Abstract
Costus pictus, belonging to the family Costaceae, is one of the valuable medicinal plants with its anti-diabetic property. Despite ever-increasing demand from the pharmaceutical industry, this species is being less exploited at molecular level. Hence, an effort has been made in the present study to characterize the 15 accessions of C. pictus collected from different geographical regions of India through random amplified polymorphic DNA (RAPD) and inter-simple sequence repeats (ISSR) markers. A total of 25 RAPD and 20 ISSR primers were used in the present study. The RAPD analysis generated 343 loci, of which 124 were polymorphic with an average of 4.96 loci per primer. While, ISSR primers produced 177 loci, of which 77 were polymorphic with an average of 3.85 loci per primer. The similarity coefficients ranged from 0.86–0.99, 0.84–0.95 and 0.86–0.96 for RAPD, ISSR and combined RAPD-ISSR, respectively. The UPGMA dendrogram generated using these data showed low level of divergence among the accessions from South and West regions. Further, accession-specific bands were also revealed by RAPD and ISSR markers which might be contributed to specific trait. This investigation was an understanding of genetic variation within the C. pictus accessions. The present finding indicates that both the marker tools RAPD and ISSR combined or individually can be used in determining the genetic relationship between the accessions. It may be concluded that data of hereditary differences appeared among the C. pictus accessions could be utilized for their conservation and reproducing programs.
Keywords: Costus pictus, Genetic diversity, RAPD, ISSR, Polymorphism
Introduction
Interest in the utilization of medicinal and aromatic plants as pharmaceuticals, perfumes and cosmetics, flavoring, herbal and alternative natural products has been significantly expanded in the recent years (Ayensu 1996). Costus pictus D. Don is an important medicinal plant that belongs to family Costaceae, normally known as Spiral flag or Insulin plant. This plant is widely used by the tribal people of Kolli hills of Tamilnadu, to treat diabetes (Elavarasi and Saravanan 2012). C. pictus recently introduced in India from the American countries as an herbal care for diabetes and, therefore, commonly called as ‘insulin plant’ (Benny 2004). The monocot family Costaceae has close to seven genera and containing about 143 known species (Christenhusz and Byng 2016). This is believed to be native of tropical climates of Asia, Central America, South America and Africa. Recently, this plant gains very high medicinal interest due to its medicinal potential and shows various pharmacological properties such as anti-diabetic, diuretic effect, anti-oxidant, anti-cancers and putative activity (Hegde et al. 2014). Particularly, in south Indian region it is grown as ornamental plant.
Various important phytochemical properties have been reported from C. pictus plant including ascorbic acid, α-tocopherol, β-carotene, steroids, triterpinoids, alkaloids, tannins, saponins and flavonoids (Devi and Urooj 2010). It is also reported that leaves of this plant contain 21.2% fibers including significant amounts of the elements K, Ca, Cr, Mn, Cu, and Zn (Hegde et al. 2014). Because of its prospective use as valuable medicine, integration of pictus species into the agricultural forestry organizations would not only make accessible to majority of the backward populace that uses it but also pay attention towards its genetic conservation.
For improvement of the breeding and cultivation of plants, understanding its genetic assortment is very important. Morphological inspection is a very simple and direct way to study diversity, from which we cannot get significant genetic diversity among accessions (Nag et al. 2015). Cataloging of the various genera and species is largely based on morphological characteristics. However, these characters may not be significantly diverse and usually require growing plants by farmers up to the maturity prior to identification and also influenced by various environmental factors. Thus, it is essential to determine genetic diversity of such species so that only promising accessions can be multiplied and preserved.
Rapid advances in molecular marker systems have proven to be powerful tools in the assessment and evaluation of genetic diversity and in the interpretation of genetic relationships within and among species could help to breeders for genetic improvement of important medicinal plants (Grover and Sharma 2016). With the molecular approach, identification of particular plant accessions/genotypes seems to be more effective as compared to traditional morphological marker system as it directly accesses the plants hereditary information and makes it possible to understand the genetic relationship between individuals (Paterson et al. 1991). Random amplified polymorphic DNA (RAPD) marker system has been extensively used to distinguish the genetic variation at molecular level in several medicinal and aromatic plants (Ganie et al. 2012; Kumar et al. 2013; Khanna et al. 2014; Prajapat et al. 2015; Bhattacharyya and Kumaria 2015).
The advantages of RAPD markers are that they do not require prior sequence information, less expensive and can efficiently differentiate taxa below the species level, since it targets both coding and non-coding regions of the genome (Ganie et al. 2015). Other marker systems, namely inter-simple sequence repeats (ISSR) are similarly PCR-based molecular marker that amplifies genomic regions flanked by the SSR sequences repeat oriented in opposite orientation. RAPD and ISSR studies have been widely used in medicinal plants for genetic diversity analysis (Patel et al. 2015; Sarwat et al. 2008; Tripathi et al. 2012). This marker is highly polymorphic, reproducible and widely used for genetic diversity, gene tagging, phylogeny, evolutionary biology and genome mapping studies (Godwin et al. 1997).
As per our knowledge and literature survey, there is a lack of information on the molecular characterization of the Costus pictus plant. Thus, the present study was carried out with different accessions of C. pictus collected from various geographical regions of India using RAPD and ISSR markers.
Materials and methods
Plant material and DNA extraction
A total of fifteen C. pictus samples were collected from different geographical regions of India (Table 1). Two grams of fresh and young leaves was collected from each accession and rapidly frozen in liquid nitrogen for DNA extraction. DNA was isolated using the CTAB method as described by Doyle and Doyle (1990) with minor modification. Further, purity and concentration were measured using the Nanodrop (Thermo Scientific, USA) software.
Table 1.
Details of Costus pictus accessions collected from different regions of India for genetic diversity analysis
| S. no. | Accessions name | Place of collection | State | Latitude (°′N) | Longitude (°′E) |
|---|---|---|---|---|---|
| 1 | CP-1 | Howrah | West Bengal | 22°33′38.4″N | 88°17′20.1″E |
| 2 | CP-2 | Bhopal | Madhya Pradesh | 23°15′35.7588″N | 77°24′45.414″E |
| 3 | CP-3 | Panji | Goa | 15°29′07.4″N | N73°49′49.0″E |
| 4 | CP-4 | Hyderabad | Telangana | 17°23′6.1584″N | 78°29′12.0156″E |
| 5 | CP-5 | Navsari | Gujarat | 20°56′48.1272″N | 72°57′7.326″E |
| 6 | CP-6 | Daang | Gujarat | 20°49′31.3608″N | 73°42′2.5524″E |
| 7 | CP-7 | Vellore | Tamilnadu | 12°55′13.8″N | 79°07′32.3″E |
| 8 | CP-8 | Thekkady | Kerala | 9°36′11.1924″N | 77°9′41.2488″E |
| 9 | CP-9 | Bangalore | Karnataka | 12°58′17.7564″N | 77°35′40.4268″E |
| 10 | CP-10 | Nashik | Maharashtra | 19°59′50.8308″N | 73°47′23.2872″E |
| 11 | CP-11 | Vansada | Gujarat | 20°45′28.5552″N | 73°21′50.5368″E |
| 12 | CP-12 | Chennai | Tamilnadu | 13°4′57.648″N | 80°16′14.5848″E |
| 13 | CP-13 | Palode | Kerala | 8°43′27.9624″N | 77°1′29.2116″E |
| 14 | CP-14 | Dharwad | Karnataka | 15°27′32.1264″N | 77°0′0.0000″E |
| 15 | CP-15 | Nagpur | Maharashtra | 21°8′44.88″N | 79°5′17.358″E |
RAPD and ISSR amplification
RAPD amplification was performed using 10-mer short oligonucleotide primers randomly from operon series (Eurofins Genomic, India). PCR amplification was carried out with 20 µl reaction volume containing 1X Taq-PCR buffer with MgCl2 (Finnzymes, Thermo scientific, USA), 5 unit Taq Polymerase (Finnzymes, Thermo scientific, USA), 30 ng genomic DNA, 10 pM of primer (Eurofins Genomic, India), 200 µM dNTPs (Finnzymes, Thermo scientific, USA), in 200 µl PCR tubes (Axygen, USA) in thermocycler instrument (Eppendorf, Germany). RAPD-PCR mixture was initially denatured at 94 °C for 6 min, followed by 40 cycles of 94 °C for 1 min, 36–38 °C (depending on primer composition) for 1 min, 72 °C for 1 min, and final extension at 72 °C for 8 min. ISSR primers used in the present study were designated from the previous reports of Mohanty et al. (2014) and Verma et al. (2015). ISSR-PCR mixture was subjected to initial denaturation at 94 °C for 10 min followed by 35 cycles of denaturation at 94 °C for 1.3 min, annealing at 40–68 °C (depending on primer composition) for 1 min and extension at 72 °C for 2 min with a final extension at 72 °C for 10 min.
Agarose gel electrophoresis
Amplified PCR products of RAPD and ISSR were fractionated on 1.5% agarose in 1X Tris–borate–EDTA buffer for 1.5 h. DNA bands were visualized by staining with ethidium bromide (EtBr) and photographed on a UV transilluminator using gel documentation system (Bio-Rad, USA). Molecular size of the amplified product was estimated using a known molecular 100 + 500 bp DNA Ladder (Thermo fisher, USA). The experiment was repeated twice with each primer and only those results were considered in which repeatable patterns (DNA fingerprint) were observed.
Data analysis
Qualitative scoring of bands was done from gel photographs obtained from RAPD and ISSR analysis. Clearly reproducible bands from each accession were scored as unit character and data were recorded in a binary matrix (‘0’ for absence and ‘1’ for presence) sheet. The binary data obtained from the marker systems were analyzed individually using Dice’s coefficient (Dice 1945) to determine pairwise comparison to estimate the genetic relationship among the accessions. Genetic similarity coefficient was computed using the Dice’s coefficient with the NTSYS-pc software (Rohlf 1998). Dendrogram was constructed using the unweighted pair group method with arithmetic averages (UPGMA) algorithms in the NYSYSpc 2.02 based on the genetic distance of Nei and Li (1979). To judge primer efficiency, polymorphism information content (PIC) was calculated according to the formula described by Garcia et al. (2004) PIC = 1 − ∑f 2, where “f” is the frequency of “ith” allele. EIGEN module of NTSYSpc 2.02 software was used to analyze principal component analysis (PCA) and results were graphically expressed in the form of 3D plots. The Mantel matrix correspondence test for RAPD and ISSR markers was carried out using the MXCOMP function of the NTSYSpc 2.02 (Mantel 1967).
Results
Performance of marker systems
RAPD marker analysis
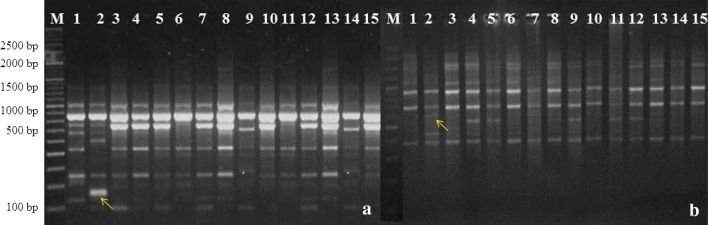
A total of 35 primers were initially screened for their ability to produce polymorphic patterns and only 25 of them were selected which gave reproducible and distinct polymorphic amplified products. The data collected from random amplification of polymorphic DNA (RAPD), detected total of 343 loci in 15 accessions, out of which 124 (35.57%) were polymorphic (Table 2). The molecular size of the amplified PCR products ranged from 110 (OPM-14; OPN-02) to 3190 bp (OPN-16). Moreover, the RAPD primer OPK-16 produced the maximum number of 24 loci, while OPN-08 produced the minimum number of 7 loci. The average polymorphic loci were found to be 4.96 per primer. The RAPD primer OPK-20 gave the highest polymorphism, while the lowest polymorphism was detected by the OPK-07 primer (Table 2). The average PIC (Polymorphism Information Content) value for RAPD was 0.91. The highest PIC value obtained was 0.95 for OPK-16 marker and the lowest PIC value was 0.85 for OPN-8. In the present study, a total of 25 RAPD primers examined, from which five primers (OPK-18; OPN-6; OPN-7; OPM-9; OPN-10) produced accession-specific amplicons (Fig. 1a; Table 4).
Table 2.
Numerical data as obtained from PCR amplification by RAPD primers among Costus pictus accessions
| Sr. no. | Primer name | Primer sequence 5′–3′ | GC content (%) | Annealing temperature | Total number of bands | Total number of loci | Monomorphic loci | Polymorphic loci | Polymorphism % | PIC | Molecular weight (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | OPK-07 | AGCGAGCAAG | 60 | 36 | 133 | 10 | 8 | 2 | 20.00 | 0.89 | 220–1500 |
| 2 | OPK-16 | GAGCGTCGAA | 60 | 36 | 283 | 24 | 15 | 9 | 37.50 | 0.95 | 160–2670 |
| 3 | OPK-18 | CCTAGTCGAG | 60 | 36 | 159 | 14 | 8 | 6 | 42.86 | 0.92 | 190–2580 |
| 4 | OPK-20 | GTGTCGCGAG | 70 | 38 | 154 | 16 | 8 | 8 | 50.00 | 0.91 | 280–2200 |
| 5 | OPM-01 | GTTGGTGGCT | 60 | 36 | 184 | 17 | 10 | 7 | 41.18 | 0.93 | 230–2640 |
| 6 | OPM-02 | ACAACGCCTC | 60 | 36 | 128 | 12 | 7 | 5 | 41.67 | 0.90 | 220–1720 |
| 7 | OPM-03 | GGGGGATGAG | 70 | 38 | 121 | 13 | 7 | 6 | 46.15 | 0.89 | 260–2930 |
| 8 | OPM-04 | GGCGGTTGTC | 70 | 38 | 144 | 11 | 8 | 3 | 27.27 | 0.91 | 160–1800 |
| 9 | OPM-05 | GGGAACGTG | 67 | 37 | 156 | 14 | 8 | 6 | 42.86 | 0.90 | 120–2200 |
| 10 | OPM-06 | CTGGGCAACT | 60 | 36 | 139 | 12 | 8 | 4 | 33.33 | 0.90 | 130–3125 |
| 11 | OPM-07 | CCGTGACTCA | 60 | 36 | 138 | 12 | 7 | 5 | 41.67 | 0.90 | 245–2065 |
| 12 | OPM-08 | TCTGTTCCCC | 60 | 36 | 151 | 13 | 7 | 6 | 46.15 | 0.91 | 240–2100 |
| 13 | OPM-09 | GTCTTGCGGA | 60 | 36 | 162 | 13 | 10 | 3 | 23.08 | 0.91 | 160–2900 |
| 14 | OPM-14 | TCTGGCGCAC | 70 | 38 | 115 | 9 | 7 | 2 | 22.22 | 0.88 | 110–1350 |
| 15 | OPN-02 | ACCAGGGGCA | 70 | 38 | 109 | 9 | 6 | 3 | 33.33 | 0.87 | 110–2200 |
| 16 | OPN-04 | GACCGACCCA | 70 | 38 | 245 | 19 | 14 | 5 | 26.32 | 0.94 | 141–1600 |
| 17 | OPN-06 | GAGACGCACA | 60 | 36 | 128 | 12 | 7 | 5 | 41.67 | 0.90 | 190–3110 |
| 18 | OPN-07 | CAGCCCAGAG | 70 | 38 | 198 | 19 | 11 | 8 | 42.11 | 0.93 | 140–1050 |
| 19 | OPN-08 | ACCTCAGCTC | 60 | 36 | 92 | 7 | 5 | 2 | 28.57 | 0.85 | 390–1410 |
| 20 | OPN-09 | TGCCGGCTTG | 70 | 38 | 154 | 15 | 9 | 6 | 40.00 | 0.91 | 240–2480 |
| 21 | OPN-10 | ACAACTGGGG | 60 | 36 | 239 | 20 | 14 | 6 | 30.00 | 0.94 | 140–1860 |
| 22 | OPN-13 | AGCGTCACTC | 60 | 36 | 102 | 9 | 6 | 3 | 33.33 | 0.87 | 330–2160 |
| 23 | OPN-15 | CAGCGACTGT | 60 | 36 | 166 | 14 | 10 | 4 | 28.57 | 0.92 | 200–2420 |
| 24 | OPN-16 | AAGCGACCTG | 60 | 36 | 149 | 14 | 8 | 6 | 42.86 | 0.91 | 250–3190 |
| 25 | OPN-18 | GGTGAGGTCA | 60 | 36 | 194 | 15 | 11 | 4 | 26.67 | 0.93 | 170–3000 |
| Total | 3943 | 343 | 219 | 124 | – | – | – | ||||
| Average/primer | 157.72 | 13.72 | 8.76 | 4.96 | 35.57 | 0.91 | 110–3190a | ||||
aRange
Fig. 1.
a RAPD profile in Costus pictus accessions generated by OPM-09 and b ISSR profile in Costus pictus accessions generated by UBC-809. (M 100 + 500 bp DNA ladder, 1 CP-1, 2 CP-2*, 3 CP-3, 4 CP-4, 5 CP-5, 6 CP-6, 7 CP-7, 8 CP-8, 9 CP-9, 10 CP-10, 11 CP-11, 12 CP-12, 13 CP-13, 14 CP-14, 15 CP-15; *accession-specific band)
Table 4.
Accession-specific amplicons as revealed by RAPD and ISSR markers
| Accession code | Characterized by the RAPD markers (bp) | Characterized by the ISSR markers (bp) |
|---|---|---|
| CP-4 | OPK-18 (494) | UBC-847 (727) |
| CP-5 | OPN-6 (166) | – |
| CP-2 | OPN-7 (331) OPM-9 (188) |
UBC-848 (1,024) UBC-808 (1391) |
| CP-1 | OPN-10 (1855) | – |
RAPD-based cluster analysis
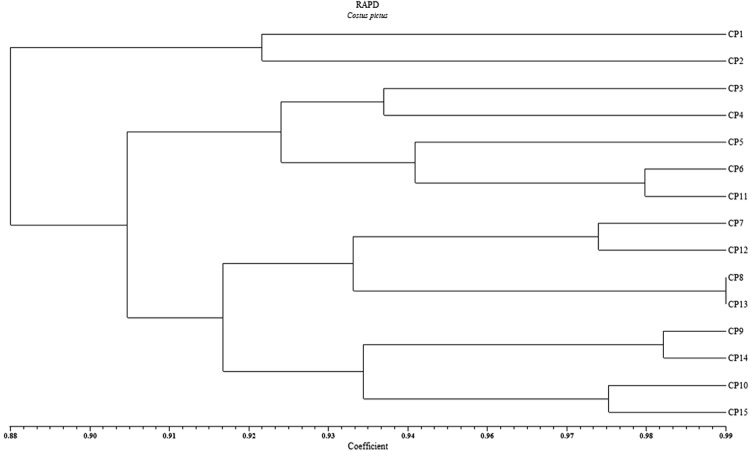
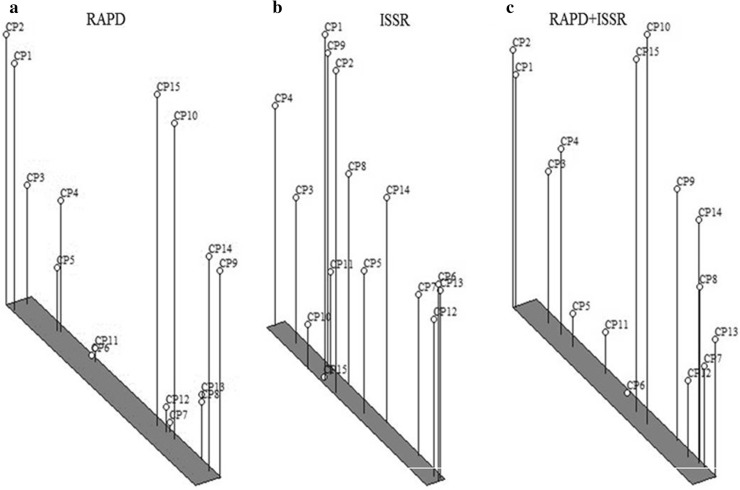
Genetic distances were obtained with UPGMA algorithm using Dice coefficient (Table 3). The results observed in the present study showed similarity coefficient ranged from 0.86 (between OC-1 and OC-9) to 0.99 (between OC-8 and OC-13) with an average value of 0.91. The UPGMA cluster analysis based on RAPD data grouped 15 accessions into two main groups (Fig. 2). The cophenetic correlation (Matrix correlation) for RAPD markers was computed and demonstrated a very good fit (r = 0.90). Group A consisted of single cluster I with two accessions CP-1 and CP-2. Group B sub divide into two clusters, cluster I consisted of 5 accessions and cluster II which enclosed 8 accessions illustrated similarity indices 92 and 91% respectively. Cluster I (B) consisted of two sub-clusters, I and II. Sub-cluster II includes CP-5, CP-6 and 11, which are of Gujarat region and CP-6 and CP-11 illustrated 97% similarity indices. Cluster II (B) include two sub-clusters each with 4 accessions, where in sub-cluster I accessions CP-8 and CP-13 showed 99% similarity coefficient.
Table 3.
Dice’s similarity coefficient based on RAPD analysis in 15 Costus pictus accessions
| Accessions | CP-1 | CP-2 | CP-3 | CP-4 | CP-5 | CP-6 | CP-7 | CP-8 | CP-9 | CP-10 | CP-11 | CP-12 | CP-13 | CP-14 | CP-15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP-1 | 1.00 | ||||||||||||||
| CP-2 | 0.92 | 1.00 | |||||||||||||
| CP-3 | 0.92 | 0.92 | 1.00 | ||||||||||||
| CP-4 | 0.91 | 0.90 | 0.94 | 1.00 | |||||||||||
| CP-5 | 0.90 | 0.89 | 0.93 | 0.94 | 1.00 | ||||||||||
| CP-6 | 0.88 | 0.88 | 0.92 | 0.93 | 0.95 | 1.00 | |||||||||
| CP-7 | 0.87 | 0.87 | 0.90 | 0.92 | 0.90 | 0.92 | 1.00 | ||||||||
| CP-8 | 0.88 | 0.87 | 0.89 | 0.90 | 0.90 | 0.92 | 0.94 | 1.00 | |||||||
| CP-9 | 0.86 | 0.87 | 0.88 | 0.90 | 0.90 | 0.91 | 0.92 | 0.94 | 1.00 | ||||||
| CP-10 | 0.89 | 0.88 | 0.89 | 0.91 | 0.90 | 0.90 | 0.91 | 0.92 | 0.94 | 1.00 | |||||
| CP-11 | 0.89 | 0.89 | 0.92 | 0.91 | 0.94 | 0.98 | 0.91 | 0.91 | 0.91 | 0.89 | 1.00 | ||||
| CP-12 | 0.88 | 0.88 | 0.90 | 0.90 | 0.89 | 0.91 | 0.97 | 0.93 | 0.92 | 0.90 | 0.92 | 1.00 | |||
| CP-13 | 0.88 | 0.87 | 0.89 | 0.90 | 0.90 | 0.91 | 0.94 | 0.99 | 0.94 | 0.91 | 0.91 | 0.94 | 1.00 | ||
| CP-14 | 0.87 | 0.87 | 0.88 | 0.89 | 0.90 | 0.90 | 0.91 | 0.93 | 0.98 | 0.94 | 0.91 | 0.92 | 0.93 | 1.00 | |
| CP-15 | 0.89 | 0.89 | 0.89 | 0.91 | 0.89 | 0.90 | 0.89 | 0.91 | 0.93 | 0.97 | 0.90 | 0.90 | 0.91 | 0.93 | 1.00 |
Bold values indicate higher and lower values of similarity index
Fig. 2.
UPGMA cluster analysis of 15 Costus pictus accessions with Dice’s similarity coefficient of RAPD
ISSR marker analysis
Among the 26 ISSR primers tested in the study, 20 primers based on di, tri or tetranucleotide repeats were further used to characterize C. pictus accessions. The present study showed that out of 177 loci, 77 (42.47%) loci were polymorphic showing an average of 3.85 polymorphic loci per primer tested. The maximum polymorphic loci were generated by primer UBC 818 and UBC 889. A total number of 177 loci, ranged from 200 bp (UBC 826) to 3070 bp (UBC 847), were produced using the 20 primers previously screened (Table 5). Among the primers studied, UBC 818 generated the highest 13 loci, while UBC 812 generated the lowest 5 loci with an average of 8.85 loci per primer. The percentage of polymorphism among tested primers was ranged from 20% for primers UBC 812 and 63.64% for primers UBC 889 with an average of 42.47% polymorphism per primer (Table 5). In this study, the highest PIC value of 0.91 for primer UBC 818 and the lowest PIC value of 0.77 for primer UBC 812, with an average value of PIC per primer 0.87, were obtained. Three ISSR primers viz., UBC-847, UBC-848 and UBC-808 produced accession-specific amplicons (Fig. 1b; Table 4).
Table 5.
Numerical data as obtained from PCR amplification by ISSR primers among Costus pictus accessions
| Sr. no | Primer name | Primer sequence 5′–3′ | GC content % | Annealing temperature | Total number of bands | Total number of loci | Monomorphic loci | Polymorphic loci | Polymorphism % | PIC value | Molecular weight (bp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | UBC 808 | AGAGAGAGAGAGAGAGC | 53 | 60 | 99 | 8 | 3 | 5 | 62.50 | 0.87 | 370–2025 |
| 2 | UBC 809 | AGAGAGAGAGAGAGAGG | 53 | 60 | 104 | 11 | 6 | 5 | 45.45 | 0.88 | 380–2190 |
| 3 | UBC 811 | GAGAGAGAGAGAGAGAC | 53 | 63 | 60 | 6 | 3 | 3 | 50.00 | 0.79 | 450–2515 |
| 4 | UBC 812 | GAGAGAGAGAGAGAGAA | 47 | 51 | 63 | 5 | 4 | 1 | 20.00 | 0.77 | 250–1700 |
| 5 | UBC 818 | CACACACACACACACAG | 53 | 53 | 253 | 13 | 6 | 7 | 53.85 | 0.91 | 260–2500 |
| 6 | UBC 826 | ACACACACACACACACC | 53 | 53 | 102 | 11 | 8 | 3 | 27.27 | 0.90 | 200–2220 |
| 7 | UBC 834 | AGAGAGAGAGAGAGAGYT | 44 | 52.5 | 113 | 10 | 6 | 4 | 40.00 | 0.88 | 320–1880 |
| 8 | UBC 835 | AGAGAGAGAGAGAGAGYC | 50 | 52.5 | 91 | 7 | 5 | 2 | 28.57 | 0.85 | 400–1790 |
| 9 | UBC 841 | GAGAGAGAGAGAGAGAY | 47 | 52.5 | 88 | 7 | 4 | 3 | 42.86 | 0.84 | 640–2830 |
| 10 | UBC 842 | GAGAGAGAGAGAGAGAYG | 50 | 53 | 122 | 9 | 4 | 5 | 55.56 | 0.89 | 355–2400 |
| 11 | UBC 847 | CACACACACACACACARC | 50 | 53 | 86 | 8 | 4 | 4 | 50.00 | 0.85 | 610–3070 |
| 12 | UBC 848 | CACACACACACACACARG | 50 | 52.5 | 101 | 8 | 6 | 2 | 25.00 | 0.86 | 335–2190 |
| 13 | UBC 852 | GTGTGTGTGTGTGTGTYC | 50 | 52 | 116 | 10 | 4 | 6 | 60.00 | 0.89 | 305–2725 |
| 14 | UBC 866 | CTCCTCCTCCTCCTCCTC | 67 | 57.5 | 94 | 9 | 4 | 5 | 55.56 | 0.87 | 305–2830 |
| 15 | UBC 880 | GGAGAGGAGAGGAGA | 60 | 55.5 | 115 | 9 | 6 | 3 | 33.33 | 0.88 | 400–2580 |
| 16 | UBC 883 | TATATATATATATABVB | 0 | 36.8 | 105 | 8 | 5 | 3 | 37.50 | 0.87 | 400–2180 |
| 17 | UBC 888 | CACACACACACACABDB | 41 | 50.5 | 105 | 8 | 6 | 2 | 25.00 | 0.87 | 390–2000 |
| 18 | UBC 889 | ACACACACACACACACDBD | 42 | 51.5 | 108 | 11 | 4 | 7 | 63.64 | 0.89 | 385–2560 |
| 19 | UBC 890 | CTCTCTCTCTCTCTVHV | 41 | 51 | 115 | 9 | 6 | 3 | 33.33 | 0.88 | 305–2770 |
| 20 | UBC 891 | TGTGTGTGTGTGTGHVH | 41 | 51 | 121 | 10 | 6 | 4 | 40.00 | 0.89 | 310–2680 |
| Total | 2161 | 177 | 100 | 77 | – | – | – | ||||
| Average/primer | 108.05 | 8.85 | 5 | 3.85 | 42.47 | 0.87 | 200–3070a | ||||
aRange
ISSR-based cluster analysis
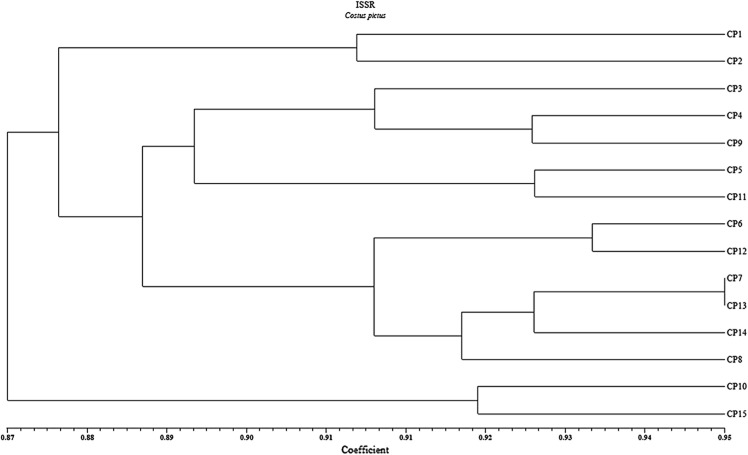
Similarity coefficient obtained by ISSR profile is shown in Table 6. Similarity coefficient was ranged from 0.84 to 0.95 with an average value of 0.88. In the present ISSR cluster analysis, the two most closely related accessions were found to be CP-8 and CP-13 (0.95), and the two most distantly related accessions were CP-1 and CP-15 with lowest similarity index (0.84). Cluster analysis of ISSR data based on the similarity matrix generated a dendrogram with two major groups (Fig. 3). The cophenetic correlation (Matrix correlation) for ISSR markers was computed and demonstrated a good fit (r = 0.74). Group B consisted of only one cluster with two populations CP-10 and CP-15 with 91% similarity indices. First group A was divided into two clusters I and II with 87% similarity; cluster I has two accessions and cluster II has 11 accessions. Second cluster (A) was divided into two sub-clusters: the first sub-cluster has 5 accessions and the second sub-cluster has 6 accessions with 91% similarity indices.
Table 6.
Dice’s similarity coefficient based on ISSR analysis in 15 Costus pictus accessions
| Accessions | CP-1 | CP-2 | CP-3 | CP-4 | CP-5 | CP-6 | CP-7 | CP-8 | CP-9 | CP-10 | CP-11 | CP-12 | CP-13 | CP-14 | CP-15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP-1 | 1.00 | ||||||||||||||
| CP-2 | 0.91 | 1.00 | |||||||||||||
| CP-3 | 0.87 | 0.86 | 1.00 | ||||||||||||
| CP-4 | 0.89 | 0.88 | 0.91 | 1.00 | |||||||||||
| CP-5 | 0.87 | 0.86 | 0.89 | 0.86 | 1.00 | ||||||||||
| CP-6 | 0.87 | 0.88 | 0.86 | 0.85 | 0.91 | 1.00 | |||||||||
| CP-7 | 0.87 | 0.87 | 0.88 | 0.88 | 0.88 | 0.92 | 1.00 | ||||||||
| CP-8 | 0.89 | 0.88 | 0.91 | 0.91 | 0.88 | 0.88 | 0.93 | 1.00 | |||||||
| CP-9 | 0.91 | 0.91 | 0.91 | 0.93 | 0.89 | 0.90 | 0.89 | 0.91 | 1.00 | ||||||
| CP-10 | 0.85 | 0.86 | 0.89 | 0.87 | 0.85 | 0.84 | 0.88 | 0.88 | 0.85 | 1.00 | |||||
| CP-11 | 0.86 | 0.85 | 0.91 | 0.91 | 0.93 | 0.88 | 0.88 | 0.89 | 0.89 | 0.87 | 1.00 | ||||
| CP-12 | 0.87 | 0.87 | 0.87 | 0.86 | 0.89 | 0.94 | 0.92 | 0.89 | 0.87 | 0.87 | 0.89 | 1.00 | |||
| CP-13 | 0.88 | 0.89 | 0.87 | 0.86 | 0.88 | 0.93 | 0.95 | 0.90 | 0.89 | 0.87 | 0.88 | 0.94 | 1.00 | ||
| CP-14 | 0.89 | 0.88 | 0.90 | 0.88 | 0.88 | 0.91 | 0.93 | 0.93 | 0.92 | 0.88 | 0.90 | 0.89 | 0.93 | 1.00 | |
| CP-15 | 0.84 | 0.85 | 0.89 | 0.89 | 0.88 | 0.87 | 0.89 | 0.88 | 0.85 | 0.92 | 0.91 | 0.88 | 0.88 | 0.87 | 1.00 |
Bold values indicate higher and lower values of similarity index
Fig. 3.
UPGMA cluster analysis of 15 Costus pictus accessions with Dice’s similarity coefficient of ISSR
Combined RAPD and ISSR analysis
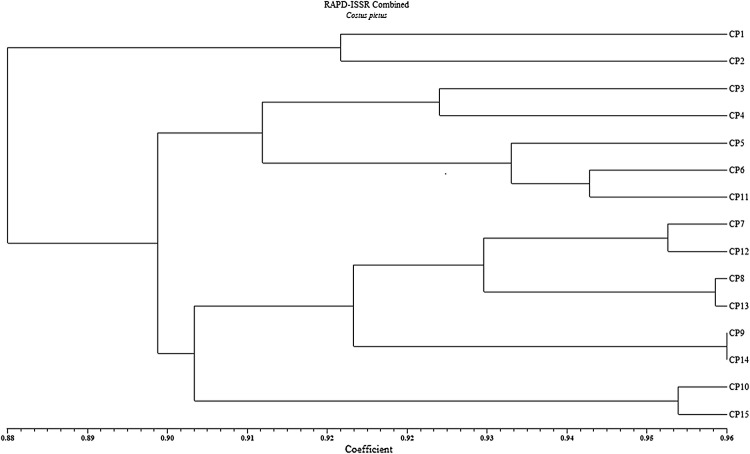
Further, to have an overview of the genetic similarities between the accessions, data of both the primers RAPD and ISSR were used to generate genetic diversity (Table 7). Based on the combined data, dendrogram indicated that all the accessions were divided into two major groups: A and B with 2 and 13 accessions, respectively (Fig. 4). The first group A had only one cluster with two accessions CP-1 and CP-2 having 92% similarities, while group B was divided into two major clusters, cluster I and cluster II with 89% similarities. Cluster I (B) and cluster II (B) both have two sub-clusters with the similarity indices, 91 and 90%. Similarity coefficient for combined RAPD-ISSR ranged from 0.86 to 0.96 with an average value of 0.90 (Table 7). The cophenetic correlation (Matrix correlation) was also calculated based on the combined data and indicated a very good fit (r = 0.90). The results showed that the two most closely related accessions were CP-9 (Karnataka) and CP-14 (Karnataka) with the highest similarity index (0.96). On the other hand, the most distinct accessions were CP-1 (Kolkata) and CP-15 (Maharashtra) at 0.86 similarity index.
Table 7.
Dice’s similarity coefficient based on combined RAPD-ISSR analysis in 15 Costus pictus accessions
| Accessions | CP-1 | CP-2 | CP-3 | CP-4 | CP-5 | CP-6 | CP-7 | CP-8 | CP-9 | CP-10 | CP-11 | CP-12 | CP-13 | CP-14 | CP-15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP-1 | 1.00 | ||||||||||||||
| CP-2 | 0.92 | 1.00 | |||||||||||||
| CP-3 | 0.90 | 0.90 | 1.00 | ||||||||||||
| CP-4 | 0.90 | 0.89 | 0.93 | 1.00 | |||||||||||
| CP-5 | 0.89 | 0.88 | 0.91 | 0.91 | 1.00 | ||||||||||
| CP-6 | 0.88 | 0.88 | 0.90 | 0.90 | 0.94 | 1.00 | |||||||||
| CP-7 | 0.87 | 0.87 | 0.89 | 0.90 | 0.90 | 0.92 | 1.00 | ||||||||
| CP-8 | 0.89 | 0.87 | 0.90 | 0.90 | 0.89 | 0.90 | 0.94 | 1.00 | |||||||
| CP-9 | 0.88 | 0.88 | 0.89 | 0.91 | 0.90 | 0.90 | 0.91 | 0.93 | 1.00 | ||||||
| CP-10 | 0.87 | 0.88 | 0.89 | 0.89 | 0.88 | 0.88 | 0.90 | 0.90 | 0.91 | 1.00 | |||||
| CP-11 | 0.88 | 0.87 | 0.91 | 0.91 | 0.93 | 0.95 | 0.90 | 0.90 | 0.90 | 0.89 | 1.00 | ||||
| CP-12 | 0.88 | 0.88 | 0.89 | 0.88 | 0.89 | 0.92 | 0.95 | 0.91 | 0.90 | 0.89 | 0.91 | 1.00 | |||
| CP-13 | 0.88 | 0.87 | 0.89 | 0.89 | 0.89 | 0.92 | 0.94 | 0.95 | 0.92 | 0.90 | 0.90 | 0.94 | 1.00 | ||
| CP-14 | 0.88 | 0.87 | 0.89 | 0.89 | 0.89 | 0.91 | 0.92 | 0.93 | 0.96 | 0.92 | 0.91 | 0.91 | 0.93 | 1.00 | |
| CP-15 | 0.86 | 0.87 | 0.89 | 0.90 | 0.89 | 0.89 | 0.89 | 0.90 | 0.90 | 0.95 | 0.90 | 0.89 | 0.90 | 0.91 | 1.00 |
Bold values indicate higher and lower values of similarity index
Fig. 4.
UPGMA cluster analysis of 15 Costus pictus accessions with Dice’s similarity coefficient combined RAPD + ISSR
Principal component analysis
Result of the PCA based on similarity coefficient of RAPD markers was comparable with its cluster analysis (Fig. 5). The PCA based on RAPD data revealed that the accessions belonging to a particular cluster were grouped together in the PCA plot (Fig. 5a). Interestingly, it showed that the accessions belong to same place state were clustered together, whereas some accessions also separated in PCA as presented by dendrogram. Similarly, ISSR-based PCA analysis revealed that the majority of accession was grouped as illustrated by the dendrogram (Fig. 5b). It is also evident from data of both the marker tools (RAPD + ISSR) that accessions were impartially distributed on PCA plots, which imitate a good genetic relatedness among the C. pictus accessions (Fig. 5c).
Fig. 5.
Three-dimensional plot of principal component analysis of Costus pictus accessions based on a RAPD, b ISSR and c combined (RAPD + ISSR) analysis
Discussion
Although work on biochemical (pharmacological properties) and morphological characterization of C. pictus plant has been attempted extensively, studies on genetic diversity are scanty. Therefore, efforts have been made in the present study to characterize fifteen accessions of C. pictus collected from different part of India, using random amplified polymorphic DNA (RAPD) and inter-simple sequence repeats (ISSR) markers.
Costus pictus is introduced from the America due to its rich indigenous medicinal properties, so that information on genetic structure and the diversity of populations is essential for their conservational program (Ganie et al. 2015). With the emergence of molecular biology techniques, DNA-based markers are now very useful tool for characterization of germplasm, assessment of genetic diversity, varietal identification, hybrid testing, clonal fidelity testing, genetic relationship, systemic and evolutionary biology, phylogenetic studies, gene tagging and marker-assisted selection (Huang and Han 2014). Molecular data can help the conservationists to investigate many tissues of concern such as gene pool fragmentation and genetic erosion (Barrett and Kohn 1991).
The present study on intrinsic genetic diversity analysis with both marker systems revealed that the higher level of polymorphism was obtained with ISSR (42.47%) than that of RAPD (35.57%). It was found that the degree of genetic diversity or polymorphism was relatively lower in C. pictus accessions, which indicates narrow gene pool. Our results are in accordance with the findings of Thomas and Palni (2016) who reported only 9.09% of polymorphism in six C. pictus accessions with RAPD markers. Another study conducted by Mandal et al. (2007) found 73.33% of polymorphism in 14 closely related accessions of Costus speciosus with RAPD markers. Moreover, low-level polymorphism in ISSR and RAPD markers was also reported in many previous studies, for examples, (26.43% RAPD; 24.36% ISSR) in Bruguiera gymnorrhiza and (14.4% RAPD; 12.73% ISSR) in Heritiera fomes (Dasgupta et al. 2015). Higher reproducibility and stability of ISSR system than RAPD were found by Zietkiewicz et al. (1994). However, it was noted in the present study that high reproducibility for both RAPD and ISSR markers can be achieved when the PCR conditions are well defined.
The present study showed low level of polymorphism in C. pictus accessions. This may be due to the less number of primers and/or accessions used in this study which may not sufficient to capture the diversity. In addition, we can assumed that C. pictus had been travelled from one location to another than shifted to longer distance, then adapted and correlated with that environmental conditions prevailed over there. For instance, adaptability and evolution of any species are based on the level of genetic diversity present in populations, which reflect the richness of diverse germplasm in a specific environment or location. The percentage of polymorphic loci can be used as indicator to measure the degree of genetic diversity in C. pictus. From the genetic perspective, a higher level of genetic diversity results in a greater ability to adapt and evolve (Li and Chen 2004). Low genetic variation could result in decreased adaptability and elevates the occurrence of less beneficial genes leading to eventual extinction of the species and thus the lower level of diversity within the populations should be paid special attention (Dasgupta et al. 2015).
The distribution of accessions to the dendrogram based on banding patterns of RAPD, ISSR and combined RAPD-ISSR may significantly differ because each technique amplifies different regions of genome. Interestingly, both the marker tools divided all C. pictus accessions into two main clusters. The combined RAPD-ISSR banding patterns increase the validity of the results as it allows to screen more segments site in the genome. Still, many studies concluded that ISSR would be a superior tool than RAPD for phylogenetic study (Marotti et al. 2007).
A close genetic similarity was observed in some of the accessions analyzed as shown by high values of similarity index. In this study, the average similarity index for Costus accessions using RAPD accessions was ranged from 0.87 to 0.99. While similarity index of the ISSRs ranged from 0.84 to 0.95. High range of similarity indices in both the samples for RAPD and ISSR markers clearly indicated that there was low genetic variability between accessions collected from different geographical regions. The similarity index for combined RAPD-ISSR markers ranged from 0.87 to 0.96, which is higher than the ISSR-based markers indicating less diversity, while its was lower than the RAPD marker. This finding is consistent with the study in Artemisia capillarys (Shafie et al. 2011).
Accessions collected from the Gujarat (CP-5, CP-6 and CP-11) regions showed very low genetic diversity and grouped into same cluster with 93% similarities. Same kind of results was observed from the accessions collected from the Tamilnadu (CP-7 and CP-12), Kerala (CP-8 and CP-13) and Karnataka (CP-9 and CP-14). Although, accessions collected from Kolkata (CP-1) and Madhya Pradesh (CP-2) were genetically distant from other accessions collected from South-West regions of India. Similar results were also reported in many plant species (Rabbani et al. 1998; Dias et al. 1993). It was noted that accession collected from South regions of India showed high similarities among them and formed single cluster II with 90% similarities.
Due to the same geographical location, a high genetic similarity was expected among C. pictus accessions in the southern part of country. But the broad genetic base observed in this study indicates an earlier introduction of this species to south, and subsequently leading to accumulation of variation. The clusters in dendrogram might be correlated some extent of the geographical distribution, as revealed in the present results. These results are in agreement with some previous study, which concluded that the accessions from diverse geographical regions can be genetically similar (Jain et al. 2003). Additionally, recent introduction of the C. pictus accessions in the collection site which are of same ancestor could be the reason for genetic similarities between accessions (Jose and Reddy 2010). Mantle matrix correspondence test revealed high matrix correlation in all the cases. Correlation values for RAPD, ISSR and combined data were 0.90, 0.74 and 0.90, respectively, which represent good to fit. The PCA was used to demonstrate the multiple dimension distribution of the accessions in a scatter-plot. Some differences were observed in the separation of individual accessions to their respective clusters as obtained from the UPGMA dendrogram as well as from PCA for both the markers. This multivariate approach was used to complement the information obtained from the cluster analysis methods because it is more informative regarding distances among major groups (Taran et al. 2005).
Because of its valuable medicinal properties, C. pictus needs more attention toward the conservation program. Sustainable management strategies can be developed by the understanding of the population structure of C. pictus. Even though there are no more molecular data to support the present analysis, it appears that identification based on the morphology alone cannot correctly designate different accessions of the same species. Studies at the molecular level are necessary to understand genetic diversity and relationships among accessions. Such discrepancies were also observed in rice (Virk et al. 1995).
Conclusion
On the basis of findings of present study, it can be concluded that genetic variability in the studied accessions of C. pictus is low, though accessions were collected from a diverse geographical regions This might be due to the deployment of a small set of markers and accessions to capture the diversity. However, accession-specific amplicons generated by RAPD and ISSR primers could be converted to co-dominant SCAR marker. As significant research has not been perform, there is a scope of research at genetic level to understand the genetic pools, to develop conservation strategies and breeding programs for this newly introduced plant. Thus, above results can be the beginning for future research to understand the genetic variability of C. pictus.
Acknowledgements
The authors are sincerely thankful to the Department of Plant Molecular Biology and Biotechnology, Navsari Agricultural University, Navsari, for providing all the necessary facilities to conduct this research.
Author contributions
AN Collected samples and conducted main experiment; PP Analyzed the data and critically reviewed the manuscript; RK Reviewed the manuscript and provided all the facilities to conduct this experiment; JMP Helped in experimental setup and analyzed the data.
Compliance with ethical standards
Conflict of interest
The authors declare that they do not have any conflict of interest.
References
- Ayensu ES. World medicinal plant resources. In: Chopra VL, Khoshoo TN, editors. Conservation for productive agriculture. New Delhi: ICAR; 1996. pp. 11–42. [Google Scholar]
- Barrett SCH, Kohn JR. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE, editors. Genetics and conservation of rare plants. Oxford: Oxford University Press; 1991. pp. 3–30. [Google Scholar]
- Benny M. Insulin plant in gardens. Nat Prod Rad. 2004;3:349–350. [Google Scholar]
- Bhattacharyya P, Kumaria S. Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Syst Evol. 2015;301(1):201–210. doi: 10.1007/s00606-014-1065-1. [DOI] [Google Scholar]
- Christenhusz MJ, Byng JW. The number of known plants species in the world and its annual increase. Phytotaxa. 2016;261(3):201–217. doi: 10.11646/phytotaxa.261.3.1. [DOI] [Google Scholar]
- Dasgupta N, Nandy P, Sengupta C, Das S. RAPD and ISSR marker mediated genetic polymorphism of two mangroves Bruguiera gymnorrhiza and Heritiera fomes from Indian Sundarbans in relation to their sustainability. Physiol Mol Biol Plants. 2015;21(3):375–384. doi: 10.1007/s12298-015-0308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi VD, Urooj A. Nutrient profile and antioxidant components of Costus specious Sm. and Costus igneus Nak. Indian J Nat Prod Resour. 2010;1:116–118. [Google Scholar]
- Dias JS, Monteiro AA, Lima MB. Numerical taxonomy of Portuguese Tronchuda cabbage and Galega Kale landraces using morphological characters. Euphytica. 1993;69:51–68. doi: 10.1007/BF00021725. [DOI] [Google Scholar]
- Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. doi: 10.2307/1932409. [DOI] [Google Scholar]
- Doyle LJ, Doyle JJ. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–14. [Google Scholar]
- Elavarasi S, Saravanan K. Ethnobotanical study of plants used to treat diabetes by tribal people of Kolli Hills, Namakkal District, Tamilnadu, Southern India. Int J Pharm Tech Res. 2012;4:404–411. [Google Scholar]
- Ganie SH, Srivastava PS, Narula A, Ali Z, Sharma MP. Authentication of shankhpushpi by RAPD markers. Eurasia J Biosci. 2012;6:39–46. doi: 10.5053/ejobios.2012.6.0.5. [DOI] [Google Scholar]
- Ganie SH, Upadhyay P, Das S, Sharma MP. Authentication of medicinal plants by DNA markers. Plant Gene. 2015;4:83–99. doi: 10.1016/j.plgene.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CL, Dsouza SAP. Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol. 2004;27(4):579–588. doi: 10.1590/S1415-47572004000400019. [DOI] [Google Scholar]
- Godwin ID, Aitken EA, Smith LW. Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis. 1997;18(9):1524–1528. doi: 10.1002/elps.1150180906. [DOI] [PubMed] [Google Scholar]
- Grover A, Sharma PC. Development and use of molecular markers: past and present. Crit Rev Biotechnol. 2016;36(2):290–302. doi: 10.3109/07388551.2014.959891. [DOI] [PubMed] [Google Scholar]
- Hegde PK, Rao HA, Rao PN. A review on Insulin plant (Costus igneus Nak) Pharmacogn Rev. 2014;8(15):67–72. doi: 10.4103/0973-7847.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- Jain N, Shasany AK, Sundaresan V, Rajkumar S, Darokar MP, Bagchi GD, Gupta AK, Kumar S, Khanuja SP. Molecular diversity in Phyllanthus amarus assessed through RAPD analysis. Curr Sci. 2003;85(10):1454–1458. [Google Scholar]
- Jose B, Reddy LJ. Analysis of the essential oils of the stems, leaves and rhizomes of the medicinal plant Costus pictus from southern India. Int J Pharm Pharm Sci. 2010;2(Suppl 2):100–101. [Google Scholar]
- Khanna PK, Chandra R, Kumar A, Dogra N, Gupta H, Gupta G, Verma V. Correlation between morphological, chemical and RAPD markers for assessing genetic diversity in Withania somnifera (L.) Dunal. J Crop Sci Biotechnol. 2014;17(1):27–34. doi: 10.1007/s12892-013-0104-8. [DOI] [Google Scholar]
- Kumar S, Sharma R, Kumar V, Vyas GK, Rathore A. Combining molecular-marker and chemical analysis of Capparis decidua (Capparaceae) in the Thar Desert of Western Rajasthan (India) Revista de Biología Trop. 2013;61(1):311–320. doi: 10.15517/rbt.v61i1.11130. [DOI] [PubMed] [Google Scholar]
- Li HS, Chen GZ. Genetic diversity of Sonneratia alba in China detected by Inter-simple Sequence Repeats (ISSR) analysis. Acta Bot Sin. 2004;46:512–521. [Google Scholar]
- Mandal AB, Thomas VA, Elanchezhian R. RAPD pattern of Costus speciosus Koen ex. Retz., an important medicinal plant from the Andaman and Nicobar Islands. Curr Sci. 2007;93(3):369–373. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209–220. [PubMed] [Google Scholar]
- Marotti I, Bonetti A, Minelli M, Catizone P, Dinelli G. Characterization of some Italian common bean (Phaseolus vulgaris L.) landraces by RAPD, semi-random and ISSR molecular markers. Genet Resour Crop Evol. 2007;54(1):175–188. doi: 10.1007/s10722-005-3133-4. [DOI] [Google Scholar]
- Mohanty S, Panda MK, Acharya L, Nayak S. Genetic diversity and gene differentiation among ten species of Zingiberaceae from Eastern India. 3 Biotech. 2014;4(4):383–390. doi: 10.1007/s13205-013-0166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Ahuja PS, Sharma RK. Genetic diversity of high-elevation populations of an endangered medicinal plant. AoB Plants. 2015;7:plu076. doi: 10.1093/aobpla/plu076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HK, Fougat RS, Kumar S, Mistry JG, Kumar M. Detection of genetic variation in Ocimum species using RAPD and ISSR markers. 3 Biotech. 2015;5(5):697–707. doi: 10.1007/s13205-014-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Tanksley SD, Sorreis ME. DNA markers in plant improvement. Adv Agron. 1991;46:39–90. doi: 10.1016/S0065-2113(08)60578-7. [DOI] [Google Scholar]
- Prajapat P, Sasidharan N, Ballani A. Assessment of Genetic Diversity in Four Brassica species using randomly amplified polymorphic DNA markers. Int J Agric Environ Biotechnol. 2015;8(4):831–836. doi: 10.5958/2230-732X.2015.00093.5. [DOI] [Google Scholar]
- Rabbani MA, Iwabuchi A, Murakami Y, Suzuki T, Takayanagi K. Phenotypic variation and the relationships among mustard (Brassica juncea L.) germplasm from Pakistan. Euphytica. 1998;101:357–366. doi: 10.1023/A:1018305201279. [DOI] [Google Scholar]
- Rohlf FJ (1998) NTSys-pc: numerical taxonomy and multivariate analysis system, version 2.02. Exter Software, Setauket
- Sarwat M, Das S, Srivastava PS. Analysis of genetic diversity through AFLP, SAMPL, SSR and RAPD markers in Tribulus terrestris, a medicinal herb. Plant Cell Rep. 2008;27(3):519–528. doi: 10.1007/s00299-007-0478-5. [DOI] [PubMed] [Google Scholar]
- Shafie MS, Hasan SM, Zain AM, Shah RM. RAPD and ISSR markers for comparative analysis of genetic diversity in wormwood capillary (Artemisia capillaris) from Negeri Sembilan, Malaysia. J Med Plants Res. 2011;5(18):4426–4451. [Google Scholar]
- Taran B, Zhang C, Warkentin T, Tullu A, Vanderberg A. Genetic diversity among varieties and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome. 2005;48:257–272. doi: 10.1139/g04-114. [DOI] [PubMed] [Google Scholar]
- Thomas SC, Palni LMS. Taxonomic, molecular and physiological evaluation of Costus pictus D. Don plants originally obtained from different parts of Kerala, India. J Appl Fund Sci. 2016;2(2):141–148. [Google Scholar]
- Tripathi N, Chouhan DS, Saini N, Tiwari S. Assessment of genetic variations among highly endangered medicinal plant Bacopa monnieri (L.) from Central India using RAPD and ISSR analysis. 3 Biotech. 2012;2(4):327–336. doi: 10.1007/s13205-012-0059-3. [DOI] [Google Scholar]
- Verma S, Singh S, Sharma S, Tewari SK, Roy RK, Goel AK, Rana TS. Assessment of genetic diversity in indigenous turmeric (Curcuma longa) germplasm from India using molecular markers. Physiol Mol Biol Plants. 2015;21(2):233–242. doi: 10.1007/s12298-015-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk PS, Ford-Lloyd BV, Jackson MT, Newbury HJ. Use of RAPD for the study of diversity within plant germplasm collections. Heredity. 1995;74(2):170–179. doi: 10.1038/hdy.1995.25. [DOI] [PubMed] [Google Scholar]
- Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20(2):176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]