |
We also investigated the impact of symmetric misspecification of the error and random effect distribution in this scenario. When the t-distribution was used for the error term (or for the random effect term) results are similar to those for the normally distributed error term (or random effect term) (Table 4). Thus in this scenario, our test was robust to symmetric misspecification of the error or random effect distribution, similar to Scenario I.
3.5. Remarks
The primary purpose of the goodness of fit tests studied in this paper is to assess the quality of the fixed-effect part of the mean response in the presence of a mixed-effect variance structure. Yet it is well known that there is ambiguity in Gaussian linear models as to which terms contribute to the fixed-effect predictors and which terms to the variance. To be specific, we consider the model
| (23) |
where X* = (X1, X2), (X*, X3) are jointly normally distributed with means 0, and , and the random error . By grouping the γX3 term together with the error ∊, we see that model (23) is equivalent to the model
| (24) |
where and ∊* are defined in terms of E(X3|X*) = MT X* and by ∊* = ∊ + γ(X3−MTX*), , and . This argument shows that the portion of a normal linear model describing is not uniquely determined, where D denotes the data-vector of covariates, that is in (23), and in (24). However, since our goodness of fit tests for adequacy of the mean structure are considered conditional on D, and are specified in terms of covariate-defined cells, these two models (23) and (24) are in fact distinguishable if cells under (23) are taken to depend non-trivially on the omitted covariate X3.
This argument also highlights the lack of power for the test in the setting of main effects (Scenarios I and III) with an omitted covariate when the cell partition was not based on the omitted covariate or a transformation of it, or for an omitted interaction term, when the cell partition is based on only one of the variables that define the interaction (Scenario II). When cell partitions are based on only on X* no lack of fit in the mean structure can be detected, as it is correctly specified with respect to X*.
4. Data example
On April 26, 1986, an accident at the Chernobyl power plant in Ukraine, close to the border with Belarus, released large amounts of radioactive materials including iodine-131 (I-131) into the atmosphere from the destroyed reactor. Deposition of these materials contaminated the territory. Radioisotopes of iodine, e.g. I-131, are concentrated in the thyroid gland. Belarusians exposed to the accident were enrolled in a cohort study to evaluate the relationship between I-131 doses and thyroid cancer risk (Stezhko et al, 2004). Investigators were also interested in studying iodine deficiency in this population, as it impacts I-131 absorption.
We therefore evaluated the relationship between levels of serum thyroglobulin (TG), a marker of iodine deficiency, and variables that might reflect or impact dietary iodine intake, including age at the time of exam, age at the time of the accident, rural or urban residence, smoking status, urinary iodine levels, serum thyroid-stimulating hormone (TSH) levels, serum anti-thyroglobulin antibody (ATG) levels, thyroid volume, presence of thyroid nodules (yes/no), presence of goiter (yes/no) and presence of any thyroid abnormality (yes/no).
We used data on m = 933 men from four of the five study regions, who had complete covariate information, whose ATG and TSH levels were measured by a luminescence assay, and who had TG ≤ 80 (to exclude those with thyroid disease). Among these men, 404 had a single TG measurement, 484 had two, 42 three and 3 four TG measurements during follow-up, resulting in N = 1510 observations. log(TG) was normally distributed (Anderson-Darling test p-value p=0.09).
We fit various models using Proc GLIMMIX, SAS 9.2. Model 1 included all the variables mentioned above, with the exception of presence of nodules, and an interaction term of ATG levels with presence of any thyroid abnormality that was marginally significant (Wald test p-value p = 0.054) and had a log-likelihood value of −1625.2. The random effect variance estimate was and the error variance estimate was . Model 2 had no interaction term, but included presence of nodules and resulted in a log-likelihood of −1621.3. The variance component estimates were similar to model 1, and . However, as models 1 and 2 are not nested, we could not compare them using a likelihood ratio test.
To assess the fit of both models, each person in the dataset was assigned to one of the L = 8 cells defined by the quartiles of ATG and the response “yes” or “no” to the question “presence of any thyroid abnormality”. There was no indication of lack of fit for either model, with p = 0.32 and p = 0.40 for models 1 and 2 respectively. We also calculated the test statistic for a second cell partition with L = 4 cells defined by “presence of nodules” (yes/no) and “presence of goiter” (yes/no), with p = 0.19 and p = 0.70 for models 1 and 2 respectively. These results suggested that both models fit the data adequately. Thus omitting the interaction term of the variable “presence of any thyroid abnormality” with ATG levels does not affect the fit to the data.
5. Discussion
Schoenfeld (1980) presented a class of omnibus chi-squared goodness of fit tests for the proportional hazards regression model. We adapted this idea and proposed a class of goodness of fit tests for testing the statistical adequacy of the mean structure of a linear mixed model, with cell partitions based on covariates. We described the asymptotic properties of the test when parameters are estimated and developed its theoretical power under local alternatives. We assessed factors that affect the power, the impact of choice of cell partitions on the test as well as the robustness of the test with respect to error distribution and distribution of random e orts in simulations. When a specific covariate associated with outcome is omitted, such as an interaction term or a covariate correlated with terms already in the model, cell partitions based on the omitted covariate result in adequate power of the test. In our simulations we studied models involving only a few covariates. In such cases, Wald testing and likelihood-based model building tools could undoubtedly be used instead. In practical settings our test would be recommended when there are many potential predictors that should in fact not appear in the model. In such circumstances, many nonlinear terms involving omitted variables would not be Wald-tested. We also found that the estimated theoretical power calculated using Le Cam’s third lemma was reliable at least when the number of clusters m was above 50. However, when m is very small, it may be advisable to rely on the empirical power computed through simulations. Our test was also robust to symmetric violations of the normality assumption of the error distribution as well as the violation of normality of the random effects distribution.
This goodness of fit test can be used to test the statistical adequacy of the fixed effects part of a finally selected LMM. It should not be used if one wants to test if a specific covariate should be included in the model, as standard tests such as the Wald test have better power for that purpose (e.g. Scenario I, Table 2). However, when a covariate is missing from the dataset, our test can detect model inadequacy when the cell partition is based on an existing covariate in the working model, which is correlated with the omitted covariate, while no Wald-type test can be applied (Scenario III). Also, the Wald test did not have power to select a variable that entered the mean model only through a quadratic term (Scenario IV), while our test clearly showed lack of fit of the finally selected model with respect to cells defined by that variable. This is particularly important in the situation when many predictors are available, and testing all possible higher order terms or interactions is not practical. In addition, investigators might not consider the inclusion of a higher order term for a variable that has no main effect. We have shown using simple examples that our proposed test has good power to detect many sorts of model inadequacies, not all of which would be tested exhaustively by other methods.
To implement the test one only needs the final model parameter estimates and their variance covariance matrix, which are standard outputs from any statistical software. As a note of caution, in applying the test one must modify the estimated variance matrix , projecting its eigenspace corresponding to extremely small eigenvalues to 0, to ensure the correct degrees of freedom for the test statistic.
Pan and Lin (2005) developed methods for checking the adequacy of generalized linear mixed models by comparing the cumulative sums of residuals over covariates or predicted values. Our proposed test has additional flexibility in defining cells based on multiple covariates, the test statistic follows a known distribution and is thus easily computed, and we present a broader class of LMMs.
Our goodness of fit test examines multiple features of the data, corresponding to residuals within each covariate cell and bears some relation to the multiaspect framework by Pesarin and Salmaso (2010), Salmaso and Solari (2005) and Marozzi (2007). Future work could attempt to adapt their permutational approaches for several populations to the goodness of fit test in a single population.
Notably, the cell partition used for our test is based on covariates, not on the response variable as for standard Pearson χ2 statistic. In future research we plan to further investigate the choice of covariate-based cells partition on the performance of our proposed test. A related issue is sparse cells. Our asymptotic results were derived letting the sample size go to infinity for a fixed cell partition and thus asymptotically cells are not sparse. However, in a real dataset the issue of sparse cells could arise. Maydeu-Olivares and Joe (2005, 2006) and Cagnone (2012) studied the impact of sparse cells when assessing the goodness of fit of latent variable models. For use with heavily cross-classified and sparse covariate-space cell decompositions the limited-information approach of Maydeu-Olivares and Joe (2005, 2006) could be used in our setting and will be part of future investigations. Other possible extensions include derivation of the distribution of the test statistic for random components with heavy tails, for example, under symmetric α-stable distributional assumption for the errors and random effects. However, these extensions of the mixed model theory presented in our paper are technically difficult and we are not aware of any related results in the literature.
Supplementary Material
Acknowledgment
We thank the investigators of the ‘US-Belarusian Study of Thyroid Cancer and Other Thyroid Diseases Following the Chernobyl Accident’ (National Cancer Institute, Columbia University, U.S.A, and Republican Research Center of Radiation Medicine and Human Ecology, Belarus) for providing the data and Jincao Wu for help with computations. We also thank the reviewers for helpful comments and suggestions. This work is part of M. Tang’s Ph.D. thesis done at the University of Maryland.
Appendix
Appendix A: Proof of Theorem 1
Let J be the limit of the sample information matrix per observation given in (3). The consistency of the MLE in model (2) follows from Miller (1977). By Taylor series expansion of the score function S(θ) = ▽ log L(θ), where L(θ) denotes the likelihood function,
| (25) |
As the Fisher information (3) is block diagonal, . Under Assumption 1.2 Y − Xβ ~ N(0, V), and the score functions for β, i.e. the first p components of S(θ), are Sβ(θ) = XTV−1(Y − Xβ). By extracting the first p components of (25), with A ≈ B denoting , we have
Thus,
which is a linear combination of Gaussian random variables.
Under Assumptions 1.4, 1.5, 1.7, which ensure the existence of components of the covariance matrix of the test statistic, we get as N → ∞,
| (26) |
Appendix B: Proof of Corollary 2
Under asymptotic normality of ,
Since is a linear combination of components of the left hand side of (26), , with .
Appendix C: Proposition 1
Proposition 1. Suppose that a sequence ZN of random q-vectors is asymptotically distributed as , where rank(Σ0) = r ≥ 1 and there exists a known ξ > 0 smaller than the minimum positive eigenvalue of Σ0, and that is a consistent covariance-matrix-valued estimator of Σ0.
Let the spectral decomposition of be given by
where ckN are the eigen values and form an orthonormal eigenbasis determined from . Define
and let be any other generalized inverse of , i.e. any matrix such that . Then
and .
and as N → ∞.
Proof of Proposition. Note first that while the eigenvectors vkN are not necessarily uniquely determined if any eigenvalues have multiplicity greater than 1, the eigenspaces spanned by {vkN: 1 ≤ k ≤ q, ckN ≤ s} are uniquely and measurably determined from for each real s > 0. Therefore and all of the random variance matrices , are well-defined, coordinate-free and measurably defined from .
Without loss of generality, let the eigenvalues ck,N of be indexed in nondecreasing order. Since the k’th smallest eigenvalue is a continuous function on the set of q × q symmetric nonnegative definite matrices (Golub and van Loan 1983, pp. 18-19), it follows from the convergence , that for arbitrarily small ∊ ∈ (0, ξ), the event
has probability converging to 1 as N → ∞. This implies that on AN(∊), the range space of is exactly the span of the eigenvectors vkN with k ≥ q − r + 1, and therefore that rank on the event AN(∊). Moreover, on the event AN(∊), for all x ∈ Rq with ||x|| = 1,
since max{|ckN|: k ≤ q − r} ≤ ∊ and . This shows the matrix sup-norm of converges in probability to 0 as N → ∞, completing the proof of (i).
By (i), the asymptotic distribution of ZN is the same as , where is independent of ZN and the matrix square-root is the symmetric square-root equal to . Therefore, by the continuous mapping theorem, the asymptotic distribution of is the same as the distribution of , which is since is symmetric and idempotent with trace r. The only feature of that has been used in this proof is the generalized-inverse property shared by . This fact about generalized inverses, which completes the proof of assertion (ii), was previously proved in detail by Rao (1973, 1b.5.(viii), 3b.4.(vii) or 3b.5.(iv)).
Appendix D: Assumptions for Theorem 3
For the rest of the Appendices we employ the notation v⊗2 = vvT for any vector v.
Assumption 3.1. The true parameter point θ0 = (β0, ψ0) is an interior point of .
Assumption 3.2. E(αi) = E(∊ij) = 0, , and there is a δ > 0, such that and .
Assumption 3.3. X is a matrix of random variables, (xi, ni) are i.i.d. with , being positive definite, and .
Appendix E: Proof of Theorem 3
The following Lemma is used in proving Theorem 3.
Lemma 1. Let {uin: n ≥ 1, 1 ≤ i ≤ n} be a triangular array of i.i.d. random variables within each row (i.e., across i) with mean 0 and finite variance , and that these variables are independent of the random array {cin: n ≥ 1, 1 ≤ i ≤ n which satisfies, as n → ∞, (a) max1≤i≤n |cin| → 0 and (b) in probability, where κ ∈ (0, ∞). Then as n → ∞.
Proof of Lemma 1: is a martingale with respect to the filtration and the Lemma follows directly from the Martingale Central Limit Theorem (Hall and Heyde, 1980).
Proof of Theorem 3: Let the ni×p covariate matrix for the i-th cluster be . Then
Then
with , i = 1, …, m, l = 1, …, L. Let , . We next show that has a limiting Gaussian distribution by using the multivariate Central Limit Theorem. For any constant vector C = (C1, …, CL)T, since the inverse of Vi is , we have
where the double index (i, j) is placed in one-to-one correspondence with the single index s. Because and are i.i.d and satisfy conditions (a) and (b) of Lemma 1, the above sums have limiting normal distributions as m → ∞. Because αi and ∊ij are independent and are conditionally independent given (xi, ni). As the two sums are jointly normal and asymptotically uncorrelated they are asymptotically independent and the limiting distribution of is normal. Moreover, for any constant vector C, its limiting variance is of the form CTΣC with the same fixed Σ,
Therefore, , and , where r = rank(Σ). We replace Σ with , the reconstructed estimated variance matrix defined as immediately following equation (11) by means of the singular value decomposition applied to any consistent estimator of Σ. One such consistent estimator of Σ is to replace all parameters in Σ with least squares and method of moments estimators. Based on Proposition 1 in Appendix C, for large N. Thus
Appendix F: Derivation of the power of the test
We derive the power of the test for LMM (1) under contiguous alternatives, based on Le Cam’s third lemma (Van der Vaart, 2000).
Lemma 2. (Le Cam’s third lemma) Let PN and QN be two measures on a measurable space, corresponding to a null distribution under investigation, and an alternative hypothesis respectively. Suppose WN is a real valued statistic for every N. If
| (27) |
then .
Let H0 : θN = θ0, and , where a is a constant vector and θN → θ0, as n → ∞. By Taylor expansion, under Theorem 5.21 in van der Vaart (2000),
where
| (28) |
and the limit of the sample Fisher information per observation is
Thus
For the special case when we fit a reduced model to the data, using instead of XN×p with p* < p, we estimate the coefficient β* corresponding to X*. The sum over the expected values under the model, e(·), in (6) has Rp as its domain. Let e* (·) denote the sum over the expected values under the reduced model, computed using instead of XN×p with domain Rp*. Let be the first vector component of (27). Under the null hypothesis PN, WN → N(0, Σ*), based on Corollary 2.
Next, we compute the variance-covariance matrix Σ in (27), which is equivalent to the variance-covariance matrix of and .
where denotes the information matrix corresponding to β*, and
Thus,
| (29) |
Under equation (28), since both and tr(V−1) are scalars, we have
Similarly, we get . Therefore (29) becomes
Since both and can be written as a matrix multiplied by the same normal vector , we obtain asymptotic joint normality of and . Because is asymptotically uncorrelated with both and as shown in the above, and aTSN(θ0) are also asymptotically jointly normal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cagnone S. A note on goodness of fit test in latent variable models with categorical variables. Commun. Stat. A Theor. 2012;41:2983–2990. [Google Scholar]
- Claeskens G, Hart JD. Goodness-of-fit tests in mixed models. Test. 2009;10:1100–1120. [Google Scholar]
- Cox DR. Tests of separate families of hypotheses; Proc. Fourth Berkeley Symp. on Math. Statist. and Prob..1961. pp. 105–123. [Google Scholar]
- Crainiceanu CM, Ruppert D. Likelihood ratio tests in linear mixed models with one variance component. J. R. Statist. Soc. B. 2004;66:165–185. D. [Google Scholar]
- Godfrey LG. Misspecification Tests in Econometrics: The Lagrange Multiplier Principle and Other Approaches (Econometric Society Monographs) Cambridge University Press; Cambridge, United Kingdom: 1988. [Google Scholar]
- Golub GH, Van Loan CF. Matrix Computations. The Johns Hopkins Studies in Mathematical Sciences, The Johns Hopkins University Press; Baltimore and London: 1983. [Google Scholar]
- Hall P, Heyde CC. Martingale Limit Theory and its Applications. Academic Press; New York: 1980. [Google Scholar]
- Jiang J. REML Estimation: Asymptotic Behavior and Related Topics. Ann. Stat. 1996;24:255–286. [Google Scholar]
- Jiang J. Goodness-of-fit tests for mixed model diagnostics. Ann. Stat. 2001;4:1137–1164. [Google Scholar]
- Khuri AI, Mathew T, Sinha BK. Statistical Tests for Mixed Linear Models. John Wiley & Sons; New York: 1998. [Google Scholar]
- Lombardía MJ, Sperlich S. Semiparametric inference in generalized mixed effects models. J. R. Statist. Soc. B. 2008;70:913–930. [Google Scholar]
- Marozzi M. Multivariate tri-aspect non-parametric testing. J. Nonparametr. Stat. 2007;19:269–282. [Google Scholar]
- Marozzi M. A modified Cucconi test for location and scale change alternatives. Colomb. J. Statist. 2012;35:369–382. M. [Google Scholar]
- Maydeu-Olivares A, Joe H. Limited- and full-information estimation and goodness-of-fit testing in 2n contingency tables: A unified framework. J. Am. Stat. Assoc. 2005;100:1009–1020. [Google Scholar]
- Maydeu-Olivares A, Joe H. Limited information goodness-of-fit testing in multidimensional contingency tables: a unified framework. Psychometrika. 2006;71:713–732. [Google Scholar]
- McCulloch CE, Searle SR. Generalized, Linear, and Mixed Models. John Wiley & Sons; New York: 2001. [Google Scholar]
- Miller JJ. Asymptotic properties of maximum likelihood estimates in the mixed model of the analysis of variance. Ann. Stat. 1977;5:746–762. [Google Scholar]
- Pan Z, Lin DY. Goodness-of-fit methods for generalized linear mixed models. Biometrics. 2005;61:1000–1009. doi: 10.1111/j.1541-0420.2005.00365.x. [DOI] [PubMed] [Google Scholar]
- Pesarin F, Fortunato L. Permutation Tests for Complex Data: Theory, Applications and Software. John Wiley & Sons; New York: 2010. [Google Scholar]
- Rao CR. Linear statistical inference and its applications. second Edition John Wiley & Sons; New York: 1973. [Google Scholar]
- Rao CR, Wu Y. A strongly consistent procedure for model selection in a regression problem. Biometrika. 1989;76(2):369–374. [Google Scholar]
- Richardson AM, Welsh AH. Asymptotic properties of restricted maximum likelihood (REML) estimates for hierarchical mixed linear models. Austral. J. Statist. 1994;36:31–43. [Google Scholar]
- Ritz C. Goodness-of-fit tests for mixed models. Board of the Foundation of Scand. J. Stat. 2004;31:443–458. [Google Scholar]
- Salmaso L, Solari A. Multiple aspect testing for case-control designs. Metrika. 2005;62:331–340. [Google Scholar]
- Schoenfeld D. Chi-squared goodness-of-fit tests for the proportional hazards regression model. Biometrika. 1980;67:145–153. [Google Scholar]
- Self SG, Liang K. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. Amer. Statist. Assoc. 1987;82:605–610. [Google Scholar]
- Shao J. An asymptotic theory for linear model selection. Stat. Sinica. 1997;7:221–264. [Google Scholar]
- Stezhko VA, Buglova EE, Danilova LI, et al. A cohort study of thyroid cancer and other thyroid diseases after the Chernobyl accident: Objectives, design and methods. Radiat. Res. 2004;161:481–492. doi: 10.1667/3148. [DOI] [PubMed] [Google Scholar]
- Tang M. Ph.D Thesis: Goodness of fit test for generalized linear mixed models. Department of Mathematics, University of Maryland; College Park: 2010. [Google Scholar]
- van der Vaart AW. Asymptotic Statistics. Cambridge University Press; Cambridge, United Kingdom: 2000. [Google Scholar]
- Wand MP. Fisher information for generalized linear mixed models. J. Multivariate. Anal. 2007;98:1412–1416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.