SUMMARY
The malaria parasite exports numerous proteins into its host red blood cell (RBC). The trafficking of these exported effectors is complex. Proteins are first routed through the secretory system, into the parasitophorous vacuole (PV), a membranous compartment enclosing the parasite. Proteins are then translocated across the PV membrane in a process requiring ATP and unfolding. Once in the RBC compartment the exported proteins are then refolded and further trafficked to their final localizations. Chaperones are important in the unfolding and refolding processes. Recently, it was suggested that the parasite TRiC chaperonin complex is exported, and that it is involved in trafficking of exported effectors. Using a parasite-specific antibody and epitope-tagged transgenic parasites we could observe no export of Plasmodium TRiC into the RBC. We tested the importance of the parasite TRiC by creating a regulatable knockdown line of the TRiC-θ subunit. Loss of the parasite TRiC-θ led to a severe growth defect in asexual development, but did not alter protein export into the RBC. These observations indicate that the TRiC proteins play a critical role in parasite biology, though their function, within the parasite, appears unrelated to protein trafficking in the RBC compartment.
Keywords: malaria, TRiC, T-complex protein, chaperonin, protein export, regulatable expression
INTRODUCTION
The most virulent species of Plasmodium that causes human malaria, P. falciparum, exports several hundred proteins into its host cell (reviewed extensively in Boddey and Cowman, 2013; Spillman et al., 2015). Some proteins are injected into the RBC during invasion, via release of the apical secretory organelles. However, the majority of exported proteins are synthesized in the parasite post-invasion and trafficked through the secretory system, where they cross the parasite plasma membrane and are deposited in the parasitophorous vacuole (PV; a membranous compartment encompassing the parasite within the RBC). A putative translocon has been implicated in the trafficking of proteins across the PV membrane (Beck et al., 2014; Elsworth et al., 2014; Mesen-Ramirez et al., 2016). The Plasmodium translocon of exported proteins (PTEX), is a macromolecular complex comprising the proteins EXP2, Heat Shock Protein 101 (HSP101), thioredoxin 2 (TRX2), PTEX88 and PTEX150 (de Koning-Ward et al., 2009; Bullen et al., 2012). The exact mechanism of translocation has not been established, but the process of crossing the PV membrane has been shown to require both ATP (Ansorge et al., 1996) and unfolding (Gehde et al., 2009; Heiber et al., 2013; Mesen-Ramirez et al., 2016). In the PV this unfolding may involve PV-resident chaperones, such as HSP101 (Beck et al., 2014), or HSP70-x (Kulzer et al., 2012; Grover et al., 2013). Following translocation, chaperones are also required, as the exported proteins need to be refolded and delivered to their final locations within the erythrocyte (Maier et al., 2009).
P. falciparum parasites export several chaperones into the RBC, including HSP70-x (Kulzer et al., 2012) and numerous co-chaperone HSP40 proteins (Kulzer et al., 2010; Kulzer et al., 2012; Petersen et al., 2016). The HSP40s are thought to modulate HSP70 function, and a direct interaction between one of the parasite HSP40 co-chaperones (PFA0660w) and HSP70-x has been demonstrated (Daniyan et al., 2016). Some of the HSP40s are targeted to chaperone-rich, mobile, membranous structures in the RBC cytoplasm, called J-dots (Kulzer et al., 2010). J-dots are only present in P. falciparum infection (Petersen et al., 2016), and have been implicated in the trafficking of the important P. falciparum virulence factor, Erythrocyte Membrane Protein 1 (PfEMP1) (Kulzer et al., 2010). In addition to these parasite chaperones, human proteins have also been implicated in trafficking of P. falciparum exported proteins (Ansorge et al., 1996; Banumathy et al., 2002; de Koning-Ward et al., 2009). The human HSP70 (HSPA1A) relocalizes to the erythrocyte plasma membrane upon parasite infection, in close proximity to areas where the PfEMP1 virulence factor is displayed (Banumathy et al., 2002). The relative importance and interactions between human and parasite chaperones is not well understood.
Recently, another chaperone class, called TRiC (T-complex protein 1 (TCP-1) ring complex; also known as chaperonin containing TCP-1 (CCT)), was implicated in the export of P. falciparum proteins (Mbengue et al., 2015). PfTRiC was identified in punctate structures throughout the infected RBC (iRBC) cytosol, and was suggested to be involved in trafficking from the PV to the Maurer’s clefts (parasite-derived membranous structures thought to be involved in protein sorting to the erythrocyte cytoskeleton/membrane) (Mbengue et al., 2015). TRiC is a class II chaperonin, which typically forms a high molecular weight heterohexadecamer, comprised of two eight-membered rings (Spiess et al., 2004; Hartl et al., 2011). Each ring consists of eight different (though very closely related) subunits, and this complexity is thought to enable the chaperonin to interact with a large range of diverse substrates, including actin (Gao et al., 1992) and tubulin (Yaffe et al., 1992). All eight subunits have been identified in the P. falciparum genome (Mbengue et al., 2015; Olshina et al., 2016). Additionally, proteomics studies have identified human TRiC in mature RBC (Wagner et al., 2004; Pasini et al., 2006).
Here, we investigate the localization, oligomerization and essentiality of the Plasmodium TRiC. We show that PfTRiC forms a large complex in the parasite cytosol, at the estimated size of the predicted heterohexadecamer. Using a PfTRiC-specific antibody and epitope tagged PfTRiC subunits we find no biochemical evidence that the PfTRiC is exported into the RBC compartment. Additionally, we show that knock-down of the PfTRiC-θ subunit is lethal, and determine that the lethality is not due to a change in protein export into the RBC. Collectively, this study is consistent with an essential role for PfTRiC within the parasite compartment.
RESULTS
Anti-human TRiC antibodies specifically detect soluble RBC TRiC subunits, but not parasite TRiC subunits
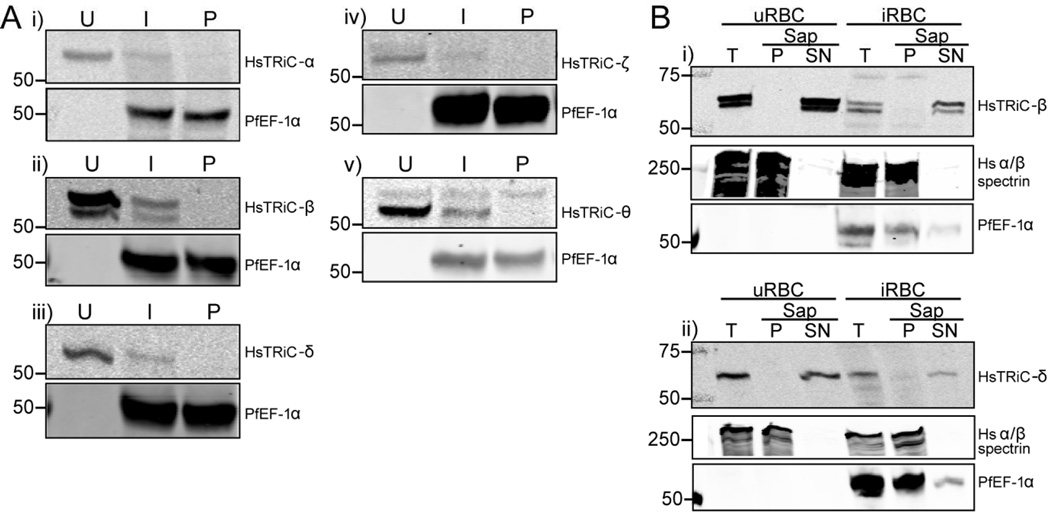
A previous RBC proteome study detected peptides against TRiC subunits in the soluble RBC fraction (Pasini et al., 2006). To confirm that human TRiC subunits are present in the RBC, total lysates were prepared from uninfected RBC (uRBC) and probed with antibodies against human TRiC subunits α (predicted size/s 60.3 kDa), β (57.5 and 52.7 kDa), δ (57.9 and 54.7 kDa), ζ (58.0 and 53.2 kDa) and θ (59.6, 57.6 and 51.6kDa)(Fig. 1A and Supplemental Fig. 1). All subunits examined were detected at their predicted full-length molecular weight, and additionally the prediction of an alternative splice isoform of the human β subunit was experimentally confirmed (sizes and splice site prediction from the UniProtKB database; UniProt, 2015). The alternative splice isoforms predicted for the δ, ζ or θ subunits were not observed in RBC. We hypothesized that the human antibodies may cross-react with the PfTRiC subunits, resulting in an increased signal in the infected RBC (iRBC) compared to uRBC. However, there was decreased signal for all five subunits examined (Fig. 1A), probably due to the extensive ingestion and degradation of the RBC cytosol during the parasite’s residence in a host RBC (Goldberg, 2005). No signal was detected in isolated parasites (Fig. 1A), further highlighting the specificity of the anti-human TRiC subunit antibodies.
Figure 1. RBC contain TRiC subunits in the cytosolic fraction.
A, Lysates from uninfected RBC (U), trophozoite-stage infected RBC (I) or saponin isolated parasites (P) were extracted with RIPA buffer and probed with antibodies against human TRiC subunits i) α, ii) β, iii) δ, iv) ζ and v) θ. PfEF-1α is a cytosolic parasite protein. B, Uninfected (uRBC) or infected (iRBC) RBC samples (T = total sample) were treated with 0.05 % saponin (20 s, room temperature), releasing the soluble contents of the RBC and parasitophorous vacuole (SN = supernatant fraction), leaving the insoluble pellet (P = pellet fraction). Human α/β spectrin is a control for the insoluble RBC fraction, and PfEF-1α is a soluble parasite protein. Equivalent fractions were loaded in all blots. Immunoblots are representative of two-four independent experiments. Marker bands are in kDa.
Fractionation, using saponin to release the soluble contents of the RBC and PV, revealed that the human TRiC-β and -δ subunits were in the soluble RBC fraction (Fig. 1B). Upon heat stress, human TRiC-α translocates from the cytosolic fraction and associates with the cytoskeleton (Wagner et al., 2004). We hypothesized that the ‘stress’ of parasite infection may lead to translocation of human TRiC subunits from the soluble to insoluble fraction. However, the human TRiC-β and -δ subunits remained in the soluble cytosolic fraction in iRBC (Fig. 1B). These results confirm that human TRiC subunits are present in the cytosol of RBC, and demonstrate that this localization is unchanged upon parasite infection.
Parasite PfTRiC-θ antibody recognizes a high molecular weight complex in the parasite cytosol
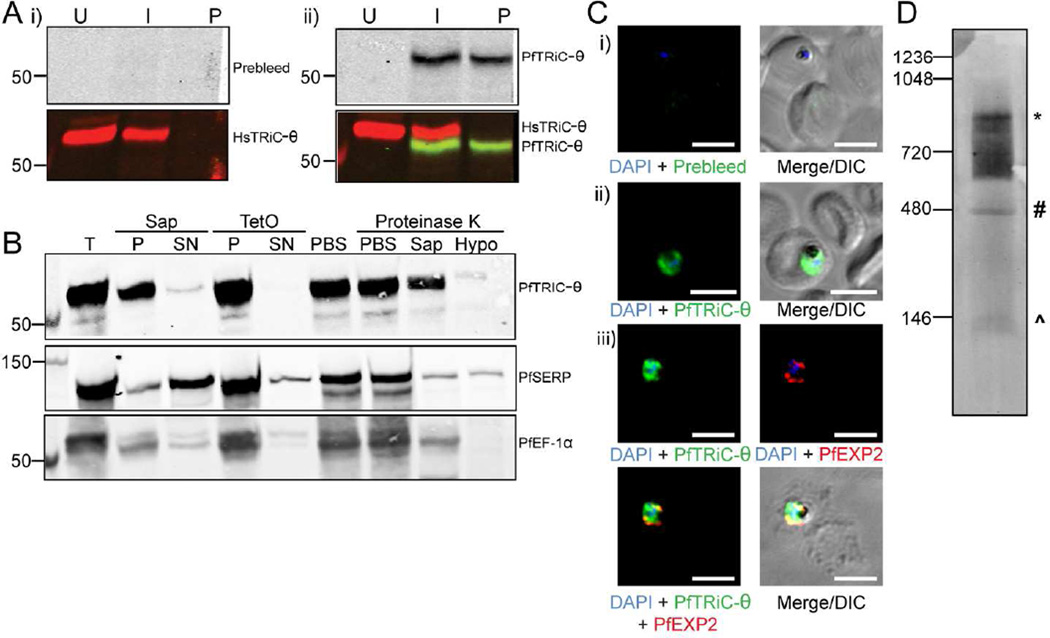
To determine whether the previously reported anti-PfTRiC-θ antibody was specifically detecting the parasite θ subunit, total lysates were prepared from uRBC, iRBC and saponin-isolated parasites (a term henceforth used to describe parasites where the RBC membrane and PV are permeabilized but the parasite membrane remains intact), and immunoblots were probed with anti-PfTRiC-θ. As previously reported (Mbengue et al., 2015), the preimmune serum did not recognize any proteins in Plasmodium lysates, and the PfTRiC-θ antiserum recognized a single band at ∼60 kDa (consistent with the predicted mass of 60.9 kDa)(Fig. 2A). The human and Plasmodium anti-TRiC-θ antibodies recognized distinct bands, highlighting the specificity of each antibody. The human antibody (expected size 59.6 kDa) recognized a band at a higher molecular weight than the Plasmodium antibody (expected size 60.9 kDa), suggesting that there may be some posttranslational modification, or alternative splicing leading to the sizes observed on the SDS-PAGE. Fractionation, using saponin to release the soluble contents of the RBC and PV, and tetanolysin O (TetO) to release the soluble contents of the RBC (leaving the PV intact) revealed that PfTRiC-θ was in the pellet fraction of both assays, consistent with a localization within the parasite compartment (Fig. 2B). Furthermore, a protease protection assay showed that PfTRiC-θ was in the parasite compartment (Fig. 2B). There was a minor component of PfTRiC-θ in the saponin-soluble fraction, and a small loss of PfTRiC-θ in the saponin-containing limb of the protease protection assay. This is likely due to some lysis of the parasite during the saponin treatment (there was also a small loss of the soluble parasite protein PfEF-1α).
Figure 2. PfTRiC-θ forms a high molecular weight complex in the parasite cytosolic fraction.
A, Lysates from uninfected RBC (U), trophozoite-stage infected RBC (I) or saponin-isolated parasites (P) were extracted with RIPA buffer and probed with antibodies against human TRiC-θ and i) prebleed and ii) immune sera generated against PfTRiC-θ. B, Infected RBC samples (T = total sample) were fractionated with 0.05 % saponin (Sap; 20 s, room temperature) or 1 HU of TetO (20 min, 37 °C). PfSERP is a soluble parasitophorous vacuole protein, released into the saponin-soluble fraction (SN= supernatant), but remaining in the TetO-insoluble fraction (P = pellet). PfEF-1α is a soluble parasite protein, remaining in the saponin- and TetO-insoluble fractions. A proteinase K accessibility assay (last four lanes of blot) was performed with TetO-insoluble fractions from trophozoite-stage infected RBC. The sample was divided into four and incubated in PBS (two samples), 0.05 % saponin, or a hypotonic lysis solution (Hypo). Samples were treated with 1 mg/mL Proteinase K (30 min, on ice) as indicated. Immunoblots, indicating that the protease gains access to PfTRiC-θ after hypotonic lysis, are representative of two independent experiments. C, Immunofluorescence assay (IFA) of paraformaldehyde-fixed (i and ii) or acetone-fixed (iii) trophozoite-infected RBC. DAPI stains the parasite nuclei and PfEXP2 delineates the parasite PV compartment. DIC = differential interference contrast. Scale bars = 5 µm. Data are representative of three independent assays. D, Saponin-isolated parasites were permeabilized with digitonin, the soluble fraction was separated by blue-native polyacrylamide gel electrophoresis and the blot was probed with anti-PfTRiC-θ. * = expected size of the heterohexadecamer. # = expected size of heterooctamer. ^ = expected size of monomers. Immunoblots are representative of three independent assays. Equivalent fractions were loaded in all blots. Marker bands are in kDa.
IFA was performed with the PfTRiC-θ antibodies in both aldehyde- and acetone-fixed iRBC. In contrast to the previous report (Mbengue et al., 2015), we never observed any fluorescence in the RBC compartment, with strong signal only present throughout the parasite (Fig. 2C). The fluorescence pattern was not always evenly distributed in the cytosol, although it was difficult to distinguish if this was due to the fixation process, or whether it represents areas of higher PfTRiC-θ distribution within the parasite.
In other organisms, TRiC proteins function in a high molecular weight complex. The oligomerization state of PfTRiC-θ was investigated in the parasite, using Blue Native PAGE analyses. Examining a soluble lysate from saponin-isolated parasites, multiple complexes were identified using the PfTRiC-θ antiserum. The highest distinct band corresponds to the size of the predicted heterohexadecamer (957.2 kDa), and an additional band at the size of the heterooctamer (478.6 kDa) was also observed (Fig. 2D). A smear < 146 kDa likely corresponds to monomeric PfTRiC-θ. There was also a smear from ∼550 kDa to the size of the heterohexadecamer. From this experiment it is clear that PfTRiC-θ can form high molecular weight complexes in the parasite soluble fraction.
C-terminally tagged PfTRiC subunits are not exported
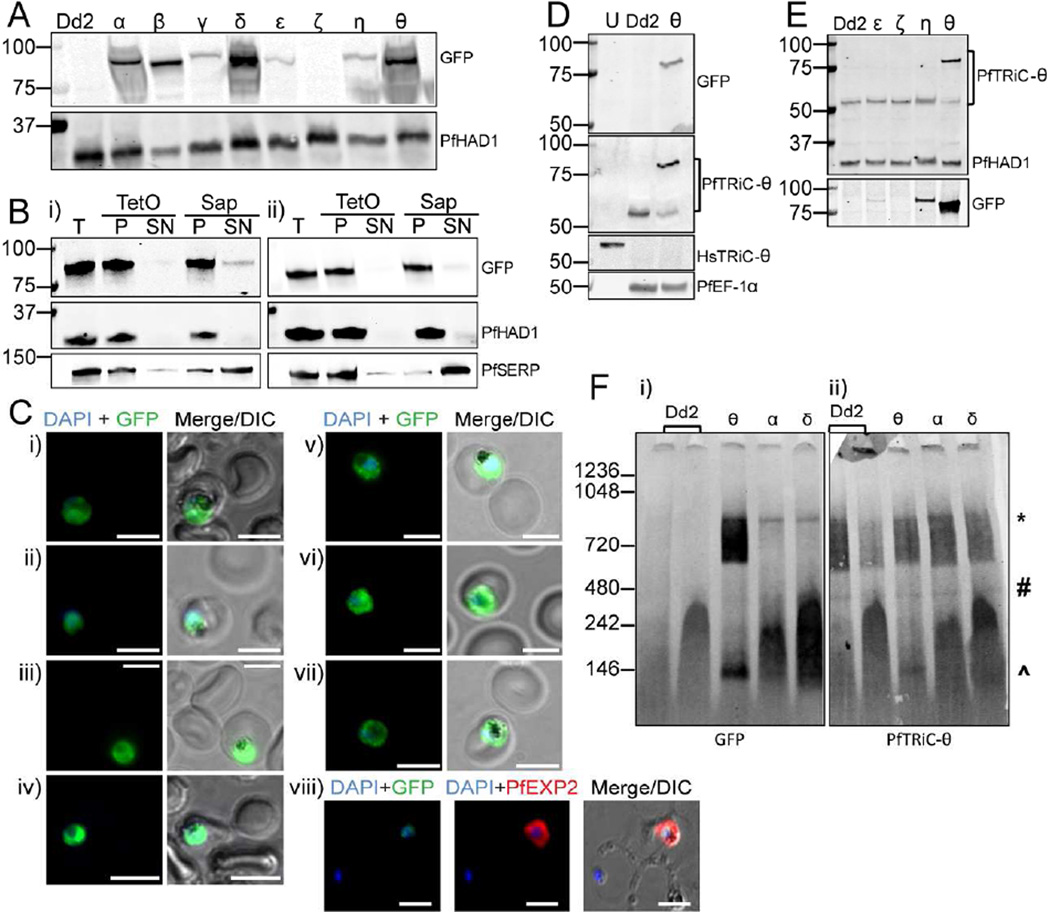
As our localization data was different to previously published results, despite using the same polyclonal PfTRiC-θ antibody, we sought to confirm the results obtained in Fig. 2 using epitope tagged PfTRiC subunits. We ectopically overexpressed each of the PfTRiC subunits, under the expression of the strong, constitutively active HSP86 promoter. Tagging the TRiC subunits at the N-termini with the large reporter GFP may result in disruption of heterohexadecamer formation (Spiess et al., 2015), thus to avoid this we C-terminally tagged the subunits, including a ten amino acid flexible linker sequence (PRPGAAHYAA) between the TRiC C-terminus and GFP (Klemba et al., 2004). All GFP-tagged subunits were successfully overexpressed, except the PfTRiC-ζ, where the parasites returned from transfection after selection (from N = 6 transfections) but GFP signal was never observed (Fig. 3A). Despite being expressed under control of the same promoter, the PfTRiC-α, -δ and –θ were more highly expressed than the PfTRiC-β, -γ, -ε or –η subunits. This may reflect mechanisms for regulation of the degree of overexpression achievable, or could be due to the constructs integrating (via piggyBac transposon-mediated integration (Balu et al., 2005)), in different genomic locations.
Figure 3. GFP-tagged PfTRiC subunits also form high molecular weight complexes in the parasite cytosolic fraction.
A, Ectopic expression of GFP-tagged PfTRiC subunits under the control of the strong, constitutive HSP86 promoter. Immunoblots show successful overexpression of all subunits, except PfTRiC-ζ. Dd2 is the parent parasite line and PfHAD1 is a loading control. B, Infected RBC samples (T = total sample) from the i) PfTRiC-η-GFP line and ii) PfTRiC-θ-GFP line were fractionated (SN = supernatant, P = pellet) with 0.05 % saponin (Sap; 20 s, room temperature) or 1 HU of TetO (20 min, 37 °C). PfSERP is an integrity marker for the parasitophorous vacuole and PfHAD1 is an integrity marker for the parasite compartment. C, IFA of paraformaldehyde-fixed RBC infected with PfTRiC-GFP lines i) α, ii) β, iii) γ, iv) δ, v) ε, vi) η and vii) θ. IFA was also performed on acetone-fixed RBC infected with PfTRiC-θ-GFP parasites probed with anti -PfTRiC-θ, and PfEXP2 to delineate the parasitophorous vacuole (viii). DAPI stains the parasite nuclei. Scale bars = 5 µm. Data are representative of two independent assays. D, Lysates from uninfected RBC (U), saponin-isolated trophozoite-stage infected RBC (Dd2; parent) or saponin-isolated PfTRiC-θ-GFP-infected RBC were probed with GFP or PfTRiC-θ antibodies. Parasite EF-1α is a parasite control marker, and HsTRiC-θ is the RBC control marker. E, Lysates from saponin-isolated PfTRiC-GFP parasites, and Dd2 parent parasites, probed with PfTRiC-θ antibody. GFP and PfHAD1 are loading controls. F, Saponin-isolated parasites (PfTRiC-α,δ and θ-GFP) were digitonin permeabilized, the soluble fraction was separated by blue-native polyacrylamide gel electrophoresis and the blot was probed with anti-GFP and anti-PfTRiC-θ. * = expected size of the heterohexadecamer. # = expected size of heterooctamer. ^ = expected size of monomers. Immunoblots are representative of three independent assays. Equivalent fractions were loaded in all blots. Marker bands are in kDa.
Fractionation of the PfTRiC-η-GFP (Fig. 3Bi) and -θ-GFP (Fig. 3Bii) lines revealed that these TRiC subunits were in the saponin and TetO pellet fractions, consistent with localization in the parasite compartment, as seen with the PfTRiC-θ antibody. IFA of all seven GFP-tagged lines was also consistent with cytoplasmic localization (Fig. 3C). Consistent with this, evaluation of the full-length immunoblot from Fig. 3A (Supplemental Fig. 2) failed to reveal a ∼ 27kDa GFP ‘core’. Exported proteins often display a proteolysis product (Boddey et al., 2016; Mesen-Ramirez et al., 2016; Spillman et al., 2016), due to endocytosis of exported proteins during residence in the PV and proteolysis down to the tightly folded GFP/tag in the digestive vacuole.
We probed lysates of the PfTRiC-θ-GFP parasites with both anti-GFP antibody (detecting the tagged protein) and the PfTRiC-θ antibody (detecting both the native untagged and GFP tagged proteins). The dominant species in the PfTRiC-θ-GFP parasite line was the GFP-tagged protein, with decreased expression of untagged PfTRiC-θ, compared to the Dd2 parent line (Fig. 3D). The level of untagged PfTRiC-θ was reduced to 38 ± 8 % (N = 3; mean ± S.E.M.) of that in the parent line and other GFP-tagged PfTRiC lines. In contrast, there was no downregulation of native PfTRiC-θ in parasite lines overexpressing other GFP-tagged subunits (Fig. 3E). These results suggest there may be a feedback mechanism regulating PfTRiC-θ protein expression from the endogenous locus. This has been observed before for the Chloroquine Resistance Transporter (CRT) locus, where overexpression of a MYC-tagged vivax CRT protein in P. falciparum reduced expression of the native falciparum untagged CRT protein; (Sa et al., 2006).
The localization of the PfTRiC-GFP-tagged lines, by biochemical fractionation and IFA, is only relevant if the GFP-tagged subunits can function like the native TRiC subunits. To investigate this, the ability of the GFP-tagged lines to form higher molecular weight complexes was examined, using blue native PAGE. The three most abundant PfTRiC-GFP-tagged lines, the -α, -δ, and –θ subunits, all formed a high molecular weight complex at the predicted size of the heterohexadecamer (Fig. 3F). As was seen with the PfTRiC-θ antibody, the PfTRiC-θ-GFP subunit formed a series of high molecular weight species, from ∼550 kDa to the size of the heterohexadecamer. This high molecular weight smear was observed with both the native and GFP-tagged protein, and observed using anti-GFP and anti-PfTRiC-θ antibodies. These results are consistent with the PfTRiC-θ forming alternative complexes compared to the other PfTRiC subunits.
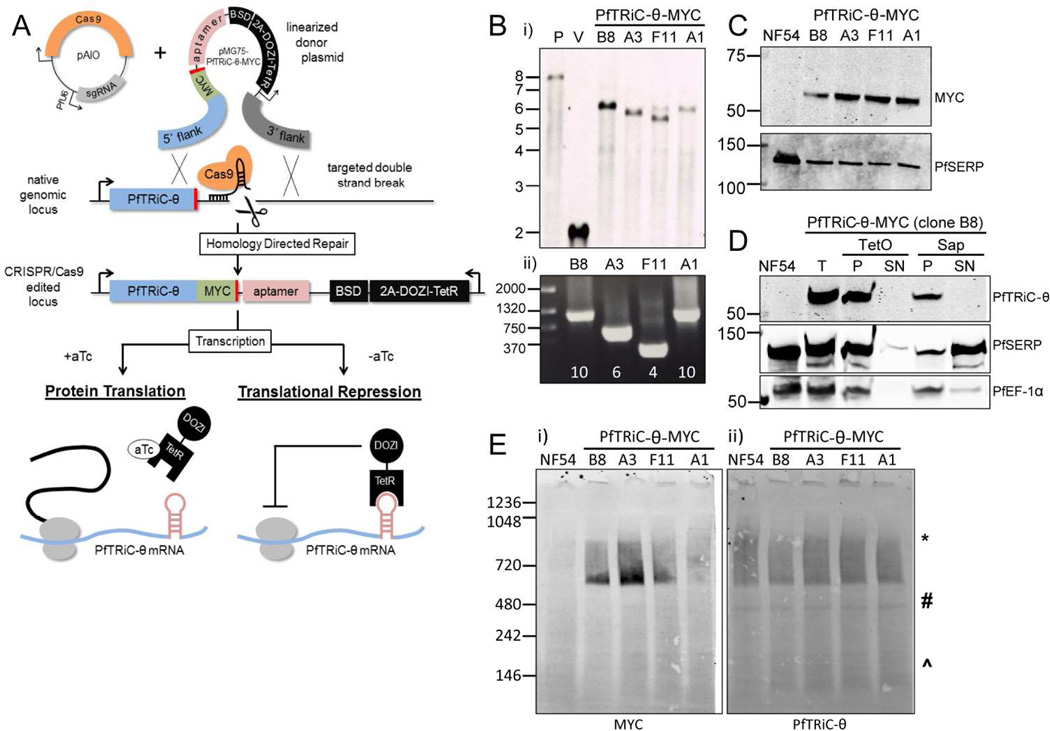
Creating a regulatable PfTRiC-θ line
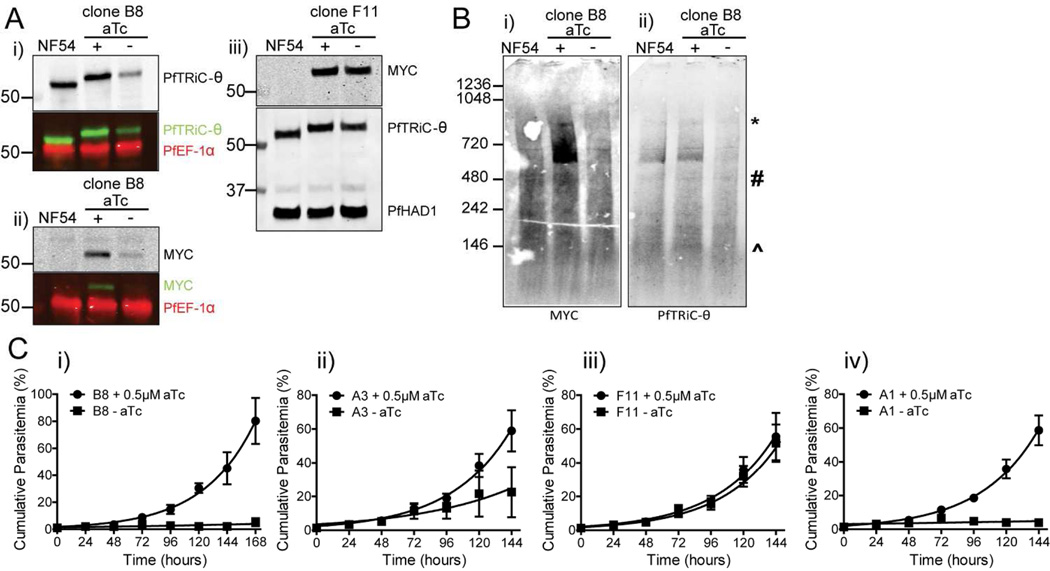
To study the importance of the PfTRiC proteins in asexual parasites we attempted to knockout PfTRiC-α and -ζ subunits using double homologous recombination. Despite using a CRISPR/Cas9 directed strategy to target the desired loci, no knockout parasites could be obtained (data not shown). As the parasites are haploid, to study essential genes it is necessary to use conditional expression approaches. A 10 x repeat Tet-repressor-binding aptamer array was introduced in the 3’ UTR of pftric-θ (Fig. 4A)(Ganesan et al., 2016). A Tet repressor (TetR)-DOZI (development of zygote inhibited) fusion is also encoded on the same plasmid. When aTc is present it binds the TetR-DOZI and expression of PfTRiC-θ is unaffected. However, when aTc is removed, TetR-DOZI binds the aptamer array and the RNA-modifying ability of DOZI can target the mRNA for sequestration/degradation (Fig. 4A)(Ganesan et al., 2016). Using CRISPR/Cas9, we successfully integrated a 3’ MYC tag/aptamer array at the PfTRiC-θ locus, confirmed by Southern blotting (Fig. 4Bi). Further genetic characterization of this locus revealed that the 10-element aptamer array had spontaneously truncated in several of our independent clones, and analytical PCR across this region suggested that the PfTRiC-θ-MYC clones retained between 4 and 10 aptamer elements (Fig. 4Bii). Western blotting confirmed that the PfTRiC-θ-MYC clones expressed a MYC-tagged protein of the correct molecular weight (Fig. 4C), and fractionation of PfTRiC-θ-MYC (using clone B8, which retains a 10-element aptamer array) demonstrated that the PfTRiC-θ-MYC protein was in the saponin and TetO pellet fractions (Fig. 4D), consistent with localization in the parasite compartment. PfTRiC-θ-MYC tagged proteins formed the same higher molecular weight complexes (Fig. 4E; ∼550 kDa to the size of the heterohexadecamer) as we observed with the wild type PfTRiC-θ, and GFP-tagged versions of the protein.
Figure 4. Generation of PfTRiC-θ-MYC aptamer-tagged lines.
A, Schematic outlining the strategy for tagging the 3’ end of PfTRiC-θ with a MYC epitope tag, and introducing an exogenous 3’ UTR containing an aptamer array. The desired integration event was achieved using CRISPR/Cas9 editing, using an “All-In-One” vector (pAIO) expressing both the sgRNA and Cas9 endonuclease. When anhydrotetracycline (aTc) is present it binds the Tet repressor (TetR), allowing translation of the PfTRiC-θ mRNA. When aTc is removed, the TetR binds the aptamer element in the 3’UTR, and the DOZI modifier targets the mRNA for sequestration/degradation, preventing translation of the PfTRiC-θ protein. B, i) Southern blot of digested vector (V) and genomic DNA from the NF54 parental line (P) and four tagged clones (probed with the 5’ homologous flank DNA), showing the expected integration event. Expected sizes are 7.9 kb for the parent, 2.0 kb for the vector and 6.0 kb for the integrants. Marker bands are in kb. ii) PRC screening using primers flanking the aptamer array, indicating the presence of a 10-element array in clones B8 and A1, 6-element array in clone A3 and 4-element array in F11. Marker bands are in bp. C, Lysates from NF54 and four PfTRiC-θ-MYC-tagged clones were prepared in RIPA buffer and immunoblots probed with anti-MYC antibody. PfSERP is a loading control. Immunoblots are representative of two independent experiments. Marker bands are in kDa. D, Infected RBC samples (T = total sample) from the PfTRiC-θ-MYC clone B8 parasite line were fractionated (SN = supernatant, P = pellet) with 0.05 % saponin (Sap; 20 s, room temperature) or 1 HU of TetO (20 min, 37 °C). PfSERP and PfEF1- α are integrity markers for the PV and parasite compartments, respectively. E, Saponin-isolated PfTRiC-θ-MYC clones were permeabilized with digitonin, the soluble fraction was separated by blue-native polyacrylamide gel electrophoresis and the blot was probed with anti-MYC and anti-PfTRiC-θ. * = expected size of the heterohexadecamer. # = expected size of heterooctamer. ^ = expected size of monomers. Equivalent fractions were loaded in all blots. Immunoblots are representative of two independent assays. Marker bands are in kDa.
The degree of regulation of expression correlates with the number of aptamers in the array, and knockdown reveals that PfTRiC-θ is essential for asexual parasite growth
Introduction of the aptamer array in the 3’ UTR may alter the basal expression of the target protein (independently of TetR-DOZI regulation)(Ganesan et al., 2016). This was not the case in the PfTRiC-θ-MYC parasites, as probing with the PfTRiC-θ antibody revealed similar protein expression between the NF54 parent line and PfTRiC-θ-MYC parasites (Fig. 5Ai-iii; expression of PfTRiC-θ-MYC in clone B8 was 105 ± 11 % of NF54, N = 2; mean ± range/2). When aTc was removed from ring-stage parasites for 24 h from the PfTRiC-θ-MYC clone B8 parasites (a clone with a 10-element array) there was a significant decrease in PfTRiC-θ-MYC expression (Fig. 5Ai,ii). PfTRiC-θ-MYC levels in clone B8 were reduced to 21 ± 5 % of NF54 levels (N = 4; mean ± S.E.M) In contrast, when aTc was removed for 24 h from the PfTRiC-θ-MYC clone F11 parasites (a clone with only a 4-element array), regulation of PfTRiC-θ-MYC levels was modest (Fig. 5Aiii). In this clone, knockdown of PfTRiC-θ-MYC levels to ∼49 ± 5 % of NF54 levels was observed (N = 4; mean ± S.E.M).
Figure 5. PfTRiC-θ is essential for parasite growth, and loss of the TRiC-θ subunit results in loss of the high molecular weight complex.
A, Lysates from NF54 and PfTRiC-θ-MYC-tagged clone B8, grown ± 0.5 µM aTc for 24 h, were prepared in RIPA buffer and immunoblots probed with i) anti- PfTRiC-θ antibody or ii) anti-MYC antibody. PfEF-1α is a loading control. In iii), lysates were prepared from the PfTRiC-θ-MYC-tagged clone F11 line. PfHAD1 is a loading control. Cell number equivalent fractions were loaded in all blots. Immunoblots are representative of four independent experiments. Marker bands are in kDa. B, Saponin-isolated NF54, or PfTRiC-θ-MYC clone B8, grown ± 0.5 µM aTc for 24 h, were digitonin permeabilized, the soluble fraction was separated by blue-native polyacrylamide gel electrophoresis and the blot was probed with i) anti-MYC and ii) anti-PfTRiC-θ. * = expected size of the heterohexadecamer. # = expected size of heterooctamer. ^ = expected size of monomers. Immunoblots are representative of two independent assays, with standard SDS-PAGE run in parallel (as in A) to confirm knockdown was achieved, and ensure equal parasite loading. Cell number equivalent fractions were loaded in all blots. Marker bands are in kDa. C, Growth assay, beginning with asynchronous parasite culture, of the four PfTRiC-θ-MYC-tagged clones, grown ± 0.5 µM aTc. Values are mean parasitemia, with error bars representing standard error of the mean from two independent experiments. The fitted line is an exponential growth equation (GraphPad Prism version 5).
Loss of one subunit in the heterohexadecamer is thought to disrupt formation of the entire complex (Spiess et al., 2004). Consistent with this, when aTc was removed from the PfTRiC-θ-MYC clone B8 parasites, the high molecular weight complexes were lost (Fig. 5B).
To investigate the effect of loss of PfTRiC-θ on asexual parasite growth, aTc was removed from the culture media and growth was measured over several cycles of intraerythrocytic development. Both clones with a 10-element array (PfTRiC-θ-MYC clones B8 and A1) showed a severe growth defect upon aTc removal (Fig. 5Ci,iv). In PfTRiC-θ-MYC clone A3, containing a 6-element array, the growth defect was intermediate (Fig. 5Cii), whereas in PfTRiC-θ-MYC clone F11, with a 4-element array, and only ∼50 % protein knockdown, there was no growth phenotype (Fig. 5Ciii). These results are consistent with PfTRiC-θ being an essential protein in asexual P. falciparum development. To investigate at what stage of asexual development the growth defect manifested, aTc was removed from the culture media at different times from synchronized PfTRiC-θ-MYC clone B8 parasites and progression through the life cycle was monitored by Giemsa smear. When aTc was removed at early ring stage, the parasites proceeded for ∼24 hrs through to trophozoite stage, however the parasites did not form normal schizonts and no rings were recovered in the next cycle. Consistent with this, when aTc was removed during trophozoite stage, there was substantial growth arrest. When aTc was removed in late schizont stage, the parasites proceeded to re-invade normally and continue through ring stage, followed by arrest at the trophozoite stage. These results suggest that PfTRiC-θ is most critical in the trophozoite stage. The growth arrest for the PfTRiC-θ-MYC parasites is very rapid, in contrast to the slower phenotype observed for regulation of PfATP4 using the aptamer array approach, which appears to take several cycles to cause a growth defect (Ganesan et al., 2016).
Loss of the PfTRiC complex does not alter export of PfSBP1 or PfREX1
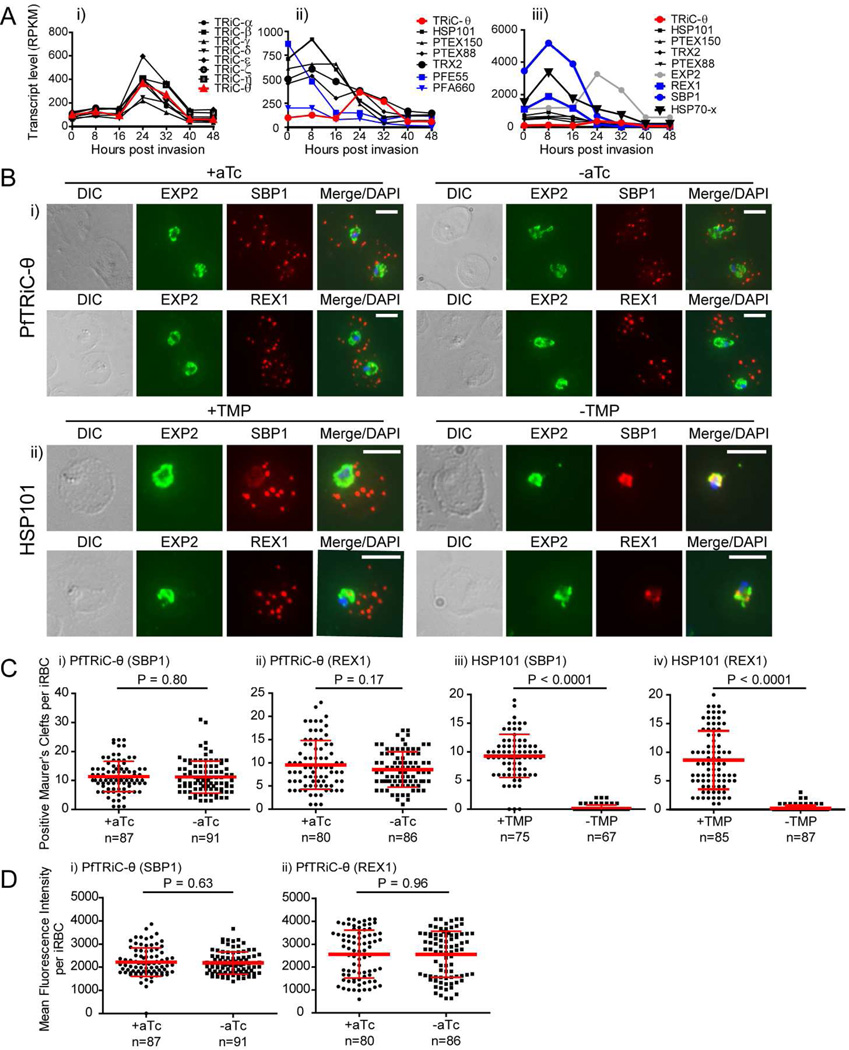
A previous study implicated the activity of the PfTRiC complex in the export of proteins (notably PfSBP1) into the RBC, particularly in export to the Maurer’s clefts (Mbengue et al., 2015). Expression of the PfTRiC subunits is synchronous, peaking ∼24 h post invasion (Fig. 6Ai). In contrast, expression of components of PTEX (HSP101, PTEX150, PTEX88 and TRX2) is high immediately after invasion (Fig. 6Aii), allowing the export of proteins into the RBC as early as ∼2 h post invasion for PfREX1(Gruring et al., 2011), and ∼ 4 h post invasion for PfSBP1(Gruring et al., 2011). Indeed, the peak expression of PfREX1 and PfSBP1 precedes that of the PfTRiC subunits by ∼16 h (Fig. 6Aiii). The expression profile of the other core component of PTEX, PfEXP2, is also high early (like the other PTEX components), though peak expression of PfEXP2 does not occur until about ∼ 24 h post invasion (Fig. 6Aiii). Expression of chaperones known to be exported and be involved in the trafficking of other exported proteins (HSP70-x and the HSP40s PFE0055c and PFA0660w) is also highest between 0–8 h post invasion (Fig. 6Aii, iii). These comparisons highlight that peak PfTRiC expression occurs later than that of export machinery, chaperones involved in protein trafficking in the RBC compartment, and (most (Pelle et al., 2015)) exported proteins.
Figure 6. Loss of PfTRiC-θ does not alter export of PfSBP1 or PfREX1.
A, Transcript levels determined using RNA sequencing, from synchronized Plasmodium falciparum 3D7 parasites, expressed as Reads Per Kilobase of exon model per Million mapped reads (RPKM; scale is different between panels). Data from Otto et al. dataset (Otto et al., 2010) as extracted from PlasmoDB (Aurrecoechea et al., 2009). i) PfTRiC subunits (black) and PfTRiC-θ (red), ii) Components of the Plasmodium translocon of exported proteins (PTEX; black), PfTRiC-θ (red) and exported HSP40s (PFE55 and PFA660 abbreviations as per (Kulzer et al., 2010; Kulzer et al., 2012); blue), iii) Exported proteins PfSBP1 and PfREX1 (blue), PfEXP2 (gray), components of the Plasmodium translocon of exported proteins (PTEX; black), PfTRiC-θ (red) and PfHSP70-x (bold black). B, IFA of acetone-fixed ring-stage infected RBC. i) PfTRiC-θ ± 0.5 µM aTc, ii) HSP101 ± 10 µM trimethoprim (TMP). DAPI stains the parasite nuclei and PfEXP2 delineates the parasite parasitophorous vacuole compartment. Scale bars = 5 µm. Data are representative of two independent assays. DIC = differential interference contrast. C, The number of PfSBP1-(i,iii) and PfREX1-(ii,iv) positive Maurer’s clefts were quantified using Volocity software in the PfTRiC-θ-MYC aptamer tagged lines grown ± 0.5 µM aTc (i,ii), and the HSP101-HA tagged lines grown ± 10 µM TMP (iii,iv). D, The mean fluorescence intensity of the total PfSBP1 (i) or PfREX1 (ii) signal was quantified in the PfTRiC-θ-MYC clone B8 aptamer-tagged line grown ± 0.5 µM aTc. Data for C and D are pooled from two independent experiments, with mean values ± standard deviation shown. P values are the results of two-tailed t-test.
To investigate the effect of loss of PfTRiC-θ on protein export, aTc was removed from the culture medium during the late trophozoite stage, and export was measured by IFA during the next intraerythrocytic cycle. Using this timing for aTc removal, PfTRiC-θ-MYC-B8 levels were reduced to 27 ± 5 % of the NF54 levels (N = 2; mean ± range/2). There were no obvious differences in export of PfSBP1 or PfREX1 upon aTc removal (Fig. 6Bi). This was in contrast to the block in export of PfSBP1 and PfREX1 observed upon TMP removal in the previously characterized PfHSP101-DDD line (Fig. 6Bii). These IFA results were quantified (Fig. 6C,D), using Volocity software to count the number of antibody-positive Maurer’s clefts per iRBC and also measure the mean fluorescence intensity of the Maurer’s clefts (total fluorescence per iRBC). In the control PfHSP101-DDD line (Beck et al., 2014), there was a significant decrease in the number of PfSBP1-/PfREX1-positive Maurer’s clefts after TMP depletion (Fig. 6Ciii,iv). However there was no difference in the number of positive Maurer’s clefts per cell in the PfTRiC-θ-MYC clone B8 upon aTc removal (Fig. 6Ci,ii). There was also no difference in the mean fluorescence intensity of PfSBP1 or PfREX1 in PfTRiC-θ-MYC clone B8 upon aTc removal (Fig. 6D). The results suggest that under these conditions PfTRiC-θ is not involved in PfSBP1/PfREX1 export.
RBC TRiC subunits interact with each other, but do not interact with PfEXP2 or PfHRPII
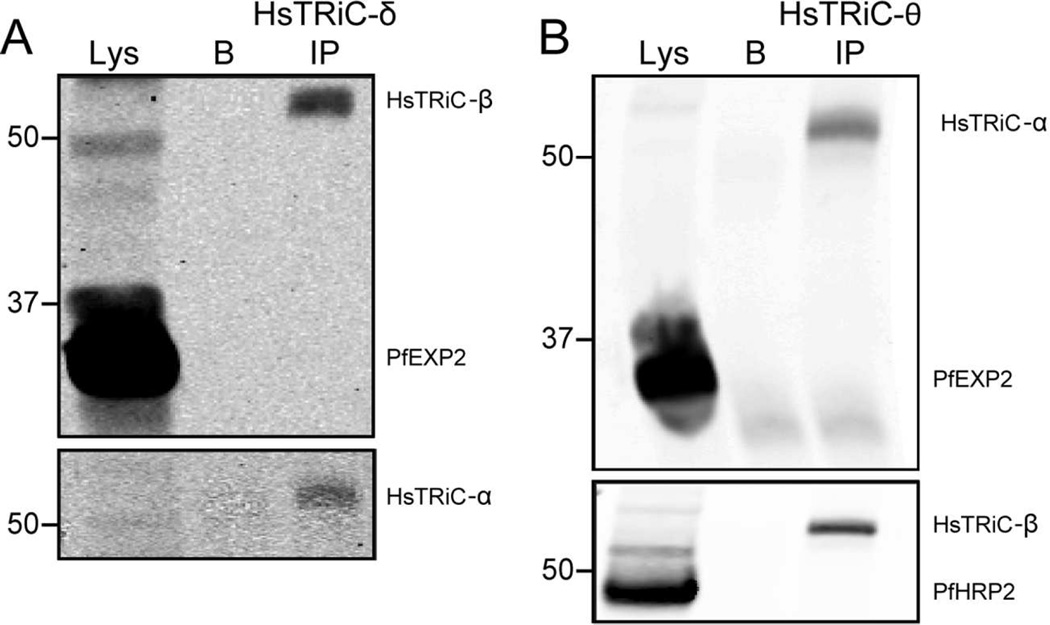
Although RBCs contain TRiC subunits (Wagner et al., 2004 and Fig. 1; Pasini et al., 2006), the ability of these subunits to form micro-complexes or higher order oligomers in RBC has not been investigated. We attempted to analyze these complexes in RBC lysates using blue native PAGE (as per Fig. 2,3). However, the high hemoglobin content of the lysate confounded our analysis, a challenge noted previously when studying RBC lysates (D’Amici et al., 2012). To examine if the human TRiC subunits could interact, human TRiC subunit -δ or -θ were immunoprecipitated from RBC lysates. Both human TRiC-δ and TRiC-θ co-immunoprecipitated human TRiC-β and TRiC-α (Fig. 7A,B).
Figure 7. Human TRiC subunits interact with each other, but do not interact with export related parasite proteins.
Lysates from trophozoite-stage infected RBC were extracted with RIPA buffer. Immunoprecipitation (IP) using A, anti-human TRiC-δ or B, anti-human TRiC-θ. Lys = lysate input, B = bead-only control. Immunoblots are representative of two independent experiments each. Equivalent fractions were loaded in all blots. Marker bands are in kDa.
The previous study examining the role of PfTRiC in export proposed a model where TRiC could both interact with exported proteins as they exited PTEX, and could interact with exported parasite proteins in the RBC (Mbengue et al., 2015). To investigate this possibility we probed whether PfEXP2 (the proposed transmembrane pore of PTEX) or PfHRPII (a soluble exported protein) were co-immunoprecipitated with the human TRiC subunit -δ or -θ. PfEXP2 and PfHRPII were robustly detected in the iRBC lysate, however under these conditions no interaction between human TRiC-δ or –θ and PfEXP2 or PfHRPII was observed (Fig. 7A,B).
DISCUSSION
The TRiC chaperonin complex is essential for protein folding in eukaryotes (Spiess et al., 2004; Hartl et al., 2011). It has been implicated in multiple important functions, including folding nascent polypeptides (Frydman et al., 1994; McCallum et al., 2000), forming multimeric protein complexes (Melville et al., 2003), preventing protein aggregate formation (Tam et al., 2006) and regulating response to proteotoxic stress (Neef et al., 2010; Neef et al., 2014). Our results suggest an important role for the PfTRiC complex during P. falciparum asexual development. Using multiple, complementary approaches, including biochemical fractionation and immunofluorescence, our data demonstrates that the P. falciparum TRiC proteins are localized within the parasite compartment, and suggest that they do not get exported into the host RBC (Fig. 2–4). Typical for class II chaperonins, the parasite TRiC subunits formed multiple high molecular weight complexes, observed using both a subunit-specific antibody and epitope-tagged subunits (Fig. 2–5). When the PfTRiC-θ subunit was knocked down, the parasites displayed a severe growth defect, and could not proceed through the subsequent intraerythrocytic growth cycle (Fig. 5). Despite this growth defect, before parasite death, the parasites continued to export proteins into their host RBC, suggesting that this chaperonin does not play an essential role in protein trafficking in the RBC compartment.
In this study we observed no export of PfTRiC, despite a previous report that PfTRiC-θ localizes to puncta in the RBC cytosol (Mbengue et al., 2015). It is difficult to explain this discrepancy, but it is possible that under certain IFA conditions the rabbit antisera could non-specifically label the Maurer’s clefts, which has been reported previously (Spielmann et al., 2006; Maier et al., 2007). Additionally, we determined that PfTRiC was in the parasite cytoplasm, whereas Mbengue and colleagues proposed that PfTRiC-θ was in a soluble RBC fraction (Mbengue et al., 2015). Sufficient controls were not provided to define their subcellular fractionation, so it is possible their soluble RBC fraction may contain some parasite material. It was also reported that GFP-fusions of the PfTRiC-α, -ζ or -θ could not be generated (Mbengue et al., 2015). GFP is a large tag, and a direct fusion at the N-termini does not allow the subunits to oligomerize and they are overexpressed in their monomeric form (Spiess et al., 2015). Thus it is possible that GFP-fusions of the PfTRiC-α, -ζ or -θ could not be generated if integration was attempted at the endogenous loci, as loss of function through disrupting oligermization ability is likely lethal. In our GFP-overexpression parasites (expressed from an ectopic locus) the native untagged subunits will still be able to form the heterohexadecamer. Additionally, we included a 10 amino acid flexible linker sequence (PRPGAAHYAA) between the TRiC subunit and the GFP, which allowed formation of the heterohexadecamer, even in the presence of this large tag (Fig. 3). Smaller tags, like the MYC epitope, can be fused to the C-termini and the TRiC subunits retain the ability to form higher molecular weight oligomers (Brackley and Grantham, 2010; Spiess et al., 2015). Consistent with this, we were able to tag the PfTRiC-θ with MYC at the endogenous locus (Fig. 4) and high molecular weight oligomers containing PfTRiC-θ were observed (Fig. 5).
A well-established function of TRiC is in the folding of actin (Gao et al., 1992). It was recently demonstrated that P. falciparum actin could bind a complex putatively identified as PfTRiC (Olshina et al., 2016). The putative complex was identified in cytosolic parasite lysates (prepared after saponin isolation of iRBCs)(Olshina et al., 2016). Actin is essential for multiple cellular functions in Plasmodium (Baum et al., 2006; Olshina et al., 2016), thus it is possible that the parasite death upon PfTRiC-θ knockdown may be a result of incorrect actin folding. Aside from this interaction with actin, PfTRiC was reported to interact with the exported protein PfSBP1 (Mbengue et al., 2015). Our results would suggest that this interaction occurs within the parasite, perhaps upon post-translational exit from the ER, during trafficking of exported proteins from the ER to the parasite plasma membrane, or potentially within the PV space. Determining the full range of substrates of PfTRiC will require further investigation.
In addition to the heterohexadecamer form, the TRiC subunits can function as monomers (Liou and Willison, 1997; Tam et al., 2006; Brackley and Grantham, 2010), other microcomplexes with two or more subunits (Liou and Willison, 1997), homo-oligomers (Bhaskar et al., 2015), and can also form homohexadecamers (Sergeeva et al., 2013). In our study we observed monomeric forms and micro-complexes for all subunits examined (Fig. 3–5) and notably the higher molecular weight complexes observed for the TRiC-θ subunit were different to those of other subunits (Fig. 4,5). One function of these smaller molecular weight complexes may be to regulate protein homeostasis by ‘sensing’ substrate levels and providing feedback to alter expression of the substrate which is then folded by the heterohexadecamer (Elliott et al., 2015). Elliot and colleagues showed that TRiC-ε interacts with the myocardin-related cotranscription factor-A (MRTF-A)(Elliott et al., 2015). MRTF-A is usually bound to actin. However, when monomeric actin levels decrease, MRTF-A can traffic to the nucleus to activate actin transcription through the serum response factor pathway (Vartiainen et al., 2007). Increasing the monomeric expression of TRiC-ε delays MRTF-A trafficking to the nucleus, highlighting a link between actin expression and actin folding (Elliott et al., 2015). Additionally, the monomeric forms may sequester misfolded protein intermediates preventing proteotoxicity. For example, the TRiC-α monomer can bind pathogenic huntingtin proteins before toxic aggregation (Tam et al., 2006). Our tagged lines generated in this study may be used for further work to characterize the function of monomers and other micro-complexes in P. falciparum.
In addition to unique interactions with specific substrates, subunits can also differ in their expression profiles and localization. Interestingly, when the mouse TRiC subunits were overexpressed in BALB 3T3 cells, the TRiC-δ subunit was uniquely associated with the plasma membrane (Spiess et al., 2015). Also, in yeast, TRiC-δ is expressed at a level several fold higher than other subunits (Matalon et al., 2014). The expression profile of the eight subunits in P. falciparum is synchronous (Fig. 6). Although not seen in the Otto dataset (Fig. 6; (Otto et al., 2010)), using the RNA sequencing dataset of Vignali and colleagues (Vignali et al., 2011) the PfTRiC-δ subunit is expressed ∼15 times higher than the average expression of the other subunits, suggesting that this subunit may warrant further investigation. We did not observe any phenotypic differences between the PfTRiC-δ-GFP line and the other overexpression lines (Fig. 3).
In this study, we confirmed the expression of the human α, β, δ, ζ and θ subunits in RBCs (Fig. 1). The human subunits could interact with each other (Fig. 7), although whether TRiC complexes are functional in protein homeostasis in the RBC, or whether their presence is vestigial from an earlier more metabolically active stage in RBC development remains to be examined. The ability of the RBC TRiC to translocate in response to heat stress suggests they may be functional (Wagner et al., 2004). Although our results suggest that PfTRiC-θ is not important for protein export, it is possible that the human TRiC may be involved. The human TRiC machinery is coopted by other microbes. TRiC subunits are recruited by viral proteins where they may play a role in viral replication (Lingappa et al., 1994; Kashuba et al., 1999; Hong et al., 2001; Inoue et al., 2011), and the human TRiC is also involved in translocation of the anthrax toxin into host cells (Slater et al., 2013). Although it is difficult to modulate TRiC expression in RBCs, the recent advances in using hematopoietic stem cell based screening to study malaria (Egan et al., 2015) may prove useful in clarifying the role of RBC TRiC in malaria protein export.
Understanding the function of PfTRiC is topical, as a recent transcriptomic study identified several TRiC subunits as being associated with reduced sensitivity to the front-line antimalarial artemisinin (Mok et al., 2015). An upregulation of TRiC (and another protein stress pathway, the Plasmodium reactive oxidative stress complex) was correlated with a delayed parasite clearance half time (Mok et al., 2015). Finding ways to selectively inhibit TRiC activity, which would likely be lethal to the parasites, may be useful in combination therapy to aid in combating artemisinin resistance. Ideally, PfTRiC-specific inhibitors, with no activity against the human enzyme could be designed. This strategy is being investigated for other parasite chaperones, with promising parasite selectivity results (Cockburn et al., 2014). The first small molecule modulator of human TRiC was recently described (Neef et al., 2010; Neef et al., 2014), and further characterization of its mode of action will prove useful in designing inhibitors against PfTRiC.
EXPERIMENTAL PROCEDURES
Parasite Culture and Transfection
Asexual P. falciparum parasites were cultured in RPMI with 0.5 (w/v) % Albumax II, as described previously (Klemba et al., 2004). Strains used in this study were 3D7 (Saint Louis, derived from MRA-102 (Bopp et al., 2013)), Dd2 and NF54attB (referred to as NF54 throughout (Adjalley et al., 2011)). Uninfected RBC (uRBC) were obtained from the Barnes-Jewish Hospital blood bank (Saint Louis, MO).
Cell Fractionation and Western Blotting
“Total” cell extracts (∼ 6 × 107 cells) were prepared by washing cells twice in phosphate-buffered saline (PBS; pH 7.4). The pellet was then dissolved in 1x SDS sample buffer. Lysates were prepared from uRBC, trophozoite-stage iRBC (enriched > 95 % parasitemia on a QuadroMACS Separator (Ribaut et al., 2008)), or saponin-isolated trophozoite-stage parasites (treated with 0.05 % saponin in PBS, 20 s, room temperature (RT)).
Saponin fractionation was performed by treating uRBC or enriched iRBC with 0.05 % saponin in PBS (20 s, RT), releasing the soluble contents of the RBC and PV. Samples were centrifuged (17,000 × g, 10 min), and the supernatant was removed to a new tube. Pellets were washed once in PBS, and the supernatant was re-centrifuged (17,000 × g, 5 min) before equivalent samples were dissolved in 1x SDS sample buffer. Tetanolysin O (TetO) fractionation was performed by treating uRBC or enriched iRBC with 1 hemolytic unit (HU) TetO in PBS (20 min, 37 °C), releasing soluble contents of the RBC. The activity of TetO was confirmed by observing release of hemoglobin into the supernatant fraction (also see immunoblot in Supplemental Fig. 3). Samples were centrifuged/prepared as for the saponin fractionation.
To obtain the total soluble fraction, cell pellets were solubilized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl (pH 7.6), 0.1 % SDS, 150 mM NaCl, 0.5 % sodium deoxycholate and 1 % NP-40) with 1x protease inhibitor cocktail (PIC; Roche) for 5 min on ice. The sample was centrifuged (17,000 × g, 5 min) and the supernatant diluted in SDS sample buffer (to 1x final concentration, from 5x stock).
For the protease protection assay (Spielmann et al., 2006), enriched iRBC were treated with TetO, as above, and the pellet fraction was divided into four samples. The pellets were incubated in 1) PBS alone, 2) PBS + 1 mg/mL Proteinase K (Sigma-Aldrich), 3) 0.05 % saponin in PBS + 1 mg/mL Proteinase K, or 4) hypotonic lysis solution (5 mM Tris-HCl, pH 8.0) + 1 mg/mL Proteinase K. Samples were incubated for 30 min, on ice, and the reaction was stopped as described previously (Thavayogarajah et al., 2015), by incubating with 5 mM phenylmethylsulfonyl fluoride and 1x PIC (5 min, RT). Samples were diluted in 1x SDS sample buffer.
Samples in SDS sample buffer were heated (5 min, 95 °C) and separated by SDS-PAGE. Proteins were transferred onto nitrocellulose (0.45 µm; BioRad) and blots were blocked in Odyssey Blocking Buffer (Licor). Primary antibodies used: mouse monoclonal anti-cMyc Antibody (9E10; ThermoScientific; 3:200), rat monoclonal anti-human TCP1-α (23c; Enzo; 1:1000), rat monoclonal anti-human TCP1-β (PK/8/4/4i/2f; Enzo; 1:1000), rabbit monoclonal anti-human-TCP-δ (EPR8495(B); EMD Millipore; 1:1000), rabbit polyclonal anti-human TCP1-ζ (ThermoScientific; 1:1000), mouse monoclonal anti-human TCP1-θ (E-7; Santa Cruz Biotechnology Inc; 1:1000), goat polyclonal anti-GFP (ab5450; Abcam; 1:1200), rabbit polyclonal anti-PfHAD1 ((Guggisberg et al., 2014); 1:24,000), rabbit polyclonal anti-PfTRiC-θ (or prebleed; (Mbengue et al., 2015); 1:1000), rabbit polyclonal anti-PfSERP ((Ragge et al., 1990); 1:1000), rabbit polyclonal anti-PfEF-1α (1363(Mamoun and Goldberg, 2001); 1:20,000), mouse monoclonal anti-PfHRPII (2G12; (Parra et al., 1991); 1:1000), mouse monoclonal anti-PfEXP2 (7.7; (Hall et al., 1983); 1:1000) and rabbit polyclonal anti-human α/β-spectrin (S1515; Sigma-Aldrich; 1:500). Blots were washed in Tris-buffered saline with 0.05% Tween 20 and incubated with IR680- or 800-conjugated secondary antibodies (1:15,000). Blots were imaged with the LI-COR Odyssey Imaging System, and quantification performed using LI-COR Odyssey Image Studio software v4.
Blue Native PAGE
Trophozoite-stage parasites were isolated from total culture by saponin treatment (0.05 % saponin in PBS, 20 s, RT) and washed three times in PBS. Samples were solubilized in 1x NativePAGE sample buffer, 1 % w/v digitonin (Invitrogen Native PAGE Sample Preparation Kit) and incubated on ice for 45 min. Each sample was centrifuged (17,000 × g, 5 min) and the supernatant was transferred to a new tube. G-250 Sample Additive was added to a final concentration of 0.25 %, and samples were separated on a NativePAGE Novex 4–16 % Bis Tris gel (1mm thickness) using a Surelock Xcell Minicell setup running on ice, with pre-chilled buffers. Nativemark Unstained protein standard (Invitrogen) was also loaded. The anode buffer was the NativePAGE Running buffer (Invitrogen) and the setup was run with Dark blue cathode buffer (Invitrogen) at 150 V for 45–60 min, and then switched to Light blue cathode buffer and run at 200 V for a further 1–1.5 h. Proteins were transferred onto polyvinylidene difluoride membrane (Immobilon-P PVDF; Sigma-Aldrich) in NuPAGE Transfer Buffer (Invitrogen), and blocking/immunoblotting performed as described above.
Immunoprecipitation (IP)
Trophozoite-stage parasites were enriched (VarioMACS) and washed twice in PBS before lysate was prepared in ice cold RIPA with 1x PIC. Lysates were nutated at 4 °C for 1 h and cleared by centrifugation (17,000 × g, 5 min). Supernatants were then nutated at 4 °C for 3 h with protein G Dynabeads, with or without the human TRiC-δ or -θ antibodies (∼ 2 µg). Immunocomplexes were separated magnetically and washed five times in RIPA with 1x PIC before lysis in SDS sample buffer. SDS/PAGE, blocking and immunoblotting was performed as described above.
Immunofluorescence Assay (IFA)
Two methods of fixation were used to perform indirect IFA (Tonkin et al., 2004; Beck et al., 2014). For the aldehyde-based method, cells were attached to concanavalin A (0.5 mg/mL) coated coverslips and fixed in 4 % (v/v) paraformaldehyde/ 0.0075 % (v/v) glutaraldehyde in PBS (30 min). Cells were permeabilized in 0.1 % Triton X100 in PBS (10 min), treated with 0.1 mg/mL NaBH4 in PBS (10 min) before blocking in 3 % (w/v) bovine serum albumin (1–16 h). Alternatively, thin smears were prepared, and slides fixed in cold 100 % acetone for two min before blocking as above. Primary antibodies used: rabbit polyclonal anti-PfTRiC-θ (or prebleed; (Mbengue et al., 2015); 1:200), rabbit polyclonal anti-GFP (ab6556; Abcam; 1:200), mouse monoclonal anti-PfEXP2 (7.7; (Hall et al, 1983); 1:500), rabbit polyclonal anti-PfSBP1 (BR28; (Blisnick et al., 2000); 1:500) and rabbit polyclonal anti-PfREX1 ((Hawthorne et al., 2004); 1:500). Secondary Alexa Fluor 488- or 594-conjugated antibodies were used at 1:2000. Coverslips were mounted in Prolong Gold Antifade Mountant with DAPI (Molecular Probes). Microscopy was performed using an AxioImager.M1 epifluorescence microscope (Zeiss) with a Hamamatsu ORCA-ER digital CCD camera, as previously described (Beck et al., 2014). AxioVision software Release 4.8.1 was used for image processing.
Molecular Cloning and Generation of Transgenic Parasites
To generate parasites that over-express PfTRiC subunits tagged with GFP at the C-terminus (ectopic expression, under the strong HSP86 promoter), full-length sequences were amplified from 3D7 genomic DNA using the primer pairs; 1+2 (pftric-α; PF3D7_1132200), 3+4 (pftric-β; PF3D7_0306800), 5+6 (pftric-γ; PF3D7_1229500), 7+8 (pftric-δ; PF3D7_1357800), 9+10 (pftric-ε; PF3D7_0320300), 11+12 (pftric-ζ; PF3D7_0608700), 13+14 (pftric-η; PF3D7_0308200) and 15+16 (pftric-θ; PF3D7_0214000). All oligonucleotide sequences are in Table 1 (Supporting Information). The resulting amplicons were inserted into the XhoI/AvrII sites of the pTEOE vector using InFusion cloning (Clontech). pTEOE is the pTyEOE vector (described in Beck et al., 2014), containing the hDHFR selection cassette in place of the yDHODH cassette. The resulting pTEOE-PfTRiC plasmids (100 µg) were transfected into ring-stage parasites (Rug and Maier, 2013). pTEOE constructs were cotransfected with 50 µg pHTH (MRA-912; deposited at MR4 by Prof. John Adams, as part of the BEI Resources Repository, NIAID, NIH). Positive selection (10 nM WR-99210) was applied 24 h post transfection.
For CRISPR/Cas9 editing, single guide RNAs (sgRNAs) were expressed from a cassette containing the 5’ and 3’ UTRs from the P. falciparum U6 gene, similar to a previously reported strategy (Ghorbal et al., 2014). The U6 locus was PCR amplified from 3D7 genomic DNA (17+18) and subcloned into pCR4-TOPO (Life Technologies). The endogenous U6 sequence was then replaced with a pre-sgRNA sequence containing BtgZI sites and a Cas9 binding sequence using the QuickChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies) and primer 19. This PfU6 pre-sgRNA cassette was then moved into the vector pHHT-TK (Duraisingh et al., 2002) at NotI. For Cas9 expression from the same vector, S. pyogenes Cas9 was PCR amplified (20+21) from the vector pMJ920 (addgene #42234) (Jinek et al., 2013) and inserted in the vector pTyEOE-6xFLAG (Beck et al., 2014). Cas9 was then PCR amplified from this plasmid (22+23) with a 3’ 2x Nuclear Localization Sequence and 1x FLAG tag. The calmodulin promoter and yDHOD selectable marker were PCR amplified (24+25) and fused to the Cas9 PCR amplicon with an intervening 2A skip peptide (Straimer et al., 2012) in a second PCR reaction (23+24). This CAM promoter-yDHOD-2A–Cas9 amplicon was inserted into the sgRNA expression vector between XbaI/XhoI, removing the hDHFR and thymidine kinase selection cassettes, and fusing Cas9 to the P. berghei dihydrofolate reductase/thymidylate synthase (DT) 3’ UTR (previously supporting expression of thymidine kinase). The resulting plasmid containing both Cas9 and sgRNA expression cassettes was named pAll-In-One (pAIO).
To generate parasites expressing a MYC-tagged, regulatable version of PfTRiC-θ, we replaced the endogenous 3’ UTR with a 10 x TetR aptamer array (Ganesan et al., 2016) using CRISPR/Cas9 editing (schematic in Figure 4A). Two guide RNA (gRNA) targets immediately upstream and downstream of the PfTRiC-θ stop codon were chosen. These gRNA seed sequences were ordered as sense and antisense primer pairs (26+27 and 28+29), annealed and inserted into the vector pAIO at BtgZI using InFusion, resulting in the plasmids pAIO-TRiC-θ gRNA1 and pAIO-TRiC-θ gRNA2. A 5’ homology donor template (up to but not including the stop codon) and a 3’ homology donor template (beginning 37 bp downstream of the stop codon just after the gRNA2 protospacer adjustment motif) were PCR amplified from P. falciparum NF54 genomic DNA (30+31 and 32+33), assembled in a second PCR reaction (31+32) and inserted between AscI/BstEII in the vector pMG75 (Ganesan et al., 2016), resulting in the vector pMG75-TRiC-θ-MYC. Primer 31 was designed to generate a synonymous shield mutation to eliminate the gRNA1 protospacer adjustment motif located within the coding sequence. This vector (100 µg) was linearized at the AflII site between the 3’ and 5’ donor templates and co-transfected with either pAIO-TRiC-θ gRNA1 or 2 (50 µg) into P. falciparum NF54attB parasites (Adjalley et al., 2011). Anhydrotetracycline (aTc; Cayman Chemical) was maintained in the culture at 0.5 µM from the time of transfection. Positive selection (2.5 µg/mL Blasticidin S) was applied 24 h post transfection. Parasites were obtained from four independent transfections, and cloning by limiting dilution resulted in four independent clones designated B8 and A3 (edited with pAIO-TRiC-θ gRNA1) and F11 and A1 (edited with pAIO-TRiC-θ gRNA2). Verification that the desired integration event was achieved was confirmed by Southern blotting. Genomic DNA (3 µg; Qiagen DNeasy Blood and Tissue Kit) was isolated from the NF54 parent and PfTRiC-θ-MYC clones. Genomic DNA and vector DNA was precipitated and triple digested overnight (HindIII, XmaI, AflII). Southern blotting using the Amersham AlkPhos Direct Labeling kit (GE Healthcare) was performed (Ponpuak et al., 2007), using the 5’ flank as the probe. To examine the repeat status of the 10 x TetR aptamer array, a diagnostic PCR was performed on B8, A3, F11 and A1 genomic DNA using primers flanking the array (34+35).
Parasite growth assays and knockdown quantification
PfTRiC-θ-MYC clones were washed once in media without aTc, and then plated in triplicate with or without 0.5µM aTc at 2% hematocrit and an initial parasitemia of ∼1%. Media changes were performed daily and parasitemia was monitored by flow cytometry (BD FACSCanto flow cytometer) using acridine orange staining (1.5 µg/mL in PBS). Each well was subcultured at 72 and 96 h to avoid overgrowth and cumulative parasitemia was back calculated based on the subculturing schedule.
To quantify whether removal of aTc resulted in PfTRiC-θ-MYC knockdown, clones B8 and F11 were tightly synchronized. Young rings (∼ < 8 h post invasion) were washed (3 × 5 min washes in media without aTc). Cultures were then plated with or without 0.5µM aTc and grown for 20–24 h. Lysates of the total soluble fraction were prepared in RIPA, and processed for WB, as described above.
Analyses of Blood Stage Transcriptome
An RNA sequencing dataset (Otto et al., 2010) from synchronized 3D7 P. falciparum parasites was retrieved from PlasmoDB (Aurrecoechea et al., 2009) (Release 28). In addition to the data for pftric subunits, data was obtained for pfhsp101 (PF3D7_1116800), pfptex150 (PF3D7_1436300), pfptex88 (PF3D7_1105600), pftrx2 (PF3D7_1345100), pfexp2 (PF3D7_1471100), pfsbp1 (PF3D7_0501300), pfrex1 (PF3D7_0935900), pfa0660w (PF3D7_0113700), pfe0055c (PF3D7_0501100.1) and pfhsp70-x (PF3D7_0831700).
Quantification of protein export
PfTRiC-θ-MYC clone B8 parasites were synchronized to a ∼5 h window. When parasites reached late trophozoite stage (∼15 h before egress), aTc was removed (3 × 5 min washes in media without aTc). Cultures were then plated with or without 0.5µM aTc and grown under shaking conditions (to minimize multiply invaded RBC (Allen and Kirk, 2010)) for 30 h. Analysis of HSP101-DDD (dihydrofolate reductase-based destabilization domain) clone 13F10 parasites (Beck et al., 2014) was carried in an identical fashion with 10 µM trimethoprim (Sigma) removed from the cultures rather than aTc. Thin smears were prepared for acetone fixation, and IFA was performed as described earlier (with 30 min blocking step and 1 h incubations with primary and secondary antibodies). The remaining culture was used to quantify the level of PfTRiC-θ-MYC knockdown (as described above). Ten images were acquired for each condition using the DAPI channel for field selection to avoid bias. To enable comparison, the same exposure times were used for each image (515 ms for PfSBP1, 150 ms for PfREX1, 500 ms for PfEXP2 and 50 ms for DAPI). Images were analyzed using Volocity 6.3 (PerkinElmer). Each single-infected RBC was cropped to an area slightly larger than the border of the RBC using the DIC channel as a guide. The PVM and parasite were marked using the “find objects” measurement tool for the PfEXP2-488 channel (automatic threshold setting with threshold offset set to 40.5 % and minimum object size set to 0 µm2). Individual Maurer’s clefts were then identified using the “find spots” measurement tool for the PfSBP1 or PfREX1-594 channel (offset minimum spot intensity set to 80 % and brightest spot within radius set to 0.5 µm). All spots within the PfEXP2 object boundary were then removed using the “subtract” measurement tool and the number and fluorescence intensity of the remaining spots in each infected cell were collected.
Supplementary Material
Acknowledgments
NS acknowledges funding from the National Health and Medical Research Council of Australia (Overseas Biomedical Fellowship 1072217). JB acknowledges funding from the National Institutes of Health (NIH F32 #AI115965). DG acknowledges funding from the Howard Hughes Medical Institute. We thank C. Braun-Breton (University Montpellier) for the anti-PfTRiC-θ and anti-PfSBP1 antibody, A. Odom (Washington University School of Medicine) for the anti-PfHAD1 antibody, J. Przyborski and K. Lingelbach (University of Marburg) for anti-PfSERP antibody, D. Taylor (University of Hawaii) for the anti-PfHRPII antibody, L. Tilley for anti-PfREX1 antibody, J. McBride, D. Cavanagh and EMRR for the anti-PfEXP2 antibody and Jacobus Pharmaceutical Company Inc. for WR-92210. We thank B. Vaupel and A. Oksman for technical assistance and Goldberg lab members for advice and suggestions.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
AUTHOR CONTRIBUTIONS
NS designed, performed and analyzed experiments, coordinated the study, and wrote the paper. JB designed, performed and analyzed experiments and wrote parts of the paper. SG and JN contributed the pMG75 vector before publication. DEG coordinated the study and wrote parts of the paper. All authors revised, read and approved the final version of the manuscript.
REFERENCES
- Adjalley SH, Johnston GL, Li T, Eastman RT, Ekland EH, Eappen AG, Richman A, et al. Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue. Proc Natl Acad Sci U S A. 2011;108:E1214–E1223. doi: 10.1073/pnas.1112037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RJ, Kirk K. Plasmodium falciparum culture: the benefits of shaking. Mol Biochem Parasitol. 2010;169:63–65. doi: 10.1016/j.molbiopara.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Ansorge I, Benting J, Bhakdi S, Lingelbach K. Protein sorting in Plasmodium falciparum-infected red blood cells permeabilized with the pore-forming protein streptolysin O. Biochem J. 1996;315(Pt 1):307–314. doi: 10.1042/bj3150307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu B, Shoue DA, Fraser MJ, Jr, Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac . Proc Natl Acad Sci U S A. 2005;102:16391–16396. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banumathy G, Singh V, Tatu U. Host chaperones are recruited in membrane-bound complexes by Plasmodium falciparum . J Biol Chem. 2002;277:3902–3912. doi: 10.1074/jbc.M110513200. [DOI] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Baum B, Speed TP, Cowman AF. Regulation of apicomplexan actin-based motility. Nat Rev Microbiol. 2006;4:621–628. doi: 10.1038/nrmicro1465. [DOI] [PubMed] [Google Scholar]
- Beck JR, Muralidharan V, Oksman A, Goldberg DE. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature. 2014;511:592–595. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar, Mitra K, Kuldeep J, Siddiqi MI, Goyal N. The TCP1gamma subunit of Leishmania donovani forms a biologically active homo-oligomeric complex. FEBS J. 2015;282:4607–4619. doi: 10.1111/febs.13521. [DOI] [PubMed] [Google Scholar]
- Blisnick T, Morales Betoulle ME, Barale JC, Uzureau P, Berry L, Desroses S, Fujioka H, et al. Pfsbp1, a Maurer’s cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol Biochem Parasitol. 2000;111:107–121. doi: 10.1016/s0166-6851(00)00301-7. [DOI] [PubMed] [Google Scholar]
- Boddey JA, Cowman AF. Plasmodium nesting: remaking the erythrocyte from the inside out. Annu Rev Microbiol. 2013;67:243–269. doi: 10.1146/annurev-micro-092412-155730. [DOI] [PubMed] [Google Scholar]
- Boddey JA, O’Neill MT, Lopaticki S, Carvalho TG, Hodder AN, Nebl T, Wawra S, et al. Export of malaria proteins requires co-translational processing of the PEXEL motif independent of phosphatidylinositol-3-phosphate binding. Nat Commun. 2016;7:10470. doi: 10.1038/ncomms10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackley KI, Grantham J. Subunits of the chaperonin CCT interact with F-actin and influence cell shape and cytoskeletal assembly. Exp Cell Res. 2010;316:543–553. doi: 10.1016/j.yexcr.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Bullen HE, Charnaud SC, Kalanon M, Riglar DT, Dekiwadia C, Kangwanrangsan N, Torii M, et al. Biosynthesis, localization, and macromolecular arrangement of the Plasmodium falciparum translocon of exported proteins (PTEX) J Biol Chem. 2012;287:7871–7884. doi: 10.1074/jbc.M111.328591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IL, Boshoff A, Pesce ER, Blatch GL. Selective modulation of plasmodial Hsp70s by small molecules with antimalarial activity. Biol Chem. 2014;395:1353–1362. doi: 10.1515/hsz-2014-0138. [DOI] [PubMed] [Google Scholar]
- D’Amici GM, Rinalducci S, Zolla L. Depletion of hemoglobin and carbonic anhydrase from erythrocyte cytosolic samples by preparative clear native electrophoresis. Nat Protoc. 2012;7:36–44. doi: 10.1038/nprot.2011.427. [DOI] [PubMed] [Google Scholar]
- Daniyan MO, Boshoff A, Prinsloo E, Pesce ER, Blatch GL. The Malarial Exported PFA0660w Is an Hsp40 Co-Chaperone of PfHsp70-x. PLoS One. 2016;11:e0148517. doi: 10.1371/journal.pone.0148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, Sanders PR, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- Egan ES, Jiang RH, Moechtar MA, Barteneva NS, Weekes MP, Nobre LV, Gygi SP, et al. Malaria. A forward genetic screen identifies erythrocyte CD55 as essential for Plasmodium falciparum invasion. Science. 2015;348:711–714. doi: 10.1126/science.aaa3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KL, Svanstrom A, Spiess M, Karlsson R, Grantham J. A novel function of the monomeric CCTepsilon subunit connects the serum response factor pathway to chaperone-mediated actin folding. Mol Biol Cell. 2015;26:2801–2809. doi: 10.1091/mbc.E15-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsworth B, Matthews K, Nie CQ, Kalanon M, Charnaud SC, Sanders PR, Chisholm SA, et al. PTEX is an essential nexus for protein export in malaria parasites. Nature. 2014;511:587–591. doi: 10.1038/nature13555. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Ganesan SM, Falla A, Goldfless SJ, Nasamu AS, Niles JC. Synthetic RNA-protein modules integrated with native translation mechanisms to control gene expression in malaria parasites. Nat Commun. 2016;7:10727. doi: 10.1038/ncomms10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Gehde N, Hinrichs C, Montilla I, Charpian S, Lingelbach K, Przyborski JM. Protein unfolding is an essential requirement for transport across the parasitophorous vacuolar membrane of Plasmodium falciparum . Mol Microbiol. 2009;71:613–628. doi: 10.1111/j.1365-2958.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Goldberg DE. Hemoglobin degradation. Curr Top Microbiol Immunol. 2005;295:275–291. doi: 10.1007/3-540-29088-5_11. [DOI] [PubMed] [Google Scholar]
- Grover M, Chaubey S, Ranade S, Tatu U. Identification of an exported heat shock protein 70 in Plasmodium falciparum . Parasite. 2013;20:2. doi: 10.1051/parasite/2012002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruring C, Heiber A, Kruse F, Ungefehr J, Gilberger TW, Spielmann T. Development and host cell modifications of Plasmodium falciparum blood stages in four dimensions. Nat Commun. 2011;2:165. doi: 10.1038/ncomms1169. [DOI] [PubMed] [Google Scholar]
- Guggisberg AM, Park J, Edwards RL, Kelly ML, Hodge DM, Tolia NH, Odom AR. A sugar phosphatase regulates the methylerythritol phosphate (MEP) pathway in malaria parasites. Nat Commun. 2014;5:4467. doi: 10.1038/ncomms5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R, McBride J, Morgan G, Tait A, Zolg JW, Walliker D, Scaife J. Antigens of the erythrocytes stages of the human malaria parasite Plasmodium falciparum detected by monoclonal antibodies. Mol Biochem Parasitol. 1983;7:247–265. doi: 10.1016/0166-6851(83)90025-7. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hawthorne PL, Trenholme KR, Skinner-Adams TS, Spielmann T, Fischer K, Dixon MW, Ortega MR, et al. A novel Plasmodium falciparum ring stage protein, REX, is located in Maurer’s clefts. Mol Biochem Parasitol. 2004;136:181–189. doi: 10.1016/j.molbiopara.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Heiber A, Kruse F, Pick C, Gruring C, Flemming S, Oberli A, Schoeler H, et al. Identification of new PNEPs indicates a substantial non-PEXEL exportome and underpins common features in Plasmodium falciparum protein export. PLoS Pathog. 2013;9:e1003546. doi: 10.1371/journal.ppat.1003546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Choi G, Park S, Chung AS, Hunter E, Rhee SS. Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J Virol. 2001;75:2526–2534. doi: 10.1128/JVI.75.6.2526-2534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Aizaki H, Hara H, Matsuda M, Ando T, Shimoji T, Murakami K, et al. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology. 2011;410:38–47. doi: 10.1016/j.virol.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashuba E, Pokrovskaja K, Klein G, Szekely L. Epstein-Barr virus-encoded nuclear protein EBNA-3 interacts with the epsilon-subunit of the T-complex protein 1 chaperonin complex. J Hum Virol. 1999;2:33–37. [PubMed] [Google Scholar]
- Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum . The Journal of cell biology. 2004;164:47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulzer S, Charnaud S, Dagan T, Riedel J, Mandal P, Pesce ER, Blatch GL, et al. Plasmodium falciparum-encoded exported hsp70/hsp40 chaperone/co-chaperone complexes within the host erythrocyte. Cell Microbiol. 2012;14:1784–1795. doi: 10.1111/j.1462-5822.2012.01840.x. [DOI] [PubMed] [Google Scholar]
- Kulzer S, Rug M, Brinkmann K, Cannon P, Cowman A, Lingelbach K, Blatch GL, et al. Parasite-encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum-infected erythrocyte. Cell Microbiol. 2010;12:1398–1420. doi: 10.1111/j.1462-5822.2010.01477.x. [DOI] [PubMed] [Google Scholar]
- Lingappa JR, Martin RL, Wong ML, Ganem D, Welch WJ, Lingappa VR. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. The Journal of cell biology. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AK, Willison KR. Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J. 1997;16:4311–4316. doi: 10.1093/emboj/16.14.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- Maier AG, Rug M, O’Neill MT, Beeson JG, Marti M, Reeder J, Cowman AF. Skeleton-binding protein 1 functions at the parasitophorous vacuole membrane to traffic PfEMP1 to the Plasmodium falciparum-infected erythrocyte surface. Blood. 2007;109:1289–1297. doi: 10.1182/blood-2006-08-043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoun CB, Goldberg DE. Plasmodium protein phosphatase 2C dephosphorylates translation elongation factor 1beta and inhibits its PKC-mediated nucleotide exchange activity in vitro. Mol Microbiol. 2001;39:973–981. doi: 10.1046/j.1365-2958.2001.02289.x. [DOI] [PubMed] [Google Scholar]
- Matalon O, Horovitz A, Levy ED. Different subunits belonging to the same protein complex often exhibit discordant expression levels and evolutionary properties. Curr Opin Struct Biol. 2014;26:113–120. doi: 10.1016/j.sbi.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Mbengue A, Vialla E, Berry L, Fall G, Audiger N, Demettre-Verceil E, Boteller D, et al. New Export Pathway in Plasmodium falciparum-Infected Erythrocytes: Role of the Parasite Group II Chaperonin, PfTRiC. Traffic. 2015;16:461–475. doi: 10.1111/tra.12266. [DOI] [PubMed] [Google Scholar]
- McCallum CD, Do H, Johnson AE, Frydman J. The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. The Journal of cell biology. 2000;149:591–602. doi: 10.1083/jcb.149.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville MW, McClellan AJ, Meyer AS, Darveau A, Frydman J. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol Cell Biol. 2003;23:3141–3151. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesen-Ramirez P, Reinsch F, Blancke Soares A, Bergmann B, Ullrich AK, Tenzer S, Spielmann T. Stable Translocation Intermediates Jam Global Protein Export in Plasmodium falciparum Parasites and Link the PTEX Component EXP2 with Translocation Activity. PLoS Pathog. 2016;12:e1005618. doi: 10.1371/journal.ppat.1005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Jaeger AM, Gomez-Pastor R, Willmund F, Frydman J, Thiele DJ. A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 2014;9:955–966. doi: 10.1016/j.celrep.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Turski ML, Thiele DJ. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 2010;8:e1000291. doi: 10.1371/journal.pbio.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshina MA, Baumann H, Willison KR, Baum J. Plasmodium actin is incompletely folded by heterologous protein-folding machinery and likely requires the native Plasmodium chaperonin complex to enter a mature functional state. FASEB J. 2016;30:405–416. doi: 10.1096/fj.15-276618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ME, Evans CB, Taylor DW. Identification of Plasmodium falciparum histidine-rich protein 2 in the plasma of humans with malaria. J Clin Microbiol. 1991;29:1629–1634. doi: 10.1128/jcm.29.8.1629-1634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- Pelle KG, Oh K, Buchholz K, Narasimhan V, Joice R, Milner DA, Brancucci NM, et al. Transcriptional profiling defines dynamics of parasite tissue sequestration during malaria infection. Genome Med. 2015;7:19. doi: 10.1186/s13073-015-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen W, Kulzer S, Engels S, Zhang Q, Ingmundson A, Rug M, Maier AG, et al. J-dot targeting of an exported HSP40 in Plasmodium falciparum-infected erythrocytes. Int J Parasitol. 2016;46:519–525. doi: 10.1016/j.ijpara.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Ponpuak M, Klemba M, Park M, Gluzman IY, Lamppa GK, Goldberg DE. A role for falcilysin in transit peptide degradation in the Plasmodium falciparum apicoplast. Mol Microbiol. 2007;63:314–334. doi: 10.1111/j.1365-2958.2006.05443.x. [DOI] [PubMed] [Google Scholar]
- Ragge K, Arnold HH, Tummler M, Knapp B, Hundt E, Lingelbach K. In vitro biosynthesis and membrane translocation of the serine rich protein of Plasmodium falciparum . Mol Biochem Parasitol. 1990;42:93–100. doi: 10.1016/0166-6851(90)90116-4. [DOI] [PubMed] [Google Scholar]
- Ribaut C, Berry A, Chevalley S, Reybier K, Morlais I, Parzy D, Nepveu F, et al. Concentration and purification by magnetic separation of the erythrocytic stages of all human Plasmodium species. Malar J. 2008;7:45. doi: 10.1186/1475-2875-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rug M, Maier AG. Transfection of Plasmodium falciparum . Methods in molecular biology. 2013;923:75–98. doi: 10.1007/978-1-62703-026-7_6. [DOI] [PubMed] [Google Scholar]
- Sa JM, Yamamoto MM, Fernandez-Becerra C, de Azevedo MF, Papakrivos J, Naude B, Wellems TE, et al. Expression and function of pvcrt-o, a Plasmodium vivax ortholog of pfcrt, in Plasmodium falciparum and Dictyostelium discoideum . Mol Biochem Parasitol. 2006;150:219–228. doi: 10.1016/j.molbiopara.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Sergeeva OA, Chen B, Haase-Pettingell C, Ludtke SJ, Chiu W, King JA. Human CCT4 and CCT5 chaperonin subunits expressed in Escherichia coli form biologically active homo-oligomers. J Biol Chem. 2013;288:17734–17744. doi: 10.1074/jbc.M112.443929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater LH, Hett EC, Clatworthy AE, Mark KG, Hung DT. CCT chaperonin complex is required for efficient delivery of anthrax toxin into the cytosol of host cells. Proc Natl Acad Sci U S A. 2013;110:9932–9937. doi: 10.1073/pnas.1302257110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T, Hawthorne PL, Dixon MW, Hannemann M, Klotz K, Kemp DJ, Klonis N, et al. A cluster of ring stage-specific genes linked to a locus implicated in cytoadherence in Plasmodium falciparum codes for PEXEL-negative and PEXEL-positive proteins exported into the host cell. Mol Biol Cell. 2006;17:3613–3624. doi: 10.1091/mbc.E06-04-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M, Echbarthi M, Svanstrom A, Karlsson R, Grantham J. Over-Expression Analysis of All Eight Subunits of the Molecular Chaperone CCT in Mammalian Cells Reveals a Novel Function for CCTdelta. J Mol Biol. 2015;427:2757–2764. doi: 10.1016/j.jmb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Spillman NJ, Beck JR, Goldberg DE. Protein export into malaria parasite-infected erythrocytes: mechanisms and functional consequences. Annu Rev Biochem. 2015;84:813–841. doi: 10.1146/annurev-biochem-060614-034157. [DOI] [PubMed] [Google Scholar]
- Spillman NJ, Dalmia VK, Goldberg DE. Exported Epoxide Hydrolases Modulate Erythrocyte Vasoactive Lipids during Plasmodium falciparum Infection. MBio. 2016;7 doi: 10.1128/mBio.01538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straimer J, Lee MC, Lee AH, Zeitler B, Williams AE, Pearl JR, Zhang L, et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat Methods. 2012;9:993–998. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavayogarajah T, Gangopadhyay P, Rahlfs S, Becker K, Lingelbach K, Przyborski JM, Holder AA. Alternative Protein Secretion in the Malaria Parasite Plasmodium falciparum . PLoS One. 2015;10:e0125191. doi: 10.1371/journal.pone.0125191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin CJ, van Dooren GG, Spurck TP, Struck NS, Good RT, Handman E, Cowman AF, et al. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol Biochem Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic acids research. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Vignali M, Armour CD, Chen J, Morrison R, Castle JC, Biery MC, Bouzek H, et al. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J Clin Invest. 2011;121:1119–1129. doi: 10.1172/JCI43457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CT, Lu IY, Hoffman MH, Sun WQ, Trent JD, Connor J. T-complex polypeptide-1 interacts with the erythrocyte cytoskeleton in response to elevated temperatures. J Biol Chem. 2004;279:16223–16228. doi: 10.1074/jbc.M310730200. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.