Phosphorylation of cortactin downstream of the EGF receptor–Src-Arg kinase cascade triggers maturation of invadopodia, actin-rich protrusions that breast cancer cells use to invade the extracellular matrix. Phosphocortactin recruits Vav2 to invadopodia to activate Rac3 and support actin polymerization, matrix degradation, and invasion.
Abstract
Breast carcinoma cells use specialized, actin-rich protrusions called invadopodia to degrade and invade through the extracellular matrix. Phosphorylation of the actin nucleation–promoting factor and actin-stabilizing protein cortactin downstream of the epidermal growth factor receptor–Src-Arg kinase cascade is known to be a critical trigger for invadopodium maturation and subsequent cell invasion in breast cancer cells. The functions of cortactin phosphorylation in this process, however, are not completely understood. We identify the Rho-family guanine nucleotide exchange factor Vav2 in a comprehensive screen for human SH2 domains that bind selectively to phosphorylated cortactin. We demonstrate that the Vav2 SH2 domain binds selectively to phosphotyrosine-containing peptides corresponding to cortactin tyrosines Y421 and Y466 but not to Y482. Mutation of the Vav2 SH2 domain disrupts its recruitment to invadopodia, and an SH2-domain mutant form of Vav2 cannot support efficient matrix degradation in invasive MDA-MB-231 breast cancer cells. We show that Vav2 function is required for promoting invadopodium maturation and consequent actin polymerization, matrix degradation, and invasive migratory behavior. Using biochemical assays and a novel Rac3 biosensor, we show that Vav2 promotes Rac3 activation at invadopodia. Rac3 knockdown reduces matrix degradation by invadopodia, whereas a constitutively active Rac3 can rescue the deficits in invadopodium function in Vav2-knockdown cells. Together these data indicate that phosphorylated cortactin recruits Vav2 to activate Rac3 and promote invadopodial maturation in invasive breast cancer cells.
INTRODUCTION
To invade surrounding tissues, invasive epithelial cancer cells must penetrate the basement membrane, a network of extracellular matrix (ECM) proteins that supports the overlying epithelium. Cancer cells can penetrate this barrier by forming protrusive actin-rich structures called invadopodia that concentrate ECM degrading activity to cell–substrate contact points. Invadopodium formation is stimulated by epidermal growth factor (EGF) released by tumor-associated macrophages (Wyckoff et al., 2004; Roh-Johnson et al., 2014). Once formed, invadopodia provide routes for metastatic cells to escape the epithelium, traverse the stroma, and enter the bloodstream (Yamaguchi et al., 2005b).
Invadopodia mature via a series of events into matrix-degrading structures. Initially, small punctate clusters form called invadopodium precursors, which are composed of actin, the actin nucleation–promoting factors cofilin, cortactin, and N-WASp, and the actin-related protein (Arp) 2/3 complex (Lorenz et al., 2004; Desmarais et al., 2009; Oser et al., 2009). These precursors have two fates: they can either disappear (Moshfegh et al., 2015) or stabilize through increased actin polymerization and recruitment of matrix metalloproteinases, developing into matrix-degrading invadopodia (Artym, 2006; Oser et al., 2009; Sharma et al., 2013; Beaty and Condeelis, 2014). We previously showed that an EGF receptor–Src-Arg kinase cascade phosphorylates cortactin to trigger actin polymerization and invadopodia stabilization (Mader et al., 2011; Beaty et al., 2013). Disruption of Arg-mediated cortactin phosphorylation in invasive human breast cancer cells compromises invadopodia function and reduces the ability of the cell to invade the ECM. Moreover, knockdown of Arg results in decreased invasion and metastasis in a mouse xenograft model of human breast cancer (Gil-Henn et al., 2013).
In invadopodium precursors, cofilin initiates the polymerization of short actin filaments that stimulate Arp2/3 complex to nucleate dendritic actin filament to drive protrusions and maturation of the invadopodium (Uruno et al., 2001; Weaver et al., 2002; Yamaguchi et al., 2005a; Oser et al., 2009; Magalhaes et al., 2011; Bravo-Cordero et al., 2013). Of importance, cortactin contains two tyrosine phosphorylation sites (Y421, Y466) that regulate cofilin activation and therefore invadopodium function (Oser et al., 2010; Magalhaes et al., 2011; Bravo-Cordero et al., 2013). In addition, phospho-cortactin (Y421, Y466) binds to the SH2 domain–containing proteins Nck1, Src, and Arg, driving invadopodium formation and function as a mature ECM degradation organelle (Okamura and Resh, 1995; Lapetina et al., 2009; Oser et al., 2009, 2010; Mader et al., 2011; Bravo-Cordero et al., 2013).
We screened a library of human SH2 domains to identify those that bind selectively to tyrosine-phosphorylated cortactin. We demonstrate that these proteins bind selectively with high affinity to phosphotyrosine-containing peptides containing cortactin residues Y421 or Y466 but not to phosphopeptides containing Y482. We report here, for the first time, that phospho-cortactin binds selectively to the Rho-family GTPase guanine nucleotide exchange factor (GEF) Vav2. Knockdown of either cortactin or Vav2 or mutational disruption of their binding interface abrogates Vav2 localization to invadopodia and decreases matrix degradation and cell invasion. We show that invadopodia require Vav2 GEF activity for maturation, although a deficit in Vav2 can be rescued by exogenous Rac3 activation. Using a fluorescence resonance energy transfer (FRET)–based biosensor for Rac3, we find “rings” of Rac3 activity around invadopodia that form just before they begin to degrade matrix. Together these data reveal that phosphorylated cortactin recruits Vav2 to invadopodia to activate Rac3 and support actin polymerization, invadopodium function, matrix degradation, and invasion.
RESULTS
Identification of Vav2 as a phospho-cortactin–binding protein
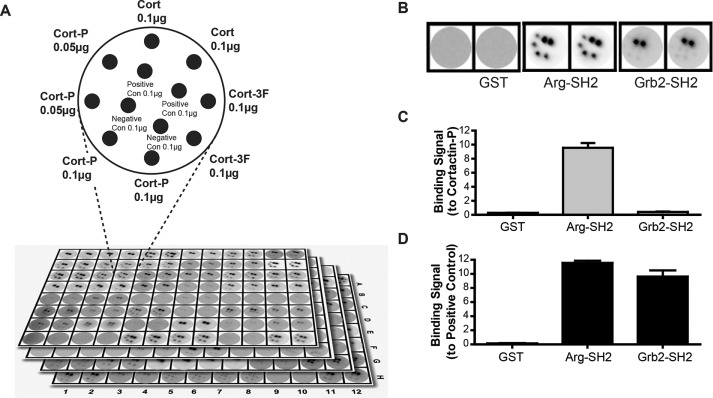
We showed previously that phospho-cortactin can bind to the SH2 domains of Arg and Nck1 and that all three proteins are key regulators of actin-interacting proteins in fibroblasts and cancer cells (Lapetina et al., 2009; Oser et al., 2010). To identify possible additional interacting partners of phospho-cortactin, we screened a set of 104 glutathione S-transferase (GST)–tagged human SH2 domains (of 115 total SH2 domain–containing proteins in the human proteome) for their ability to selectively bind tyrosine phosphorylated cortactin. Replicate samples of nonphosphorylated wild-type cortactin (Cort), a nonphosphorylatable cortactin mutant with three key tyrosine residues mutated to phenylalanine (Cort3F), or phosphorylated cortactin (CortP) were spotted on nitrocellulose membranes, along with pervanadate-treated or phosphatase-treated cell extracts as positive and negative controls for tyrosine phosphorylation. The membranes were placed into 96-well plates, and each well was incubated with a distinct GST-SH2 domain–horseradish peroxidase (HRP) conjugate (Machida et al., 2003, 2007; Gifford et al., 2014). Binding to the pervanadate-treated positive control verified that each SH2 domain was able to bind phosphotyrosine-containing proteins. No SH2 domain bound to the phosphatase-treated cell-lysate negative control. We identified 39 SH2 domains that bound selectively to phospho-cortactin but did not bind nonphosphorylated cortactin or cortactin 3F (Figures 1, A–D, and 2, A and B, and Supplemental Figure S1).
FIGURE 1:
SH2 domains in the human proteome against phosphorylated cortactin identify Vav2 as a phospho-cortactin–binding protein. (A) Schematic of dot blot membranes used in the SH2 domain–binding screen. Nonphosphorylated cortactin (Cort), a nonphosphorylatable cortactin mutant (Cort-3F), and phosphorylated cortactin (Cort-P) were spotted on nitrocellulose membrane disks. Pervanadate- or phosphatase-treated cell lysates were used as positive and negative controls, respectively. Each disk was probed with a separate GST-SH2 domain fusion in quadruplicate. (B) Selected examples of control, positive, and negative binding results. A negative control GST-only probe did not bind to any sample. The Arg SH2 domain bound to both the positive control and Cort-P but not to nonphosphorylated Cort, Cort-3F, or the negative control, demonstrating specific binding to phosphorylated cortactin. The Grb2 SH2 domain bound only to the positive control, demonstrating that the probe was active but does not bind to phosphorylated cortactin. (C) Quantification of binding signal to Cort-P from examples in B. (D) Quantification of binding signal to positive control from examples in B.
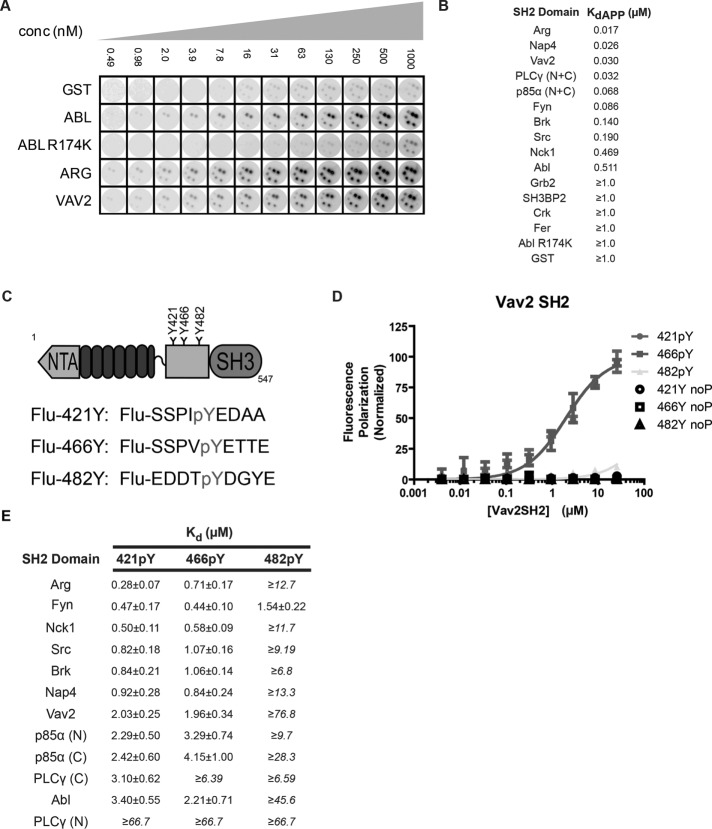
FIGURE 2:
Validation of SH2 domain screen for phospho-cortactin–binding proteins and identification of the Vav2 SH2 domain as a specific binder of the cortactin 421 and 466 phosphotyrosine residues by fluorescence anisotropy. (A) Cortactin-spotted nitrocellulose disks were probed with increasing concentrations of the top interacting SH2 domains. Selected noninteracting SH2 domains were also tested as negative controls (GST, the AblR174K SH2 point mutant). (B) Each SH2 domain identified in the original screen bound to phosphorylated cortactin in a concentration-dependent manner with submicromolar affinities, whereas none of the negative control SH2 domains bound with appreciable affinity. The maximal concentration tested was 1.0 µM, and therefore we place a lower limit on the negative control binding affinities. Note that p85α and PLCγ contain two tandem SH2 domains. For these experiments these probes contained both SH2 domains (N + C). It is important to note that these apparent affinities are for GST-SH2 fusions, which can dimerize and likely lower the reported KdAPP values. (C) Cortactin domains (NTA; N-terminal acidic region; SH3; Src homology 3) and construction of fluoresceinated (Flu-) peptides corresponding to each cortactin phosphorylation site. (D) Example fluorescence polarization curves for the Vav2 SH2 domain and phosphorylated cortactin peptides. Vav2 SH2 domain binds in a concentration-dependent manner to phosphorylated sites 421 (421pY; circles) and 466 (466pY; squares) but not to 482 (482pY; triangles) or nonphosphorylated controls of each site (noP). (E) Calculated KD values from fluorescence anisotropy experiments as described in C and D for the top 10 binding candidates from initial screen in Figure 1. Note that p85α and PLCγ contain two tandem SH2 domains. Here, N and C refer to either the individual N-terminal or C-terminal SH2 domains, respectively. All experiments were done in triplicate. Under conditions in which saturation was not achieved, a lower limit for binding affinity is given.
The 10 most strongly detected phospho-cortactin–binding SH2 domains included several known cortactin-binding proteins, such as Arg, Src, Nck1, and Abl, and we detected novel interactions with the SH2 domains of the Rho-family GTPase activator Vav2, Nap4/SOCS7, the p85α subunit of phosphoinositide 3-kinase, phospholipase Cγ, and the Src-family kinase Fyn. We repeated the screen using increasing concentrations of these top 10 hits to determine their relative apparent binding affinities. Each SH2 domain probe bound phospho-cortactin in a concentration-dependent manner with submicromolar affinity. The Arg SH2 domain bound with the tightest apparent affinity, followed by the Nap4, Vav2, PLCγ, p85α, Fyn, Brk, Src, Nck1, and Abl SH2 domains (Figure 2B and Supplemental Figure S1). Note that p85α and PLCγ contain two tandem SH2 domains, and the SH2 probe contained both SH2 domains. This screen demonstrated that phospho-cortactin binds selectively and with high affinity to discrete groups of SH2 domain–containing proteins.
The Vav2 SH2 domain binds phospho-cortactin in vitro and in vivo
Cortactin was initially reported to be phosphorylated on three tyrosine sites (Y421, Y466, Y482; Zhan et al, 1993), but mutation of only two of these sites (Y421 and Y466) abrogates cortactin tyrosine phosphorylation by both Src and Abl family kinases (Boyle et al., 2007). We incubated 5 nM of each fluorescently labeled nonamer cortactin phosphopeptide (Figure 2C) with increasing concentrations of monomeric SH2 domains cleaved from GST and monitored the change in fluorescence polarization to measure the affinities and binding specificities of SH2 domain binding. Each SH2 domain bound to phospho-Y421 and phospho-Y466 peptides with submicromolar to low-micromolar affinities (Figure 2E) but bound with significantly weaker affinity (KD > 5 μM), if at all, to the phospho-Y482 peptide. In the few cases in which saturated binding was not achieved, we report the lower limits of KD. The Arg SH2 domain showed a 2.5-fold greater affinity for the phospho-Y421 peptide relative to the phospho-Y466 peptide, whereas other SH2 domains bound with similar affinities to both sites (Figure 2, C–E, and Supplemental Figure S2). The Vav2 SH2 domain bound with similar low-micromolar affinity to both the phospho-Y421 and phospho-Y466 peptides (Figure 2D). These data indicate that the identified SH2 domains bind selectively and with high affinity to the phospho-Y421 and phospho-Y466 peptides.
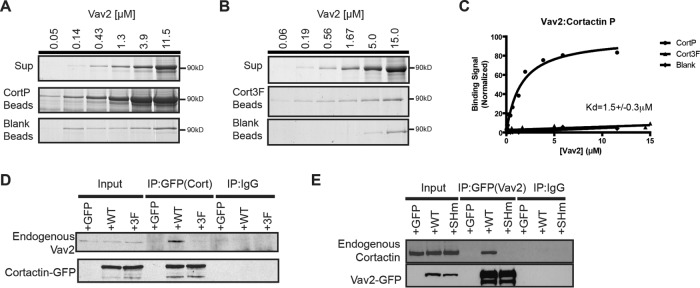
Given Vav2’s involvement in promoting Rac-mediated cellular protrusions (Liu and Burridge, 2000; Marignani and Carpenter, 2001), we decided to see whether a Vav2–cortactin interaction was involved in breast cancer invadopodium function. As a first step, we tested whether purified recombinant full-length Vav2 and phosphorylated cortactin interact directly in vitro and in cells. We incubated agarose beads covalently coupled to phosphorylated cortactin or nonphosphorylatable cortactin (Cort-3F) with increasing concentrations of full-length Vav2. Vav2 binds to phosphorylated cortactin with low-micromolar affinity (KD = 1.5 ± 0.3 μM) but does not detectably bind nonphosphorylatable cortactin (Figure 3, A–C). We also examined the phospho-cortactin–Vav2 interaction in cells by transiently transfecting HEK293T cells with control green fluorescent protein (GFP), wild-type cortactin-GFP (Cort-WT-GFP), or Cort-3F-GFP and treated with pervanadate to promote cortactin phosphorylation. Endogenous Vav2 coimmunoprecipitated with wild-type cortactin but not with GFP only or with Cort-3F-GFP (Figure 3D). We also found that cortactin coimmunoprecipitated with wild-type Vav2 after pervanadate treatment but not with GFP or with Vav2 bearing a point mutation (R698K) in its SH2 domain that disrupts binding to phosphotyrosine (Vav2-SH2m; Figure 3E). Together with the binding experiments, these results indicate that Vav2 interacts directly with phospho-cortactin in an SH2 domain–dependent manner and that this interaction can occur in cells.
FIGURE 3:
Phospho-cortactin binds to Vav2. Representative Coomassie blue–stained SDS–PAGE gel showing binding of Vav2 to Tyr-phosphorylated cortactin (CortP) (A) beads but not to nonphosphorylatable cortactin (Cort3F) beads (B) nor to beads blocked with an excess of Tris. (C) Fitted single-site-binding curves demonstrating Vav2 binding to CortP (1.5 ± 0.3 µM) but not to nonphosphorylated cortactin or control beads. (D) Vav2 coimmunoprecipitates only with phosphorylated cortactin. GFP control, wild type cortactin-GFP (WT-GFP), or nonphosphorylatable cortactin-GFP (3F-GFP) was expressed in HEK293T cells and immunoprecipitated using anti-GFP or immunoglobulin G (IgG) control, and immunoblots were probed for endogenous coimmunoprecipitating Vav2 and cortactin. (E) Cortactin coimmunoprecipitates with only Vav2 containing a functional SH2 domain. GFP control, wild-type vav2-GFP (WT-GFP), or SH2 mutant Vav2 (R698K) (SHm-GFP) was expressed in HEK293T cells and immunoprecipitated using anti-GFP or IgG control, and immunoblots were probed for endogenous coimmunoprecipitating Vav2 and cortactin.
Vav2 localizes to invadopodia and is required for matrix degradation
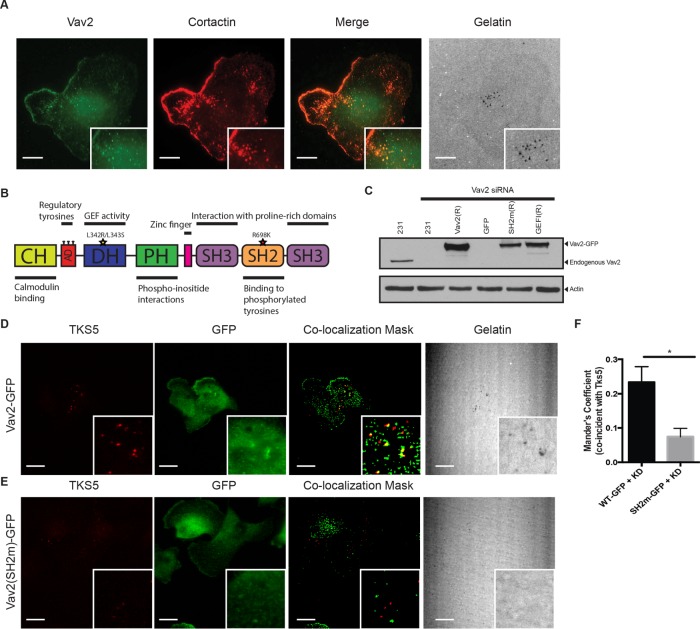
Although phospho-cortactin interacts with Vav2 in vitro and in cells (Figures 2 and 3), it was important to test whether this interaction occurs at invadopodia, which comprise a small fraction of the total cell volume. To test this hypothesis, we used the highly invasive MDA-MB-231 triple-negative (Her2–/ER–/PR–) breast cancer cell line, which produces invadopodia in culture and exhibits aggressive invasive behavior in vivo (Artym, 2006; Gil-Henn et al., 2013; Patsialou et al., 2013). MDA-MB-231 cells (231 cells) were incubated on a fluorescently labeled gelatin matrix and immunostained for Vav2 and cortactin, which is known to localize to invadopodia (Bowden et al., 2006). Vav2 and cortactin colocalized prominently at sites of matrix degradation (Figure 4A).
FIGURE 4:
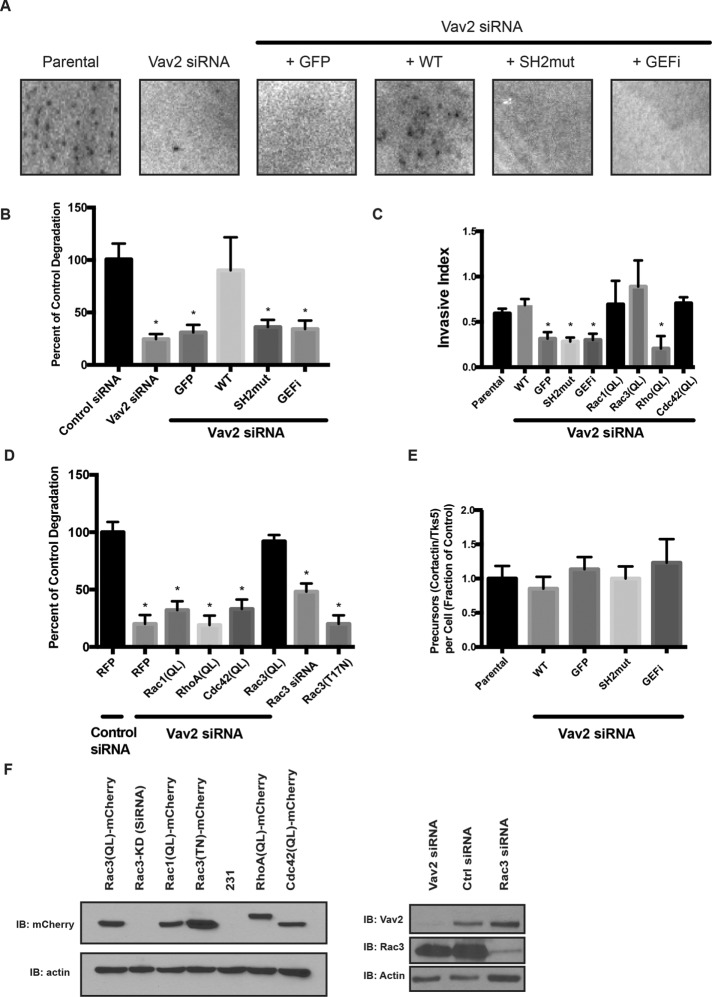
Localization of Vav2 to invadopodia is mediated by its SH2 domain. (A) Colocalization of Vav2 and cortactin at sites of gelatin degradation. Insets, blow-ups of sites of invadopodia action. Scale bars, 10 μm. (B) Domain diagram of Vav2 indicating relevant mutations: SH2mut (R698K) and GEFi (L342R/L343S). (C) Western blot analysis of MDA-MB-231 cells transfected with control or Vav2 siRNA and cell lines stably expressing RNAi-resistant rescue constructs with wild-type Vav2-GFP (Vav2(R)), Vav2SH2m-GFP (SH2m(R)), or Vav2GEFi-GFP (GEFI(R)). Blots were immunoblotted for Vav2 and for β-actin as a loading control. (D–F) Endogenous Vav2 was knocked down in cells stably expressing RNAi-resistant Vav2-GFP (D) or Vav2SH2m-GFP (E). Cells were plated on fluorescent gelatin and immunostained for TKS5 and GFP. Scale bars, 10 μm. (F) Quantification of colocalization of WT and SH2m Vav2 with Tks5 at invadopodia.
We next asked whether Vav2 localization to invadopodia required binding of the SH2 domain of Vav2 to phospho-cortactin. To accomplish this, we generated stable cell lines expressing RNA interference (RNAi)–resistant Vav2-GFP, Vav2-SH2m-GFP, or GFP alone, in which we knocked down >98% of the endogenous Vav2 using small interfering RNA (siRNA; Figure 4C). After plating on matrix for 4 h, we fixed the cells and immunofluorescently labeled GFP and Tks5, a scaffolding protein found in invadopodia. Vav2-GFP colocalized with puncta of Tks5 and matrix degradation (Figure 4, D–F), indicating functional invadopodia, whereas Vav2-SH2m-GFP did not. Together, these results suggest that Vav2 requires SH2 domain–mediated binding to phosphorylated cortactin to localize to invadopodia.
ECM degradation is the hallmark of fully mature invadopodia (Chen, 1989; Barsky et al., 1983; Artym, 2006; Oser et al., 2009). We found that Vav2-knockdown cells were deficient in degradation, as revealed by the total area of matrix degradation observed over 18 h (Figure 5, A and B). Expression of RNAi-resistant Vav2-GFP rescued these deficits, whereas Vav2-SH2m-GFP could not rescue this phenotype (Figure 5, A and B, and Supplemental Figure S4).
FIGURE 5:
Degradation of ECM requires both the SH2 domain and the GEF function of Vav2, and a constitutively active form of Rac3 can rescue Vav2 deficiency. Parental MDA-MB-231 cells or cells stably expressing GFP-tagged, RNAi-resistant WT or mutants of Vav2 were treated with control siRNA or Vav2 siRNA (Vav2 KD), plated on a fluorescent matrix, and allowed to degrade for 18 h. (A) Representative images of degradation area formed by the different cell lines. (B) Quantification of invadopodia matrix degradation area per cell. Ten fields were averaged in triplicate experiments. Vav2-knockdown and mutant constructs degrade significantly less matrix than wild-type constructs or control (p < 0.05). (C) Parental and mutant MDA-MB-231 cells were plated on a Matrigel-coated Transwell insert and allowed to migrate toward serum-containing medium. Relative invasion through Matrigel was quantified and normalized to proteolysis-independent migration. Vav2-knockdown and mutant constructs have a significant defect in Transwell invasion, but constitutively active Rac1 (Q61L), Rac3 (Q61L), RhoA (Q63L), or Cdc42 (Q61L; QL mutants) rescue the Vav2-knockdown phenotype (p < 0.05). Experiments performed in triplicate (n = 10 per condition). (D) Cells were treated with a control siRNA, Vav2 siRNA, or Rac3 siRNA or transfected with a dominant-negative (T17N) form of Rac3. Quantification of invadopodia matrix degradation area per cell for each of the cell types normalized to control. Ten fields were averaged in triplicate experiments. (E) Quantification of invadopodium formation (as measured by cortactin/Tks5 double-positive puncta). Cells were serum starved overnight in the presence of MMP inhibitor and treated with EGF (n = 20 per condition). (F) Western blot analysis of MDA-MB-231 cells transfected with control siRNA, Rac3 siRNA, and Vav2 siRNA and cell lines stably expressing rescue constructs with mCherry-tagged dominant-negative Rac3(TN), constitutively active Rac1(QL), Rac3(QL), RhoA(QL), or Cdc42(QL). Samples were immunoblotted for Rac3, Vav2, or mCherry and for β-actin as a loading control.
Vav2 knockdown does not affect the assembly of cortactin and Tks5-positive invadopodial precursors, but Vav2 is required for EGF-stimulated actin polymerization at invadopodia
We next determined which step in invadopodium maturation was affected by the loss of Vav2 function. We serum starved control 231 cells or Vav2-knockdown 231 cells, with or without Vav2-GFP and Vav2-SH2m-GFP rescue, overnight while inhibiting matrix degradation with the matrix metalloproteinase inhibitor GM6001. After 12 h, we washed out the inhibitor and stimulated with EGF to synchronously induce invadopodia formation (Mader et al., 2011). Each cell type formed statistically indistinguishable numbers of cortactin- and Tks5-positive puncta, which may be on the path to making invadopodia (Figure 5E). Together with the finding that matrix degradation is compromised in Vav2-knockdown cells, these results suggests that whereas Vav2 does not appear to be required for cortactin:Tks5 puncta formation, Vav2 does regulate the maturation of invadopodium precursors into fully mature matrix-degrading invadopodia.
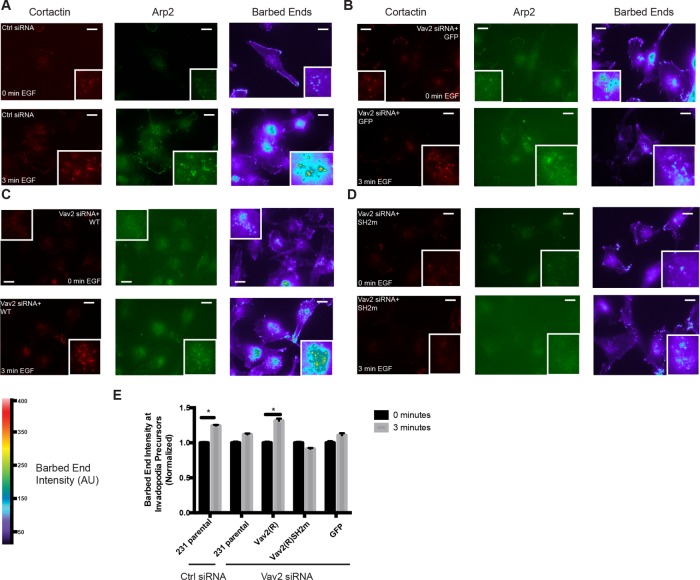
Cofilin and Arp2/3 complex–dependent actin polymerization is required for invadopodial maturation (Yamaguchi et al., 2005a; Desmarais et al., 2009; Oser et al., 2009, 2010; Magalhaes et al., 2011). To investigate whether Vav2 has a role in this process, we measured the relative assembly of actin at invadopodia as described previously (Oser et al., 2009, 2010; Mader et al., 2011). Briefly, we serum starved cells overnight, stimulated them for 3 min with EGF, and visualized the relative incorporation of biotinylated G-actin monomers at barbed ends of preexisting actin by measuring the average biotin-actin intensity at Arp2-positive puncta. We found that the intensity of actin was increased by 25% in control siRNA-treated cells after EGF treatment (Figure 6, A and E). This EGF-induced increase in actin barbed-end labeling was not observed in Vav2-knockdown cells and could be restored by reexpression of wild-type Vav2-GFP but not Vav2SH2m-GFP (Figure 6, B–E). These data suggest that proper Vav2 localization to invadopodia is critical for EGF-induced actin polymerization at invadopodia.
FIGURE 6:
EGF-induced formation of actin barbed ends at invadopodia requires Vav2. Parental and Vav2-rescued MDA-MB-231 cells were knocked down using control or Vav2 siRNA and either left untreated (0 min EGF) or stimulated with EGF for 3 min. Cells were fixed, gently permeabilized, and allowed to incorporate biotin-actin, followed by fixation and staining for biotin actin and Arp2 to label invadopodia. Representative micrographs for control scrambled siRNA (A), Vav2 siRNA knockdown with GFP rescue alone (B), an RNAi-resistant Vav2-GFP construct (C), or an RNAi-resistant Vav2-GFP with the SH2m mutation (D). Scale bars, 10 μm. (E) Quantification of free actin barbed-end formation as measured by average biotin-actin intensity in Arp2-positive puncta.
Vav2 is required for invasion through extracellular matrix
To determine whether Vav2 affects invasive migration toward a chemotactic cue, we plated MDA-MB-231 cells in 0.5% serum and measured their ability to invade through Matrigel (Gaggioli et al., 2007) toward 10% serum–containing medium. Vav2-knockdown cells exhibited significantly reduced invasion compared with control cells and were rescued by reexpression of Vav2-WT but not Vav2-SH2m, indicating that Vav2 is required for invasive cell migration through ECM and the SH2 domain of Vav2 is essential for mediating this function (Figure 5C).
To account for possible migratory deficits resulting from altered Vav2 knockdown, we also measured the ability of cells to chemotax in a Dunn chamber using serum as an attractant (Zicha et al., 1997; Allen et al., 1998). Serum-starved cells were plated in the center of the chamber, and migration toward a chemotactic gradient of 10% serum was visualized by videomicroscopy over 4 h. We found that Vav2-knockdown cells have a deficit in directional migration, whereas their migration speed was similar to that of wild-type cells (Supplemental Figure S3).
Vav2 acts through Rho-family GTPases to regulate invadopodium function
Depletion of Vav2 in MDA-MB-231 cells induces a severe invadopodial maturation defect that is rescued by wild-type Vav2-GFP but not by Vav2-SH2m-GFP. Because Vav2 is a GEF, we also asked whether Vav2 GEF activity is required for its role in promoting invadopodia function. To address this, we mutated the catalytic residues of the Vav2 Dbl domain to render it inactive (L342R/L343S; hereafter denoted Vav2GEFi; Marcoux and Vuori, 2003) and developed a cell line stably expressing Vav2GEFi-GFP (Figure 4C). After knockdown of endogenous Vav2, we found that Vav2GEFi-GFP–expressing cells have impaired ability to degrade matrix, similar to Vav2-knockdown cells or cells expressing Vav2-SH2m-GFP (Figure 5B). Vav2GEFi-GFP was also unable to rescue the defects in Matrigel invasion of Vav2-knockdown cells (Figure 5C).
Active Rac3 can rescue the Vav2-knockdown phenotype in invadopodia
The finding that Vav2 GEF activity is required for invadopodium function raised the question of which Rho-family GTPase is targeted by Vav2 in this context. Despite repeated attempts, we did not observe significant changes in the activities of Rac1, RhoA, or Cdc42 in whole-cell extracts of EGF-stimulated cells (unpublished data), possibly because invadopodia represent a small fraction of the total cell volume. As an alternative approach, we tested whether constitutively active (generated by mutation of a conserved glutamine to leucine; hence QL) mutants of Rho-family GTPases (Figure 5F) could rescue invadopodial function in Vav2-knockdown cells, an approach previously used by others (Razidlo et al., 2014). MDA-MB-231 cells in which Vav2 was knocked down via siRNA were transfected with constitutively active mutants of RhoA, Rac1, Rac3, and Cdc42. We found that constitutively active Rac3 alone fully rescued matrix degradation ability in Vav2-knockdown cells (Figure 5D and Supplemental Figure S4). Although Rac1 and Rac3 have substantial amino acid sequence homology (92%) and are both expressed in MDA-MB-231 cells, previous work suggested that Rac3 has a distinct role in promoting cancer aggressiveness (Gest et al., 2013).
Rac3 is a Vav2 GEF substrate
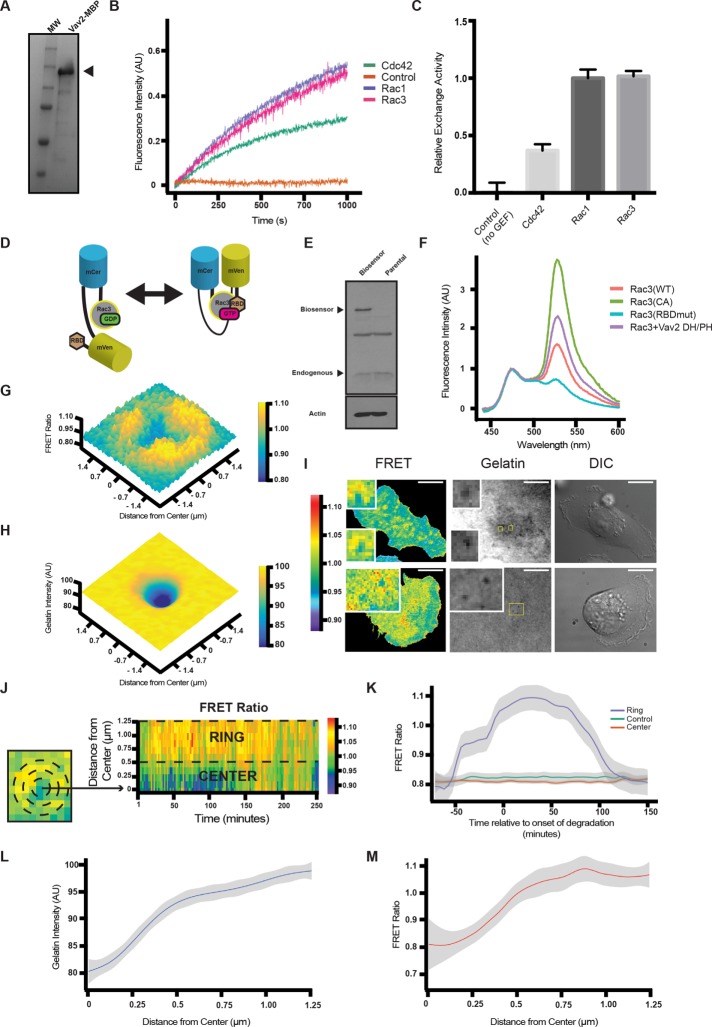
Rac3 has not previously been identified as a Vav2 substrate. We performed in vitro GTP exchange assays with purified proteins to test whether Vav2 is capable of promoting nucleotide exchange on purified Rac3. Purified recombinant Vav2 promoted GTP loading onto recombinant Rac1 and Rac3, but its activity was significantly reduced using Cdc42 as a substrate (Figure 7, A–C). The relative activities we measured for Vav2 on Rac1 and Cdc42 are consistent with a previous study (Abe et al., 2000). These data indicate that Vav2 is a GEF for Rac3.
FIGURE 7:
Rac3 GTP/GDP exchange is stimulated by Vav2. (A) Coomassie blue–stained gel showing a prominent band at the expected (150 kDa) molecular weight for Vav2-MBP. (B) Representative fluorescence traces from mant-GTP exchange assays performed with Vav2 GEF and the indicated Rho-family GTPase. The control trace represents Rac1 with no Vav2 added. Experiments were run in triplicate, and maximum slope was used to quantify exchange activity according to the kit manufacturer’s instructions. (C) Quantification of relative GEF activity for the various GTPases normalized to Rac1. (D) Diagram of FRET Rac3 biosensor. (E) Western blot analysis of parental MDA-MB-231 cells or cells stably expressing the Rac3 biosensor. Blots were immunoblotted for Rac (top) and β-actin (bottom) as a loading control. (F) Emission spectrum of Rac3 biosensor activity in cell lysates of cells expressing WT, CA mutant, RBD mutant, or Rac3 + Vav2 (DH/PH). (G) A 3D plot of maximum intensity projection over time (50 min) of FRET ratio from a representative invadopodium. (H) A 3D plot of minimum intensity projection over time (50 min) of gelatin intensity from a representative invadopodium corresponding to G. (I) Still frames from movies of MDA-MB-231 cells expressing the Rac3 FRET biosensor plated on fluorescent matrix. Insets, ring of Rac3 activity around actively degrading invadopodia. Scale bars, 10 μm. (J) Radial heat-map kymograph representation of invadopodia. To depict Rac3 FRET activity, mean FRET values in concentric annuli centered on a degradation puncta identified in the fluorescent gelatin channel were recorded and plotted as a function of time as a heat map. A boundary between the “ring” and “center” was determined by maximizing the mean difference in FRET signal between the “ring” and “center” among the pooled data. (K) LOWESS smoothed fits of the FRET signal from the “ring” and “center” regions, with 95% confidence intervals, along with averaged FRET data from a control area of the cell that is not participating in matrix degradation. Time zero is the onset of degradation observed in the fluorescent gelatin channel. As the invadopodium degrades gelatin, Rac3 activity increases in a ring around the invadopodium core over a time scale of minutes to hours and then returns to baseline. In contrast, control areas of the cell and the center of degradation puncta do not change significantly from baseline. (L) LOWESS smoothed fit of the gelatin intensity at invadopodia, averaged over time from 25 to 75 min, as a function of distance from the center of matrix degradation activity, with 95% confidence intervals. (M) LOWESS smoothed fit of the FRET ratio at invadopodia, averaged over time from 25 to 75 min, as a function of distance from the center of matrix degradation activity, with 95% confidence intervals.
We knocked down Rac3 and, in separate cells, expressed a dominant-negative (T17N) mutant that is incapable of GTP binding (Figure 5F). We found that both treatments disrupted the ability of cells to degrade matrix, consistent with a central role for Rac3 in allowing invadopodia to become functional (Figure 5D). The dominant-negative Rac3 construct appeared to be more potent in disrupting matrix degradation than Rac3 knockdown and approached a level that was indistinguishable from that of Vav2 knockdown. The elevated inhibition could be due to residual wild-type Rac3 after knockdown. Alternatively, the dominant-negative mutant of Rac3 may block Vav2 from activating other Rho GTPases, including Rac1 and Cdc42.
A Rac3 FRET biosensor reveals “rings” of Rac3 activity that surround active invadopodia and precede matrix degradation
To pinpoint where and when Rac3 becomes activated during matrix degradation, we developed a new FRET-based Rac3 biosensor (Dora-Rac3). Constructed similarly to previous RhoA and Rac1 biosensors (Pertz et al., 2006; Timmerman et al., 2015; van Unen et al., 2015; Kedziora et al., 2016), a full-length Rac3 was placed at the C-terminal end of the biosensor (see Materials and Methods for details). The design preserved an intact C-terminal hypervariable region and a CAAX box (for prenylation) in Rac3, thereby minimizing perturbation of its interaction with plasma membrane and RhoGDI. The sensor also consists of a bright FRET pair, Cerulean and Venus, and a p21-binding domain (PBD) of Pak1 that binds to active Rac3. When GTP is loaded onto the Rac3 moiety, the modules form a closed conformation and increase FRET between the two fluorescent proteins (Figure 7D).
To validate the Dora-Rac3 biosensor, we expressed it in HEK293 cells and measured its fluorescence emission spectrum (Figure 7F, normalized to Cerulean emission) upon donor excitation at 430 nm. A constitutively active mutant of Rac3 (Q61L) was constructed in the biosensor and showed pronounced FRET emission (FRET/CFP ratio of 3.65). As a negative control, a biosensor mutant bearing point mutations (H83,86D) in the PBD that disrupts Rac3 binding produced minimal FRET (ratio of 0.74). The high dynamic range (4.93-fold) of the Dora-Rac3 suggests a sensitive response of the biosensor to the activation of Rac3. Overexpression of the wild type Dora-Rac3 under growth conditions (10% serum) yielded elevated FRET (ratio of 1.59) in comparison with the negative control, consistent with a “hyperactivation” state of Rac3 as previously described (Mira et al., 2000). The elevated FRET is also sensitive to coexpression of RhoGDI (unpublished data), indicating that the expression level of the sensor could be important. Nevertheless, coexpression of the DH/PH domain of Vav2 significantly increased FRET, demonstrating a catalytic activity of Vav2 toward Rac3 in vivo (Figure 7F).
We generated an MDA-MB-231 cell line stably expressing low levels of the Dora-Rac3 biosensor to limit off-target effects often caused by overexpression (Figure 7E). These cells were serum starved overnight and then plated in serum-containing medium on a fluorescent gelatin matrix. The sensor imaging was conducted using total internal reflection fluorescence (TIRF) microscopy to focus on the Rac3 activation events in invadopodia while rejecting the vast background signal contributed from the cell volume. In contrast to previous studies of Rac1 in invadopodia, we failed to detect Rac3 activation in the center of invadopodia during prolonged imaging. Instead, we observed an increase of Rac3 activity (FRET/CFP ratio increase of 0.2–0.3 above baseline) in ring-like structures surrounding invadopodia and coincident with the onset of gelatin degradation as observed in the fluorescent matrix channel (shown as time-averaged three-dimensional [3D] plots in Figure 7, G and H, and at a single time point in Figure 7I). To analyze these structures quantitatively, we measured the mean FRET signal in concentric circles centered on degradation puncta observed in the gelatin channel as a function of time from the onset of degradation. The FRET signal from each circle was averaged and is presented as a radial kymograph in Figure 7J and as a radial cross section in Figure 7M (Figure 7L shows the corresponding gelatin intensity plot). These “activity rings” of Dora-Rac3 appeared before gelatin degradation was detected, increased coincident with the first detection of degradation, and persisted for up to 1 h during matrix degradation before gradually decaying to baseline (Figure 7K). In contrast, the center of the ring remained at baseline throughout the time period of active degradation at a similar FRET ratio to control areas of the cell where matrix degradation was not observed. These sensor studies, for the first time, revealed tightly regulated spatiotemporal dynamics of Rac3 activation in relation to invadopodia and suggest a potential role of Rac3 activation in invadopodia maturation and matrix degradation.
DISCUSSION
Invadopodia are actin-rich, subcellular structures capable of degrading extracellular matrix that are required for tumor metastasis in a number of different cancer cell types (Artym, 2006; Dedes et al., 2010; Bravo-Cordero et al., 2011). We investigated the consequences of cortactin phosphorylation downstream of the EGF-Src-Arg kinase cascade, which was previously described as a key molecular switch promoting the maturation of invadopodia into degradative structures (Mader et al., 2011; Magalhaes et al., 2011). To accomplish this, we performed a high-throughput screen of a nearly complete human SH2 domain library to identify the SH2 domains that selectively interact with phosphorylated cortactin. Among the strongest hits identified in this screen was the SH2 domain of a Rho-family GEF, Vav2.
We investigated the role of Vav2 in invadopodia function in metastatic breast cancer cells downstream of its recruitment by phosphorylated cortactin via its SH2 domain. We showed that Vav2 localizes to invadopodia in MDA-MB-231 cells and that the Vav2 SH2 domain is required for this localization. Although Vav2 is not required for invadopodium precursor formation, it does appear to be essential for matrix degradation—the hallmark of mature invadopodia—as well as for invasion through ECM toward a chemotactic cue. We show that Vav2 is required for actin polymerization and that its GEF activity is critical for its function at invadopodia. Moreover, we showed that the target of Vav2 GEF activity is Rac3. Blockade of Rac3 activity by knockdown or expression of a dominant-negative mutant of Rac3 recapitulates the Vav2 knockdown phenotype.
Vav-family GEFs play general roles in cancer invasiveness
Vav2 is one of three Rho GTPase GEFs that couple a Dbl homology domain with SH2 and SH3 binding domains (Schuebel et al., 1996). Vav2 has been relatively understudied in cancer, although there is some clinical evidence suggesting that it may participate in metastasis in cancer patients. Jiang et al. (2014) showed that Vav2 overexpression can predict a more aggressive subtype of breast cancer. In melanoma cells, knockdown of Vav2 reduced invasiveness in response to a gradient of CXCL12 (Bartolome, 2006). In contrast, in non–small cell lung cancer samples, as well as in head and neck squamous cell carcinoma samples, hyperphosphorylation of Vav2 is correlated with more aggressive tumors (Patel et al., 2007; Ahn et al., 2013). Finally, in a model of hepatocellular cancer, a metastasis-suppressive microRNA was found to exert its effects, in part, by down-regulating Vav2 (Wang et al., 2013). Razidlo et al. (2015) implicated Vav1, a closely related family member of Vav2, in the invasive behavior of pancreatic tumor cells. In their system, Vav1 activates Cdc42 downstream of activation by Src, promoting invasive behavior. Knockdown of Vav1 dramatically reduces the invasive behavior of pancreatic tumor cells.
Vav2 is required for invadopodia-mediated matrix degradation
Because invadopodia are dynamic structures and comprise only a small fraction of the total cell volume, we used a novel Rac3 biosensor to directly demonstrate that Rac3 activity is distributed in a ring-like structure at mature, matrix-degrading invadopodia. This result echoes previous findings in which Rho-family GTPases were found to exhibit extraordinarily tightly regulated ring activity patterns at invadopodia related to the regulation of actin polymerization by cofilin (Bravo-Cordero et al., 2011).
Taken together, our results indicate that Vav2 functions to promote invadopodia maturation. The function of active Rac3 at invadopodia is unclear; possibilities include modulating actin polymerization or assisting with the recruitment of matrix metalloproteinases (MMPs) because MMP recruitment cannot occur without proper actin polymerization at invadopodia (Murphy and Courtneidge, 2011). It is also possible that in addition to its GEF activity, the SH3 domains of Vav2 are important for its function at invadopodia. For example, Trio, a different Rho-family GTPase, requires its SH3 domain to interact with Rac1 for promoting invadopodia disassembly (Moshfegh et al., 2015).
Rac3 is required for matrix degradation by invadopodia and is the target of Vav2
An unresolved question is how Rac3 activity is confined to a ring around actively degrading invadopodia. Previous work suggested that such a tight spatiotemporal pattern of Rho GTPase activation requires interplay between GEFs and GAPs (Moshfegh et al., 2015). Note that we did not detect restricted Rac3 diffusion at invadopodia (unpublished data), suggesting that sustained localization of Rac3 to invadopodia is unlikely and that a gradient of active Rac3 in a more homogeneous distribution of total Rac3 is more probable.
Rac3 is hyperactive in a number of highly aggressive human cancer cell lines, and this is associated with increased membrane localization and up-regulated Pak kinase activity (Mira et al., 2000). Introducing constitutively active Rac3 increases the invasiveness of the MDA-MB-435 melanoma cell line, whereas a dominant-negative version of Rac3 reduces its aggressiveness (Baugher et al., 2005). There is also evidence that Rac3 depletion is correlated with a reduction in MMP-9 secretion via a Pak-dependent mechanism (Gest et al., 2013). Clinical studies have found Rac3 to be overexpressed in some types of human breast cancer tumors (Morris et al., 2000). Moreover, increased Rac3 expression is correlated with decreased disease-free survival in prostate cancer patients (Engers et al., 2007).
GEFs as pharmacologic targets for the disruption of cancer invasion
Because of their role in promoting cancer progression, Rho-family GTPases are tempting pharmacologic targets. In fact, Rho GTPase inhibition decreases metastasis in some models without affecting normal cell viability (Montalvo-Ortiz et al., 2012; Menhofer et al., 2014). Unfortunately, these inhibitors have pleiotropic effects that can cause collateral damage to the vasculature, nervous system, and immune system. Instead, the inhibition of GEF activity—for example, a selective Vav2 inhibitor—may be a more promising target, as Vav2-knockout mice are generally viable and have no gross deficits (Doody et al., 2001; Vigil et al., 2010). Alternatively, direct inhibitors of the Vav2 SH2–phospho-cortactin interaction may be worth pursuing; indeed, specific inhibitors of the SH2 domains of Grb2, Grb7, and STAT3 have been developed and show promising results in breast cancer models (Morlacchi et al, 2014).
MATERIALS AND METHODS
Molecular cloning and protein purification
Human Vav2 and Vav2 mutant cDNAs (kind gifts from Christopher McCulloch, University of Toronto) were cloned into pFastBac expression vectors (Qiagen), pLXSN, pBabe, or pN1 vectors as appropriate. Mutants were introduced by PCR and site-directed mutagenesis. Expression levels of Vav2 mutants were determined by Western blotting of puromycin-selected (0.5–1.0 μg/ml) or G418-selected (200–800 μg/ml) cells.
Purified recombinant Vav2 was generated using baculovirus produced in Sf9 cells and used to infect Hi5 cells according to the manufacturer’s protocol (Bac-to-Bac expression system; ThermoFisher). Hi5 cells were pelleted 48 h after baculovirus infection, resuspended in lysis buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 5% glycerol, and protease and phosphatase inhibitors), dissociated by being passed through a French press, lysed with 1% Triton X-100, and bound to glutathione beads. Protein was cleaved with PreScission Protease as previously described (Warren et al., 2012), and buffer exchanged over a G-25 column into assay buffer: 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.35, 100 mM NaCl, 5% glycerol, 0.01% Triton X-100, and 1 mM dithiothreitol.
For imaging experiments, 100,000 cells were plated in six-well plates and transfected with 1 μg of plasmid DNA using JetPrime reagent as per the manufacturer’s instructions 24–48 h before imaging. siRNA target sequences used for Vav2 and Rac3 knockdowns were 5′-CUGACGUCUUUCUGAUCUG-3′ (Rac3) and 5′-AAAGUCCGGUCCAUAGUCA-3′ (Vav2) (Dharmacon, GE Life Sciences). Nonsilencing control siRNA was obtained from Qiagen.
Screening assay
Recombinant phosphorylated cortactin was expressed in insect cells using Arg- and His-tagged cortactin. Lysates were incubated with nickel beads, and high-salt washes (0.5, 1.25, 0.5 M KCl) were used to wash impurities and cortactin-bound Arg. Elution was performed with 200 mM imidazole, and phosphorylation was confirmed by Western blotting using anti-phosphotyrosine antibodies.
Recombinant SH2 domain binding was measured on cortactin affixed to nitrocellulose as described previously (Machida et al., 2007). Briefly, dots (0.4 μg in 1 μl) of phosphorylated cortactin, cortactin 3F, pervanadate-treated cell lysate, and phosphatase-treated cell lysate were spotted on nitrocellulose membrane and dried overnight. Membranes were soaked in transfer buffer (20% MeOH, 12.5 mM Tris-HCl, pH 8.0, 100 mM glycine) for 30 min, rinsed twice with TBST (150 mM NaCl, 10 mM Tris-HCl, pH 8.0, and 0.05% Tween-20), and blocked at room temperature for 1 h in TBST with 10% nonfat dry milk, 1 mM Na3VO4, and 1 mM EDTA. Recombinant SH2 domains conjugated to GSH horseradish peroxidase were added at a concentration of 1 μg/ml and incubated for 1 h at room temperature, washed with TBST, dried, incubated with ECL solution, and exposed to film (Nollau and Mayer, 2001; Machida et al., 2007).
5-Carboxyfluorescein–labeled peptide synthesis
Peptides corresponding to tyrosine phosphorylation sites Y421 (SSPIpYEDAA), Y466 (SSPVpYETTE), and Y482 (EDDTpYDGYE) were synthesized on a 30-µmol scale using microwave-assisted reactions and standard Fmoc chemistry. After removal of the final Fmoc protecting group, the resin was washed alternatively with dimethylformamide (DMF) and methylene chloride for a total of 16 washes and dried for 20 min under N2. The peptides were treated overnight with a cocktail composed of 5-carboxyfluorescein, succinimidyl ester (8 mg, 0.017 mmol), and diisopropylethylamine (24 µl, 17.8 mg, 0.138 mmol) in 1.5 ml DMF.
Completed peptides were treated with a cleavage cocktail of 2.5% (vol/vol) 3,6-dioxa-1,8-octanedithiol, 2.5% (vol/vol) H2O, 2.5% (vol/vol) triisopropylsilane in trifluoroacetic acid (50% power at 400-W maximum, 38°C ramp for 2 min, hold for 30 min; followed by treatment with fresh cocktail at 50% power, 400-W maximum, 38°C, ramp 2 min, hold 5 min). Crude peptides were concentrated by rotary evaporation and reconstituted in acetonitrile/water (1:1) for purification by reverse-phase high-performance liquid chromatography (HPLC).
The efficiency of each synthesis was assessed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis of the crude reaction mixture followed by purification to homogeneity by reverse-phase HPLC. Peak samples were reconfirmed by a second round of MALDI-TOF. After purification, peptides were lyophilized and stored in the dark at −20°C.
Fluorescence polarization measurements
SH2 domains were dialyzed into binding buffer (50 mM HEPES, pH 7.25, 150 mM NaCl, 0.01% NP40, 5% glycerol) and 30 µl volume serially diluted into prechilled 384-well low-flange black, flat-bottom nonbinding microplate (Corning) while on ice. A 5-µl amount of 35 nM fluorescent peptide dissolved in binding buffer was added to each well (final concentration 5 nM) and mixed five times by pipetting, and plates were incubated at 4°C for 40 min. Fluorescence polarization experiments were performed with an Analyst AD (Molecular Devices, Sunnyvale, CA) spectrofluorimeter. Each well was excited using 485-nm light, and emission was read at 530 nm. Fluorescence polarization was measured 1 mm from the bottom of each well with an integration time of 560 ms. All experiments were conducted in triplicate. The change in fluorescence polarization was normalized such that maximum change for each condition was set at an arbitrary value of 100 for the ease of comparison. Normalized values were graphed and fit to a single-binding-site hyperbola using GraphPad Prism.
Binding assay
Recombinant protein was covalently coupled to AminoLink beads at a concentration of 1 mg/ml gel bed as per the manufacturer’s protocol and stored as 50% slurry in 50 mM HEPES, pH 7.25, 125 mM NaCl, 0.01% NP40, and 5% glycerol. Input protein was diluted in a serial manner with 20 μl saved as input sample. Beads were blocked with Tris and 5% bovine serum albumin, and the reaction was incubated at 4°C with rotation for 2 h. Supernatant was removed and beads rapidly washed with 1 ml of binding buffer (3.65× phosphate-buffered saline [PBS]). Bound protein was recovered with 4× SDS–PAGE buffer and analyzed by SDS–PAGE gel stained with Coomassie Blue R-250. Densitometry was performed with Quantity One software. Background-subtracted and binding curves were generated by GraphPad Prism according to Specific binding = (Bmax*[Ligand]/[Ligand]) + KD + Nonspecific*[Ligand].
Coimmunoprecipitation and Western blotting
Cells were transiently transfected using JetPrime. After 48 h, cells were incubated with 150 μM sodium pervanadate for 15 min, washed with PBS, and lysed with immunoprecipitation buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 5% glycerol, and 1% Triton X-100 with protease and phosphatase inhibitors. Lysates were centrifuged at 14,000 rpm in an Eppendorf table-top centrifuge for 10 min at 4°C and precleared with protein A/G beads for 20 min with rotation. Protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce Chemical), and an equal amount of total protein was used for immunoprecipitation using 2 μg of antibody. Lysates were then incubated with antibody overnight at 4°C before protein A/G beads were added for 1 h. Beads were washed and then boiled in SDS–PAGE buffer run on an SDS–PAGE gel. Protein was then transferred to a nitrocellulose membrane and immunoblotted.
Cell lines and culture
The MDA-MB-231 cell line (American Type Culture Collection [ATCC]) and the HEK 293 cell line (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (FBS), antibiotics, and l-glutamine. Stable cell lines were produced by first transfecting the Phoenix Amphotrophic packaging cell line with a retroviral construct (pLXSN or pBabe) using Lipofectamine or JetPrime according to the manufacturer’s instructions. After 48 h, virus-containing supernatant was collected, filtered, and combined with concentration reagent (42.5% PEG 6000, 1.5 M NaCl, 40% PBS) in a 1:4 PEG-to-supernatant ratio. Solution was incubated for 24 h and centrifuged, and the pellet was resuspended in PBS and immediately used to infect MDA-MB-231 cells. Cells were selected with 600–800 μg/ml G418 or 0.5 μg/ml puromycin to generate stable lines.
Serum starvation was performed in 0.5% FBS and 0.5% BSA in DMEM overnight. Cells were starved in 0.345% BSA in L15 medium for 10 min and then stimulated with 2.5 nM EGF. Live-cell imaging was performed in phenol red–free DMEM supplemented with 20 mM HEPES and 10% FBS.
Antibodies and reagents
Cortactin (sc-30771), Tks5 (sc-30122), and Arp2 (sc-H-84) antibodies were from Santa Cruz Biotechnology. Cortactin (ab33333) was from Abcam. Anti-actin (Millipore), anti-GFP (Rockland), Rac1/3 (Upstate), specific anti-Rac3 (ProteinTech Group), and anti-Rac1 (Upstate) were also used. All Alexa Fluor–conjugated secondary antibodies were obtained from Molecular Probes (Life Technologies). GM 6001 was from Enzo Life Sciences.
Matrix degradation assay and immunofluorescence
The invadopodia matrix degradation assay was performed as previously described (Mader et al., 2011). In summary, gelatin was conjugated with either Alexa 405 or Alexa 568 dye (Molecular Probes). MatTek dishes or coverslips were treated with 1 M HCl and coated with 50 μg/ml poly-l-lysine. A 2.5% gelatin/2.5% sucrose solution was made in PBS supplemented with a 1:50 mix of Alexa-labeled gelatin, and unlabeled gelatin was warmed to 37°C before addition to poly-l-lysine–coated dishes. Gelatin was cross-linked with 0.5% glutaraldehyde and quenched with 5 mg/ml sodium borohydride. We plated 200,000 MDA-MB-231 cells on the gelatin overnight. Cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.1% Triton X-100, and then blocked with 2% FBS and 3% BSA in PBS. All antibodies were diluted in blocking buffer.
Invadopodium precursors were identified as previously described (Mader et al., 2011). Briefly, cells were plated on fluorescent dye–conjugated gelatin in the presence of the broad-spectrum MMP inhibitor GM 6001, serum starved overnight, and stimulated with 2.5 nM EGF. Precursors were identified as cortactin-Tks5–rich puncta not colocalizing with degradation puncta, whereas mature invadopodia colocalize with degradation puncta. Degradation area was calculated as total area per field in thresholded images and normalized to the total number of cells in each field in ImageJ. Images were acquired on a Nikon TE2000 inverted microscope with a Retiga cooled charge-coupled device (CCD) camera using Nikon Elements and processed using ImageJ.
Colocalization analysis
MDA-MB-231 cells were plated on fluorescently labeled gelatin, fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and fluorescently labeled as described in Oser et al. (2010). Images were acquired by a Nikon TE2000 inverted microscope with a Retiga cooled CCD camera using Nikon Elements and processed using ImageJ and the JaCop plug-in.
Transwell invasion assay
Transwell invasion assays were performed as described (Oser et al., 2010). Briefly, the bottom surface of 8.0-μm Transwell supports (CoStar) were treated with 10 μg/ml fibronectin for 1 h and dried. Upper surfaces were coated with 50 μl of reduced growth factor Matrigel (2.5 mg/ml) for 1 h at 37 C. Excess Matrigel was gently removed and the chamber equilibrated in DMEM at 37°C for 1 h. After equilibration, the bottom chamber medium was replaced with 10% FBS in DMEM. A total of 50,000 cells were resuspended in 200 μl of 0.5% FBS/DMEM and plated in the upper chamber. As a control, cells were also plated in Transwell membranes without Matrigel and allowed to invade for 24 h, followed by fixation in 4% PFA. Cells that did not invade were scraped off the upper surface of the membrane with cotton swabs, and cells at the bottom were stained with 4′,6-diamidino-2-phenylindole and counted under a fluorescence microscope.
Actin barbed-ends assay
The measurement of actin incorporation into barbed ends in invadopodia was performed as described previously (Oser et al., 2009). In summary, cells were transfected and serum starved overnight. Cells were stimulated with 2.5 nM EGF in L15 medium and permeabilized in 20 mM HEPES (pH 7.5), 138 mM KCl, 4 mM MgCl2, 3 mM ethylene glycol tetraacetic acid, 0.2 mg/ml saponin, 1 mM ATP, and 1% BSA. Cells were incubated with 0.5 μM biotin-actin (Cytoskeleton) for 1 min and fixed with 4% PFA, blocked with 1% FBS with 1% BSA and 3 μM phalloidin in BSA, and immunostained with fluorescein isothiocyanate (FITC)–anti-biotin, cortactin, and Arp2 to identify invadopodium precursors. Barbed-end intensity at each invadopodium was determined by measuring FITC-biotin mean fluorescence intensity in background-subtracted images and were normalized to the no-stimulation control condition.
Immunoblotting
Cells were washed with PBS, lysed in SDS–PAGE sample buffer, and boiled. Samples were subjected to SDS–PAGE, transferred to nitrocellulose, blocked with 4% milk or 5% BSA (4G10 antibody only), and processed/immunoblotted as previously described (Mader et al., 2011).
GEF activity assay
Recombinant baculovirus expressing MBP-Vav2 was purified from Hi5 insect cells as previously described in Tanis et al. (2003). The MBP tag was cleaved off with PreScission protease before use. Glutathione S-transferase (GST)–tagged Rac3 was produced from bacterial cells using the pGEX-6P1 plasmid, and the GST tag was cleaved with PreScission protease. Before use in assays, proteins were exchanged into the manufacturer’s recommended buffer. Other Rho GTPases used in the assay were purchased from the manufacturer (Cytoskeleton). We determined Vav2 exchange activity on Rac3 and other Rho GTPases using a fluorophore-based RhoGEF exchange assay kit according to the manufacturer’s instructions (BK100; Cytoskeleton).
Fluorescence spectroscopic analysis of N-methylanthraniloyl (mant)–GTP incorporation into purified Rho-family GTPase was carried out at 25°C. Exchange reaction assay mixtures containing 20 mM Tris (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 50 μg/ml BSA, 0.75 μM mant-GTP, and 2 μM Rac1, Cdc42, or Rac3 GTPase were prepared and allowed to equilibrate. After equilibration, the mixtures were placed into the fluorescence spectrophotometer, and fluorescence measurements were taken approximately every 10 s with excitation and emission wavelengths of 360 and 440 nm, respectively, and 10-nm bandwidth. After ∼120 s to establish a baseline, purified Vav2 was added to 0.8 μM, and the relative mant fluorescence was measured. Experiments were performed in triplicate.
FRET imaging
The construction of the dimerization-optimized reporter for activation (Dora)–Rac1 sensor and its negative control was described previously (Timmerman et al., 2015). The Dora-Rac3 biosensor and its control were generated by replacing the coding sequences of Rac1 with those of Rac3. The Dora-Rac3 biosensor was subcloned into pLXSN retroviral plasmid. Fluorimetry on cell extracts was performed after transfection of the indicated the plasmids into HEK293 cells using JetPrime. Cells were fixed with formaldehyde in PBS and fluorescence spectra measured with excitation at 433 nm and emission spectrum between 40 and 600 nm as described previously (Pertz et al., 2006). Live-cell FRET imaging was performed on a custom Nikon Ti-E microscope with a 60× TIRF objective (numerical aperture 1.49), an iXon Ultra electron-multiplying CCD, and MetaMorph software. Cells were maintained in a heated chamber supplemented with 5% CO2. Excitation was performed with a 442-nm laser, and TIRF FRET images were acquired with 500-ms exposure for the donor and acceptor channels. Images of the Alexa 567–labeled gelatin substrate were also obtained. Images were analyzed with custom-written software in ImageJ and R. Rac3 activation was monitored by measuring yellow fluorescent protein FRET and cyan fluorescent protein donor intensities. For spatiotemporal analysis of Rac3 activity at invadopodia, areas of new degradation were identified in the fluorescent gelatin channel. Using custom-written software in R, Matlab, and ImageJ, we assessed FRET activity in concentric annuli centered on the degradation spot. FRET intensity for each annulus was averaged. Then the time of the onset of degradation was determined separately for each new degradation spot, and the FRET data for each spot were synchronized to the timing of degradation initiation. A boundary between the “ring” and the “center” was then determined by finding a radial distance value that maximized the mean difference in FRET between the center and ring among the pooled experimental data, and the data were then plotted as a function of time. A control area of the cell that did not cause degradation in the gelatin channel was analyzed analogously.
Statistical analysis
Analysis was performed with unpaired, two-tailed t-test or ANOVA with Newman–Keuls posttest, as appropriate. Significance was defined as p < 0.05. Error bars represent SEM. Calculations were performed using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Javier Bravo-Cordero for invaluable advice during the initial stages of this project and are grateful to Xianyun Ye for technical support. This work was funded by National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) Grant T32GM007205 (B.J.R.), the Israel Science Foundation and Israel Cancer Research Fund (to H.G.H.), NIH/National Cancer Institute (NCI) Grants CA150344 and CA1154966 (J.C.), LLS Quest for CURES (K.M.), NIH/NIGMS Grant GM117061 (Y.I.W.), and NIH/NCI Grant CA133346 (A.J.K.).
Abbreviations used:
- Arp
actin-related protein
- ECM
extracellular matrix
- EGF
epidermal growth factor
- FRET
fluorescence resonance energy transfer
- GEF
guanine nucleotide exchange factor
- HRP
horseradish peroxidase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-12-0885) on March 29, 2017.
REFERENCES
- Abe K, Rossman KL, Liu B, Ritola KD, Chiang D, Campbell SL, Burridge K, Der CJ. Vav2 is an activator of Cdc42, Rac1, and RhoA. J Biol Chem. 2000;275:10141–10149. doi: 10.1074/jbc.275.14.10141. [DOI] [PubMed] [Google Scholar]
- Ahn J, Truesdell P, Meens J, Kadish C, Yang X, Boag AH, Craig AWB. Fer protein-tyrosine kinase promotes lung adenocarcinoma cell invasion and tumor metastasis. Mol Cancer Res. 2013;11:952–963. doi: 10.1158/1541-7786.MCR-13-0003-T. [DOI] [PubMed] [Google Scholar]
- Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym VV. Dynamic interactions of cortactin and membrane Type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- Barsky SHS, Siegal GPG, Jannotta FF, Liotta LAL. Loss of basement membrane components by invasive tumors but not by their benign counterparts. Lab Invest. 1983;49:140. [PubMed] [Google Scholar]
- Bartolome RA. Activation of Vav/Rho GTPase signaling by CXCL12 controls membrane-type matrix metalloproteinase-dependent melanoma cell invasion. Cancer Res. 2006;66:248–258. doi: 10.1158/0008-5472.CAN-05-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugher PJ, Krishnamoorthy L, Price JE, Dharmawardhane SF. Rac1 and Rac3 isoform activation is involved in the invasive and metastatic phenotype of human breast cancer cells. Breast Cancer Res. 2005;7:R965–R974. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty BT, Sharma VP, Bravo-Cordero JJ, Simpson MA, Eddy RJ, Koleske AJ, Condeelis J. β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol Biol Cell. 2013;24:1661–1675. doi: 10.1091/mbc.E12-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty BT, Condeelis J. Digging a little deeper: the stages of invadopodium formation and maturation. Eur J Cell Biol. 2014;93:438–444. doi: 10.1016/j.ejcb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden E, Onukoyi E, Slac R, Myoui A, Yoneda T, Yamada K, Mueller S. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312:1240–1253. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Boyle SN, Michaud GA, Schweitzer B, Predki PF, Koleske AJ. A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr Biol. 2007;17:445–451. doi: 10.1016/j.cub.2007.01.057. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A novel spatiotemporal RhoC activation pathway locally regulates cofilin activity at invadopodia. Curr Biol. 2011;21:635–644. doi: 10.1016/j.cub.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Magalhaes MAO, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J Exp Zool. 1989;251:167–185. doi: 10.1002/jez.1402510206. [DOI] [PubMed] [Google Scholar]
- Dedes KJ, Lopez-Garcia M-A, Geyer FC, Lambros MBK, Savage K, Vatcheva R, Wilkerson P, Wetterskog D, Lacroix-Triki M, Natrajan R, et al. Cortactin gene amplification and expression in breast cancer: a chromogenic in situ hybridisation and immunohistochemical study. Breast Cancer Res Treat. 2010;124:653–666. doi: 10.1007/s10549-010-0816-0. [DOI] [PubMed] [Google Scholar]
- Desmarais V, Yamaguchi H, Oser M, Soon L, Mouneimne G, Sarmiento C, Eddy R, Condeelis J. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil Cytoskeleton. 2009;66:303–316. doi: 10.1002/cm.20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doody GM, Bell SE, Vigorito E, Clayton E, McAdam S, Tooze R, Fernandez C, Lee IJ, Turner M. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nat Immunol. 2001;2:542–547. doi: 10.1038/88748. [DOI] [PubMed] [Google Scholar]
- Engers R, Ziegler S, Mueller M, Walter A, Willers R, Gabbert HE. Prognostic relevance of increased Rac GTPase expression in prostate carcinomas. Endocr Relat Cancer. 2007;14:245–256. doi: 10.1677/ERC-06-0036. [DOI] [PubMed] [Google Scholar]
- Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- Gest C, Joimel U, Huang L, Pritchard L-L, Petit A, Dulong C, Buquet C, Hu C-Q, Mirshahi P, Laurent M, et al. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer. 2013;13:63. doi: 10.1186/1471-2407-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Liu W, Mader CC, Halo TL, Machida K, Boggon TJ, Koleske AJ. Two amino acid residues confer different binding affinities of Abelson family kinase SRC homology 2 domains for phosphorylated cortactin. J Biol Chem. 2014;289:19704–19713. doi: 10.1074/jbc.M114.556480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Henn H, Patsialou A, Wang Y, Warren MS, Condeelis JS, Koleske AJ. Arg/Abl2 promotes invasion and attenuates proliferation of breast cancer in vivo. Oncogene. 2013;32:2622–2630. doi: 10.1038/onc.2012.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Prabakaran I, Wan F, Mitra N, Furstenau DK, Hung RK, Cao S, Zhang PJ, Fraker DL, Guvakova MA. Vav2 protein overexpression marks and may predict the aggressive subtype of ductal carcinoma in situ. Biomark Res. 2014;2:22. doi: 10.1186/2050-7771-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedziora KM, Leyton-Puig D, Argenzio E, Boumeester AJ, van Butselaar B, Yin T, Wu YI, van Leeuwen FN, Innocenti M, Jalink K, et al. Rapid remodeling of invadosomes by Gi-coupled receptors: dissecting the role of Rho GTPases. J Biol Chem. 2016;291:4323–4333. doi: 10.1074/jbc.M115.695940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapetina S, Mader CC, Machida K, Mayer BJ, Koleske AJ. Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J Cell Biol. 2009;185:503–519. doi: 10.1083/jcb.200809085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BP, Burridge K. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol Cell Biol. 2000;20:7160–7169. doi: 10.1128/mcb.20.19.7160-7169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Machida K, Mayer B, Nollau P. Profiling the global tyrosine phosphorylation state. Mol Cell Proteomics. 2003;2:215–233. doi: 10.1074/mcp.R300002-MCP200. [DOI] [PubMed] [Google Scholar]
- Machida K, Thompson CM, Dierck K, Jablonowski K, Kärkkäinen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P. High-throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- Mader CC, Oser M, Magalhaes MAO, Bravo-Cordero JJ, Condeelis J, Koleske AJ, Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes MAO, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M, Chen X, Koleske AJ, Condeelis J. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J Cell Biol. 2011;195:903–920. doi: 10.1083/jcb.201103045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux N, Vuori K. EGF receptor mediates adhesion-dependent activation of the Rac GTPase: a role for phosphatidylinositol 3-kinase and Vav2. Oncogene. 2003;22:6100–6106. doi: 10.1038/sj.onc.1206712. [DOI] [PubMed] [Google Scholar]
- Marignani PA, Carpenter CL. Vav2 is required for cell spreading. J Cell Biol. 2001;154:177–186. doi: 10.1083/jcb.200103134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menhofer MH, Kubisch R, Schreiner L, Zorn M, Foerster F, Mueller R, Raedler JO, Wagner E, Vollmar AM, Zahler S. The actin targeting compound Chondramide inhibits breast cancer metastasis via reduction of cellular contractility. PLoS One. 2014;9:e112542. doi: 10.1371/journal.pone.0112542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci USA. 2000;97:185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo-Ortiz BL, Castillo-Pichardo L, Hernández E, Humphries-Bickley T, la Mota-Peynado De A, Cubano LA, Vlaar CP, Dharmawardhane S. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J Biol Chem. 2012;287:13228–13238. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlacchi P, Robertson FM, Klostergaard J, McMurray JS. Targeting SH2 domains in breast cancer. Future Med Chem. 2014;6:1909–1926. doi: 10.4155/fmc.14.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CM, Haataja L, McDonald M, Gough S, Markie D, Groffen J, Heisterkamp N. The small GTPase RAC3 gene is located within chromosome band 17q25.3 outside and telomeric of a region commonly deleted in breast and ovarian tumours. Cytogenet Cell Genet. 2000;89:18–23. doi: 10.1159/000015583. [DOI] [PubMed] [Google Scholar]
- Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2015;17:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollau P, Mayer BJ. Profiling the global tyrosine phosphorylation state by Src homology 2 domain binding. Proc Natl Acad Sci USA. 2001;98:13531–13536. doi: 10.1073/pnas.241215998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Resh MD. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270:26613–26618. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- Oser M, Mader CC, Gil-Henn H, Magalhaes M, Bravo-Cordero JJ, Koleske AJ, Condeelis J. Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J Cell Sci. 2010;123:3662–3673. doi: 10.1242/jcs.068163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V, Rosenfeldt HM, Lyons R, Servitja J-M, Bustelo XR, Siroff M, Gutkind JS. Persistent activation of Rac1 in squamous carcinomas of the head and neck: evidence for an EGFR/Vav2 signaling axis involved in cell invasion. Carcinogenesis. 2007;28:1145–1152. doi: 10.1093/carcin/bgm008. [DOI] [PubMed] [Google Scholar]
- Patsialou A, Bravo-Cordero JJ, Wang Y, Entenberg D, Liu H, Clarke M, Condeelis JS. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital. 2013;2:e25294. doi: 10.4161/intv.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Razidlo GL, Magnine C, Sletten AC, Hurley RM, Almada LL, Fernandez-Zapico ME, Ji B, McNiven MA. Targeting pancreatic cancer metastasis by inhibition of Vav1, a driver of tumor cell invasion. Cancer Res. 2015;75:2907–2915. doi: 10.1158/0008-5472.CAN-14-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razidlo GL, Schroeder B, Chen J, Billadeau DD, McNiven MA. Vav1 as a central regulator of invadopodia assembly. Curr Biol. 2014;24:86–93. doi: 10.1016/j.cub.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M, Bravo-Cordero JJ, Patsialou A, Sharma VP, Guo P, Liu H, Hodgson L, Condeelis J. Macrophage contact induces RhoA GTPase signaling to trigger tumor cell intravasation. Oncogene. 2014;33:4203–4212. doi: 10.1038/onc.2013.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel KE, Bustelo XR, Nielsen DA, Song BJ, Barbacid M, Goldman D, Lee IJ. Isolation and characterization of murine vav2, a member of the vav family of proto-oncogenes. Oncogene. 1996;13:363–371. [PubMed] [Google Scholar]
- Sharma VP, Eddy R, Entenberg D, Kai M, Gertler FB, Condeelis J. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr Biol. 2013;23:2079–2089. doi: 10.1016/j.cub.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanis KQ, Veach D, Duewel HS, Bornmann WG, Koleske AJ. Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol Cell Biol. 2003;23:3884–3896. doi: 10.1128/MCB.23.11.3884-3896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman I, Heemskerk N, Kroon J, Schaefer A, van Rijssel J, Hoogenboezem M, van Unen J, Goedhart J, Gadella TWJ, Yin T, et al. A local VE-cadherin and Trio-based signaling complex stabilizes endothelial junctions through Rac1. J Cell Sci. 2015;128:3041–3054. doi: 10.1242/jcs.168674. [DOI] [PubMed] [Google Scholar]
- Uruno T, Liu J, Zhang P, Yx Fan, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- van Unen J, Reinhard NR, Yin T, Wu YI, Postma M, Gadella TWJ, Goedhart J. Plasma membrane restricted RhoGEF activity is sufficient for RhoA-mediated actin polymerization. Sci Rep. 2015;5:14693. doi: 10.1038/srep14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy. Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zhao N, Li S, Fang J-H, Chen M-X, Yang J, Jia W-H, Yuan Y, Zhuang S-M. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology. 2013;58:642–653. doi: 10.1002/hep.26373. [DOI] [PubMed] [Google Scholar]
- Warren MS, Bradley WD, Gourley SL, Lin Y-C, Simpson MA, Reichardt LF, Greer CA, Taylor JR, Koleske AJ. Integrin β1 signals through Arg to regulate postnatal dendritic arborization, synapse density, and behavior. J Neurosci. 2012;32:2824–2834. doi: 10.1523/JNEUROSCI.3942-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Heuser JE, Karginov AV, Lee W-L, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005a;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005b;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Zhan X, Hu X, Hampton B, Burgess WH, Friesel R, Maciag T. Murine cortactin is phosphorylated in response to fibroblast growth factor-1 on tyrosine residues late in the G1 phase of the BALB/c 3T3 cell cycle. J Biol Chem. 1993;268:24427–24431. [PubMed] [Google Scholar]
- Zicha D, Dunn G, Jones G. Analyzing chemotaxis using the dunn direct-viewing chamber. In: Pollard JW, Walker JM, editors. Basic Cell Culture Protocols. Totowa, NJ: Humana Press; 1997. pp. 449–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.