Abstract
F-box proteins are part of one of the largest families of regulatory proteins that play important roles in protein degradation. In plants, F-box proteins are functionally very diverse, and only a small subset has been characterized in detail. Here, we identified a novel F-box protein FBX92 as a repressor of leaf growth in Arabidopsis. Overexpression of AtFBX92 resulted in plants with smaller leaves than the wild type, whereas plants with reduced levels of AtFBX92 showed, in contrast, increased leaf growth by stimulating cell proliferation. Detailed cellular analysis suggested that AtFBX92 specifically affects the rate of cell division during early leaf development. This is supported by the increased expression levels of several cell cycle genes in plants with reduced AtFBX92 levels. Surprisingly, overexpression of the maize homologous gene ZmFBX92 in maize had no effect on plant growth, whereas ectopic expression in Arabidopsis increased leaf growth. Expression of a truncated form of AtFBX92 showed that the contrasting effects of ZmFBX92 and AtFBX92 gain of function in Arabidopsis are due to the absence of the F-box-associated domain in the ZmFBX92 gene. Our work reveals an additional player in the complex network that determines leaf size and lays the foundation for identifying putative substrates.
Keywords: Cell cycle, F-box protein, Leaf development
Introduction
The leaf is the major photosynthetic organ of plants, and as such provides the basis for food, feed and bio-energy production by humans. Because leaf size, shape and number strongly influence photosynthetic capacity, understanding the molecular networks underneath is pivotal for future food security. The largest part of leaf development occurs post-embryonically: leaf primordia are initiated at the flanks of the shoot apical meristem in a position that is characteristic for the species. During a first phase after leaf initiation, cells are only proliferating because cell division and expansion are balanced (Donnelly et al. 1999, Andriankaja et al. 2012, Gonzalez et al. 2012), which is followed by a phase of cell expansion starting at the tip of the leaf, thereby establishing a cell cycle arrest front that remains in both Arabidopsis and maize at a constant position for a few days, and then rapidly declines (Andriankaja et al. 2012, Avramova et al. 2015). From then on, leaves enlarge solely because of an increase in average cell size due to cell expansion and because of meristemoid divisions, generating extra pavement cells while forming stomata, until the leaf reaches its final size (Gonzalez et al. 2012). Both leaf initiation and its further growth are under control of a complex set of intrinsic signals, such as phytohormones, that regulate cell proliferation and cell expansion to elaborate the final shape and size of the leaf (Gonzalez et al. 2012, Powell and Lenhard 2012, Gonz�lez and Inz� 2015).
The cell division cycle is precisely controlled to duplicate the DNA correctly during the S-phase and to generate two daughter cells during mitosis. Complexes formed by cyclin-dependent kinase (CDK) and cyclin are the core cell cycle regulators that play crucial roles at both the G1 to S and the G2 to M phase transitions (Harashima et al. 2013). The activity of these complexes is regulated by (de)phosphorylation, interaction with inhibitors and targeted protein degradation (De Veylder et al. 2007). Targeted protein degradation of cell cycle regulators happens through the ubiquitin–proteasome pathway, a highly precise post-translational regulatory process that ensures irreversibility of cell cycle progression (Genschik et al. 2014). The ubiquitin–proteasome pathway involves covalent attachment of polyubiquitin chains to targeted substrate proteins through the consecutive action of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3) (Hotton and Callis 2008). Ubiquitinated proteins are subsequently recognized and degraded via the 26S proteasome (Hershko and Ciechanover 1998). Substrate specificity of the pathway is defined by the E3 ligase, which binds to specific target proteins and stimulates in this way the conjugation of ubiquitin to this target protein. In Arabidopsis thaliana (Arabidopsis), >1,400 genes or approximately 5% of the proteome encode elements of the ubiquitin–proteasome pathway (Smalle and Vierstra 2004). Several mutants and plants transgenic for the ubiquitin–proteasome pathway with altered leaf size have been identified, illustrating that controlled proteolysis is an important layer of regulation during development. For instance, a mutation in the E3 ubiquitin ligases BIG BROTHER (BB or EOD1) and DA2 prolongs the timing of cell proliferation in different organs (Disch et al. 2006, Xia et al. 2013). BB/EOD1 and DA2 act synergistically with the ubiquitin receptor DA1, and inactivation of DA1 causes the formation of larger plants (Li et al. 2008). Similarly, loss of function of a subunit of the 19S proteasome, RTP2A, results in larger organs due to an increased cell size partially compensated by a reduced cell number (Kurepa et al. 2009). Also the PEAPOD transcription factors, negative regulators of meristemoid activity, are subjected to F-box-mediated proteolysis by STERILE APETAL (SAP) (Wang et al. 2016). Another important system that enables ubiquitin-mediated degradation of proteins important for growth is the multiprotein E3 ubiquitin ligase anaphase-promoting complex/cyclosome (APC/C) (De Veylder et al. 2007, Eloy et al. 2015). The subunits APC10 (Eloy et al. 2011) and CELL DIVISION CYCLE PROTEIN 27 HOMOLOG A (CDC27a) (Rojas et al. 2009) both promote cell proliferation and an increased leaf size, whereas SAMBA is a plant-specific negative regulator of the APC/C and its inactivation increases organ size (Eloy et al. 2012).
A major type of E3 ligases that is involved in cell cycle control are the SCF E3 ligases, which consist of four components, Cullin1/Cdc53, Rbx1/Roc1/Hrt1, Skp1 (ASK1 in plants) and an F-box protein (Cardozo and Pagano 2004). The scaffold protein Cullin1 interacts at its C-terminus with Rbx1/Roc1/Hrt1, which binds to the E2 loaded with ubiquitin, and at its N-terminus with Skp1, which binds to the F-box protein that interacts with the target proteins for degradation (Bai et al. 1996). The SCF complex has an important function in the proteolysis of cell cycle regulatory proteins, although it can also mark other proteins for destruction (Genschik et al. 2014). More precisely, the SCF complex plays a critical role during the G1 to S phase transition, which requires the degradation of CDK inhibitors (CKIs, also known as ICK/KRP proteins) to release CDK activity (Verkest et al. 2005, Ren et al. 2008, Noir et al. 2015). For example, the F-BOX-LIKE17 (FBL17) is essential to maintain normal cell proliferation by mediating the degradation of the CDK inhibitor KIP-RELATED PROTEIN2 (KRP2) known to switch off CDKA;1 kinase activity (Noir et al. 2015).
F-box proteins identify the target proteins for degradation, recruit them and position them in proximity to E2 for ubiquitination (Skaar et al. 2013). Via their F-box domain, a structural motif consisting of approximately 50 conserved amino acids (Xiao and Jang 2000), they are anchored to the SCF complex, whereas the C-terminal domain binds the target proteins for ubiquitination and degradation usually via protein interaction motifs. Examples of these protein interaction motifs occurring in plants are kelch repeats, WFBX920, LRR and tubby (Gagne et al. 2002, Kuroda et al. 2002, Jain et al. 2007, Jia et al. 2013). F-box proteins in plants belong to a large family: about 700 F-box proteins have been identified in Arabidopsis (Risseeuw et al. 2003) and Oryza sativa (rice) (Jain et al. 2007) and about 350 in Zea mays (maize) (Jia et al. 2013), numbers that are much larger than the number of F-box proteins identified in most other eukaryotes (Gagne et al. 2002, Schumann et al. 2011). They are involved in a large variety of biological processes, including flower development, hormone perception and signaling, circadian rhythms, defense responses, senescence, embryogenesis and seedling development (Lechner et al. 2006, Hua et al. 2011, Schumann et al. 2011). Evidence is coming to light that E3 ligases might act as receptors of hormones or other molecules for signal transduction, suggesting a novel mechanism to link internal and external cues to cell division (Jurado et al. 2008, Achard and Genschik 2009, del Pozo and Manzano 2014). However, the specific function of the majority of the F-box proteins currently remains unclear (Schumann et al. 2011).
Here, we identified a maize F-box protein, ZmFBX92, for which ectopic expression in Arabidopsis resulted in plants with larger leaves, although maize plants overexpressing ZmFBX92 showed no obvious phenotypes. In contrast, gain-of-function mutants of the Arabidopsis FBX92 homologous gene, AtFBX92, developed smaller leaves than the wild type (WT) due to a reduced cell number. In concert, Arabidopsis plants with reduced AtFBX92 expression levels had larger leaves due to an increased cell division rate and consequential cell number. The opposite effects of ZmFBX92 and AtFBX92 gain of function in Arabidopsis can be explained by the presence of an F-box-associated domain in the AtFBX92 gene that is lacking in the ZmFBX92 gene. Overexpression of a truncated AtFBX92 allele lacking the F-box-associated domain resulted in a larger leaf size phenotype than in plants with reduced levels of AtFBX92 or which ectopically expressed ZmFBX92. In addition, the increase in expression levels of several cell cycle genes in plants with reduced levels of AtFBX92 indicates that this newly identified F-box protein AtFBX92 acts as a repressor of leaf growth by affecting cell division.
Results
Ectopic ZmFBX92 expression in Arabidopsis positively affects leaf size
In the maize genome, about 350 F-box proteins have been identified and a small subset has previously been reported to respond to multiple stress treatments, such as salt, drought, cold and heat (Kakumanu et al. 2012, Jia et al. 2013). We investigated whether expression of one of these F-box genes, ZmFBX92, in Arabidopsis would alter plant growth under mild osmotic stress. Therefore, ZmFBX92 was expressed in Arabidopsis under control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter and five independent, homozygous, single-locus lines were selected to analyze their phenotype. The results were very similar for all lines, thus only the results of line ZmFBX92OE11, hereafter ZmFBX92OE, are presented.
Phenotypic analysis of ZmFBX92OE and WT [Columbia-0 (Col-0)] plants revealed that the leaf area was significantly larger in the transgenic plants, while no other phenotypic abnormalities were observed (Fig. 1; Supplementary Fig. S1A). The projected rosette areas (PRAs) of WT and ZmFBX92OE plants grown in vitro in standard and mild osmotic stress conditions (25 mM mannitol) were determined from 6 until 21 days after stratification (DAS) (Fig. 1A). On average, osmotic stress reduced the rosette area by about 60% at 21 DAS. ZmFBX92OE plants had a significantly increased rosette area compared with the WT under both conditions (Fig. 1A). At 21 DAS, this difference was about 35% and about 50% under control and mild stress conditions, respectively. The increased rosette area in ZmFBX92OE occurred already very early during development: at 6 DAS the rosette size was about 50% larger in transgenic plants than in the WT under both control and mild stress conditions (Fig. 1A, inset). There was no significant interaction effect [‘three-way’ analysis of variance (ANOVA)], indicating that the effect of ZmFBX92OE expression on plant growth was comparable under both control and mild osmotic stress conditions. Hence, for further analysis, we focused on rosette growth and leaf development under control conditions.
Fig. 1.
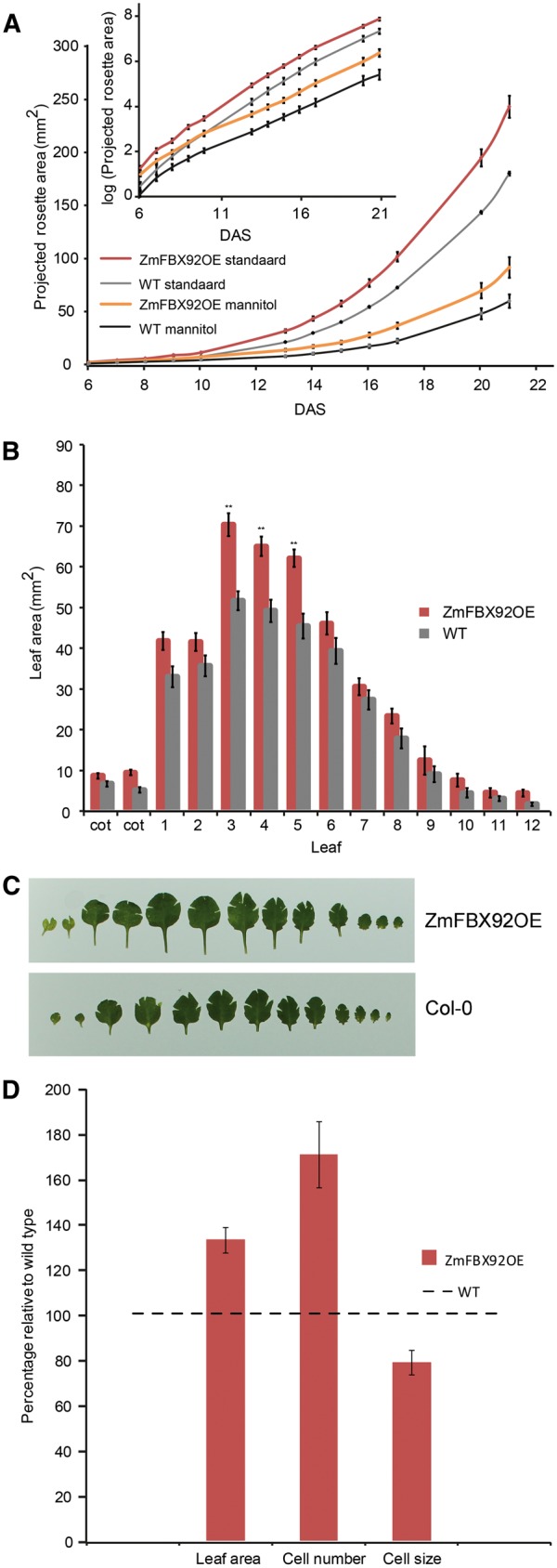
Effect of ZmFBX92 expression on rosette and leaf growth in Arabidopsis and cellular basis of the leaf size differences. (A) PRA of a ZmFBX92OE transgenic line and the WT over time from 6 until 21 DAS. Plants were grown in vitro on standard medium or medium containing 25 mM mannitol. Inset: PRA in log scale. Values represent the mean � SE (ntransgenic line = 25–27, nWT = 39). (B) Individual leaf size of 22-day-old WT and ZmFBX92OE plants grown in vitro. Values represent the mean � SE (n = 7). Significant differences (Student’s t-test): *P < 0.05; **P < 0.01 relative to the WT. (C) Representative pictures from the measurements shown in (B). (D) Average area, pavement cell number and pavement cell size of leaf 3 at 22 DAS of ZmFBX92OE plants relative to the WT. Values represent the mean � SE (n = 3).
The positive effect of ZmFBX92OE on leaf size in Arabidopsis was corroborated by determining the individual leaf areas at 22 DAS (Fig. 1B, C). Both mature and juvenile leaves were larger in ZmFBX92OE plants. To examine to what extent a difference in cell proliferation and/or cell expansion was responsible for the increased leaf size, the number and size of abaxial epidermal cells were compared in WT and ZmFBX92OE leaves. The fully mature (22 DAS) third leaf was approximately 30% larger in ZmFBX92OE plants due to a highly increased cell number (approximately 70%), which was partially compensated by a reduction in cell size of approximately 20% (Fig. 1D). Thus, ectopic expression of ZmFBX92 in Arabidopsis resulted in larger leaves primarily due to an increased cell number.
ZmFBX92 overexpression in maize has no effect on leaf size
To investigate if variation in ZmFBX92 expression levels also affects plant development in maize, we generated three independent, single-locus lines overexpressing ZmFBX92 under control of the Brachypodium distachyon elongation factor1α promoter (pBdEF1α) (Coussens et al. 2012), showing an approximately 50- to 800-fold higher expression level than the control inbred line B104 (Supplementary Fig. S2A). Leaf growth under control and mild drought conditions was monitored in the two lines with the highest overexpression level, ZmFBX92OE1 and ZmFBX92OE2. Several final leaf size-related parameters were determined, i.e. area, width, length and weight, and these measurements were complemented with the kinetic parameters ‘leaf elongation rate’ and ‘leaf elongation duration’ (Voorend et al. 2014) and shoot-related parameters at seedling stage, i.e. fresh weight, V-stage and leaf number (Supplementary Fig. S2B–K) (Baute et al. 2015). Mild drought stress negatively impacted all these parameters, except leaf elongation duration, which was prolonged (Supplementary Fig. S2D), implying that mild drought stress primarily reduces growth rate. When comparing growth of ZmFBX92OE and control B104 plants, no significant differences were observed, for any of the measured parameters, under control conditions or under mild drought stress (Supplementary Fig. S2B–K).
Altered AtFBX92 expression level influences leaf size by affecting cell number
The putative orthologous gene of ZmFBX92 in Arabidopsis was identified using PLAZA (Proost et al. 2015), and is further designated AtFBX92 (At3g07870). To examine the function of this gene in Arabidopsis, we generated plants with altered AtFBX92 expression levels. Transgenic Arabidopsis plants were generated that ectopically overexpressed AtFBX92 under the control of the constitutive CaMV 35S promoter. Four independent transformation events with a single-insertion locus and varying expression levels of AtFBX92 were selected for further analysis (Supplementary Fig. S1B). Because the phenotypes of the lines with the weakest overexpression were similar, we present here only the analysis of AtFBX92OE7 together with that of AtFBX92OE2 showing a stronger increase in expression level.
In contrast to what was observed for plants that ectopically expressed ZmFBX92, AtFBX92OE plants showed a reduction in the rosette area compared with WT plants (Fig. 2A; Supplementary Fig. S1C). This reduction was comparable under mild osmotic stress (Supplementary Fig. S3A). No other obvious phenotypes were observed in AtFBX92OE plants. The decreased rosette size was visible already very early during development (Fig. 2A, inset), from 6 DAS onwards. The negative effect of AtFBX92 overexpression on leaf growth was confirmed by determining the individual leaf areas of 22-day-old plants grown in vitro. The areas of the mature leaves were significantly smaller in the AtFBX92OE7 plants compared with the WT, whereas for AtFBX92OE2 plants, all leaves were significantly smaller, including the younger leaves (Fig. 2B, C). To explore the cellular basis of the leaf size decrease, leaf development of AtFBX92OE and WT plants grown in vitro was analyzed at the cellular level. Similar to the case for ZmFBX92OE, cell number and cell size of the abaxial epidermis of the third leaf were determined at 21 DAS, when this leaf is fully matured (Fig. 2D). The mature third leaf of AtFBX92OE2 and AtFBX92OE7 was 45% and 16%, respectively, smaller than that of the WT, due to a strong reduction in cell number (52% and 22% for AtFBX92OE2 and AtFBX92OE7, respectively), which was partially compensated by an increased cell size (15% and 7% for AtFBX92OE2 and AtFBX92OE7, respectively).
Fig. 2.
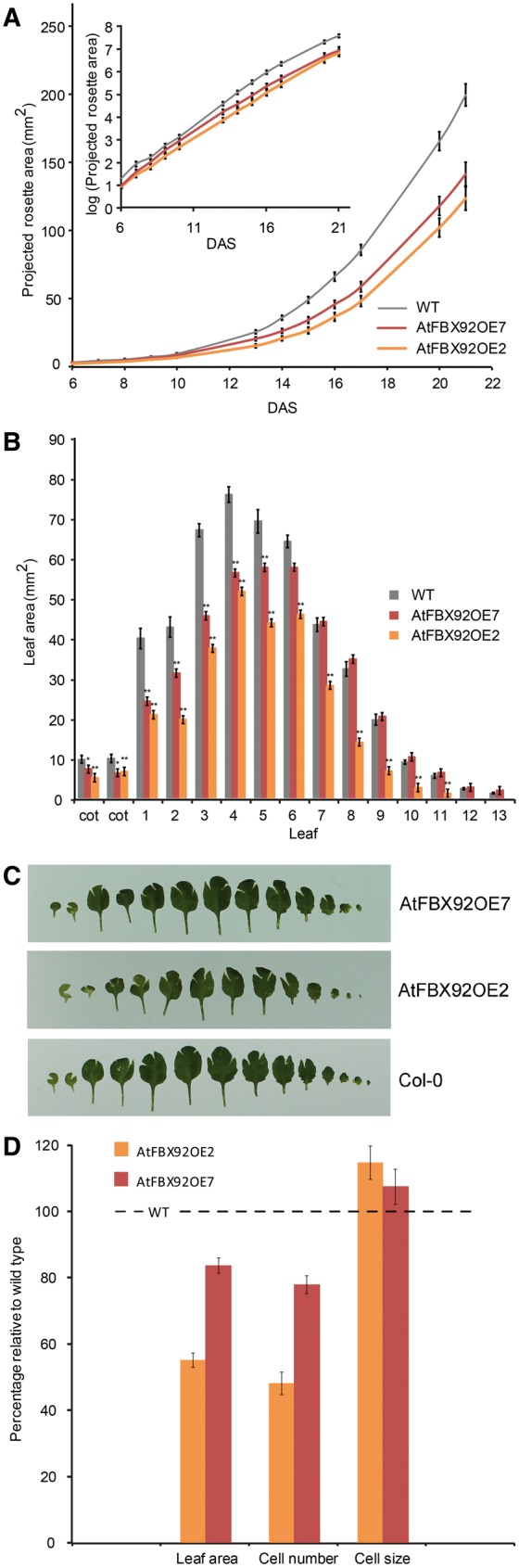
Effect of AtFBX92 ectopic expression on rosette and leaf growth under standard conditions in vitro and cellular basis of leaf size differences. (A) PRA of AtFBX92OE transgenic lines and the WT over time from 6 until 21 DAS. Plants were grown in vitro on standard medium. Inset: PRA in log scale. Values represent the mean � SE (ntransgenic lines = 26–32, nWT = 36). (B) Individual leaf size of 22-day-old WT, AtFBX92OE7 and AtFBX92OE2 plants grown in vitro. Values represent the mean � SE (n = 7). Significant differences (Student’s t-test): *P < 0.05; **P < 0.01 relative to the WT. (C) Representative pictures from the measurements shown in (B). (D) Average area, pavement cell number and pavement cell size of leaf 3 at 21 DAS of AtFBX92OE7 and AtFBX92OE2 plants relative to the WT. Values represent the mean � SE (n = 3).
In parallel with the plants overexpressing AtFBX92, transgenic plants with reduced expression levels were generated by designing an artificial microRNA (amiRNA) targeting AtFBX92 using the tool at http://wmd3.weigelworld.org/cgi-bin/webapp.cgi (Ossowski et al. 2008). Three homozygous, independent, single-locus lines with reduced expression levels (Supplementary Fig. S1D) were analyzed for their leaf phenotype and, because they were very similar, only the results of amiFBX92-4, hereafter designated amiFBX92, is presented (Fig. 3; Supplementary Fig. S1E). The PRA was determined from 5 until 21 DAS and shown to be larger in amiFBX92 compared with the WT from the first day of analysis onwards (Fig. 3A, inset). This increase in PRA was comparable under mild osmotic stress (Supplementary Fig. S3B). Next, the individual leaf areas were determined at 21 DAS. All leaves except leaf 3 of amiFBX92 were significantly larger than those of the WT (Fig. 3B, C). Cellular analysis of the fully matured first leaf pair at 21 DAS showed that the increase in leaf area (24%) was due to an increased cell number (47%), partially compensated by a decreased cell size (16%) (Fig. 3D). Taken together, our data indicate that altering AtFBX92 expression levels influences leaf size in opposing ways, which is primarily the result of a difference in cell number.
Fig. 3.
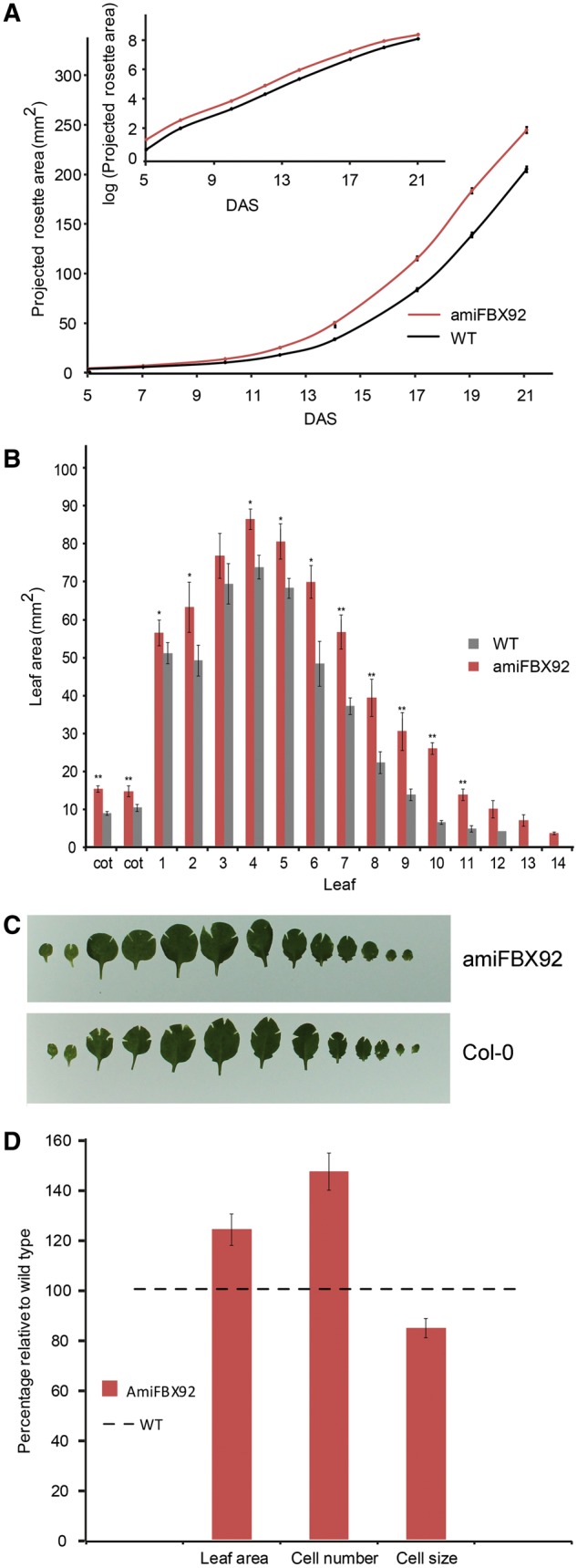
Effect of AtFBX92 down-regulation on rosette and leaf growth under standard conditions in vitro and cellular basis of the leaf size differences. (A) PRA of amiFBX92 and the WT over time from 5 until 21 DAS. Plants were grown in vitro on standard medium. Inset: PRA in log scale. Values represent the mean � SE (transgenic line= 22, nWT = 30). (B) Individual leaf size of 21-day-old WT and amiFBX92 plants grown in vitro. Values represent the mean � SE (n = 7). Significant differences (Student’s t-test): *P < 0.05; **P < 0.01 relative to the WT. (C) Representative pictures from the measurements shown in (B). (D) Average area, pavement cell number and pavement cell size of leaves 1 and 2 at 21 DAS of amiFBX92 plants relative to the WT. Values represent the mean � SE (n = 3).
Opposing phenotypes in ZmFBX92- and AtFBX92-overexpressing plants
Unexpectedly, ectopic expression of ZmFBX92 and overexpression of AtFBX92 in Arabidopsis resulted in opposite leaf phenotypes. A sequence search using Pfam (Finn et al. 2014) and InterPro revealed that AtFBX92 harbors an F-box-associated domain, type 3, C-terminally of the F-box domain, whereas this F-box-associated domain is lacking in ZmFBX92 (Supplementary Fig. S4). To analyze if overexpression of AtFBX92 without the F-box-associated domain has a phenotype comparable with ectopic expression of ZmFBX92, a deletion mutant construct AtFBX92del was generated, expressing the N-terminal part of the gene including the F-box domain but not the F-box-associated domain, under the control of the constitutive CaMV 35S promoter (Supplementary Fig. S5). Three independent, single-locus lines with high expression levels of AtFBX92del (Supplementary Fig. S1F) were selected for further phenotypic characterization. Because the leaf phenotypes of the three lines were very similar in vitro, only the results for AtFBX92del12 with the lowest expression level, hereafter named AtFBX92del, are shown. Quantitative image analysis of the PRA over time, from 6 DAS until 24 DAS, indicated that AtFBX92del plants were larger than the WT (Fig. 4A). Additionally, the PRA increase started very early during development (Fig. 4A, inset), similar to the effect observed in ZmFBX92OE and amiFBX92 lines. In agreement with this, the individual leaf areas determined at 20 DAS were significantly larger than those of the WT (Fig. 4B, C). Consistently, this increase in size of the first leaf pair (27%) at 20 DAS was due to an increased cell number (29%), while there was no significant effect on cell size (Fig. 4D).
Fig. 4.

Effect of AtFBX92del expression on rosette and leaf growth under standard conditions in vitro and cellular basis of the leaf size differences. (A) PRA of AtFBX92del and the WT over time from 6 until 24 DAS. Plants were grown in vitro on standard medium. Inset: PRA in log scale. Values represent the mean � SE (ntransgenic line= 46, nWT = 58). (B) Individual leaf size of 20-day-old WT and AtFBX92del plants grown in soil. Values represent the mean � SE (n = 12). Significant differences (Student’s t-test): **P < 0.01 relative to the WT. (C) Representative pictures from the measurements shown in (B). (D) Average area, pavement cell number and pavement cell size of leaves 1 and 2 at 20 DAS of AtFBX92del plants relative to the WT. Values represent the mean � SE (n = 3).
AtFBX92 expression pattern
To analyze the spatiotemporal expression pattern of AtFBX92, we engineered a construct consisting of the 1.3-kb fragment upstream of the ATG codon of AtFBX92 fused to a green fluorescent protein (GFP)–β-glucuronidase protein (GUS) reporter cassette in a FAST vector (Shimada et al. 2010). This construct, named pAtFBX92:GFP:GUS, was subsequently introduced into Arabidopsis plants. Histochemical analyses of three independent Arabidopsis transgenic lines showed a similar expression pattern (Fig. 5). In general, AtFBX92 was widely expressed in young seedlings, although expression seemed somewhat lower in the hypocotyl (Fig. 5A), and strongly in the whole root, except for the most distal region of the basal meristem (Fig. 5B). AtFBX92 expression was observed as leaf development progressed from proliferative to mature tissue, although expression was somewhat lower in younger than in older leaves (Fig. 5C). This is in agreement with the expression levels in the transcriptomics data set of leaf 3 during the subsequent phases of proliferation, expansion and maturation, showing that AtFBX92 expression was low in fully proliferative tissue and increased gradually towards maturity (Supplementary Fig. S6) (Andriankaja et al. 2012). In 6-week-old plants, GUS expression was also detected in the stems, sepals, style, the most distal part of carpels and in pollen grains of reproductive tissues (Fig. 5D). GUS expression was also detected in developing siliques, in the valves but not in seeds (Fig. 5E).
Fig. 5.
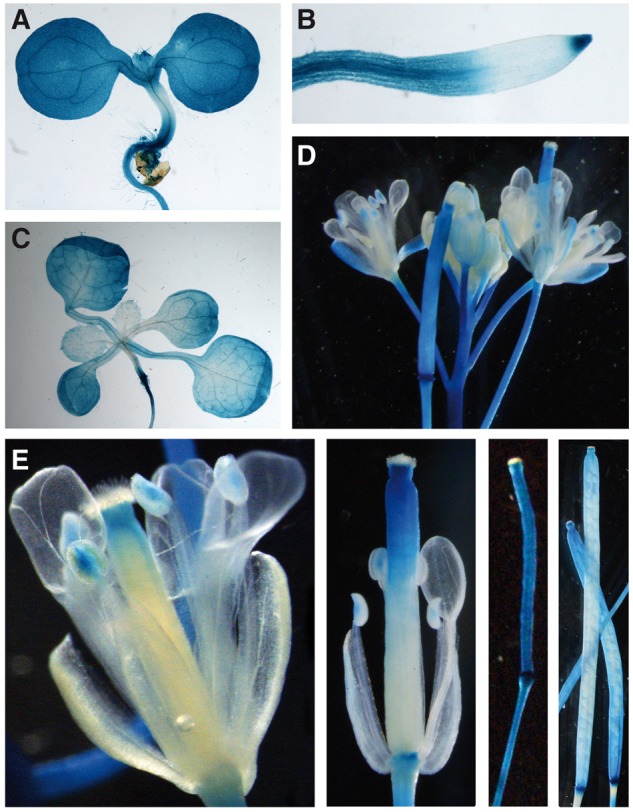
Expression of the pAtFBX92:GFP:GUS reporter gene at different developmental stages. (A) Seedling shoot at 6 DAS. (B) Main root. (C) Seedling shoot at 13 DAS. (D) Flower cluster of a 6-week-old plant. (E) Carpels and developing siliques of a 6-week-old plant.
Rosette growth in soil
To validate the in vitro observed differences in rosette size of plants with altered AtFBX92 expression levels, we grew AtFBX92OE, amiFBX92, AtFBX92del and the corresponding WT plants in soil on the automated imaging platform WIWAM, which allows the PRA to be followed over time (Skirycz et al. 2011b). Two lines of each construct were analyzed. We could confirm that also in soil, AtFBX92OE plants were smaller than the WT, whereas amiFBX92 and AtFBX92del plants were larger, although for AtFBX92del plants, this could only be confirmed for one of the two lines (Fig. 6A; Supplementary Fig. S7). In general, the effects in soil seemed to be less pronounced than in vitro, because the absolute percentage difference from the WT was always larger in vitro than in soil. At 20 DAS, for instance, the PRA of amiFBX92 grown in vitro was 25% larger than that of the WT vs. 15% when grown in soil; 32% for AtFBX92del grown in vitro vs. 18% when grown in soil; and −38% and −29% for AtFBX92OE2 and AtFBX92OE7, respectively, grown in vitro vs. −14% and −16%, respectively, when grown in soil. The PRAs of amiFBX92 and AtFBX92del plants grown in soil were significantly larger than those of the WT already very early during development (6 DAS), similar to plants grown in vitro (Supplementary Fig. S7B, C, insets). However, AtFBX92OE lines grown in soil were only significantly smaller than the WT from 18 DAS onwards (Supplementary Fig. S7A), whereas in vitro a significant reduction was also observed from 6 DAS onwards (Fig. 2A, inset). The stronger reduction in rosette growth for AtFBX92OE2 than for AtFBX92OE7 observed in vitro was completely lacking in soil; both lines showed an equal decrease of PRA (Supplementary Fig. S7A). Remarkably, growth of AtFBX92OE2 and AtFBX92OE7 in soil resulted in epinastic leaves (Fig. 6A), an effect that was not seen in vitro (Supplementary Fig. S1C).
Fig. 6.

Effect of AtFBX92 misexpression on leaf size of plants grown in soil under standard and drought stress conditions. Representative pictures of 20-day-old amiFBX92, AtFBX92del, AtFBX92OE and Col-0 plants grown in soil under standard conditions (A) and under mild drought stress (B).
The effect of AtFBX92 on tolerance to mild drought stress
The reduction in PRAs of AtFBX92OE and amiFBX92 plants grown under osmotic stress conditions by adding mannitol to the medium was not significantly different from the reduction in PRA for WT plants grown under these conditions (Supplementary Fig. S3). Addition of mannitol to the growth medium is often used in lab settings as a proxy for osmotic stress. Because the response of soil-grown plants is closer to natural conditions, we also evaluated the effect of mild drought stress on soil-grown plants with altered AtFBX92 levels and WT plants (Fig. 6B; Supplementary Fig. S8). We grew these plants on the WIWAM platform that allows, in addition to automated imaging, automated weighing and watering to control the applied water regime (see the Materials and Methods for details). In WT plants, the rosette size at 20 DAS was 34% reduced under mild drought conditions compared with well-watered conditions (Supplementary Fig. S7, S8). For the two amiFBX92 lines, the reduction was comparable with the reduction in WT plants (35% and 33% reduction in PRA at 20 DAS for amiFBX92-4 and amiFBX92-13, respectively). Overall, there was no significant difference in the PRA of amiFBX92 and the WT in response to mild drought stress over time (Supplementary Fig. S8B), a result similar to that for the response to osmotic stress (Supplementary Fig. S3A). In contrast, the PRA of AtFBX92del plants under mild drought stress was not significantly different from the PRA of WT plants (Supplementary Fig. S8C), whereas under standard conditions AtFBX92del plants were significantly larger than the WT (Supplementary Fig. S7C). In contrast to the effect of down-regulating AtFBX92 in standard conditions (Supplementary Fig. S7B), the PRA of AtFBX92OE plants under mild drought stress was significantly larger (AtFBX92OE2) or equal to (AtFBX92OE7) the PRA of WT plants (Supplementary Fig. S8A). This is in contrast to what was found under osmotic stress (Supplementary Fig. S3A), for which there was no difference in response on PRA between WT and AtFBX92OE plants.
Down-regulation of AtFBX92 increases the cell division rate
To investigate the effect of the reduction in FBX92 expression at the cellular level over time, leaf growth was analyzed kinematically (De Veylder et al. 2001). The first two initiated leaves of amiFBX92 and WT plants grown in vitro were harvested daily from 5 until 21 DAS for quantitative image analysis of leaf blade area, and cell number and cell size of the abaxial epidermis (Fig. 7). The leaf size of amiFBX92 plants was not significantly different from that of the WT until 7 DAS, when the difference became significant (Fig. 7A, inset). At maturity, amiFBX92 leaves were approximately 30% larger than those of the WT. The cell area remained constant until 8 DAS, i.e. during the period of cell division, and then increased exponentially (Fig. 7C). Transgenic and WT plants followed the same trend of cell area increase over time, suggesting that the down-regulation of AtFBX92 did not affect cell expansion. The cell number per leaf, however, was strongly increased in amiFBX92 plants compared with the WT (Fig. 7B); at maturity the difference was approximately 25%, supporting our previous data. Average cell division rates of the whole leaf were estimated from the exponential increase in cell number. Cell cycle duration was higher in amiFBX92 plants (16 � 2 h) than in the WT (20 � 1 h) from 5 until 7 DAS, after which they followed the same trend, reaching zero at day 14 in the first leaf pair (Fig. 7D). Thus, the increased final leaf size in amiFBX92 is most probably due to augmented cell division rates during very early stages of leaf development.
Fig. 7.
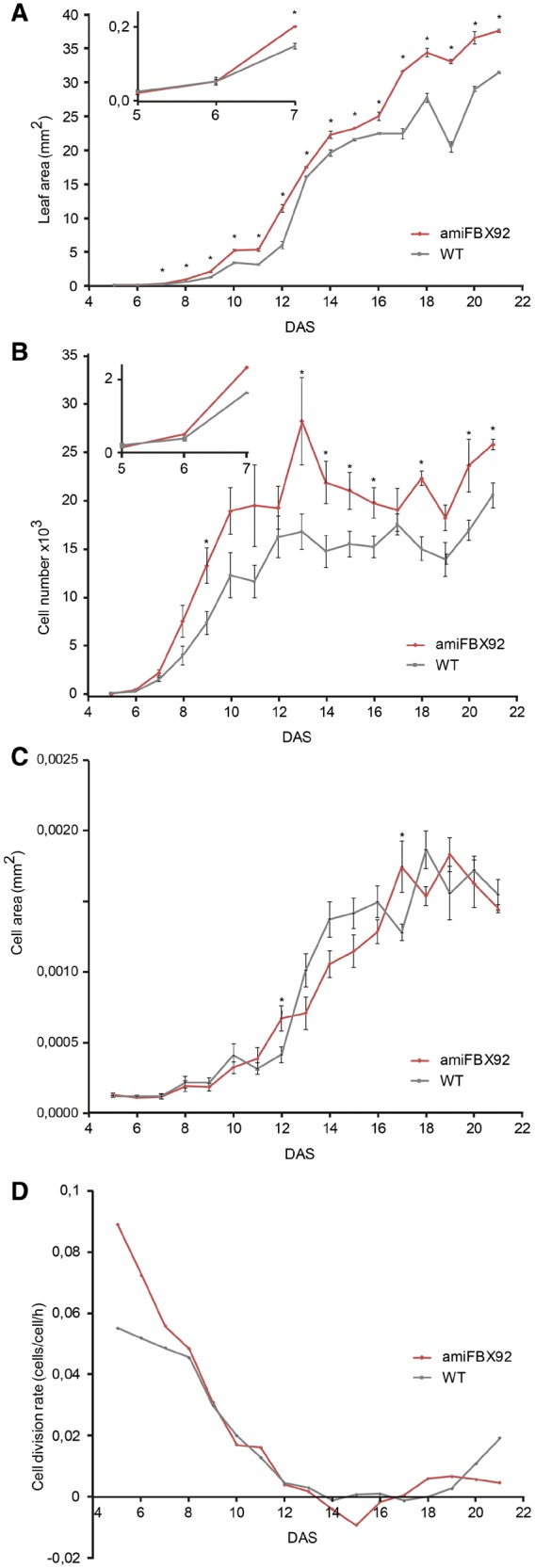
Kinematic analysis of the first leaf pair of amiFBX92 and WT plants grown in vitro from 4 to 22 DAS. (A) Leaf area. Inset: measurements at 5–7 DAS. (B) Cell number. Inset: measurements at 5–7 DAS. (C) Cell area. (D) Cell division rate. Values represent the mean � SE (n = 4–6) for (A), (B) and (C); significant differences (Student’s t-test): *P < 0.05; **P < 0.01 relative to the WT.
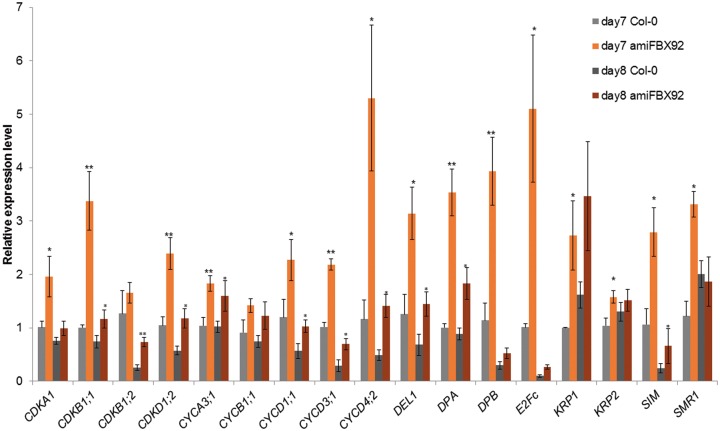
Quantitative reverse transcription–PCR (qRT–PCR) of cell cycle genes during early phases of development in amiFBX92
Because the cell proliferation rate is affected in amiFBX92 plants, we examined the expression of several cell cycle genes. Total RNA was isolated from the first leaf pair of WT and amiFBX92 plants at 7 and 8 DAS, the first time points at which cell numbers were significantly different, and subjected to qRT–PCR. The two major cell cycle phase transitions, the G1 to S and G2 to M, are controlled by the consecutive action of cyclin–CDK complexes (Inz� and De Veylder 2006). We found that transcript levels of genes involved in the G1 to S phase transition, such as the D-type cyclins, and genes involved in the G2 to M transition, such as CYCB1;1, CDKB1;1 and CDKB1;2, were increased in amiFBX92 lines (Fig. 8). Surprisingly, in addition to these positive cell cycle regulators, the expression levels of some of the negative cell cycle regulators—ICK/KIP/CIP-RELATED PROTEINS (ICK/KRPs), SIAMESE (SIM) and SIM-related (SMR) proteins—were also up-regulated in amiFBX92 leaves (Fig. 8). We could confirm this in an additional amiFBX92 line, amiFBX92-13 (Supplementary Fig. S9A). Moreover, expression levels of both positive and negative cell cycle genes were down-regulated and up-regulated in proliferating AtFBX92OE and AtFBX92del leaves, respectively, compared with WT leaves, supporting the increase in expression of cell cycle genes in proliferative leaves with reduced AtFBX92 activity and/or expression levels (Supplementary Fig. S9B, C).
Fig. 8.
Relative expression levels of cell cycle genes in amiFBX92 in the first leaf pair compared with the WT at 7 and 8 DAS as determined by qRT–PCR. Values were normalized against the expression level of the housekeeping gene and represent the main expression levels of the indicated transcripts of three biological repeats � SE. Significant differences (Student’s t-test): *P < 0.10; **P < 0.01 relative to the WT.
Discussion
In this study, we present the characterization of a new plant-specific F-box-containing protein we designated FBX92. Reducing AtFBX92 expression levels using an amiRNA approach resulted in plants that developed larger cotyledons and leaves, whereas an opposite effect was seen in plants overexpressing AtFBX92. We showed that this difference in leaf size was primarily due to an effect on cell number, compensated slightly by an effect on cell size. The cell number in a plant organ is determined by three things: first, the cell number in the leaf primordia recruited from the shoot apical meristem (SAM); secondly, the cell proliferation rate; and thirdly, the cell proliferation duration (Gonzalez et al. 2012). Because the leaf primordia of amiFBX92 plants were equal in size to those of WT plants at the first time point of analysis (5 DAS), we can assume that the number of cells recruited from the SAM for leaf initiation is not altered. Detailed kinematic analysis of the first amiFBX92 leaf pair indicated that the observed increase in leaf size was specifically due to an enhanced rate of cell division during the first days of leaf development, which was maintained until maturity. Moreover, the average duration of the cell cycle decreased from 20 h in the WT to about 16 h in amiFBX92. There are only a few genes known to increase the rate of cell division in developing leaves. Overexpression of the APC/C subunit APC10 accelerates the average cell cycle duration from 21 to 19 h (Eloy et al. 2011). It is likely that CDC27a, another regulator of APC/C, also affects the rate of cell division (Rojas et al. 2009). The observation that cell division is enhanced in the amiFBX92 plants was further supported by the enhanced expression level of several cell cycle genes in amiFBX92 and their decreased expression levels in AtFBX92OE plants during this early leaf development. In agreement with our observations, CDKA levels in the roots of different Arabidopsis ecotypes are positively correlated with cell division rates (Beemster et al. 2002). Also in cell suspension cultures, the expression levels of A-, B- and D-type cyclins and CDKB1;1 coincided with cell division rates (Richard et al. 2001). For several genes shown to function in organ size determination by altering cell division rates, it has been shown that, when altering their expression, the expression of cell cycle-regulating genes was also affected (Achard and Genschik 2009, Lee et al. 2009, Rojas et al. 2009, Eloy et al. 2011). We found both positive and negative cell cycle regulators up-regulated in amiFBX92 proliferative leaves, although it has been shown that constitutive expression of the negative regulators KRP and SMR results in growth retardation (Verkest et al. 2005, Churchman et al. 2006, Hudik et al. 2014). This discrepancy might be due to the specific time points of our analysis at which the leaf was fully proliferative. It is very possible that in later stages of leaf development, the levels of these positive and negative regulators are not affected or show different trends in amiFBX92 or AtFBX92OE. Also, in cell suspension cultures, the expression levels of negative cell cycle regulators peak with the highest cell division rate (Richard et al. 2001), and in animal systems some of the KRPs have a role in assembling CDK–cyclin complexes (Sherr and Roberts 1999). In agreement with the kinematic analysis, the up-regulation of both positive and negative regulators in proliferative leaves might suggest that there is no effect on the timing of the transition from cell division to cell expansion. Moreover, because genes involved in the G1 to S phase transition as well as genes involved in the G2 to M transition show differential expression, we can speculate that reducing AtFBX92 activity results in a faster progression through both S-phase and mitosis. In agreement with this, AtFBX92 expression levels seem not to be specific for one of the phases of the cell cycle in cell cultures (Menges et al. 2003). Possibly, the effect of altered AtFBX92 levels on the expression of cell cycle genes is indirect; it might be that AtFBX92 targets a currently unknown growth-regulating factor for proteolysis. For instance, the SCF complex containing SLEEPY1 (SLY1) is an F-box protein that affects cell division indirectly by gibberellin-mediated degradation of the growth-repressing DELLA proteins (Achard et al. 2009).
Although the cell cycle and the proteolytic processes are conserved between yeast, animals and plants, only a few plant F-box proteins have been identified to modulate the G1 to S phase transition of the cell cycle (reviewed by Genschik et al. 2014). SKP2A and SKP2B act as positive and negative regulators, respectively, of root cell division by targeting specific cell cycle regulators for degradation (del Pozo et al. 2006, Ren et al. 2008). Plants with reduced SKP2A and SKP2B expression levels exhibit only mild phenotypes (del Pozo et al. 2006, Manzano et al. 2012). Also the effect of AtFBX92 perturbation on plant development is rather mild. This might be due to only limited variation of expression levels in the amiFBX92 and AtFBX92OE lines compared with WT plants. Additionally, it is possible that this F-box protein is also regulated at the post-transcriptional level. For instance, it has been shown that many F-box proteins are intrinsically unstable because they are often themselves targeted for degradation by the ubiquitin–proteasome pathway (Jurado et al. 2008, Marrocco et al. 2010). This would be in agreement with the fact that we could not stably express a tagged version of this F-box protein, either in cell cultures or in seedlings (data not shown). Also, it is likely that there are redundant mechanisms governing the different steps of cell division; potentially there might be a partial compensation by other proteins and protein complexes, which has also been suggested for other F-box proteins (Dharmasiri et al. 2005, Qiao et al. 2009, An et al. 2010, Schumann et al. 2011, Manzano et al. 2012). Recently, an F-box protein, FBL17, was identified as an important regulator of the cell cycle at different stages of plant development (Gusti et al. 2009, Zhao et al. 2012, Noir et al. 2015). Loss of FBL17 function drastically impaired plant development during the sporophytic life cycle as well as during gametogenesis by reducing cell proliferation, due to an increased stability of the cell cycle inhibitor KRP2, although the drastic phenotype suggests that this F-box protein also targets other substrates for degradation (Noir et al. 2015).
The expression of AtFBX92 was examined in a large number of published microarray data sets using the Arabidopsis eFP Browser (www.bar.utoronto.ca) and Genevestigator (https://www.genevestigator.com), showing that expression was rather weak in most tissues except in pollen. Analysis of GUS reporter lines confirmed that AtFBX92 is expressed in almost all sporophytic tissues, in tissues with low proliferation rates, but not in root meristems. GUS expression was also visible in reproductive tissues, i.e. in sepals, style, carpels, developing siliques and pollen grains, although gametogenesis and seed set were not affected in amiFBX92, AtFBX92OE and AtFBX92del plants.
Ectopic expression of ZmFBX92 in Arabidopsis and of the deletion mutant AtFBX92del, both lacking the F-box-associated interaction domain, resulted in plants with larger leaves, comparable with the phenotype of amiFBX92. We hypothesize that these ectopic or mutant proteins bind to the SCF complex and inhibit binding of the native AtFBX92 protein, in that way preventing the ubiquitination of the target proteins, because the F-box-associated interaction domain that most probably recruits the target is lacking in these ectopic or mutant proteins. Overexpression of ZmFBX92 in maize, however, has no apparent phenotype. Because the native ZmFBX92 lacks the F-box-associated interaction domain, it is possible that this protein must interact with a protein containing an F-box-associated interaction domain to exert its function. In maize, there are 17 genes with an F-box-associated interaction domain, whereas there are 283 in Arabidopsis (bioinformatics.psb.ugent.be/plaza/). However, only two of the 17 genes in maize, GRMZM2G055789 and GRMZM2G083000, lack the F-box domain and thus are potential candidates for interaction with ZmFBX92 and possibly other F-box domain-ontaining proteins. This might explain why the number of F-box proteins in Arabidopsis is much larger, about double, than the number of F-box proteins identified in maize (Risseeuw et al. 2003, Jia et al. 2013). Moreover, more than half of the maize F-box proteins contain only the F-box domain without other known motifs (Jia et al. 2013), whereas in Arabidopsis only 14% of the F-box proteins have no additional domains (Risseeuw et al. 2003). Perhaps the lower number of F-box proteins in maize is partially compensated by the possibility of different combinations of F-box proteins and F-box-interacting domain proteins to target other proteins for degradation or play a role under other conditions.
The effects of altering AtFBX92 levels on leaf size were largely comparable in vitro and in soil under well-watered conditions. However, opposite effects on growth upon AtFBX92 overexpression were obtained under osmotic stress and drought stress. Under mild drought stress applied in soil, leaves of AtFBX92OE plants were larger than those of WT plants, whereas under standard or mild in vitro osmotic stress conditions, they grew more slowly than the WT. Although osmotica such as mannitol are often used to mimic drought stress responses, their relevance is debatable (Verslues et al. 2006, Lawlor 2013). Our data suggest that AtFBX92 might be involved in the mild drought stress response, but not in the osmotic stress response. It has often been seen that enhancing drought tolerance by altering gene expression levels, as shown here for AtFBX92OE plants, results in growth inhibition and a significant yield penalty (Yang et al. 2010). Leaves of amiFBX92 plants were significantly larger than those of the WT under standard conditions in vitro and in soil, and in stress conditions when grown on mannitol or under mild drought conditions. There was no significant difference in effect of both stresses compared with standard conditions, implying that AtFBX92 plays no role in the osmotic or drought stress response, in contrast to what is suggested by the results for AtFBX92OE plants. Expression levels of AtFBX92 do not vary significantly under mild drought stress and osmotic stress (Skirycz et al. 2011a, Clauw et al. 2015, Dubois et al. 2017), supporting that the observed enhanced drought tolerance in AtFBX92OE plants is rather an indirect effect of changed AtFBX92 levels. Alternatively, functionally redundant proteins may exist that shield the effect that reduced AtFBX92 levels have on drought tolerance.
Strikingly, leaves of AtFBX92OE plants grown in soil, under both standard and mild drought stress conditions, were folded downwards, whereas the shape of AtFBX92OE and WT leaves grown in vitro was indistinguishable. Possibly, epinasty of the leaf lamina in AtFBX92OE plants only appears in specific conditions, for instance under lower relative humidity or specific light conditions (Takemiya et al. 2005, de Carbonnel et al. 2010). Epinastic leaves result from a difference in cell division rate between abaxial and adaxial cells (Romano et al. 1995). In amiFBX92 plants, although no effect on leaf flattening was observed, we could show that the increased leaf size was due to an increased cell division rate. Also other genes affecting leaf development and final leaf size have been identified that influence leaf curling, e.g. BREVIS RADIX (Beuchat et al. 2010), several members of the TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factor family (Schommer et al. 2008), jaw-D (Palatnik et al. 2003), PEAPOD (Gonzalez et al. 2015) and TCP Interactor containing EAR motif protein1 (Tao et al. 2013), often linked to hormone biosynthesis or signaling. Potentially, AtFBX92 does not affect cell cycle genes directly, but by regulating hormone signaling.
In conclusion, we identified a new F-box gene that is important for vegetative growth. This gene acts as a negative regulator of growth, as indicated by the effect on leaf size when altering AtFBX92 levels, by affecting cell division rates and expression levels of cell cycle genes. Future experiments are needed to identify putative substrates that might have a role in the regulation of organ growth.
Materials and Methods
Cloning and generation of transgenic plants
The coding region of maize F-box protein GRMZM2G059799_T02 (ZmFBX92) was amplified with Phusion High-Fidelity DNA polymerase (Thermo Fischer Scientific) from cDNA generated from leaf tissue of the maize inbred line B73. The PCR fragment was introduced by recombination via the attBattP recombination sites into pDONR™221 using the Gateway system (Invitrogen Life Technologies). Next, the ZmFBX92 coding region and pBdEF1α (Coussens et al. 2012) were transferred to the binary vector pBbm42GW7 (Anami et al. 2010) (https://gateway.psb.ugent.be) using a multisite Gateway approach (pBdEF1α:ZmFBX92). Additionally, the ZmFBX92 coding region was also introduced in the binary vector pK7GW2 (https://gateway.psb.ugent.be), under control of the CaMV 35S promoter (pCaMV35S:ZmFBX92) for the generation of Arabidopsis transgenic plants (Karimi et al. 2007).
The closest orthologous gene of ZmFBX92 in Arabidopsis was identified using PLAZA (Proost et al. 2015) (https://plaza.psb.ugent.be) and the coding region of this gene, At3g07870 (AtFBX92), was amplified with Phusion High-Fidelity DNA polymerase (Thermo Fischer Scientific) from cDNA generated from leaf tissue of Arabidopsis ecotype Col-0. In addition, a fragment consisting of the first 492 bp starting from the ATG start codon, containing the F-box domain, was amplified by PCR for further cloning. The Gateway system was used to introduce the obtained PCR fragments via recombination into pDONr221 (Invitrogen Life Technologies), followed by recombination via the attLattR sites into binary vector pK7GW2 (https://gateway.psb.ugent.be) into which a cassette containing the seed-specific napin promoter (Ellerstr�m et al. 1996) driving GFP was introduced, further indicated as pK7GW2napin, to allow the selection of transgenic seeds based on GFP expression in the seed. The generated constructs, pCaMV35S:AtFBX92 and pCaMV35S:AtFBX92del, were subsequently transformed into Arabidopsis.
For silencing of AtFBX92, a pCaMV35S:AtFBX92-amiRNA construct was designed using the pRS300 plasmid as described before (Ossowski et al. 2008), inserted in pDONR™221 (Invitrogen Life Technologies) and sequenced. Next, the DNA construct was transferred to pK7GW2napin by recombination.
For analysis of the AtFBX92 promoter, a 1,362 bp fragment upstream of the ATG start codon was amplified with Phusion High-Fidelity DNA polymerase (Thermo Fischer Scientific) from Arabidopsis Col-0 genomic DNA, cloned into pDONR™221 (Invitrogen Life Technologies) and transferred to the pFAST-G04 binary vector (Shimada et al. 2010) (https://gateway.psb.ugent.be) to generate the pAtFBX92:GFP:GUS construct. Primers used for cloning are summarized in Supplementary Table S1.
pBdEF1α:ZmFBX92 was introduced into maize cultivar B104 by Agrobacterium tumefaciens transformation of immature embryos as described before (Coussens et al. 2012).
pCaMV35S:ZmFBX92, p35S:AtFBX92, p35S:AtFBX92del, p35S:AtFBX92-amiRNA and pAtFBX92:GFP:GUS constructs were transformed into A. tumefaciens strain C58C1 RifR harboring the plasmid pMP90, followed by transformation into Arabidopsis Col-0 using the floral dip protocol (Clough and Bent 1998).
Maize growth analysis
Maize plants were grown in controlled growth chamber conditions (24 �C, 55% relative humidity, light intensity of 170 mmol m−2 s−1 photosynthetic active radiation, in a 16 h/8 h day/night cycle). Pot weight was determined daily and water was added to 100% of the initial water content under well-watered conditions, and to 70% of the initial water content under mild drought stress conditions. Leaf size-related phenotypic observations were performed as described before (Baute et al. 2015).
Arabidopsis growth analysis
Arabidopsis plants were grown in vitro on round Petri dishes containing half-strength Murashige and Skoog medium supplemented with 1% (w/v) sucrose at 21 �C, light intensity of 70 mmol m−2 s−1 photosynthetic active radiation, in a 16 h/8 h day/night cycle. To subject plants to mild osmotic stress, seeds were germinated on medium containing 25 mM d-mannitol (Sigma-Aldrich) (Claeys et al. 2014).
Rosette growth over time was determined for plants grown in vitro at a density of one plant per 4 cm2 by photographing the plates three times a week from 5 to 6 DAS until 21–24 DAS (dependent on the experiment) and calculating the PRA with IMAGEJ software version 1.46 (http://rsb.info.nih.gov/ij/). Relative growth rates were calculated as the log of the PRA over time.
Arabidopsis growth analysis in soil was performed on the automated phenotyping platform WIWAM (Skirycz et al. 2011b) in a growth chamber under controlled conditions (21 �C, 55% relative humidity, light intensity of 100–120 mmol m−2 s−1 photosynthetic active radiation, in a 16 h/8 h day/night cycle). The water content of the soil was kept constant at 2.19 g water g−1 dry soil for control plants during the entire experiment. For mild drought-treated plants, from 10 DAS on, water was withheld until a soil water content of 1.19 g−1 water g−1 dry soil was reached, and kept at this level until 21 DAS. Images of the rosettes were taken daily from 6 DAS until 20 DAS, and PRA and relative growth rates were determined as explained before.
For rosette leaf area measurements, seven seedlings grown in vitro or in soil for 21 or 22 d (dependent on the experiment) were dissected and spread on agar plates according to their position in the rosette. Agar plates were photographed and individual leaf area was determined using IMAGEJ software.
Kinematic analysis
Kinematic analysis was performed as described before (De Veylder et al. 2001) on the first true leaf pair of 12 amiFBX92 and Col-0 plants grown in vitro from 5 DAS until 21 DAS. Briefly, leaves 1 and 2 were harvested daily, cleared in 100% ethanol, mounted in lactic acid on microscope slides and photographed. Leaf area of each leaf was measured using IMAGEJ. To determine the average cell area at each time point, 50–100 abaxial epidermal cells of 3–5 leaves were drawn with a Leica microscope fitted with a drawing tube and a differential interference contrast objective, and the average cell area was determined with IMAGEJ. From the leaf area and average cell area, cell numbers per leaf were calculated. Average cell division rates for the whole leaf were determined as the slope of the log 2-transformed cell number.
Using the same protocol, we determined the size, average cell number and average cell size of mature leaves 1 and 2 (amiFBX92, AtFBX92del and the respective Col-0) or leaf 3 (ZmFBX92OE, AtFBX92OE and respective the Col-0).
RNA extraction and qRT–PCR
Tissue for RNA extraction was flash-frozen in liquid nitrogen immediately upon harvest to avoid degradation. To analyze expression levels of the cell cycle genes in fully proliferative tissue, about 50 amiFBX92 and WT plants were sampled and flash-frozen at 7 and 8 DAS, after which cooled RNAlater-ICE (Ambion) was added to the samples. Samples were kept at −20 �C for 1 week to allow the RNAlater-ICE to penetrate the tissue. Leaves 1 and 2 were dissected under a binocular microscope while the samples were kept on dry ice before grinding.
Total RNA was extracted from frozen material with TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA samples were treated with RNase-free DNase I (Healthcare) to eliminate residual genomic DNA possibly present after RNA extraction. First-strand cDNA was synthesized starting from 1 μg of total RNA using iScript™ (Biorad) according to the manufacturer’s instructions. cDNA was amplified on a LightCycler480 (Roche Diagnostics) in 384-well plates with LightCycler 480 SYBR Green I Master (Roche) as described by the manufacturer. Gene-specific primers were designed with the Beacon Designer™ software and are summarized in Supplementary Table S1. The specificity of the amplification was determined by analyzing the melting curves. Normalization was done against the maize 18S rRNA or Arabidopsis ACTIN1 gene, and PCR efficiency was taken into account using geNorm (Vandesompele et al. 2002). Relative expression levels were calculated based on the ΔΔ cycle threshold method (Livak and Schmittgen 2001). Data presented for testing expression levels of AtFBX92 in AtFBX92OE, amiFBX92, AtFBX92del and ZmFBX92 in Arabidopsis and maize, respectively, were from triplicates. Data presented for cell cycle gene expression levels were from duplicates of three biological repeats. Values for the three biological repeats were used for statistical analysis.
Histochemical staining of GUS activity
pAtFBX92:GFP:GUS and pCYCB1;1:DB-GUS Arabidopsis plants grown in vitro were harvested daily from 5 to 13 DAS, followed by incubation in heptane for 5 min. After removal of the heptane, plants were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronide (X-Gluc) buffer [100 mM sodium phosphate pH 7, 10 mM EDTA, 0.5 mM K3Fe(CN)6,0.5 mM K4Fe(CN)6, 0.5 g l−1 X-gluc, 1% dimethylsulfoxide (DMSO)] and incubated for 6 h at 37 �C after vacuum infiltration for 10 min . Plants were cleared in 100% (v/v) ethanol until Chl was removed, and kept in 90% lactic acid. Samples were photographed under a differential interference contrast microscope (Leica).
Supplementary data
Supplementary data are available at PCP online.
Funding
The research leading to these results has received funding from the European Research Council (ERC) [under the European Community’s Seventh Framework Programme (FP7/2007–2013) under ERC grant agreement No. 339341-AMAIZE11]; from Ghent University [‘Bijzonder Onderzoeksfonds Methusalem project’ No. BOF08/01M00408]; and from the Interuniversity Attraction Poles Program [IUAP P7/29 ‘MARS’] initiated by the Belgian Science Policy Office.
Supplementary Material
Acknowledgments
We thank Dr. Annick Bleys and Karel Spruyt for help in preparing the manuscript. D.I. conceived and co-ordinated the study. J.B. designed the experiments. J.B., J.D., S.P. and J.Bl. carried out the experiments and interpreted the results. M.V. was responsible for maize transformation. J.B. wrote the manuscript with input from the other authors. All authors read and approved the final manuscript.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- amiRNA
artificial microRNA
- APC/C
anaphase-promoting complex/cyclosome
- BB
BIG BROTHER
- pBdEF1α
Brachypodium distachyon elongation factor1α promoter
- CaMV
Caulifower mosaic virus
- CDC27a
CELL DIVISION CYCLE PROTEIN 27 HOMOLOG A
- CDK
cyclin-dependent kinase
- CKI
cyclin-dependent kinase inhibitor
- DAS
days after stratification
- ICK/KRP
ICK/KIP/CIP-RELATED PROTEIN
- FBL17
F-BOX-LIKE17
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- KRP
KIP-RELATED PROTEIN
- PRA
projected leaf area
- qRT–PCR
quantitative reverse transcription–PCR
- SAM
shoot apical meristem
- SIM
SIAMESE
- SMR
SIM-related
- TCP
TEOSINTE BRANCHED1/CYCLOIDEA/PCF
- WT
wild type
References
- Achard P., Genschik P. (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60: 1085–1092. [DOI] [PubMed] [Google Scholar]
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19: 1188–1193. [DOI] [PubMed] [Google Scholar]
- An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anami S.E., Mgutu A.J., Taracha C., Coussens G., Karimi M., Hilson P., et al. (2010) Somatic embryogenesis and plant regeneration of tropical maize genotypes. Plant Cell Tissue Organ Cult. 102: 285–295. [Google Scholar]
- Andriankaja M., Dhondt S., De Bodt S., Vanhaeren H., Coppens F., De Milde L., et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev. Cell 22: 64–78. [DOI] [PubMed] [Google Scholar]
- Avramova V., Sprangers K., Beemster G.T.S. (2015) The maize leaf: another perspective on growth regulation. Trends Plant Sci. 20: 787–797. [DOI] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J.W., et al. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- Baute J., Herman D., Coppens F., De Block J., Slabbinck B., Dell’Acqua M., et al. (2015) Correlation analysis of the transcriptome of growing leaves with mature leaf parameters in a maize RIL population. Genome Biol. 16: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T.S., De Vusser K., De Tavernier E., De Bock K., Inz� D. (2002) Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 129: 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuchat J., Scacchi E., Tarkowska D., Ragni L., Strnad M., Hardtke C.S. (2010) BRX promotes Arabidopsis shoot growth. New Phytol. 188: 23–29. [DOI] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5: 739–751. [DOI] [PubMed] [Google Scholar]
- Churchman M.L., Brown M.L., Kato N., Kirik V., H�lskamp M., Inz� D., et al. (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18: 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H., Van Landeghem S., Dubois M., Maleux K., Inz� D. (2014) What is stress? Dose–response effects in commonly used in vitro stress assays. Plant Physiol. 165: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P., Coppens F., De Beuf K., Dhondt S., Van Daele T., Maleux K., et al. (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167: 800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coussens G., Aesaert S., Verelst W., Demeulenaere M., De Buck S., Njuguna E., et al. (2012) Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J. Exp. Bot. 63: 4263–4273. [DOI] [PubMed] [Google Scholar]
- de Carbonnel M., Davis P., Roelfsema M.R.G., Inoue S.-i., Schepens I., Lariguet P., et al. (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 152: 1391–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin–SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Manzano C. (2014) Auxin and the ubiquitin pathway. Two players–one target: the cell cycle in action. J. Exp. Bot. 65: 2617–2632. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T.S., Krols L., Terras F., Landrieu I., et al. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Inz� D. (2007) The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8: 655–665. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16: 272–279. [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Dubois M., Claeys H., Van den Broeck L., Inz� D. (2017) Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 40: 180–189. [DOI] [PubMed] [Google Scholar]
- Ellerstr�m M., St�lberg K., Ezcurra I., Rask L. (1996) Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol. Biol. 32: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Eloy N., de Freitas Lima M., Ferreira P.C.G., Inz� D. (2015) The role of the anaphase-promoting complex/cyclosome in plant growth. Crit. Rev. Plant Sci. 34: 487–505. [Google Scholar]
- Eloy N.B., de Freitas Lima M., Van Damme D., Vanhaeren H., Gonzalez N., De Milde L., et al. (2011) The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68: 351–363. [DOI] [PubMed] [Google Scholar]
- Eloy N.B., Gonzalez N., Van Leene J., Maleux K., Vanhaeren H., De Milde L., et al. (2012) SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proc. Natl. Acad. Sci. USA 109: 13853–13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., et al. (2014) Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes B.P., Shiu S.-H., Durski A.M., Vierstra R.D. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P., Marrocco K., Bach L., Noir S., Criqui M.-C. (2014) Selective protein degradation: a rheostat to modulate cell-cycle phase transitions. J. Exp. Bot. 65: 2603–2615. [DOI] [PubMed] [Google Scholar]
- Gonz�lez N., Inz� D. (2015) Molecular systems governing leaf growth: from genes to networks. J. Exp. Bot. 66: 1045–1054. [DOI] [PubMed] [Google Scholar]
- Gonzalez N., Pauwels L., Baekelandt A., De Milde L., Van Leene J., Besbrugge N., et al. (2015) A repressor protein complex regulates leaf growth in Arabidopsis. Plant Cell 27: 2273–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Vanhaeren H., Inz� D. (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 17: 332–340. [DOI] [PubMed] [Google Scholar]
- Gusti A., Baumberger N., Nowack M., Pusch S., Eisler H., Potuschak T., et al. (2009) The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H., Dissmeyer N., Schnittger A. (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23: 345–356. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67: 425–479. [DOI] [PubMed] [Google Scholar]
- Hotton S.K., Callis J. (2008) Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 59: 467–489. [DOI] [PubMed] [Google Scholar]
- Hua Z., Zou C., Shiu S.-H., Vierstra R.D. (2011) Phylogenetic comparison of F-box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS One 6: e16219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudik E., Yoshioka Y., Domenichini S., Bourge M., Soubigout-Taconnat L., Mazubert C., et al. (2014) Chloroplast dysfunction causes multiple defects in cell cycle progression in the Arabidopsis crumpled leaf mutant. Plant Physiol. 166: 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inz� D., De Veylder L. (2006) Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Jain M., Nijhawan A., Arora R., Agarwal P., Ray S., Sharma P., et al. (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143: 1467–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F., Wu B., Li H., Huang J., Zheng C. (2013) Genome-wide identification and characterisation of F-box family in maize. Mol. Genet. Genomics 288: 559–577. [DOI] [PubMed] [Google Scholar]
- Jurado S., D�az-Trivi�o S., Abraham Z., Manzano C., Gutierrez C., del Pozo C. (2008) SKP2A, an F-box protein that regulates cell division, is degraded via the ubiquitin pathway. Plant J. 53: 828–841. [DOI] [PubMed] [Google Scholar]
- Kakumanu A., Ambavaram M.M.R., Klumas C., Krishnan A., Batlang U., Myers E., et al. (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 160: 846–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007) Recombinational cloning with plant Gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Wang S., Li Y., Zaitlin D., Pierce A.J., Smalle J.A. (2009) Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 150: 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Takahashi N., Shimada H., Seki M., Shinozaki K., Matsui M. (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43: 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lawlor D.W. (2013) Genetic engineering to improve plant performance under drought: physiological evaluation of achievements, limitations, and possibilities. J. Exp. Bot. 64: 83–108. [DOI] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. (2006) F-box proteins everywhere. Curr. Opin. Plant Biol. 9: 631–638. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Ko J.-H., Lee S., Lee Y., Pak J.-H., Kim J.H. (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151: 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008) Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Manzano C., Ramirez-Parra E., Casimiro I., Otero S., Desvoyes B., De Rybel B., et al. (2012) Auxin and epigenetic regulation of SKP2B, an F-box that represses lateral root formation. Plant Physiol. 160: 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco K., Bergdoll M., Achard P., Criqui M.-C., Genschik P. (2010) Selective proteolysis sets the tempo of the cell cycle. Curr. Opin. Plant Biol. 13: 631–639. [DOI] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A.H. (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53: 423–442. [DOI] [PubMed] [Google Scholar]
- Noir S., Marrocco K., Masoud K., Thomann A., Gusti A., Bitrian M., et al. (2015) The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. Plant Cell 27: 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., et al. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Powell A.E., Lenhard M. (2012) Control of organ size in plants. Curr. Biol. 22: R360–R367. [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inz� D., Mueller-Roeber B., et al. (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43: D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Chang K.N., Yazaki J., Ecker J.R. (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23: 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Santner A., del Pozo J.C., Murray J.A.H., Estelle M. (2008) Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 53: 705–716. [DOI] [PubMed] [Google Scholar]
- Richard C., Granier C., Inz� D., De Veylder L. (2001) Analysis of cell division parameters and cell cycle gene expression during the cultivation of Arabidopsis thaliana cell suspensions. J. Exp. Bot. 52: 1625–1633. [PubMed] [Google Scholar]
- Risseeuw E.P., Daskalchuk T.E., Banks T.W., Liu E., Cotelesage J., Hellmann H., et al. (2003) Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34: 753–767. [DOI] [PubMed] [Google Scholar]
- Rojas C.A., Eloy N.B., Lima M.d.F., Rodrigues R.L., Franco L.O., Himanen K., et al. (2009) Overexpression of the Arabidopsis anaphase promoting complex subunit CDC27a increases growth rate and organ size. Plant Mol. Biol. 71: 307–318. [DOI] [PubMed] [Google Scholar]
- Romano C.P., Robson P.R.H., Smith H., Estelle M., Klee H. (1995) Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27: 1071–1083. [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Ch�telat A., Cubas P., Farmer E.E., et al. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann N., Navarro-Quezada A., Ullrich K., Kuhl C., Quint M. (2011) Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol. 155: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J., Roberts J.M. (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Skaar J.R., Pagan J.K., Pagano M. (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Claeys H., De Bodt S., Oikawa A., Shinoda S., Andriankaja M., et al. (2011a) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Vandenbroucke K., Clauw P., Maleux K., De Meyer B., Dhondt S., et al. (2011b) Survival and growth of Arabidopsis plants given limited water are not equal. Nat. Biotechnol. 29: 212–214. [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590. [DOI] [PubMed] [Google Scholar]
- Takemiya A., Inoue S.-i., Doi M., Kinoshita T., Shimazaki K.-i. (2005) Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., et al. (2013) The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002) Accurate normalization of real-time quantitative RT–PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: research0034–research0034.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A., Weinl C., Inz� D., De Veylder L., Schnittger A. (2005) Switching the cell cycle. Kip-related proteins in plant cell cycle control. Plant Physiol. 139: 1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Agarwal M., Katiyar-Agarwal S., Zhu J., Zhu J.-K. (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45: 523–539. [DOI] [PubMed] [Google Scholar]
- Voorend W., Lootens P., Nelissen H., Rold�n-Ruiz I., Inz� D., Muylle H. (2014) LEAF-E: a tool to analyze grass leaf growth using function fitting. Plant Methods 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li N., Jiang S., Gonzalez N., Huang X., Wang Y., et al. (2016) SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat. Commun. 7: 11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., et al. (2013) The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Jang J.-C. (2000) F-box proteins in Arabidopsis. Trends Plant Sci. 5: 454–457. [DOI] [PubMed] [Google Scholar]
- Yang S., Vanderbeld B., Wan J., Huang Y. (2010) Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol. Plant 3: 469–490. [DOI] [PubMed] [Google Scholar]
- Zhao X.A., Harashima H., Dissmeyer N., Pusch S., Weimer A.K., Bramsiepe J., et al. (2012) A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genet. 8: e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.