Abstract
Epileptic encephalopathies (EE) are a group of severe childhood epilepsy disorders characterized by intractable seizures, cognitive impairment and neurological deficits. Recent whole-exome sequencing (WES) studies have implicated significant contribution of de novo mutations to EE. In this study, we utilized WES for identifying causal de novo mutations in 4 parent-offspring trios affected by West syndrome. As a result, we found two deleterious de novo mutations in DYNC1H1 and RTP1 in two trios. Expression profile analysis showed that DYNC1H1 and RTP1 are expressed in almost all brain regions and developmental stages. Interestingly, co-expression and genetic interaction network analyses suggested that DYNC1H1 and RTP1 are tightly associated with known epilepsy genes. Furthermore, we observed that the de novo mutations of DYNC1H1 were identified in several different neuropsychiatric disorders including EE, autism spectrum disorders and intellectual disabilities by previous studies, and these mutations primarily occurred in the functional domain of the protein. Taken together, these results demonstrate DYNC1H1 as a strong candidate and RTP1 as a potential candidate on the onset of EE. In addition, this work also proves WES as a powerful tool for the molecular genetic dissection of children affected by sporadic EE.
Introduction
Epileptic encephalopathies (EE) are typically defined as a devastating group of severe childhood epilepsy disorders characterized by early onset of seizures associated with ongoing epileptic activity1. West syndrome (MIM 308350) is one of the most common form of infantile epileptic encephalopathy, characterized by tonic spasms with clustering, arrest of psychomotor development, and hypsarrhythmia in electroencephalography (EEG)1, 2. Approximately 17% of all cases can evolve into Lennox-Gastaut syndrome (LGS) which is characterized by polymorphic intractable seizures and paroxysms of fast activity in EEG3.
In the past few years, several genes, such as ARX 4, CDKL5 5 and SPTAN1 6, have been implicated in West syndrome based on a candidate gene sequencing approach7, 8. Furthermore, several recent exome-sequencing studies have shown that West syndrome probands exhibit an excess of loss of function de novo mutations, which plays an important role in West syndrome9. Indeed, in a study in which exome sequencing was performed in 264 patient-parent trios with EE, de novo mutations were recurrently observed in probands within known causative genes for West syndrome, such as STXBP1 (n = 4) and CDKL5 (n = 2)9. In other recent studies, de novo mutations in GRIN2B 10, GNAO1 11, KCNT1 12 and SPTAN1 13 found in probands have been recognized as being associated with West syndrome. These findings showed that West syndrome is a genetically heterogeneous condition, and de novo mutations play a significant role in the onset of West syndrome.
Recent advances in the whole-exome sequencing (WES) approach have offered a cost-effective method for investigating single-nucleotide variants (SNVs) across all gene-coding regions in the genome. WES has been successfully applied to identify de novo mutations associated with neurodevelopmental disorders for autism spectrum disorders (ASDs)14, 15, intellectual disabilities (IDs)16, 17, schizophrenia (SCZ)18, 19 and EE20, 21. To obtain insight into the characterization of de novo mutations in West syndrome, we conducted WES of 4 unrelated Chinese parent-offspring trios affected by West syndrome. The results revealed two novel de novo mutations in DYNC1H1 and RTP1 which were predicted to be deleterious based on the concordance of generic damage prediction tools. Furthermore, expression, co-expression and genetic interaction network analyses provided supporting evidences for the role of DYNC1H1 and RTP1 in EE. In particular, the de novo mutations in DYNC1H1 were found to be shared among different neuropsychiatric disorders of EE, ASD and ID, which further implicated DYNC1H1 in the onset of sporadic neuropsychiatric disorders.
Results
Detection of de novo mutations
WES of 4 EE trios with West syndrome (one affected child and both unaffected parents) was performed. After the removal of sequencing adapters and trimming of low-quality bases, approximately 2.12~4.72 Gb of cleaned sequencing data were obtained for each sample (Supplemental Table 1). More than 99.42% of the sequencing reads were aligned to the human reference genome (hg19), with 56.38% effective reads from target regions being obtained after the removal of PCR duplications. The average sequencing depth for each sample was 70.25-fold, with more than 86.00% of target regions having at least a 10-fold coverage. As a result, 21,756 SNVs or InDels were identified and 3 de novo SNVs in coding regions were identified and validated in the 4 trios. We also attempted to identify rare (new or with allele frequency <0.001 based on Exome Aggregation Consortium (ExAC)22, 1000 Genomes23 and the NHLBI Exome Sequencing Project (ESP)9) inherited mutations in known or related epilepsy genes, but no rare inherited mutations were detected for the 4 trios.
Analysis of the association of de novo gene mutations with EE
Recent WES studies based on parent-offspring trios are increasingly demonstrating that de novo mutations play a prominent role in a substantial number of EE, although the extent of this contribution is not yet known24. In the present study, we observed that two of the three missense de novo mutations were clearly predicted to be functionally deleterious based on four mutation effect prediction tools (SIFT, VEST3, LRT and SiPhy) (Table 1).
Table 1.
Summary of DNMs detected by trios-based WES of EE.
| Trio | Chr | Position(hg19) | Gene | Mutation | Transcript | Protein change | SIFT | VEST3 | LRT | SiPhy |
|---|---|---|---|---|---|---|---|---|---|---|
| E1 | chr6 | 158318012 | SNX9 | missense | NM_016224 | p.Q152K | tolerable | damaging | neutral | conserved |
| E2 | chr3 | 186917389 | RTP1 | missense | NM_153708 | p.I108N | damaging | damaging | deleterious | conserved |
| E3 | chr14 | 102499496 | DYNC1H1 | missense | NM_001376 | p.M3392V | damaging | damaging | deleterious | conserved |
One novel de novo mutation was detected in DYNC1H1 in E3P proband. This mutation was nonsynonymous (c.10174A > G) and caused a methionine (Met) to valine (Val) substitution at amino acid 3392 (p.M3392V) in the conserved MT domain (microtubule-binding stalk of dynein motor) (Fig. 1a,b and Table 1). This mutation was predicted as deleterious by the four types of prediction tools employed in this study (Table 1). Furthermore, DYNC1H1 might be considered as an intolerant gene based on residual variation intolerance score (RVIS)25, which was developed to estimate functional variants deviations based on the ESP6500 dataset and Z score for missense variants from ExAC22 (Supplemental Table 2).
Figure 1.
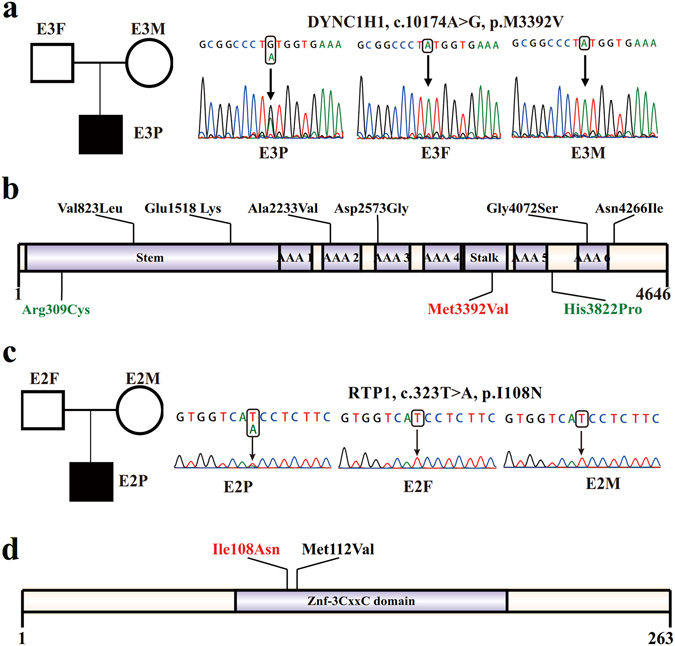
De novo mutations identified in DYNC1H1 and RTP1. (a) The de novo mutation of DYNC1H1 was confirmed in trio E3 using Sanger sequencing. (b) Schematic representation of the DYNC1H1 protein and de novo mutations of DYNC1H1 identified in EE, ASD and ID. Protein changes are shown in black for ASD, green for ID and red for EE. (c) The de novo mutation of RTP1 was confirmed in trio E2 using Sanger sequencing. (d) Schematic representation of the RTP1 protein and de novo mutations of RTP1 identified in EE. Protein change identified in the present study is shown in red, and those identified in the previous study are shown in black.
We also validated a novel missense mutation of T to A substitution (c.323T > A) in RTP1 (MIM 609137) in E2P proband (Fig. 1c and Table 1). The single nucleotide change on RTP1 produced a nonsynonymous substitution (p.I108N) (Fig. 1d). Similarly, this mutation was considered to be deleterious according to prediction software tools (Table 1). Although RTP1 tends to be not evolutionarily constrained based on RIVS or Z score for missense from ExAC, its relatively higher haploinsufficient probability26 indicates its functional importance (Supplemental Table 2). In addition to these above mentioned deleterious mutations, a single nucleotide substitution (c.454C > A) on the SNX9 gene (MIM 605952) was observed in a different proband, which result in an amino acid change (p.Q152K) (Supplementary Figure 1 and Table 1). However, this mutation was considered to be tolerable or neutral based on the predictions of the SIFT and LRT software tools, therefore, it was not considered as a possible cause.
Taken together, two de novo mutations in DYNC1H1 and RTP1 genes identified from two probands may result in severe disruption of protein function.
Expression profile of DYNC1H1 and RTP1 in the developmental human brain
To investigate the potential contribution of DYNC1H1 and RTP1 genes to human brain development and functions, we firstly assessed the expression profiles of the two genes for different brain regions and developmental stages. Gene expression levels measured using micro-arrays were obtained from the Human Brain Transcriptome database (HBT, http://hbatlas.org/) for 6 human brain regions, including the cerebellar cortex (CBC), mediodorsal nucleus of the thalamus (MD), striatum (STR), amygdala (AMY), hippocampus (HIP) and neocortex (NCX), and 15 human developmental periods, from early embryonic stages to late adulthood. Both DYNC1H1 and RTP1 were stably expressed at high levels in all 6 brain regions analyzed throughout the human lifespan (Fig. 2). In addition, DYNC1H1 expression was approximately two-fold higher than that of RTP1. Further analysis of the expression data for the two genes in 11 areas of the neocortex region based on the HBT database revealed similar expression patterns in 6 different brain regions (Supplementary Figure 2). Therefore, we concluded that DYNC1H1 and RTP1 may be important for early brain development and normal brain functions considering their stable high expression levels, and are potential candidate genes for brain function related disorders.
Figure 2.
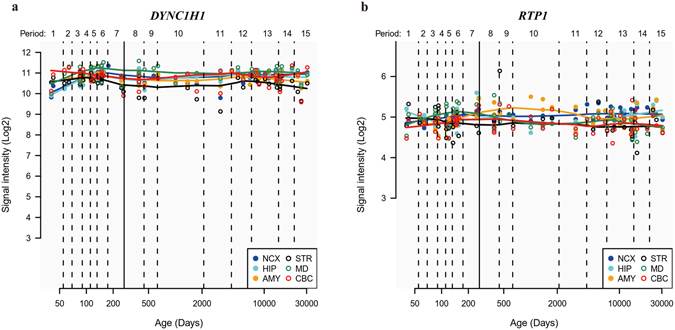
Expression analysis of DYNC1H1 and RTP1 in six human brain regions. Expression profiles are shown for DYNC1H1 (a) and RTP1 (b). The expression levels of DYNC1H1 and RTP1 are shown for the developmental stages from the embryonic development stage at 50 days to late adulthood (e.g., 30,000 days). A solid line between periods 7 and 8 separates prenatal periods from postnatal periods. CBC, cerebellar cortex; MD, mediodorsal nucleus of the thalamus; STR, striatum; AMY, amygdala; HIP, hippocampus; NCX, neocortex.
Co-expression and genetic interaction network analyses of DYNC1H1 and RTP1
To further explore the relationship of DYNC1H1 and RTP1 expression with other genes related to epilepsy, we performed a co-expression network analysis based on gene expression data for the human brain throughout development from the BrainSpan database (http://www.brainspan.org/). An obvious co-expression relationship was observed between each of the two genes and epilepsy-associated genes obtained from the EpilepsyGene database (Fig. 3). DYNC1H1 was co-expressed (Pearson correlation coefficients ≥0.6) with 46 associated epilepsy genes among all co-expressed genes (significance p = 6.6 × 10−4, Fisher’s exact test), and in addition, 15 genes (significance p = 0.024, Fisher’s exact test) were considered high-confidence genes based on their relevance to epilepsy according to gene prioritization in the EpilepsyGene database (Fig. 3). In addition, we detected a co-expression relationship between RTP1 and 17 genes associated with epilepsy (significance p = 0.025, Fisher’s exact test), almost half of which belong to the high-confidence genes (significance p = 0.012, Fisher’s exact test).
Figure 3.
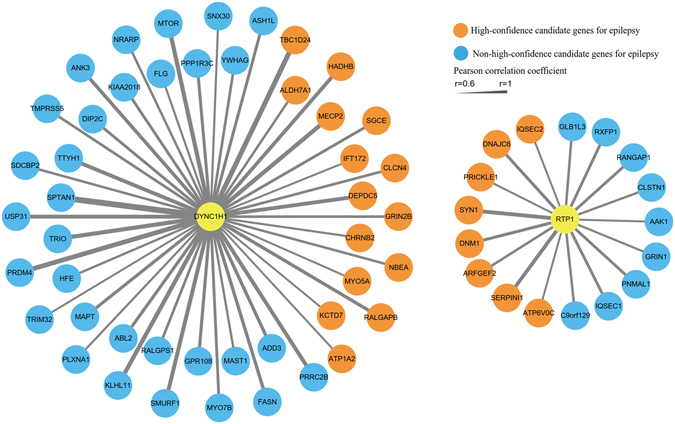
Co-expression network analysis of DYNC1H1 and RTP1. Gene co-expression levels were estimated using the Pearson correlation coefficients (r) between each pair of genes. Candidate epilepsy genes were extracted from the EpilepsyGene database.
Furthermore, to investigate the functional association between the two genes and other genes associated with epilepsy, we implemented a genetic interactions network analysis based on the interaction dataset collected from the GeneMANIA database. The results revealed that DYNC1H1 was associated with 41 epilepsy-associated genes obtained from the EpilepsyGene database to different extents (Fig. 4). Similarly, RTP1 displayed genetic interactions with 22 genes. Within this genetic interaction network, 12 high-confidence genes associated with epilepsy were detected in the network analysis of DYNC1H1, in addition to 9 genes for RTP1 (Fig. 4). Furthermore, five shared genes existed between the genetic interaction networks for DYNC1H1 and RTP1. In particular, CBL and KCNQ3, two shared genes, were considered high-confidence genes associated with epilepsy (Fig. 4). Overall, the co-expression and genetic interactions network analyses indicated that DYNC1H1 and RTP1 may share a similar role with several candidate epilepsy genes.
Figure 4.
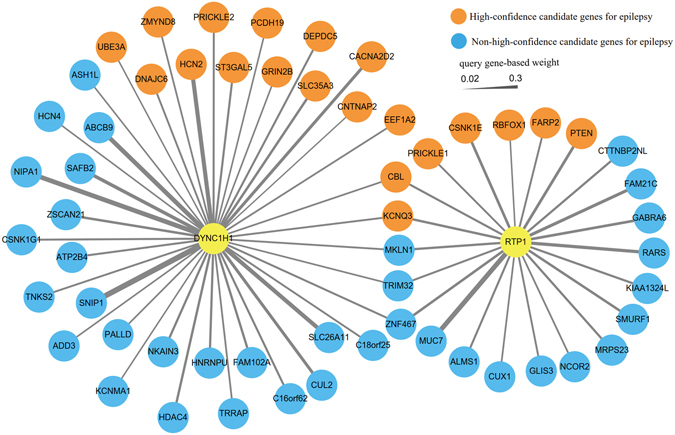
Genetic interaction network analysis of DYNC1H1 and RTP1. A node represents genes, and an edge represents interactions between each pair of genes. Candidate epilepsy genes were extracted from the EpilepsyGene database.
Discussion
The DYNC1H1 gene is located in chromosome 14q32 and was previously implicated in both central and peripheral neuronal dysfunction27, associated with syndromes involving spinal nerve degeneration and brain developmental problems28. In a previous study, frequent recurrence of mutations in DYNC1H1 (8 different de novo mutations in 16 unrelated probands) was described in a cohort of individuals affected by the malformations of cortical development (MCD) disorder using WES29. Interestingly, among the probands with de novo mutations in the DYNC1H1 gene, one proband had clinical symptoms of EE (LGS), and 4 additional probands also possessed epileptic features. Another recent study reported a novel de novo mutation in DYNC1H1 presenting with the clinical phenotype of sporadic congenital Spinal Muscular Atrophy-Lower Extremity Dominant (SMA-LED) and EE30. In addition, the DYNC1H1 gene tends to be intolerant to functional genetic variation based on RIVS and Z score for missense from ExAC. Furthermore, the detected de novo mutation (c.10174A > G, p.M3392V) in DYNC1H1 was predicted as “deleterious” based on multiple bioinformatics algorithms and was not observed in any large control population variation databases, including ExAC, 1000 Genomes, dbSNP 142 and ESP databases. These observations suggested that the de novo mutation in DYNC1H1 may play an important role in EE.
Notably, de novo mutations in the DYNC1H1 are recurrently detected based on WES studies in probands with different sporadic neuropsychiatric disorders, such as EE, ID and ASD31. In a recent study, in 2 unrelated patients with severe ID syndromes, 2 different de novo mutations were identified in the DYNC1H1 gene (Fig. 1b and Table 2)27. Notably, one 51-year-old patient with severe ID who developed generalized epileptic seizures at the age of 3 years27. Moreover, other 6 different nonsynonymous de novo mutations were detected in DYNC1H1 in 6 different trios from 3 different cohorts associated with ASD (Fig. 1b and Table 2)15, 32, 33. Indeed, the shared genetic etiology between EE and other neuropsychiatric disorders, including ASD, and ID, has previously been described34, 35. These results suggest that the de novo mutations of DYNC1H1 are shared among EE, ASD and ID, which indicate its possible contribution to these different genetic disorders.
Table 2.
The shared de novo mutations of DYNC1H1 among ASD, ID and EE.
| Chr | Mutation | Protein change | Mutation type | Disorder | Method | Reference |
|---|---|---|---|---|---|---|
| Chr14 | c.2467G > T | p.Val823Leu | SNV | ASD | WES | Rubeis S et al. Nature33 |
| Chr14 | c.6698C > T | p.Ala2233Val | SNV | ASD | WES | Rubeis S et al. Nature33 |
| Chr14 | c.7718A > G | p.Asp2573Gly | SNV | ASD | WES | Rubeis S et al. Nature33 |
| Chr14 | c.12797A > T | p.Asn4266Ile | SNV | ASD | WES | Rubeis S et al. Nature33 |
| Chr14 | c.12214G > A | p.Gly4072Ser | SNV | ASD | WES | Iossifov I et al. Nature33 |
| Chr14 | c.925C > T | p.Arg309Cys | SNV | ASD | WES | Krumm N et al. Nat Genet32 |
| Chr14 | c.11465A > C | p.His3822Pro | SNV | ID | WES | Willemsen MH et al. J Med Genet27 |
| Chr14 | c.4552C > T | p. Glu1518 Lys | SNV | ID | WES | Willemsen MH et al. J Med Genet27 |
| Chr14 | c.10174A > G | p.Met3392Val | SNV | EE | WES | In this study |
According to previous research, most of the de novo mutations in DYNC1H1 were identified in functional domains in ASD, ID and EE15, 17, 32, 33, with the exception of one variant in ID (c.11465A > C, p.H3822P) and one variant in ASD (c.12797A > T, p.N4266I). These results indicate that these domains play an important role in the function of DYNC1H1. The de novo Met3392Val mutation in DYNC1H1 identified in this study is located in the stalk domain derived from two of the coiled-coil domains and is considered to maintain dynein function and neuronal physiology. Four unrelated patients with MCD disorder carrying de novo mutations in the stalk domain and showing obvious clinical symptoms of EE and early onset epilepsy were recently reported29. This finding indicated that disruption of the stalk domain might affect cortical development associated with the occurrence of epilepsy. We are unsure about the exact mechanisms for different genetics variants in DYNC1H1 underlying the different phenotypes as mentioned above. However, location and nature of the different mutations, genetic background, and other complex factors including environmental influence, mutation timing and epigenetic factors could affect the genotype to phenotype relationship36–39.
One deleterious de novo missense mutation (c.323T > A) in the RTP1 gene was identified in another proband. RTP1 encodes a member of the receptor transporter protein family and can specifically induce functional cell surface expression of odorant receptors in human embryonic kidney cells40. Thus far, this gene has not been clearly associated with any neurodevelopmental disorder according to our knowledge. Nevertheless, one de novo mutation (c.334A > G; p.M112V) in RTP1 was recently described in an EE proband in a cohort of 264 trios based on WES9. Although RTP1 does not tend to be evolutionarily constrained based on RIVS or Z score for missense variant from ExAC, RTP1 is most likely a haploinsufficient gene. Furthermore, this mutation in RTP1 (c.323T > A) was not observed in any large control population through searching public variation database including ExAC, ESP and 1000 Genomes. These observations indicated that RTP1 may be a potential candidate gene for EE.
In recent years, WES has proved to be an efficient diagnostic approach for Mendelian diseases41. However, some limitations in WES technique impede high-efficiency detection for INDELs and CNVs, and de novo mutations from noncoding regions of UTR, introns, or intergenic regions, and synonymous mutations are usually ignored42. In addition, for some GC-rich regions, it is difficult to obtain effective sequencing coverage and depth of sequencing for variant detection43. These factors are likely to prevent the detection of possible deleterious mutations. The solution of these problems generally includes algorithm improvement, increase of read depth in target regions and application of whole-genome sequencing (WGS)42, 43. Nevertheless, it should be noted that approximately 85% of known disease-causing variants are in the coding regions44, 45. WES still represents a powerful tool for diagnosis and is adopted by many studies41, 42.
Methods
Patient recruitment
We collected samples from 4 unrelated West syndrome trios of Han Chinese ancestry from the Second Affiliated Hospital & Yuying Children’s Hospital of Wenzhou Medical University. This study protocol was approved by the Hospital Ethics Committee, and was carried out in accordance with the approved guidelines. Written informed consent was obtained from all participants or their caregivers prior to peripheral blood and clinical data collection. These cases comprised 3 males and 1 females with obvious clinical symptoms of West syndrome, characterized by refractory seizures and cognitive arrest or regression. All epilepsy diagnoses were evaluated based on an assessment of clinical seizure presentation and EEG recorded by a pediatric neurologist with experience in the clinical diagnosis of epileptic encephalopathies.
Whole-exome sequencing
Genomic DNA samples were extracted from 1 ml of peripheral blood leukocytes obtained from the parents and probands using the QIAGEN DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). DNA quality and quantity were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Approximately 2 µg of high-quality genomic DNA from each sample was prepared as the starting material for generating the sequencing library using the Agilent SureSelect Library Prep Kit according to the manufacturer’s instructions. Through target enrichment of DNA samples and construction of a hybridization library, the whole exome was captured using the Agilent SureSelect Human All Exon v5 Kit (Agilent Technologies, Santa Clara, CA, USA). In addition, all sheared DNA samples were tagged through PCR using an index (barcode) sequence. After the purification and quality assessment of the sample libraries, the samples were pooled according to mass, followed by multiplex sequencing using an Illumina HiSeq2000 sequencer (San Diego, CA, USA.) with 101 bp paired-end reads.
Data processing and de novo mutation detection
The raw sequencing reads generated through WES for the 12 individual samples were processed to remove sequence adapters and low-quality reads using the Trim_Galore program. Only sequencing reads with Phred-scaled quality scores greater than 30 and read lengths greater than 80 bp were retained for further analysis. The sequencing reads were aligned to the human reference genome (GRCH37/hg19) using the BWA program (version 0.7.12). After read mapping, duplicated reads and reads mapped to multiple genome locations were removed with the Picard software tool. GATK tools were used for the read realignment, quality score recalibration and SNP/InDel variant calling. Two software tools (ForestDNM and mirTrios) were used for de novo mutation detection, and mutations indicated to be de novo using both tools were considered as reliable de novo mutations for further analysis in the present study.
De novo mutation annotation and evaluation of the tolerance of genes
All variants were annotated using the ANNOVAR software tool and in-house codes. The consequences of mutations for protein products were inferred based on RefSeq gene annotations (http://www.ncbi.nlm.nih.gov/refseq/). Functional predictions for missense variants were obtained using 4 software tools: SIFT, LRT, SiPhy, and VEST3. A missense variant was considered deleterious based on the concordance of four generic damage prediction tools. For each sequence variant, the allele frequency (AF) was obtained from normal population variant databases, including dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), 1000 Genomes (http://www.1000genomes.org/), ESP6500 (http://evs.gs.washington.edu/EVS/), ExAC (http://exac.broadinstitute.org/), CG69 (http://www.completegenomics.com/public-data/69-genomes/) and GWAS Catalog (https://www.ebi.ac.uk/gwas/). Variants with AF <0.001 according to 1000 Genomes, ESP6500 and ExAC were defined as rare variants.
To assess the intolerance of genes to functional genetic variation, we utilized the predicted haploinsufficient probability26, residual variation intolerance score (RVIS) based on the ESP650025 dataset and Z score for missense variants from ExAC22.
Primers and Sanger sequencing validation
To validate each potential de novo mutation detected in the present study, 100 ng of genomic DNA from each sample was used to amplify the target mutation region through conventional PCR and Sanger sequencing. All primers in these assays are listed in Supplemental Table 3.
Gene co-expression and genetic interaction network
Gene expression data were obtained from the BrainSpan (http://www.brainspan.org/) database, which shows gene expression levels for specific brain regions obtained through RNA sequencing and exon microarray techniques. Gene co-expression levels were estimated based on the Pearson correlation coefficients (r) between each pair of genes. We extracted each pair of genes exhibiting high co-expression correlation coefficient (absolute r ≥ 0.6) compared with target genes and candidate epilepsy genes in the EpilepsyGene database46. Enrichment analysis of co-expressed epilepsy genes by using one-sided Fisher’s Exact test. To generate a human genetic interaction network, the direct protein-protein interaction dataset was collected from GeneMANIA47, based on the InWeb database or the Cytoscape program plugin48. In addition, GeneMANIA was employed to calculate the number of nodes and edges between the target genes and candidate epilepsy genes from the EpilepsyGene database. In the genetic interaction network, a node represents genes, and an edge represents interactions between two genes. The thickness of the edge represents the degree of genetic interaction. Both gene co-expression and the genetic interaction network were visualized using the Cytoscape program.
Electronic supplementary material
Acknowledgements
This work was financially supported through grants from the National Natural Science Foundation of China (31401110/C060605 and 81501824), and Natural Science Foundation of Zhejiang Province (LY15C090004).
Author Contributions
Z.D.L., Z.W.L., Z.W. and Y.L. designed the experiments; Z.D.L., X.C.L. and F.L. performed the experiments; Z.W.L. Y.H. and B.Y.C. analyzed the data; F.L. and Z.W. contributed reagents/materials/analysis tools; Z.D.L., Z.W.L., and Y.L. contributed to the writing of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Zhongdong Lin and Zhenwei Liu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00208-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhen Wang, Email: yuchen6046@163.com.
Yong Liu, Email: liuy5599@mail.xjtu.edu.cn.
References
- 1.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 2.Kato M, et al. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome) Am J Hum Genet. 2007;81:361–366. doi: 10.1086/518903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caraballo R, et al. Long-term follow-up of the ketogenic diet for refractory epilepsy: multicenter Argentinean experience in 216 pediatric patients. Seizure. 2011;20:640–645. doi: 10.1016/j.seizure.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Stromme P, et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 5.Weaving LS, et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saitsu H, et al. Dominant-negative mutations in alpha-II spectrin cause West syndrome with severe cerebral hypomyelination, spastic quadriplegia, and developmental delay. Am J Hum Genet. 2010;86:881–891. doi: 10.1016/j.ajhg.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamien BA, Cardamone M, Lawson JA, Sachdev R. A genetic diagnostic approach to infantile epileptic encephalopathies. J Clin Neurosci. 2012;19:934–941. doi: 10.1016/j.jocn.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Paciorkowski AR, Thio LL, Dobyns WB. Genetic and biologic classification of infantile spasms. Pediatr Neurol. 2011;45:355–367. doi: 10.1016/j.pediatrneurol.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemke JR, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol. 2014;75:147–154. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura K, et al. De Novo mutations in GNAO1, encoding a Galphao subunit of heterotrimeric G proteins, cause epileptic encephalopathy. Am J Hum Genet. 2013;93:496–505. doi: 10.1016/j.ajhg.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohba C, et al. De novo KCNT1 mutations in early-onset epileptic encephalopathy. Epilepsia. 2015;56:e121–128. doi: 10.1111/epi.13072. [DOI] [PubMed] [Google Scholar]
- 13.Writzl K, et al. Early onset West syndrome with severe hypomyelination and coloboma-like optic discs in a girl with SPTAN1 mutation. Epilepsia. 2012;53:e106–110. doi: 10.1111/j.1528-1167.2012.03437.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Lin MC, Kornblum HI, Papazian DM, Nelson SF. Exome sequencing identifies de novo gain of function missense mutation in KCND2 in identical twins with autism and seizures that slows potassium channel inactivation. Hum Mol Genet. 2014;23:3481–3489. doi: 10.1093/hmg/ddu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 17.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 18.Xu B, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nava C, et al. De novo mutations in HCN1 cause early infantile epileptic encephalopathy. Nat Genet. 2014;46:640–645. doi: 10.1038/ng.2952. [DOI] [PubMed] [Google Scholar]
- 21.Veeramah KR, et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia. 2013;54:1270–1281. doi: 10.1111/epi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand MS, et al. Recent advances in the molecular genetics of epilepsy. J Med Genet. 2013;50:271–279. doi: 10.1136/jmedgenet-2012-101448. [DOI] [PubMed] [Google Scholar]
- 25.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willemsen MH, et al. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet. 2012;49:179–183. doi: 10.1136/jmedgenet-2011-100542. [DOI] [PubMed] [Google Scholar]
- 28.Fiorillo C, et al. Novel dynein DYNC1H1 neck and motor domain mutations link distal spinal muscular atrophy and abnormal cortical development. Hum Mutat. 2014;35:298–302. doi: 10.1002/humu.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirier K, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh J, et al. Spinal Muscular Atrophy-Lower Extremity Dominant (SMA-LED), with bilateral perisylvian polymicrogyria and infantile epileptic encephalopathy, due a novel DYNC1H1 mutation. Neuromuscular Disorders. 2015;25:S222–S223. doi: 10.1016/j.nmd.2015.06.139. [DOI] [Google Scholar]
- 31.Lee BH, Smith T, Paciorkowski AR. Autism spectrum disorder and epilepsy: Disorders with a shared biology. Epilepsy Behav. 2015;47:191–201. doi: 10.1016/j.yebeh.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. A Cross-Disorder Method to Identify Novel Candidate Genes for Developmental Brain Disorders. JAMA Psychiatry. 2016;73:275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21:290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo, S. et al. Identification of Novel Compound Mutations in PLA2G6-Associated Neurodegeneration Patient with Characteristic MRI Imaging. Mol Neurobiol (2016). [DOI] [PubMed]
- 37.Miceli F, et al. Genotype-phenotype correlations in neonatal epilepsies caused by mutations in the voltage sensor of K(v)7.2 potassium channel subunits. Proc Natl Acad Sci USA. 2013;110:4386–4391. doi: 10.1073/pnas.1216867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobow K, El-Osta A, Blumcke I. The methylation hypothesis of pharmacoresistance in epilepsy. Epilepsia. 2013;54(Suppl 2):41–47. doi: 10.1111/epi.12183. [DOI] [PubMed] [Google Scholar]
- 39.Zweier C, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Fogel BL, Satya-Murti S, Cohen BH. Clinical exome sequencing in neurologic disease. Neurol Clin Pract. 2016;6:164–176. doi: 10.1212/CPJ.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tetreault M, Bareke E, Nadaf J, Alirezaie N, Majewski J. Whole-exome sequencing as a diagnostic tool: current challenges and future opportunities. Expert Rev Mol Diagn. 2015;15:749–760. doi: 10.1586/14737159.2015.1039516. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X. Exome sequencing greatly expedites the progressive research of Mendelian diseases. Front Med. 2014;8:42–57. doi: 10.1007/s11684-014-0303-9. [DOI] [PubMed] [Google Scholar]
- 44.Chong JX, et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am J Hum Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahdieh N, Rabbani B. An overview of mutation detection methods in genetic disorders. Iran J Pediatr. 2013;23:375–388. [PMC free article] [PubMed] [Google Scholar]
- 46.Ran X, et al. EpilepsyGene: a genetic resource for genes and mutations related to epilepsy. Nucleic Acids Res. 2015;43:D893–899. doi: 10.1093/nar/gku943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


