Abstract
Several aspects of meiosis are impacted by the absence of centrosomes in oocytes. Here we review four aspects of meiosis I that are significantly affected by the absence of centrosomes in oocyte spindles. One, microtubules tend to assemble around the chromosomes. Two, the organization of these microtubules into a bipolar spindle is directed by the chromosomes. Three, chromosome bi-orientation and attachment to microtubules from the correct pole require modification of the mechanisms used in mitotic cells. Four, chromosome movement to the poles at anaphase cannot rely on polar anchoring of spindle microtubules by centrosomes. Overall, the chromosomes are more active participants during acentrosomal spindle assembly in oocytes, compared to mitotic and male meiotic divisions where centrosomes are present. The chromosomes are endowed with information that can direct the meiotic divisions and dictate their own behavior in oocytes. Processes beyond those known from mitosis appear to be required for their bi-orientation at meiosis I. As mitosis occurs without centrosomes in many systems other than oocytes, including all plants, the concepts discussed here may not be not limited to oocytes. The study of meiosis in oocytes has revealed mechanisms that are operating in mitosis and will probably continue to do so.
Keywords: meiosis, microtubule, spindle assembly, chromosome segregation, oocyte, acentrosomal
Introduction
A germ cell with two homologous alleles of each gene, one inherited from each parent, progresses through meiosis to produce four haploid cells with only one allele each. The haploid state is reached by replicating the genome once followed by two divisions. It is the first meiotic, or reductional, division that separates the two parental alleles into two daughter cells and is critical for Mendel’s laws. Both meiotic divisions should be flawless because mistakes are usually lethal to the zygote. The success of meiosis usually depends on crossover formation between homologs to maintain a physical link between pairs of homologous chromosomes (bivalents) until anaphase I onset. That is why there is an inverse correlation between crossover frequency and nondisjunction of homologs (Baker and Hall 1976), and mutations that cause decreases in crossing over also increase nondisjunction. In addition, there is an increase in frequency of non-exchange chromosomes associated with human aneuploidy (Hassold and Hunt 2001; Nagaoka, et al. 2012).
The physical linkage of homologs that results from a crossover is called a chiasma, however, this linkage only directs segregation in combination with a bipolar spindle that orients homologs to opposite poles. Oocytes in many organisms have an added complication: spindle assembly occurs in the absence of centrosomes (referred to here as acentrosomal), the centriole-containing microtubule-organizing centers (MTOCs). The reason why centrioles are often eliminated prior to meiosis I in oocytes is not known but has been discussed elsewhere (Pimenta-Marques, et al. 2016; Severson, et al. 2016). The consequences of this centrosome elimination on spindle assembly, however, are dramatic. Instead of the chromosomes acting as passengers or cargo for the spindle, the chromosomes have the ability to organize meiotic spindles in a variety of systems (McKim and Hawley 1995).
The premise of this review is that chromosomes are active participants in the spindle assembly process. That the chromosomes carry information that can direct the meiotic divisions is long standing. For example, meiosis I chromosomes physically removed from a cell and placed into a meiosis II spindle still divide reductionally (Nicklas 1977; Paliulis and Nicklas 2000), showing the chromosomes dictate their own behavior. This review focuses on four aspects of meiosis I that are significantly affected by the absence of centrosomes in oocyte spindles: 1) assembly of microtubules around the chromosomes, 2) organization of these microtubules into a bipolar spindle, 3) chromosome bi-orientation and attachment to microtubules from the correct pole, and 4) chromosome movement to the poles at anaphase. Mitosis can and does occur without centrosomes in many systems other than oocytes, including all plants. Indeed, several reviews of cell division by acentrosomal mitosis have been published (Gatti, et al. 2012; Meunier and Vernos 2016; Moutinho-Pereira, et al. 2013; Wadsworth and Khodjakov 2004). Therefore, the knowledge gained from the study of oocytes is also relevant for many other cell types in a large range of organisms. In addition, for additional perspective on meiotic acentrosomal spindle assembly, a couple reviews have recently been published (Dumont and Desai 2012; Severson, et al. 2016).
Oocyte spindle assembly is dominated by chromosome-based mechanisms
Given the large size of oocytes (e.g. 120 μm diameter in humans) compared to most mitotically dividing cells (e.g. 40 μm diameter HeLa cell), nucleating and organizing microtubules must be combined with a mechanism to restrict spindle assembly to the vicinity of the chromosomes. Chromosome-directed spindle assembly has been observed in several oocyte systems. For example, early work in Drosophila and Xenopus oocytes and recent work in human oocytes show that spindle assembly begins with organization of microtubules around the chromosomes (Gard 1992; Holubcová, et al. 2015; Theurkauf and Hawley 1992). In some Drosophila mutants, oocytes chromosomes are ejected from the main spindle and form their own spindles (Cullen, et al. 2005; Theurkauf and Hawley 1992). These results show that oocyte chromosomes can organize a bipolar spindle.
Enucleated mouse oocytes are unable to form spindles in a timely manner (<12 hrs) (Schuh and Ellenberg 2007). While other studies have reported spindle assembly in bisected (Brunet, et al. 1998) or enucleated oocytes (Yang, et al. 2007), these observations were made after a substantial period of time (18 hrs) and most spindles formed were grossly abnormal. In a result strikingly similar to what has been observed in Drosophila, dispersed clusters of mouse oocyte metaphase chromosomes, induced by nocodazole treatment, each organize a meiotic spindle (Maro, et al. 1986). Thus, it appears in mouse, and also pig oocytes (Sun, et al. 2001), that chromosomes organize microtubules, but other nuclear factors may also be critical for spindle assembly (Polanski, et al. 2005). In C. elegans, meiosis I oocyte spindle assembly appears to initiate with a microtubule “cage” that forms on the inside of the nuclear envelope (Wolff, et al. 2016) (although at meiosis II, the microtubules assemble closely around the chromosomes like the other systems). Observations like these raise the possibility that spindle assembly is coupled to the release of nuclear factors caused by disassembly of the nuclear envelope. This is a mechanism that oocytes could use to restrict spindle assembly to the vicinity of the chromosomes.
The first insights into identifying the molecules required for chromatin-based spindle assembly mechanisms came from work in Xenopus egg extracts. DNA-coated beads (Heald, et al. 1996) or sperm nuclei (Nachury, et al. 2001; Wilde and Zheng 1999) promote de novo spindle assembly in a process that depends on two mechanisms: a gradient of RanGTP centered on the chromosomes and the chromosomal passenger complex (CPC) (Carazo-Salas, et al. 1999; Sampath, et al. 2004). RanGTP, whose conversion from RanGDP is stimulated by chromosome-localized RCC1, causes the release of spindle assembly factors from the inhibitory effect of importins (Clarke and Zhang 2008; Meunier and Vernos 2016). Surprisingly, expression of a dominant-negative form of Ran (RanT24N) in human, mouse, and Drosophila oocytes demonstrated that the RanGTP pathway is not essential for assembly of the first meiotic spindle in vivo (Cesario and McKim 2011; Dumont, et al. 2007; Holubcová, et al. 2015). However, RanT24N caused delayed and disorganized assembly of the first meiotic spindle (Cesario and McKim 2011; Dumont, et al. 2007; Holubcová, et al. 2015). These data suggest that RanGTP contributes to the speed and efficiency of meiosis I spindle assembly in oocytes, but that other essential mechanisms are also present. Instead, the Xenopus, mouse and Drosophila studies suggest Ran becomes critical after meiosis I, during meiosis II and subsequent embryonic mitoses. For example, RanGTP appears to be essential for establishing the mouse metaphase II spindle (Dumont et al 2007).
Chromosome-based spindle assembly and the CPC
In the absence of the RanGTP gradient, the CPC promotes spindle assembly around sperm nuclei (Maresca, et al. 2009). The CPC is composed of four proteins: INCENP, Survivin, Borealin, and Aurora B (or C) kinase (Carmena, et al. 2012). The chromatin-focused enrichment of the CPC may facilitate local kinase activation, satisfying the spatial component to spindle assembly to be around the chromosomes (Kelly, et al. 2007). Two studies in Drosophila have supported a role for the CPC in acentrosomal spindle assembly in oocytes. Partial loss of the CPC component INCENP resulted in spindle assembly delay (Colombié, et al. 2008) while the absence of the CPC components Aurora B kinase or INCENP prevented spindle assembly (Radford, et al. 2012).
In Xenopus egg extracts, both the INCENP centromere-targeting and microtubule-targeting domains are required to support chromatin-mediated spindle assembly. Therefore, it appears that the initiation of spindle assembly depends on simultaneous interactions between the CPC, the chromosomes and the microtubules (Tseng, et al. 2010). A partner for the CPC in this context could be motor proteins that bundle microtubules such as the Drosophila kinesin-6 Subito, which colocalizes with the CPC on the metaphase spindle in oocytes (Jang, et al. 2005). The bundling activity of the Subito appears to be activated only in the presence of the chromosomes after nuclear envelope breakdown (NEB) (Jang, et al. 2007). Thus, enforcement of spindle assembly around the chromosomes may also depend on the localized activation of motor proteins and their bundling activity.
The features of the chromatin that interact with the CPC to promote spindle assembly are not known. Sites that recruit the CPC include the centromeres, which may result in assembly of the kinetochores (Emanuele, et al. 2008; Kim and Yu 2015; Radford, et al. 2015; Rago, et al. 2015). Microtubules nucleated at or near kinetochores contribute to spindle assembly in somatic cells (Maiato, et al. 2004; Torosantucci, et al. 2008), but oocyte chromosomes lacking kinetochores still initiate spindle assembly (Brunet, et al. 1999; Deng, et al. 2009; Dumont, et al. 2010; Radford, et al. 2015). Therefore, there must be other chromosomal sites that make a significant contribution to oocyte spindle assembly mechanisms, although it is not yet clear what structures or molecules are important. In Drosophila mitotic cells, a chromosome-based spindle assembly pathway depends on the conserved Misato protein and the tubulin chaperone complex (Palumbo, et al. 2015). The role of these proteins in oocytes is not known.
Evidence of a single CPC-based pathway promoting initiation of spindle assembly in Drosophila oocytes has not been replicated in all organisms. For example, knocking down CPC components such as INCENP does not cause spindle assembly failure in mouse (Sharif, et al. 2010). The CPC may not be required for the initiation of spindle assembly in C. elegans, although it is required during meiosis because oocytes lacking the CPC have disorganized spindles (Schumacher, et al. 1998; Wignall and Villeneuve 2009). It may be that the existence of multiple Aurora homologs provides redundancy. For example, in mouse, like other mammals, there are three Auroras – AURKA, AURKB, and AURKC – that may compensate for each other (Balboula and Schindler 2014; Fernández-Miranda, et al. 2011; Schindler, et al. 2012). Many eukaryotes express at least two Aurora kinases, one that is pole-associated (Aurora A) and one that transits from chromosomes to the central spindle (Aurora B/C). However, the designation of an Aurora kinase as either A or B/C does not always reflect their evolutionary relationships. For example, while mammal AURKB and AURKC are more similar to one another than they are to AURKA, AURKA is phylogenetically more closely related to AURKB and AURKC than to Aurora A in Drosophila (Brown, et al. 2004) (Figure 1).
Figure 1. Evolutionary relationships between Aurora kinases.
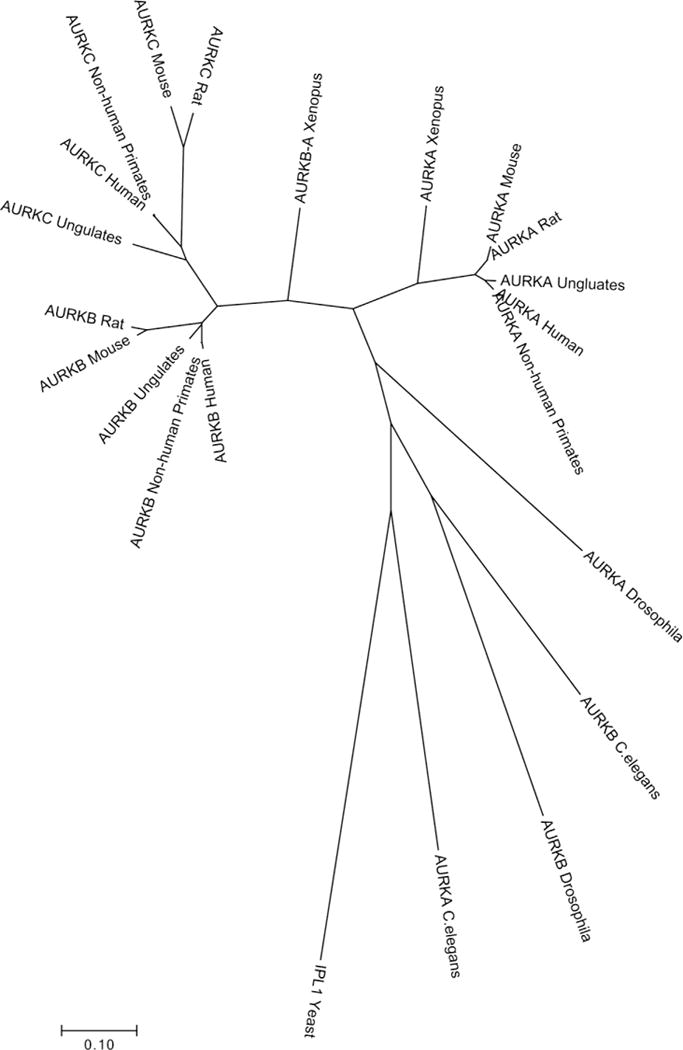
Unrooted phylogenetic tree, assembled using the Maximum Likelihood method based on the JTT matrix-based model (Felsenstein 1992; Jones, et al. 1992). The tree with the highest log likelihood is shown. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 22 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 261 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar, et al. 2016). All three mammalian Auroras cluster together, indicating independent evolution of Aurora A/B in mammals, Drosophila and C. elegans.
In mouse oocytes, AURKC is the predominant catalytic subunit in the CPC (Balboula and Schindler 2014; Sharif, et al. 2010; Yang, et al. 2010). AURKC localizes to centromeres and chromosome arms during metaphase I and is restricted to centromeres at metaphase II, whereas AURKB localizes to microtubules (Balboula and Schindler 2014; Sharif, et al. 2010; Shuda, et al. 2009). A recent finding is that AURKC is recruited to spindle poles by Haspin kinase and is required for MTOC clustering (Balboula, et al. 2016). However, in oocytes from Aurkc−/− mice, AURKB compensates, adopting AURKC localization and activity (Balboula and Schindler 2014; Schindler, et al. 2012). Conversely, embryos lacking Aurkb develop to the blastocyst stage because of AURKC (Fernández-Miranda, et al. 2011). Because of this compensation between AURKB and AURKC, establishing a requirement for initiating spindle assembly as a conserved function of Aurora kinase in oocytes has been challenging. Despite the evidence obtained from Xenopus and Drosophila, the role of the Aurora kinases in meiotic spindle assembly, and, more generally, the chromosome-based signal(s) that regulates spindle assembly continue to be an important topic of study.
Microtubule nucleation, bundling, and stabilization in oocyte spindle assembly
Microtubules may be nucleated at many cytoplasmic locations such as membranes in Drosophila (Cha, et al. 2001; Theurkauf, et al. 1992), or at dispersed, small-sized cytoplasmic foci of pericentriolar material (PCM) also known as MTOCs in mouse oocytes (Luksza, et al. 2013). Mouse meiosis I oocytes contain a large number of acentriolar MTOCs (Szollosi, et al. 1972) that assemble microtubules at a rate similar to centriole-containing centrosomes in mitotic cells (Schuh and Ellenberg 2007). These MTOCs are also observed in preimplantation mouse embryos, which are also acentrosomal (Courtois, et al. 2012). Prior to NEB, an oocyte has 1–3 MTOCs, and this number increases to ~30 upon NEB, possibly due to MTOC fragmentation rather than de novo synthesis (Clift and Schuh 2015). Their biogenesis may require AURKA, which is localized at the poles, because oocytes from transgenic mice overexpressing AURKA showed an increase in MTOC numbers (Solc, et al. 2012).
Depletion of Pericentrin, an important component of acentriolar MTOCs, in mouse oocytes causes misaligned chromosomes and perturbs, but does not eliminate, bipolar spindle formation and maturation to metaphase II (Ma and Viveiros 2014). Despite a lack of γ-tubulin at these MTOCs, microtubules still nucleate, albeit the total amount of α-tubulin incorporated into the spindle is reduced. This reduction is similar to that of oocytes expressing dominant-negative RanT24N (Dumont, et al. 2007; Schuh and Ellenberg 2007). These results suggest that the Ran and MTOC pathways are not essential for spindle assembly in mouse oocytes. Similarly, in Drosophila sas-4 mutant brain cells, centrosomes are absent and acentriolar MTOCs form, but they do not increase the efficiency of acentrosomal spindle assembly (Baumbach, et al. 2015).
Microtubules may also be nucleated within an existing spindle, which is usually associated with the Augmin pathway (Meunier and Vernos 2016; Sanchez-Huertas and Luders 2015). Augmin is not required to initiate mitotic spindle assembly, but instead enhances spindles by building on pre-existing microtubules. In Drosophila oocytes, the Augmin pathway contributes to acentrosomal spindle function but is not required for assembly (Meireles, et al. 2009). Proteins like Dgp71WD/Nedd1, which recruit microtubule nucleating factors such as γ-TuRC, have also been implicated in microtubule nucleation within spindles (Reschen, et al. 2012). Drosophila oocytes lacking Dgp71WD have reduced density of tubulin and spindle width. It is possible these pathways are downstream targets of the CPC that contribute to specific populations of microtubules within the context of acentrosomal spindle assembly.
In the previous section, we emphasized the role of the chromosomes in stabilizing and bundling microtubules. In this section, we emphasized microtubule nucleation, which may have several mechanisms. To integrate these concepts, we propose a model with three distinct phases required in all acentrosomal systems: microtubule nucleation, bundling of microtubules, and stabilization around the chromosomes (Meunier and Vernos 2016) (Figure 2). In all systems, before NEB, the cytoplasm contains diffuse microtubules. At some point in meiotic progression, which may differ between organisms, there is a profound reorganization of cytoplasmic microtubules that may reflect enhanced bundling activity. This reorganization seems most dramatic in mouse oocytes with the appearance of MTOCs prior to NEB. However, in other cases this phase may be linked to NEB, which would be consistent with observations in human, Drosophila and C. elegans oocytes. Microtubule reorganization results in the bundling of the previously dispersed microtubules and depends on microtubule motors, such as the mouse kinesin-5 Kif11 (Clift and Schuh 2015). Such bundling may help restrict the microtubules to the vicinity of the chromosomes and, as described in the next section, may contribute to the assembly of a bipolar spindle.
Figure 2. Model for acentrosomal spindle assembly.
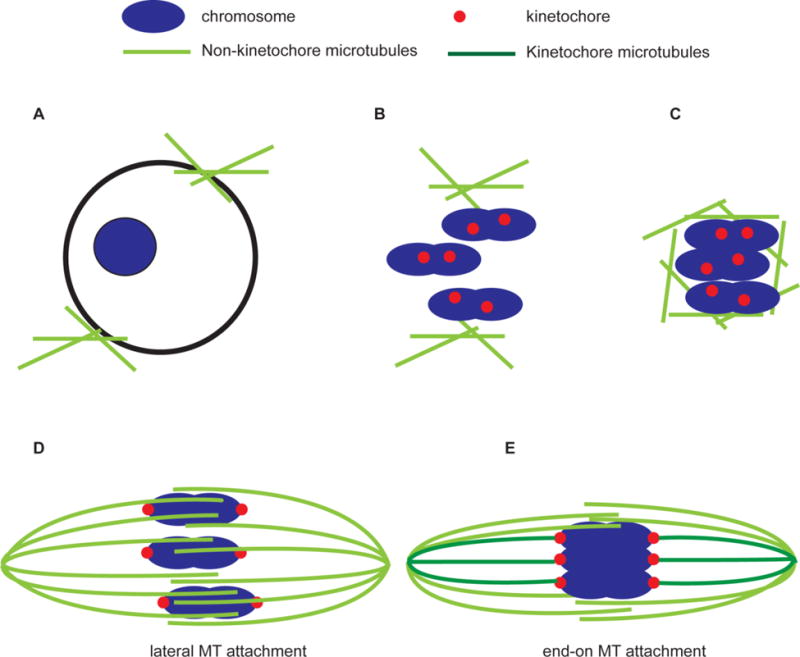
The emphasis of this model is that the accumulation and bundling of microtubules are regulated to occur in close proximity to the chromosomes. A) Oocyte before nuclear envelope breakdown. In most organisms, microtubules are visible outside the nuclear envelope. In mouse, these cluster in MTOCs. B) After nuclear envelope breakdown, there may be bundles or clusters of microtubules in close proximity, but distinct from, the chromosomes. This is seen in mouse and nematodes but not in fly or human. C) In all systems, the initial interactions between the chromosomes and microtubules may appear quite disorganized or appear as a ball of microtubules. D) A bipolar spindle forms and then lateral attachments predominate. E) Prior to anaphase, end-on attachments are stabilized. This last step is not clear in nematodes which lack point centromeres. In B and C, the kinetochores (red) are drawn relative to the chromosomes (blue) to show they are not stretched by the microtubules (green). In D and E, the kinetochores are drawn relative to the chromosomes to indicate they are stretched towards the poles.
Once NEB occurs, bundles of microtubules appear to be attracted to or stabilized by the chromosomes. Future research will be important to uncover how chromosome-based spindle assembly mechanisms are coordinated with non-chromosome-based mechanisms to form a functional oocyte spindle. While cytoplasmic elements may nucleate microtubules, and bundling may even occur prior to NEB, paramount is a mechanism that forces spindles to form around chromosomes. In all these cases, spindles reach a similar state after NEB, with a disorganized accumulation of microtubules near the chromosomes.
Bipolarity is established through a central spindle
Centrosomes function not only as a source of microtubule nucleation, but they also establish spindle bipolarity. An important question, therefore, is how oocytes accomplish bipolarity in the absence of centrosomes. Although oocytes lack centrosomes, the poles contain PCM components. The composition and assembly of these pole structures has been recently reviewed (Severson, et al. 2016). Some of these studies have shown that pole-localized proteins, and by inference pole focusing, are important for chromosome segregation. For example, proteins known to associate with acentrosomal poles include TACC, MSPS (Cullen and Ohkura 2001) and ASP in Drosophila (Riparbelli, et al. 2002) and C. elegans (van der Voet, et al. 2009; Wignall and Villeneuve 2009). Defective spindle poles and a pole-focusing defect were found in mouse oocytes lacking NuMA (Kolano, et al. 2012). The spindles were longer than normal and initially lacked focused poles. Despite kinetochore attachments occurring normally, the spindle pole defects resulted in aneuploidy. These results suggest that pole focusing is important for chromosome segregation, but is not required for spindle bipolarity.
Results in Drosophila show that pole focusing is not sufficient to ensure only two poles. In Drosophila oocytes, a central spindle, composed of the antiparallel overlap of non-kinetochore microtubules, forms at metaphase I. The kinesin-6 motor protein Subito localizes to the central spindle (Jang, et al. 2005). In subito mutants, the central spindle is missing and oocyte spindles are frequently monopolar or tripolar, suggesting the central spindle is required to maintain a bipolar spindle (Giunta, et al. 2002) (Figure 3). Several other proteins that localize to the central spindle at anaphase in somatic cells also localize to the central spindle at metaphase in Drosophila oocytes, including the CPC and the centralspindlin complex (Das, et al. 2016; Jang, et al. 2005). Interestingly, loss of centralspindlin does not lead to the same spindle polarity defects seen in subito mutants (Das, et al. 2016), suggesting that Subito itself may provide the antiparallel microtubule crosslinking function required for oocyte spindle bipolarity.
Figure 3. Defects in spindle bipolarity in Drosophila and mouse oocytes.
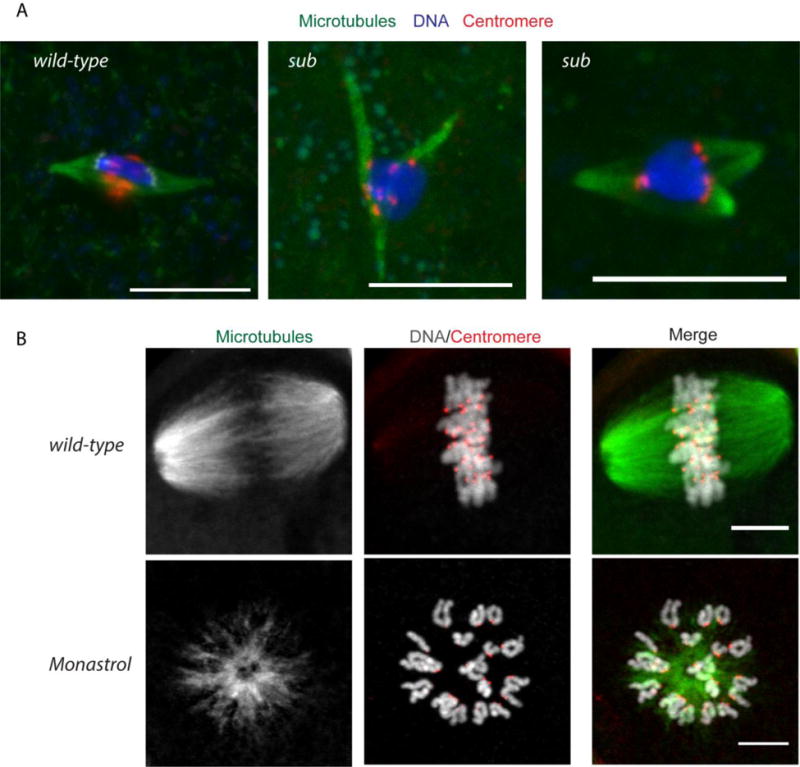
A) In Drosophila oocytes lacking the Subito protein, tripolar spindles form. Microtubules are in green, DNA in blue, and centromeres (CENP-C) in red. Scale bar is 10 μM. B) Metaphase II eggs from a control with bipolar spindle, and monastrol-treated, which inhibits kinesin-5, with a monopolar spindle. Microtubules are in green, DNA in white, and centromeres (CREST) in red. Scale bar is 10 μm.
These results are consistent with computational models that simulate key features of acentrosomal spindle assembly through sliding and bundling of microtubules by a plus-end-directed motor and clustering of minus ends by a minus-end-directed motor (Burbank, et al. 2007) or through microtubule crosslinking and microtubule depolymerization at minus ends (Janson, et al. 2007; Loughlin, et al. 2010). These models rely heavily on the antiparallel arrangement of microtubules that is typical of non-kinetochore microtubules. Non-kinetochore microtubules are a prominent feature of oocyte spindles, with estimates of their prevalence as high as 95% (compared to 5% kinetochore microtubules) in Xenopus egg extract spindles (Ohi, et al. 2007). The known properties of the kinesin-6 family are consistent with these models. Kinesin-6 proteins, which include the Subito mammalian homolog MKLP2, bundle antiparallel microtubules, an activity best known for its role in assembling the midzone at anaphase (Cesario, et al. 2006; Gruneberg, et al. 2004; Mishima, et al. 2002; Neef, et al. 2003; Nislow, et al. 1992; Tao, et al. 2016).
The role of Subito in other organisms in organizing the meiotic central spindle is not known. Other microtubule-bundling kinesins, for example kinesin-5, may have a similar function in mouse oocytes. Antiparallel microtubule crosslinking and sliding can be performed by kinesin-5 (known as Eg5, KIF11, BimC, or KLP61F). Inhibition of kinesin-5 in mouse oocytes with the small molecule inhibitor monastrol induces monopolar spindles (Mailhes, et al. 2004) (Figure 3). This phenotype is, however, distinct from that observed in subito mutants. In the absence of Subito, spindle poles undergo continuous cycles where they collapse into existing poles and then re-form, resulting in continual transitions between tripolar, bipolar, and monopolar spindles (Colombié, et al. 2008). In contrast, inhibition of kinesin-5 simply results in collapse of the spindle poles into a monopole (Mailhes, et al. 2004). These differences suggest that kinesin-5 and kinesin-6 have distinct roles in generating the bipolarity of oocyte spindles. In C. elegans, kinesin-5 is not essential for viability. A different microtubule-bundling kinesin, the kinesin-12, KLP-18, is required for spindle bipolarity (Segbert, et al. 2003). Much like Drosophila, at least conceptually, there is a ring containing the CPC and a motor protein (KLP-19, a kinesin-4) that forms around the chromosomes during metaphase I (Wignall and Villeneuve 2009). C. elegans KLP-18 may have a role like kinesin-5 and enforce bipolarity by sliding microtubules and sorting the minus ends towards the poles (Wolff, et al. 2016).
Although the role of the central spindle in promoting spindle bipolarity has not been confirmed in organisms other than Drosophila, some studies suggest it is important. As described below, bi-orientation in mouse may depend on a “prometaphase belt”, which may be similar to the Drosophila meiotic central spindle. HURP, a conserved microtubule-associated protein, localizes to mouse oocyte central spindles in a kinesin-5-dependent manner and promotes microtubule stability in this region. Importantly, HURP is required for establishment and maintenance of a bipolar meiotic spindle through MTOC sorting and clustering (Breuer, et al. 2010). Like Subito, in the absence of HURP, there is a reduction in microtubule density specifically in the region of antiparallel overlap. These results are consistent with the idea that acentrosomal spindles require a robust central spindle to establish spindle bipolarity.
Bi-orientation without “search and capture” – the importance of converting lateral to end-on attachments
Formation of a stable, bipolar spindle is a critical step in cell division, but the ultimate goal is for this structure to direct the accurate segregation of the chromosomes. For accurate chromosome segregation to occur, chromosomes must become bi-oriented. One model for how bi-orientation occurs is “search and capture” – where microtubules growing from the poles randomly make end-on attachments to kinetochores (Kirschner and Mitchison 1986; Nicklas 1997). This process, however, may not be efficient enough to find and orient all the chromosomes in a typical cell. Cells containing centrosomes probably require additional mechanisms to supplement search and capture (Heald and Khodjakov 2015; Wollman, et al. 2005). This problem is only magnified in acentrosomal spindles with the absence of defined microtubule growth from the poles. One of these additional mechanisms supporting chromosome alignment is congression, where prometaphase chromosomes are collected to the center of the spindle (Cai, et al. 2009; Kapoor, et al. 2006). An important contributor to this process is lateral attachments between the chromosomes and the microtubules.
Several studies show that lateral attachments are important for the process of homolog bi-orientation on acentrosomal spindles (Figure 4). During prometaphase in mouse oocytes, chromosomes move towards the outside edges of the developing spindle and then congress via lateral attachments with microtubules to a ring around the central part of the spindle, or the “prometaphase belt” (Kitajima, et al. 2011; Magidson, et al. 2011). There are several important features of the “prometaphase belt” model. One, it highlights an important role for the central spindle, which is a prominent feature of oocytes in several systems. Not only an organizer of bipolarity, the central spindle and its structure of antiparallel microtubule overlaps is a rich source of microtubule plus ends. In this context, the “prometaphase belt” facilitates and enhances the rate of bi-orientation by bringing kinetochores into the vicinity of a high density of microtubule plus ends, which leads to stable kinetochore-microtubule attachments. Another important feature of the “prometaphase belt” model is the prominent role of lateral chromosome-microtubule attachments. Oocytes experience a prolonged period during which stable end-on kinetochore-microtubule attachments are not observed (Brunet, et al. 1999). However, during a stage (2–4 hrs after NEB) when end-on attachments are not observed, the bivalents appear to stretch, indicating that there is enough stability in lateral attachments to provide force on the chromosomes and promote bi-orientation (Yoshida, et al. 2015).
Figure 4. Lateral and end-on kinetochore attachments in Drosophila and mouse oocytes.
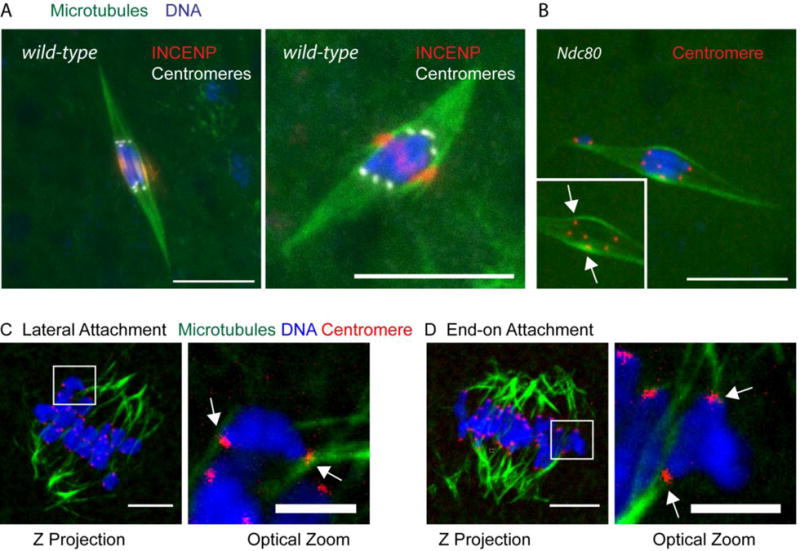
A) Wild-type Drosophila oocytes showing all kinetochores making end-on attachments, with DNA in blue, microtubules in green, central spindle protein INCENP in red and centromeres (CENP-C) in white. B) Oocyte lacking the kinetochore protein NDC80, which is required for end-on attachments, shows evidence of lateral attachments. DNA is in blue, microtubules in green, and centromeres (CENP-C) in red. Inset is a 10 μm region with the DNA removed showing most of the centromeres. C,D) Cold-treated mouse oocytes at metaphase I, showing examples of lateral (C) and end-on (D) kinetochore attachments. The boxed regions are shown at higher magnification. DNA is in blue, microtubules in green and centromeres (CREST) in red and the arrows point to lateral or end-on attachments. In all images, the scale bar is 10 μm.
The predominance of lateral attachments and absence of end-on kinetochore-microtubule attachments has also been observed in C. elegans oocytes (Wignall and Villeneuve 2009). C. elegans is an interesting case because the chromosomes are holocentric and form cup-shaped kinetochores. Lateral attachments sandwich the chromosomes between parallel bundles of microtubules, and this constrains and bi-orients the chromosomes (Wignall and Villeneuve 2009). The kinetochore protein KNL-1 is required for chromosome alignment in C. elegans oocytes (Dumont, et al. 2010), suggesting that these lateral attachments between chromosomes and microtubules are mediated by the kinetochore. Work in Drosophila oocytes has demonstrated that kinetochores are indeed essential for prometaphase chromosome movements and alignment (Radford, et al. 2015). Importantly, lateral kinetochore-microtubule attachments are sufficient to drive prometaphase chromosome movements in Drosophila oocytes (Radford, et al. 2015), consistent with the “prometaphase belt” model. Thus, in nematode, Drosophila and mouse, lateral attachments are important for bi-orientation and sufficient for substantial chromosome movement and interkinetochore stretch. End-on attachments may only be required to stabilize bi-oriented homologs (Radford, et al. 2015).
Bi-orientation, error correction and establishment of end-on attachments
Lateral attachments between kinetochores and microtubules, likely involving antiparallel bundles of microtubules in the central spindle, appear to be a conserved feature of acentrosomal meiosis. How these attachments lead to bi-orientation is not known. Several observations suggest bi-orientation in oocytes involves a mechanism of error correction. Homologous chromosomes undergo multiple rounds of stretching and relaxation in mouse oocytes (Kitajima, et al. 2011), reminiscent of “error correction”. Similarly, real-time imaging of achiasmate Drosophila chromosomes suggest they also undergo multiple rounds of attachment to each spindle pole (Hughes, et al. 2009). Genetic studies have shown that mutants in checkpoint genes, such as in the Drosophila homolog of Mps1, display chromosome segregation errors in oocytes (Gilliland, et al. 2007; Gilliland, et al. 2005; O’Tousa 1982). Mps1 is also required for chromosome segregation in mouse oocytes (Hached, et al. 2011), although further studies are required to determine if Mps1 is required for error correction.
Homologous chromosome bi-orientation is often defined as the establishment of stable end-on kinetochore-microtubule attachments, with sister chromatids (mitosis or meiosis II) or homologous chromosomes (meiosis I) making connections to opposite spindle poles. This places the chromosomes under tension, which can be observed as the stretching apart of sister or homologous centromeres. Errors are identified by a lack of end-on kinetochore-microtubule attachment or a lack of the tension provided by these attachments. Surprisingly, during meiosis I in mouse oocytes, homologous centromeres become stretched prior to the establishment of stable end-on kinetochore-microtubule attachments (Yoshida, et al. 2015). This is corroborated by data from Drosophila oocytes that shows homologous centromeres stretch apart in the absence of end-on kinetochore-microtubule attachments (Radford, et al. 2015). Similarly, C. elegans pairs of homologous chromosomes stretch towards opposite poles in the absence of end-on attachments (McNally, et al. 2016). These results suggest that end-on attachment and tension are separated in oocytes. Further research is needed to determine how incorrect attachments are identified, what defines bi-orientation and how it is monitored in meiosis I oocytes.
Does Aurora B/C have a role in error correction?
As in mitotic cells, Aurora B/C kinase could have a pivotal role in error correction. The spatial separation model for error correction suggests that incorrectly attached kinetochores are in close proximity to Aurora B kinase, which destabilizes the microtubule attachments, while correctly bi-oriented chromosomes are pulled away from Aurora B (Kalantzaki, et al. 2015; Lampson, et al. 2004). There are two problems with this model in meiosis. First, unlike mitosis and meiosis II, the sister centromeres of meiosis I orient towards the same pole; therefore, tension would not be predicted to displace kinetochores from Aurora B at inner centromeres (Watanabe 2012). Furthermore, this model probably does not apply to lateral kinetochore-microtubule attachments, which are resistant to Aurora B-mediated destabilization (Kalantzaki, et al. 2015). Because lateral attachments may have a role in oocyte bi-orientation, how error correction operates on lateral kinetochore-microtubule attachments to lead to the establishment of correctly bi-oriented chromosomes that are primed for the stabilization of end-on attachments is an important question for further study.
Additionally, evidence from the mouse suggests that Aurora B/C may not be regulated by attachment status. The conversion from lateral to stable end-on attachments in mouse oocytes depends on the timing of PP2A and CDK1 activity, rather than bi-orientation or tension/stretching status (Yoshida, et al. 2015). Even stretched apart and correctly bi-oriented chromosomes are treated like errors and “corrected” in an Aurora B-dependent manner in mouse oocytes (Yoshida, et al. 2015). During prometaphase, active Aurora B/C is located at the centromeres of stretched homologous chromosomes, which may explain the lack of stable end-on kinetochore-microtubule attachments (Yoshida, et al. 2015). Late in prometaphase, CDK1 activity increases (Davydenko, et al. 2013) and the phosphatase PP2A-B56 is recruited to centromeres (Yoshida, et al. 2015), which may promote the formation of stable end-on kinetochore-microtubule attachments, regardless of bi-orientation status. These observations help explain the failure to observe end-on attachments during most of mouse meiosis I (Brunet, et al. 1999).
These results appear to challenge the view that Aurora B/C is required to prevent incorrect attachments in oocytes. Numerous studies, however, have found that Aurora B is required for bi-orientation (Davydenko, et al. 2013; Kitajima, et al. 2011; Lane, et al. 2010; Rattani, et al. 2013; Shuda, et al. 2009; Yang, et al. 2010; Yoshida, et al. 2015). One possibility is that Aurora B/C destabilizes premature end-on attachments that occur during early prometaphase that may be at a high risk of forming stable improper attachments. Later, Aurora B/C activity decreases, allowing the stabilization of end-on attachments. Interestingly, in Drosophila sentin mutant oocytes (Sentin is an EB1 interacting protein), end-on attachments are stabilized prematurely and the result is bi-orientation errors (Głuszek, et al. 2015). Thus, oocytes may use a bi-orientation mechanism that depends on destabilizing all attachments until there is a high probability that all are correct (Figure 2). In addition, pole-localized Aurora A activity can promote error correction of chromosomes that have wandered close to the poles (Chmátal, et al. 2015; Ye, et al. 2015), but whether this represents the primary mechanism for error correction in oocytes is not known. More research is needed to understand how such a system works.
Consistent with the idea that the emphasis for oocyte bi-orientation is making correct attachments rather than having to correct errors, the presence of one or even several univalents does not cause progression delays (Gui and Homer 2012; Kolano, et al. 2012; Lane, et al. 2012; LeMaire-Adkins, et al. 1997; Nagaoka, et al. 2012; Nagaoka, et al. 2011; Sebestova, et al. 2012). In some cases, univalents bi-orient (Kouznetsova, et al. 2007), which could satisfy the checkpoint. One interpretation is that oocytes lack a robust mechanism to correct erroneous attachments. Another possibility is that the checkpoint doesn’t monitor chromosomes, but instead controls progression (i.e. the checkpoint as a timer hypothesis). This second possibility is consistent with the results described above from mouse oocytes in which stabilization of end-on attachments results from an increase throughout prometaphase in PP2A activity at centromeres, which silences the destabilizing activity of the CPC and depends on CDK1 and BubR1 (Davydenko, et al. 2013; Yoshida, et al. 2015). Therefore, in oocytes, the checkpoint and error correction machinery may function within an inflexible timescale, which could be tied to the developmental restrictions associated with oocyte maturation.
Segregation at anaphase
The bulk of studies into meiosis in oocytes (and also the bulk of this review) have been dedicated to understanding spindle assembly and homolog bi-orientation. It may be assumed that once these events have occurred, anaphase merely executes the pulling of end-on kinetochore-microtubule attachments to drive chromosome poleward movement. One problem with this simplistic view is that, in the absence of centrosomes, how are the poles anchored? Indeed, surprising results were found in C. elegans, where the poles are observed to move towards the chromosomes. At metaphase, the C. elegans meiotic spindle shortens until the kinetochores become embedded in the spindle poles (McNally, et al. 2016). The chromosomes then separate with sliding of the microtubules between them. This resembles Anaphase B (spindle elongation) and does not depend on kinetochore proteins (Dumont, et al. 2010). C. elegans meiosis also shows evidence of Anaphase A (chromosome movement), based on the observation that the chromosome move through channels in the microtubule array (Muscat, et al. 2015).
Pushing the chromosomes apart with microtubule sliding may also operate in mouse, where in meiosis II, anaphase B has been proposed to occur before anaphase A (FitzHarris 2012). This mechanism depends on spindle elongation driven by kinesin-5. The existence of these forces may explain observations in mouse that chromatin-coated beads move poleward at anaphase (Deng, et al. 2009). Similar mechanisms may be operating in Drosophila based on elongation of the spindle observed in precocious anaphase, whereas chromosome to pole distance may not decrease (McKim, et al. 1993). These results suggest that anaphase includes a combination of A- and B-type chromosome movements, although relative to mitosis, possibly in reverse order or at the same time.
A final issue when considering anaphase is the concept and consequences of the asymmetric cell division in oocytes. Although meiosis can generate four haploid products, only one of these becomes the large oocyte, while the rest are retained in much smaller polar bodies. In a variety of organisms, the oocyte spindle is positioned asymmetrically prior to the division. Not only is the spindle located close to the cortex, but it may also rotate from an initial parallel orientation to perpendicular at anaphase I or II (McNally 2013). The mechanism of spindle positioning involves interactions with the cortex (“cortical pulling”) mediated by actin and/or tubulin (Almonacid, et al. 2014; Brunet and Verlhac 2011). While the asymmetric division may only reflect elements in the cytoplasm, it is also possible that, as a consequence or cause, the meiotic spindle is asymmetric, with consequences on segregation. Indeed, the asymmetry in C. elegans meiosis causes a bias in the inheritance of univalents (Cortes, et al. 2015).
Because only one of the four meiotic products is incorporated into the zygote, an asymmetric spindle could have significant consequences on Mendelian segregation. If a pair of homologs have kinetochores that interact unequally with the spindle, then there is the potential for biased inheritance (Ross and Malik 2014). While this mechanism is plausible and attractive, it has proven difficult to observe. One study in mouse oocytes has revealed a link between asymmetric segregation and a property of the chromosomes (Chmátal, et al. 2014). This study demonstrated that some chromosomes (i.e. fusion chromosomes in natural populations) have stronger centromeres than other chromosomes (i.e. telocentric chromosomes in the same populations). These stronger centromeres appear to load more kinetochore proteins, and therefore interact with more microtubules, than weaker centromeres. It needs to be reiterated that such a difference in centromere strength only results in biased inheritance if there is an asymmetric spindle. Given the scarcity of such evidence (Hewitt 1976), more study is needed on the basis and consequences of the asymmetric spindle.
Summary
Chromosomes are the focal point of acentrosomal spindle assembly in oocytes. Amazingly, despite intensive knowledge about various spindle assembly pathways and proteins that mediate microtubule dynamics, there is still much to learn about what recruits the microtubules and what are the chromosome-based signals. For example, although the CPC is required in Drosophila and Xenopus, the generality of these findings is not clear. Most targets for acentrosomal spindle assembly come from mitotic candidates, which puts a limitation on finding factors involved in meiotic spindle assembly. New factors may be identified in screens, as has been reported in mouse oocytes, for spindle assembly factors (Pfender, et al. 2015). In addition to the central role of the chromosomes, the central spindle has a function at metaphase that is critical for spindle organization and bi-orientation. It replaces the centrosomes with an array of sliding antiparallel microtubules that can establish bipolarity. The central spindle may also provide a kinetochore-independent source of microtubules.
This review has emphasized the importance of the chromosomes because most studies have focused on the trans-acting spindle assembly factors. Almost nothing is known about the chromatin features that promote acentrosomal spindle assembly. The kinetochores are clearly neither sufficient nor necessary. There could be epigenetic factors such as histone modifications (Kelly, et al. 2010; Wang, et al. 2010; Yamagishi, et al. 2010) or proteins like HP1 (Abe, et al. 2016; Ainsztein, et al. 1998), which are known to recruit the CPC. An interesting recent development is that noncoding RNAs may have a role in chromatin-mediated spindle assembly (Blower 2016; Chen, et al. 2015; Grenfell, et al. 2016; Jambhekar, et al. 2014).
In general, there are more examples of conserved concepts (eg. chromatin-mediated spindle assembly and the central spindle) than conserved proteins that carry out these functions. This could represent differences in mechanisms, such as in the case of the CPC and spindle assembly. In other cases, it could be due to lack of studies. For example, while Subito is required for bipolarity of the Drosophila oocyte spindle, C. elegans lacks a true orthologue of Subito and there is a lack of studies on the mouse orthologue MKLP2. It is possible, however, that a similar role to Subito is taken by kinesin-12 (in C. elegans) or kinesin-5 (in mouse). Similarly, there are few studies on the motors that regulate dynamics, although depolymerizing kinesins (kinesin-13) are important for regulating spindle length and microtubule attachments in Drosophila (Radford, et al. 2012) and C. elegans (Connolly, et al. 2015). It is not known if a shift from dynamic to stable microtubules accompanies the transition from prometaphase to metaphase in meiosis as it does in mitosis (Godek, et al. 2015; Kabeche and Compton 2013).
Lateral microtubule attachments with the kinetochore, probably involving the central spindle, are a conserved element of the mechanism for bi-orientation. The stability provided by the central spindle is important in this context when, early in meiosis, there are few end-on attachments and ill-defined poles. Lateral attachments probably predominate over any search and capture type of mechanism. However, much remains to be learned about how bi-orientation is measured, especially if this occurs at kinetochores that are making lateral attachments with microtubules. Indeed, the observation that end-on attachments are stabilized late and in a temporal manner, and that some lateral attachments may be stable, suggests that bi-orientation is established using mechanisms that do not depend on end-on attachments. The models from mitosis are insufficient to explain bi-orientation at meiosis I, and more interestingly, lessons learned from meiosis may reveal mechanisms that are operating in mitosis.
Acknowledgments
We thank members of the McKim and Schindler labs for discussions and comments on the manuscript, and Christian Lehner and two anonymous reviewers for suggestions leading to significant improvements in the manuscript. We apologize to the many authors, whose papers we were not able to cite.
Writing of this review was supported by National Institutes of Health grants GM101955 to KMcK and GM112801 to KS.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Informed consent: “Informed consent was obtained from all individual participants included in the study.”
References
- Abe Y, Sako K, Takagaki K, Hirayama Y, Uchida KS, Herman JA, DeLuca JG, Hirota T. HP1-Assisted Aurora B Kinase Activity Prevents Chromosome Segregation Errors. Dev Cell. 2016;36:487–497. doi: 10.1016/j.devcel.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–1774. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Terret M, Verlhac MH. Actin-based spindle positioning: new insights from female gametes. J Cell Sci. 2014;127:477–483. doi: 10.1242/jcs.142711. [DOI] [PubMed] [Google Scholar]
- Baker BS, Hall JC. Meiotic mutants: genetic control of meiotic recombination and chromosome segregation. In: Ashburner M, Novitski E, editors. The Genetics and Biology of Drosophila. Academic Press; New York: 1976. pp. 351–434. [Google Scholar]
- Balboula AZ, Nguyen AL, Gentilello AS, Quartuccio SM, Drutovic D, Solc P, Schindler K. Haspin kinase regulates microtubule-organizing center clustering and stability through Aurora kinase C in mouse oocytes. J Cell Sci. 2016 doi: 10.1242/jcs.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula AZ, Schindler K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 2014;10:e1004194. doi: 10.1371/journal.pgen.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J, Novak ZA, Raff JW, Wainman A. Dissecting the function and assembly of acentriolar microtubule organizing centers in Drosophila cells in vivo. PLoS Genet. 2015;11:e1005261. doi: 10.1371/journal.pgen.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD. Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell reports. 2016;15:1624–1633. doi: 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer M, Kolano A, Kwon M, Li CC, Tsai TF, Pellman D, Brunet S, Verlhac MH. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol. 2010;191:1251–1260. doi: 10.1083/jcb.201005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Koretke KK, Birkeland ML, Sanseau P, Patrick DR. Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol Biol. 2004;4:39. doi: 10.1186/1471-2148-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Maria AS, Guillaud P, Dujardin D, Kubiak JZ, Maro B. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol. 1999;146:1–12. doi: 10.1083/jcb.146.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Polanski Z, Verlhac MH, Kubiak JZ, Maro B. Bipolar meiotic spindle formation without chromatin. Curr Biol. 1998;8:1231–1234. doi: 10.1016/s0960-9822(07)00516-7. [DOI] [PubMed] [Google Scholar]
- Brunet S, Verlhac MH. Positioning to get out of meiosis: the asymmetry of division. Hum Reprod Update. 2011;17:68–75. doi: 10.1093/humupd/dmq044. [DOI] [PubMed] [Google Scholar]
- Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- Cai S, O’Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario J, McKim KS. RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J Cell Sci. 2011;124:3797–3810. doi: 10.1242/jcs.084855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario JM, Jang JK, Redding B, Shah N, Rahman T, McKim KS. Kinesin 6 family member Subito participates in mitotic spindle assembly and interacts with mitotic regulators. J Cell Sci. 2006;119:4770–4780. doi: 10.1242/jcs.03235. [DOI] [PubMed] [Google Scholar]
- Cha BJ, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106:35–46. doi: 10.1016/s0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O’Neill RJ, Mellone BG. Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Dev Cell. 2015;34:73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Gabriel SI, Mitsainas GP, Martínez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L, Yang K, Schultz RM, Lampson MA. Spatial Regulation of Kinetochore Microtubule Attachments by Destabilization at Spindle Poles in Meiosis I. Curr Biol. 2015;25:1835–1841. doi: 10.1016/j.cub.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Clift D, Schuh M. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun. 2015;6:7217. doi: 10.1038/ncomms8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié N, Cullen CF, Brittle AL, Jang JK, Earnshaw WC, Carmena M, McKim K, Ohkura H. Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development. 2008;135:3239–3246. doi: 10.1242/dev.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AA, Sugioka K, Chuang CH, Lowry JB, Bowerman B. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J Cell Biol. 2015;210:917–932. doi: 10.1083/jcb.201412010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes DB, McNally KL, Mains PE, McNally FJ. The asymmetry of female meiosis reduces the frequency of inheritance of unpaired chromosomes. eLife. 2015;4:e06056. doi: 10.7554/eLife.06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;198:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Brittle AL, Ito T, Ohkura H. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol. 2005;171:593–602. doi: 10.1083/jcb.200508127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nature Cell Biol. 2001;3:637–642. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- Das A, Shah SJ, Fan B, Paik D, DiSanto DJ, Hinman AM, Cesario JM, Battaglia RA, Demos N, McKim KS. Spindle Assembly and Chromosome Segregation Requires Central Spindle Proteins in Drosophila Oocytes. Genetics. 2016;202:61–75. doi: 10.1534/genetics.115.181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydenko O, Schultz RM, Lampson MA. Increased CDK1 activity determines the timing of kinetochore-microtubule attachments in meiosis I. J Cell Biol. 2013;202:221–229. doi: 10.1083/jcb.201303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Gao J, Suraneni P, Li R. Kinetochore-independent chromosome poleward movement during anaphase of meiosis II in mouse eggs. PloS one. 2009;4:e5249. doi: 10.1371/journal.pone.0005249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol. 2010;12:894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet Res. 1992;60:209–220. doi: 10.1017/s0016672300030962. [DOI] [PubMed] [Google Scholar]
- Fernández-Miranda G, Trakala M, Martín J, Escobar B, González A, Ghyselinck NB, Ortega S, Cañamero M, Pérez de Castro I, Malumbres M. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 2011;138:2661–2672. doi: 10.1242/dev.066381. [DOI] [PubMed] [Google Scholar]
- FitzHarris G. Anaphase B precedes anaphase A in the mouse egg. Curr Biol. 2012;22:437–444. doi: 10.1016/j.cub.2012.01.041. [DOI] [PubMed] [Google Scholar]
- Gard DL. Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev Biol. 1992;151:516–530. doi: 10.1016/0012-1606(92)90190-r. [DOI] [PubMed] [Google Scholar]
- Gatti M, Bucciarelli E, Lattao R, Pellacani C, Mottier-Pavie V, Giansanti MG, Somma MP, Bonaccorsi S. The relative roles of centrosomal and kinetochore-driven microtubules in Drosophila spindle formation. Exp Cell Res. 2012;318:1375–1380. doi: 10.1016/j.yexcr.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Gilliland WD, Hughes SE, Cotitta JL, Takeo S, Xiang Y, Hawley RS. The multiple roles of mps1 in Drosophila female meiosis. PLoS Genet. 2007;3:e113. doi: 10.1371/journal.pgen.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland WD, Wayson SM, Hawley RS. The meiotic defects of mutants in the Drosophila mps1 gene reveal a critical role of Mps1 in the segregation of achiasmate homologs. Curr Biol. 2005;15:672–677. doi: 10.1016/j.cub.2005.02.062. [DOI] [PubMed] [Google Scholar]
- Giunta KL, Jang JK, Manheim EA, Subramanian G, McKim KS. subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics. 2002;160:1489–1501. doi: 10.1093/genetics/160.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głuszek AA, Cullen CF, Li W, Battaglia RA, Radford SJ, Costa MF, McKim KS, Goshima G, Ohkura H. The microtubule catastrophe promoter Sentin delays stable kinetochore-microtubule attachment in oocytes. J Cell Biol. 2015;211:1113–1120. doi: 10.1083/jcb.201507006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godek KM, Kabeche L, Compton DA. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol. 2015;16:57–64. doi: 10.1038/nrm3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell AW, Heald R, Strzelecka M. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J Cell Biol. 2016;214:133–141. doi: 10.1083/jcb.201604029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Homer H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development. 2012;139:1941–1946. doi: 10.1242/dev.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hached K, Xie SZ, Buffin E, Cladiere D, Rachez C, Sacras M, Sorger PK, Wassmann K. Mps1 at kinetochores is essential for female mouse meiosis I. Development. 2011;138:2261–2271. doi: 10.1242/dev.061317. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Heald R, Khodjakov A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J Cell Biol. 2015;211:1103–1111. doi: 10.1083/jcb.201510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Meiotic drive for B-chromosomes in the primary oocytes of Myrmeleotettix maculatus (Orthopera: Acrididae) Chromosoma. 1976;56:381–391. doi: 10.1007/BF00292957. [DOI] [PubMed] [Google Scholar]
- Holubcová Z, Blayney M, Elder K, Schuh M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science. 2015;348:1143–1147. doi: 10.1126/science.aaa9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Gilliland WD, Cotitta JL, Takeo S, Collins KA, Hawley RS. Heterochromatic threads connect oscillating chromosomes during prometaphase I in Drosophila oocytes. PLoS Genet. 2009;5:e1000348. doi: 10.1371/journal.pgen.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Emerman AB, Schweidenback CT, Blower MD. RNA stimulates Aurora B kinase activity during mitosis. PloS one. 2014;9:e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JK, Rahman T, Kober VS, Cesario J, McKim KS. Misregulation of the Kinesin-like Protein Subito Induces Meiotic Spindle Formation in the Absence of Chromosomes and Centrosomes. Genetics. 2007;177:267–280. doi: 10.1534/genetics.107.076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JK, Rahman T, McKim KS. The kinesinlike protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Mol Biol Cell. 2005;16:4684–4694. doi: 10.1091/mbc.E04-11-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loïodice I, Fu C, Brunner D, Nédélec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantzaki M, Kitamura E, Zhang T, Mino A, Novák B, Tanaka TU. Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nat Cell Biol. 2015;17:421–433. doi: 10.1038/ncb3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol. 2015;208:181–196. doi: 10.1083/jcb.201407074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Kolano A, Brunet S, Silk AD, Cleveland DW, Verlhac MH. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci U S A. 2012;109:E1858–1867. doi: 10.1073/pnas.1204686109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouznetsova A, Lister L, Nordenskjöld M, Herbert M, Höög C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Lane SI, Chang HY, Jennings PC, Jones KT. The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction. 2010;140:521–530. doi: 10.1530/REP-10-0223. [DOI] [PubMed] [Google Scholar]
- Lane SI, Yun Y, Jones KT. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development. 2012;139:1947–1955. doi: 10.1242/dev.077040. [DOI] [PubMed] [Google Scholar]
- LeMaire-Adkins R, Radke K, Hunt PA. Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J Cell Biol. 1997;139:1611–1619. doi: 10.1083/jcb.139.7.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nédélec F. A computational model predicts Xenopus meiotic spindle organization. J Cell Biol. 2010;191:1239–1249. doi: 10.1083/jcb.201006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luksza M, Queguigner I, Verlhac MH, Brunet S. Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol. 2013;382:48–56. doi: 10.1016/j.ydbio.2013.07.029. [DOI] [PubMed] [Google Scholar]
- Ma W, Viveiros MM. Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol Reprod Dev. 2014;81:1019–1029. doi: 10.1002/mrd.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailhes JB, Mastromatteo C, Fuseler JW. Transient exposure to the Eg5 kinesin inhibitor monastrol leads to syntelic orientation of chromosomes and aneuploidy in mouse oocytes. Mutat Res. 2004;559:153–167. doi: 10.1016/j.mrgentox.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Webb M, Flach G. Mechanism of polar body formation in the mouse oocyte: an interaction between the chromosomes, the cytoskeleton and the plasma membrane. J Embryol Exp Morphol. 1986;92:11–32. [PubMed] [Google Scholar]
- McKim KS, Hawley RS. Chromosomal control of meiotic cell division. Science. 1995;270:1595–1601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- McKim KS, Jang JK, Theurkauf WE, Hawley RS. Mechanical basis of meiotic metaphase arrest. Nature. 1993;362:364–366. doi: 10.1038/362364a0. [DOI] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, Panzica MT, Kim T, Cortes DB, McNally FJ. A Novel Chromosome Segregation Mechanism During Female Meiosis. Mol Biol Cell. 2016 doi: 10.1091/mbc.E16-05-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles AM, Fisher KH, Colombie N, Wakefield JG, Ohkura H. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol. 2009;184:777–784. doi: 10.1083/jcb.200811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier S, Vernos I. Acentrosomal Microtubule Assembly in Mitosis: The Where, When, and How. Trends Cell Biol. 2016;26:80–87. doi: 10.1016/j.tcb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S, Stuurman N, Afonso O, Hornsveld M, Aguiar P, Goshima G, Vale RD, Maiato H. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc Natl Acad Sci U S A. 2013;110:19808–19813. doi: 10.1073/pnas.1320013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 2015;4 doi: 10.7554/eLife.06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol. 2011;21:651–657. doi: 10.1016/j.cub.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. Chromosome distribution: experiments on cell hybrids and in vitro. Philosophical Transcripts of the Royal Society of London B. 1977;277:267–276. doi: 10.1098/rstb.1977.0017. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- O’Tousa J. Meiotic chromosome behavior influenced by mutation-altered disjunction in Drosophila melanogaster females. Genetics. 1982;102:503–524. doi: 10.1093/genetics/102.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr Biol. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Paliulis LV, Nicklas RB. The reduction of chromosome number in meiosis is determined by properties built into the chromosomes. J Cell Biol. 2000;150:1223–1232. doi: 10.1083/jcb.150.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo V, Pellacani C, Heesom KJ, Rogala KB, Deane CM, Mottier-Pavie V, Gatti M, Bonaccorsi S, Wakefield JG. Misato Controls Mitotic Microtubule Generation by Stabilizing the TCP-1 Tubulin Chaperone Complex [corrected] Curr Biol. 2015;25:1777–1783. doi: 10.1016/j.cub.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfender S, Kuznetsov V, Pasternak M, Tischer T, Santhanam B, Schuh M. Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature. 2015;524:239–242. doi: 10.1038/nature14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta-Marques A, Bento I, Lopes CA, Duarte P, Jana SC, Bettencourt-Dias M. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science. 2016;353:aaf4866. doi: 10.1126/science.aaf4866. [DOI] [PubMed] [Google Scholar]
- Polanski Z, Hoffmann S, Tsurumi C. Oocyte nucleus controls progression through meiotic maturation. Dev Biol. 2005;281:184–195. doi: 10.1016/j.ydbio.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Harrison AM, McKim KS. Microtubule-depolymerizing Kinesin KLP10A Restricts the Length of the Acentrosomal Meiotic Spindle in Drosophila Females. Genetics. 2012;192:431–440. doi: 10.1534/genetics.112.143503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Hoang TL, Głuszek AA, Ohkura H, McKim KS. Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Drosophila Oocytes. PLoS Genet. 2015;11:e1005605. doi: 10.1371/journal.pgen.1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Jang JK, McKim KS. The Chromosomal Passenger Complex is required for Meiotic Acentrosomal Spindle Assembly and Chromosome Bi-orientation. Genetics. 2012;192:417–429. doi: 10.1534/genetics.112.143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattani A, Wolna M, Ploquin M, Helmhart W, Morrone S, Mayer B, Godwin J, Xu W, Stemmann O, Pendas A, Nasmyth K. Sgol2 provides a regulatory platform that coordinates essential cell cycle processes during meiosis I in oocytes. eLife. 2013;2:e01133. doi: 10.7554/eLife.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschen RF, Colombie N, Wheatley L, Dobbelaere J, St Johnston D, Ohkura H, Raff JW. Dgp71WD is required for the assembly of the acentrosomal Meiosis I spindle, and is not a general targeting factor for the gamma-TuRC. Biol Open. 2012;1:422–429. doi: 10.1242/bio.2012596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G, Glover DM, Avides MC. A requirement for the Abnormal Spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J Cell Sci. 2002;115:913–922. doi: 10.1242/jcs.115.5.913. [DOI] [PubMed] [Google Scholar]
- Ross BD, Malik HS. Genetic conflicts: stronger centromeres win tug-of-war in female meiosis. Curr Biol. 2014;24:R966–968. doi: 10.1016/j.cub.2014.08.053. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sanchez-Huertas C, Luders J. The augmin connection in the geometry of microtubule networks. Curr Biol. 2015;25:R294–299. doi: 10.1016/j.cub.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A. 2012;109:E2215–2222. doi: 10.1073/pnas.1120517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestova J, Danylevska A, Novakova L, Kubelka M, Anger M. Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle. 2012;11:3011–3018. doi: 10.4161/cc.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, von Dassow G, Bowerman B. Oocyte Meiotic Spindle Assembly and Function. Curr Top Dev Biol. 2016;116:65–98. doi: 10.1016/bs.ctdb.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, Zernicka-Goetz M. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci. 2010;123:4292–4300. doi: 10.1242/jcs.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev. 2009;76:1094–1105. doi: 10.1002/mrd.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solc P, Baran V, Mayer A, Bohmova T, Panenkova-Havlova G, Saskova A, Schultz RM, Motlik J. Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo. Biol Reprod. 2012;87:85. doi: 10.1095/biolreprod.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun QY, Lai L, Wu GM, Park KW, Day BN, Prather RS, Schatten H. Microtubule assembly after treatment of pig oocytes with taxol: correlation with chromosomes, gamma-tubulin, and MAP kinase. Mol Reprod Dev. 2001;60:481–490. doi: 10.1002/mrd.1113. [DOI] [PubMed] [Google Scholar]
- Szollosi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- Tao L, Fasulo B, Warecki B, Sullivan W. Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat Commun. 2016;7:11182. doi: 10.1038/ncomms11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Smiley S, Wong ML, Alberts BM. Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development. 1992;115:923–936. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual Detection of Chromosomes and Microtubules by the Chromosomal Passenger Complex Drives Spindle Assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M, Lorson MA, Srinivasan DG, Bennett KL, van den Heuvel S. C. elegans mitotic cyclins have distinct as well as overlapping functions in chromosome segregation. Cell Cycle. 2009;8:4091–4102. doi: 10.4161/cc.8.24.10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P, Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13:370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wolff ID, Tran MV, Mullen TJ, Villeneuve AM, Wignall SM. Assembly of C. elegans acentrosomal spindles occurs without evident MTOCs and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol Biol Cell. 2016 doi: 10.1091/mbc.E16-05-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Yang JW, Lei ZL, Miao YL, Huang JC, Shi LH, OuYang YC, Sun QY, Chen DY. Spindle assembly in the absence of chromosomes in mouse oocytes. Reproduction. 2007;134:731–738. doi: 10.1530/REP-07-0149. [DOI] [PubMed] [Google Scholar]
- Yang KT, Li SK, Chang CC, Tang CJ, Lin YN, Lee SC, Tang TK. Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol Biol Cell. 2010;21:2371–2383. doi: 10.1091/mbc.E10-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye AA, Deretic J, Hoel CM, Hinman AW, Cimini D, Welburn JP, Maresca TJ. Aurora A Kinase Contributes to a Pole-Based Error Correction Pathway. Curr Biol. 2015;25:1842–1851. doi: 10.1016/j.cub.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kaido M, Kitajima TS. Inherent Instability of Correct Kinetochore-Microtubule Attachments during Meiosis I in Oocytes. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.04.020. [DOI] [PubMed] [Google Scholar]


