Abstract
High myopia (HM) is a leading cause of mid-way blindness with a high heritability in East Asia. Although only a few disease genes have been reported, a small proportion of patients could be identified with genetic predispositions. In order to expand the mutation spectrum of the causative genes in Chinese adult population, we investigated three genes, SLC39A5, LEPREL1 and LRPAP1, in a cohort of 187 independent Chinese patients with high myopia. Sanger sequencing was used to find possible pathogenic mutations, which were further screened in normal controls. After a pipeline of database and predictive assessments filtering, we, thereby, identified totally seven heterozygous mutations in the three genes. Among them, three novel missense mutations, c.860C > T, p.Pro287Leu and c.956G > C, p.Arg319Thr in SLC39A5, c.1982A > G, p.Lys661Arg in LEPREL1, were identified as potentially causative mutations. Additionally, the two heterozygous mutations (c.1582G > A, p.Ala528Thr; c.1982A > G, p.Lys661Arg) in one patient in LEPREL1 gene were reported in this study. Our findings will not only augment the mutation spectrum of these three genes, but also provide insights of the contribution of these genes to adult high myopia in Chinese. However, further studies are still needed to address the pathogenicity of each of the mutations reported in this study.
Introduction
High myopia is one of the most severe eye disorders with a strong genetic component1, 2. This disease is resulted primarily from excessive axial elongation of the eyeball (longer than 26 mm)3, concomitant with obvious refractive error (greater than 6 diopters4). It can also predispose the affected individuals to several ocular comorbidities, such as retinal detachment5, 6, macular degeneration7, 8 and glaucoma9. On the other hand, myopia prevalence rates vary and have been increasing worldwide. Multiple studies have shown that its prevalence ranges between 30% to 50% in American, European and Australian populations10–12, and is as high as about 71–96% in Asian countries, particularly in China, Singapore and Japan13–15.
High myopia has been widely accepted as a complex disorder. Both genetic and environmental factors have been shown to involve in the etiology of myopia16, 17. Family and twin studies have indicated that genetic factor, in particular, plays a significant role in the development of high myopia18, 19.
Despite intensive study on myopia, its exact molecular mechanism remains unclear, and it is mostly regarded as a polygenic disorder. Genome-wide association studies (GWAS) have mapped several genomic loci associated with myopia to chromosomes 11q24.120, 15q1421, 15q2522, 5p1523, 4q2524, 13q12.1225in large cohorts. On the other hand, at least 39 susceptibility loci have been identified by linkage analysis for nonsyndromic monogenic myopia26. In addition, mutations in six genes associated with high myopia have been detected by next generation sequencing. Of the six, three genes, including ZNF644 (c.2156A > G, p. Ser672Gly; c.725C > T, p.Thr242Met; c.821A > T, p.Gln274Trp; c.2014A > G, p.Ser672Gly)27, 28; SCO2 (c.157C> T; p. Gln53*)29 and SLC39A5 (c.141C > G; p.Tyr47*, c.T911C; p.Met304Thr)30, have been reported for autosomal dominant high myopia, and three other genes have been reported for autosomal recessive high myopia, including LEPREL1 (c.13C > T, p.Gln5X; c.1523C > T, p.Gly508Val)31, 32; LRPAP1 (c.605delA, p.Asn202Thrfs*8; c.863_864del, p.Ile288Argfs*118)33 and CTSH (c.485_488del, p. Leu162Profs*66)33. Recently, Jiang et al.34 has identified five novel mutations in several disease-causing genes in 298 unrelated Chinese patients with high myopia, including three heterozygous mutations (p.Lys369Met, p.Ala55OThr and p.Asp851His) inZNF644, a frame shift mutation (p.Gln67Sfs*8) in LRPAP1 and a heterozygous mutation (p.Gly413Ala) in SLC39A5. Up to now, limited mutations in the six causative genes have been confirmed, which contributed to very few high myopia cases being genetically deciphered. In summary of these previous studies, we proposed that those genes mutation exert their characteristics in different regions. SLC39A5, LEPREL1 and LRPAP1 were more likely to associate with Chinese high myopia patients. Therefore, we screened mutations in the three HM associated genes, SLC39A5, LEPREL1 and LRPAP1, and discovered additional mutations in a group of 187 unrelated Chinese patients with high myopia.
Materials and Methods
Patient recruitment
A total of 187 patients were enrolled in this study. All patients were clinically diagnosed with high myopia greater than −6.0 D. We selected totally 200 subjects as healthy controls, who met the following criteria: aged more than 60, with no systemic diseases and no high myopia and other known ocular diseases. Written informed consents were obtained from all the participants or their statutory guardians prior to the collection of their genomic DNA. The study was conducted in adherence to the tenets of the Declaration of Helsinki, and was approved by the Ethics Committee of the Eye Hospital of Wenzhou Medical University.
Mutational screening
Sanger sequencing was used as a direct and rigorous method to identify potential mutations in all three genes, the reported novel variants were further screened in matched controls. In detail, genomic DNA was extracted from leukocytes in the subjects’ peripheral venous blood using the Blood DNA Mini Kit (Simgen, Hangzhou, China) according to the manufacturer’s recommendations, and finally dissolved in TE buffer. PCR primer pairs were designed using the online program ExonPrimer35 (http://genome.ucsc.edu/cgi-bin/hgBlat) to amplify all coding regions and intron boundary of SLC39A5, LEPREL1 and LRPAP1. The seven primer pairs amplified sequences harboring the mutation were provided in Table 1. All amplified products were separated with polyacrylamide gel electrophoresis. Sequencing was performed with ABI 3730XL automated DNA sequencer (Thermo Fisher Scientific, Carlsbad, CA, USA). The sequences were compared against the known reference sequences obtained from the UCSC genome browser hg1936 (http://genome.ucsc.edu/cgi-bin/hgGateway) in order to retrieve and identify SNPs, insertions or deletions. All mutations (Table 2) were screened in 200 healthy controls.
Table 1.
PCR Primers for Sequences Harboring the Mutations in Present Study.
| Primer name | Forward | Reverse |
|---|---|---|
| Chr3:189972991 | TCAATGCAAGCTAGTGCCTG | TTTGCCTTGTTTCATTTCCC |
| Chr3:189681799 | AGCCAGAGAAGCAGGAGTTG | TTCTTTTCCTCAGACGAAGC |
| Chr3:190120600 | GAGGGAAGGTGGGAGAGG | ACTGAACAGAGATGACGGGG |
| Chr12:56231524 | GATGTTTCGGGGAGAATAGGAG | ATTTGTAACTCCAGGGATCTCG |
| Chr12:56629399 | AGTAGAGCATATGAGCGAAGGC | CAGTTCTTGACTGGGACTCTGG |
| Chr12:56630190 | GTGGAACCAGGTGTTCATCTTC | CAGCTGATAACTAGGAGCCCTG |
| Chr4:3514801 | GTCCTTGCAGTTCACCCG | CGGCCTCATCTTTCCTGC |
Table 2.
Summery of Mutations in LEPREL1, SLC39A5 and LRPAP1.
| Chr.position | Gene | Exon | Mutation | Status | Patient | Mutation | SIFT | PolyPhen2 | PROVEAN | Mutation | Note | ExAC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taster | Assessor | |||||||||||
| Chr12:56231524 | SLC39A5 | 4 | c.250C > T (p.Arg84Trp) | Het | HM_h16 | DC (0.935) | D (0.004) | Pr.D (0.995) | N (−1.97) | L (1.61) | rs199681035 | 0.0001771 |
| Chr12:56629399 | SLC39A5 | 8 | c.860C > T (p.Pro287Leu) | Het | HM_95 | DC (0.995) | D (0.047) | B (0.231) | N (−0.57) | N (0.555) | Novel | 0.0001895 |
| Chr12:56630190 | SLC39A5 | 9 | c.956G > C (p.Arg319Thr) | Het | HM_71 | P (0.283) | D (0.022) | B (0.174) | N (−0.84) | L (1.09) | Novel | — |
| Chr3:190120600 | LEPREL1 | 1 | c.132C > A (p.Phe44Leu) | Het | HM_101 | DC (0.996) | T (0.191) | B (0.002) | N (−1.58) | L (1.59) | rs367659257 | 0.0001129 |
| Chr3:189972991 | LEPREL1 | 11 | c.1582G > A (p.Ala528Thr) | Het | HM_68 | DC (0.999) | D (0.014) | Pr.D (0.977) | D (−3.55) | M (2.83) | rs199877373 | 0.000132 |
| Chr3:189681799 | LEPREL1 | 14 | c.1982A > G (p.Lys661Arg) | Het | HM_68 | DC (1) | T (0.594) | B (0.000) | N (1.06) | N (−0.22) | Novel | 3.30E-05 |
| Chr4:3514801 | LRPAP1 | 7 | c.962 G > A (p.Arg321His) | Het | HM_h32 | P (0.016) | T (0.17) | Pos.D (0.884) | N (−0.60) | M (2.2) | rs140947105 | 0.00015 |
Notes: Het, heterozygous; Mutation taster (DC, disease-causing; P, polymorphism); SIFT (D, damaging; T, tolerated); PolyPhen2 (Pr.D, probably damaging; pos.D, possible damaging; B, benign); PROVEAN (D, deleterious; N, neutral); MutationAssessor (M, medium; L, low; N, neutral).
Bioinformatics analysis
We evaluated all identified mutations using the following software and online database37. Polymorphism Phenotyping v238 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2), Sorting Intolerant from Tolerant39 (SIFT, http://sift.jcvi.org) and Mutation Taster40 (http://www.mutationtaster.org/) were employed to assess protein structure/function and evolutionary conservation. PROVEAN41 (http://provean.jcvi.org/index.php) was used to align and measure the similarity between mutation sequence and protein sequence homologs. Mutations detected in potential splice-sites were analyzed by Human Splice Finder42 (HSF, www.umd.be/HSF). SNPs minor allele frequency (MAF) was evaluated by 1000 Human Genome Project43 (ftp://1000genomes.ebi.ac.uk/vol1/ftp) and Exome Aggregation Consortium44 (ExAC, http://exac.broadinstitute.org/). Mutation assessor45(http://mutationassessor.org/) was used to predict the effect of evolutionary conservation.With Clustal Omega46 (http://www.ebi.ac.uk/Tools/msa/clustalo/), we also acquired multiple-sequence alignment of SLC39A5, LEPREL1 and LRPAP1 in different species, including Homo sapiens, Pan Troglodytes, Macacamulatta, Bostaurus, Feliscatus, Mus musculus, Gallus gallss and Danio rerio. SMART47 (http://smart.embl-heidelberg.de/) was used to simulate the topological model of the relative genes polypeptide. Furthermore, associated crystal structures of mutant and wild-type proteins were predicted by Phyre248 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi? id=index) and then visualized by Pymol Molecular Graphics System (Pymol)49.
Mutation criteria
Mutations identified in the three genes from all subjects with high myopia were filtered by the following criteria34, 50:
Variants in noncoding region that did not affect splicing sites based on prediction of the Berkeley Drosophila Genome Project (http://www.fruitfly.org/) were excluded;
Synonymous mutations in genes that did not alter splicing sites were subtracted;
Mutations with minor allele frequency (MAF) less than or equal to 0.01 in the Exome Aggregation Consortium (ExAC) were extracted;
Nonsynonymous single nucleotide mutations predicted to be benign by three commonly used silico tools (Mutation Taster, SIFT and Polyphen-2) were excluded;
Mutations were verified using dbSNP146 and those without rs number, were regarded as novel rare mutations.
Results
We screened for mutations in SLC39A5, LEPREL1 and LRPAP1 in a cohort of 187 high myopia patients with Sanger sequencing. A total of seven heterozygous mutations from six subjects were confirmed (Fig. 1) by applying the filtering criteria described in the Methods section. All mutations were located in the functional domains, except for the c.250C > T, p.Arg84Trp in SLC39A5, according to the prediction of SMART (Fig. 1A–C). None of these mutant alleles were detected in the control population.
Figure 1.
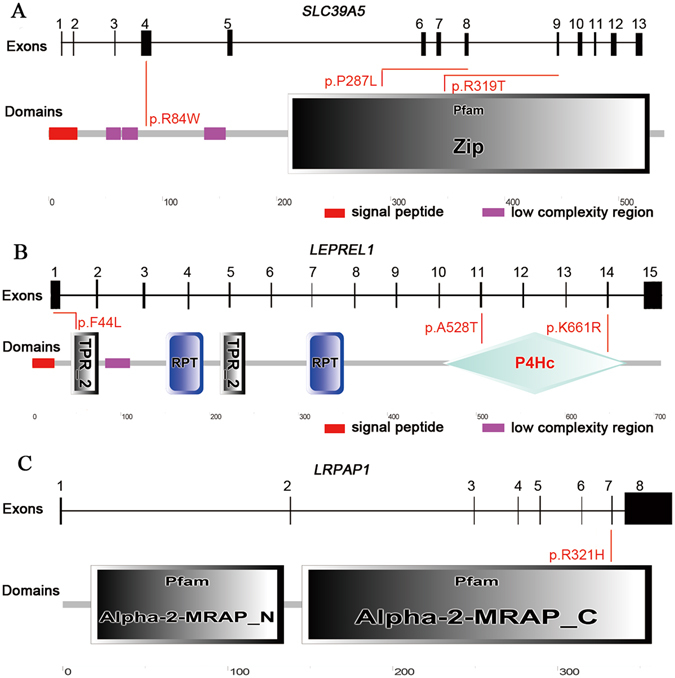
Location of the identifiedmutations in SLC39A5, LEPREL1 and LRPAP1. Exons of human SLC39A5, LEPREL1 and LRPAP1 (upper), and positions of mutated residues corresponding to the topological model of the polypeptides (under). A total of seven missense mutations colored red were identified in this study. All mutations were located in the functional domains, except for the heterozygous mutation c.250C > T (p.Arg84Trp) in SLC39A5 (A,B,C). Pfam ZIP domain is responsible for metal ion transmembrane transporter activity (A). Proteins containing TPRs are involved in many biological processes, such as cell cycle regulation, mitochondrial and peroxisomal protein transport, neurogenesis and protein folding, RPT is an internal repeat, P4Hc domain participatesin inoxidoreductase activity (B). Alpha-2-MRAP isa Pfam domain that binds to the alpha-2-macroglobulin receptor (C).
SLC39A5Mutations
Three heterozygous mutations were detected in SLC39A5 (c.860C > T, p.Pro287Leu; c.956G > C, p.Arg319Thr; c.250C > T, p.Arg84Trp) from three sporadic cases (Fig. 2A–C), among which p.Pro287Leu and p.Arg319Thr were novel (Table 2).The substitution p.Pro287Leu was predicted to be pathogenic by both SIFT and Mutation Taster at a low allele frequency. Besides that, mutated amino acid is evolutionarily highly conserved among all the tested species apart from danio after multiple orthologous sequence alignment (Fig. 3B), illustrating that it is important for protein function. Consequently, structural modeling demonstrated the absence of bonds between the mutated residue 287 leucine and residue 284 asparticacid, 290 serine, 291 valine (Fig. 4B). The mutation p.Arg319Thr was predicted to be damaging by SIFT (Table 2). In addition, 3D modeling demonstrated a newly formed bond between residue 319 and residues 320, 322 (Fig. 4C). The mutation p.Arg84Trp caused a substitution of arginine to tryptophan at position 84. This was predicted as damaging by 3 different in silico tools (PolyPhen-2, SIFT and Mutation Taster). This mutation was very rare in the ExAC database (Table 2). Furthermore, R84Wwas shown to affect highly evolutionarily conserved amino acid residues (Fig. 3A). In addition, 3D structural modeling revealed the absence of a bond between the mutated tryptophan at residue 84 and glycine at residue 83 (Fig. 4A).
Figure 2.
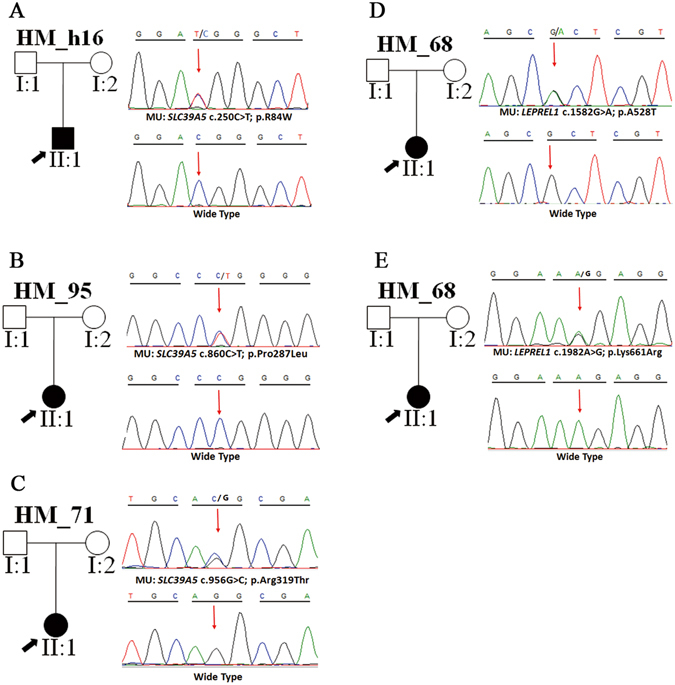
Potentially pathogenic mutations detected in this study. Pedigree plots of mutations. The black arrow represents the patient (left). Sequence profiles of identified mutations and wild type (right) were also shown (A–E).
Figure 3.
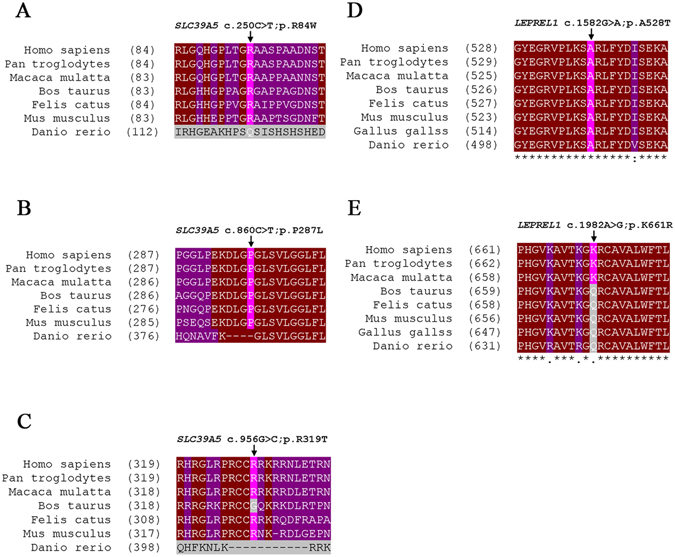
Conservation analysis revealed evolutionary conservation of the mutations. Clustal Omega results showing multiple alignments of the amino acids from different species. The arrow indicates the location of the mutations (A–E).
Figure 4.
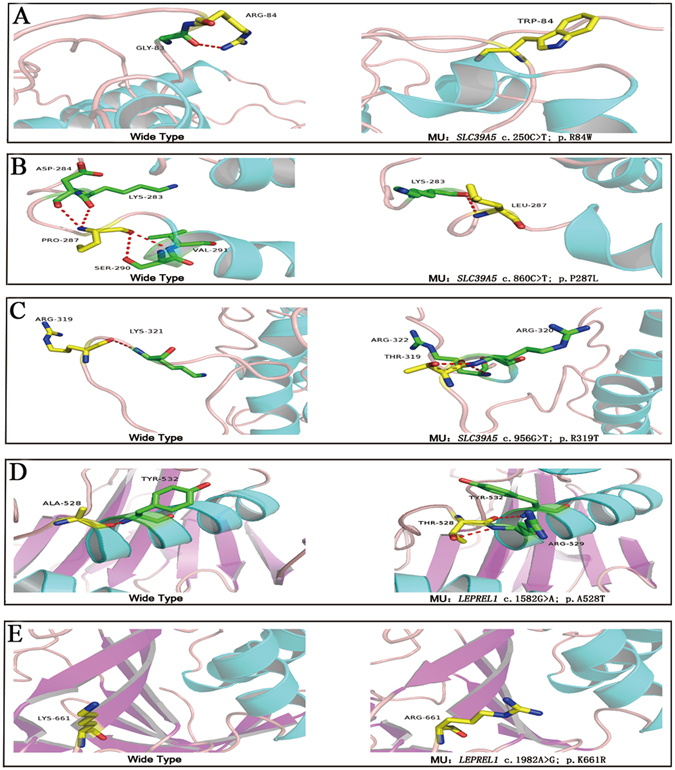
Predicted three-dimensional structure of proteins. Predicted crystal structures of wild type (left) and mutant (right) proteins. Yellow represents residue of wild type (left) and mutant (right), while green indicates residues that interact with wild-type (left) and mutant residue (right) (A–E).
LEPREL1 Mutations
In LEPREL1, three heterozygous mutations were detected in two isolated individuals. Among them, two heterozygous mutations were found in one patient (Fig. 2D,E). The c.1982A > G, p.Lys661Arg and c.1582G > A, p.Ala528Thr located in exon14 and exon11, respectively. The heterozygous p.Lys661Arg was a novel conservative mutation (Table 2) (Fig. 3E) that was predicted to be disease-causing by Mutation Taster. The p.Ala528Thr has been annotated as an exceedingly rare SNP (rs199877373), and it was predicted as a pathogenic mutation by all 5 pathogenicity prediction tools used in this study (PolyPhen-2, SIFT, Mutation Taster, PROVEAN and Mutation Assessor). Furthermore, the A528T mutation occurred at a remarkably conserved region in various species (Fig. 3D, Table 2), and was projected to produce a substituted bond between the mutated residues 528 and 529, in the 3D protein model (Fig. 4D). The third mutation c.132C > A, p.Phe44Leu was highly conserved among the different species (Figure S1A–C). Nevertheless, it was excluded based on the inheritance pattern of the gene. As expected, the mutation p.Phe44Leu was estimated as benign by both Polyphen-2 and SIFT (Table 2), and no noticeable fundamental changes were observed in protein modeling (Figure S1D, E).
LRPAP1 Variant
A heterozygous variant c.962G > A, p.Arg321His was detected in a HM patient (Figure S2A,B). It was predicted to be a benign polymorphism by both SIFT and Mutation Taster (Table 2). Likewise, R321H was excluded according to the inheritance pattern of the gene and its resulting change in residues showed no effect on crystal structure modeling (Figure S2 C–E).
Discussion
To date, tremendous efforts have been made to better understand the genetics of high myopia. However, only a few studies that explored mutations in the six causative genes (SLC39A5 30, 34, LEPREL1 31, 32, LRPAP1 33, 34, CTSH 33, SCO2 29, 34 and ZNF644 27, 34) have been reported. In this study, we attempted to replicate previous results and broaden the mutation spectrum of HM associated genes in a Chinese high myopia cohort. Based on previous reports, we proposed that SLC39A5, LEPREL1 and LRPAP1were more likely to associate with Chinese high myopia patients. We regarded them as preferential genes, and set out to screen mutations in these three genes using Sanger sequence. We identified a total of seven mutations that were predicted to influence the functional residues. These included three heterozygous mutations in SLC39A5 (p.Pro287Leu; p.Arg319Thr and p.Arg84Trp) from three sporadic cases, three heterozygous mutations (p.Lys661Arg; p.Ala528Thr and p.Phe44Leu) in LEPREL1 in two isolated individuals, a heterozygous mutation p.Arg321His in LRPAP1. Among these seven mutations, three mutations, p.Pro287Leu and p.Arg319Thr in SLC39A5 and p.Lys661Arg in LEPREL1, have not been previously reported. Our results were consistent with those reported in previous study in that missense mutations accounted for the largest proportion of the four mutation types in reported monogenic high myopia patients50.
SLC39A5 encodes the solute carrier family 39, member 5, which is a key member of the ZIP transporters for metal ions, especially in mammalian zinc omeostasis51. SLC39A5 has also been shown to express in all developmental stages of mouse ocular tissues, and is especially abundant in the sclera and in several layers of the retina. In addition, SLC39A5 might be involved in high myopia by regulating BMP/TGF-β, and the disruption of this pathway may be the underlying mechanisms of high myopia30. Guo et al. first identified that mutations in SLC39A5 were associated with autosomal dominant high myopia in two Chinese family, which included a nonsense mutation (c.141C > G, p.Tyr47*) and a missense mutation (c.911T > C, p.Met304Thr)30. Jiang et al. subsequently reported another heterozygous missense mutation c.1238G > C, p.Gly413Ala in a sporadic case34. In this study, three missense mutations p.Arg84Trp; p.Pro287Leu and p.Arg319Thr were detected in three isolated patients, and two of them have never been reported. Through secondary structure modeling, we found a novel mutation p.Pro287Leu, which was locatzed in the Pfam ZIP domain of the SLC39A5 (Fig. 1A). We modeled the 3D structure of SLC39A5 protein by applying Phyre2 program52. The structural modeling showed an absence of bond between the mutated residue 287 leucine and the residues 284 asparticacid, 290 serine and 291 valine (Fig. 4B). Since residue 290 was predicted to be a phosphorylation site, the p.Pro287Leu mutation may affect phosphorylation events. Further functional studies are needed to elucidate the molecular mechanism of high myopia as related to SLC39A5, however, our results may provided additional genetic evidence for potential contribution of SLC39A5in high myopia.
LEPREL1encodes a member of the prolyl 3-hydroxylase subfamily of 2-oxo-glutarate-dependent dioxygenase (P3H2). Expression of LEPREL1 has been detected in various collagen fibril-containing tissues, including the eye53. As reported, P3h2 n/nmouse suffered from a significant defect in collagen prolyl 3-hydroxylation compared with their wild type littermates, which led to result in structural abnormalities in multiple eye tissues. These findings suggested that altered collagen hydroxylation caused by loss of LEPREL1 can potentially contribute to the myopia54.Mordechai et al.31 previously identified a homozygous mutation c.1523G > T, p.Gly508Val in LEPREL1from affected individuals in a Bedouin kindred. Co-segregation of one homozygous nonsense mutation in LEPREL1 (c.13C > T, p.Gln5*) with autosomal-recessive high myopia was reported in Chinese family by Guo et al.32. In this study, we identified two heterozygous mutations p.Ala528Thr and p.Lys661Arg in one high myopia patient. Of note, we also found a heterozygous mutation p.Phe44Leu in LEPREL1, which was excluded in this study due to the inheritance pattern of the gene.
LRPAP1encodes a Low Density Lipoprotein (LDL) Receptor-Related Protein Associated Protein 1, a 357 amino acid protein that binds and protects the LDL receptor-related protein (LRP1), which is known to influence transforming growth factor-βactivity33, 55, 56. Furthermore, TGF-β signal pathway has been highlighted as the responsible factor for regulation of scleral metabolism in myopia57. Therefore, LRPAP1 may affect the formation of myopia by regulating TGF-β signaling. So far, three homozygous frame shift mutations (Asn202Thrfs*8; Ile288Argfs*118; Gln67Serfs*8) inLRPAP1have been reported to associate with autosomal recessive high myopia in Arabic and Chinese families33, 34. Here, we detected a heterozygous mutation p.Arg321His in LRPAP1. However, p.Arg321His already existed in the database (rs140947105) and was excluded based on the inheritance pattern of this gene. Nevertheless, co-segregation analysis could not be performed due to the unavailability of familiar samples. In addition, functional consequences of these mutations need to be investigated in future studies.
In conclusion, we carried out mutational screening in three causative genes in a large cohort of high myopia patients. We identified seven mutations, including three novel mutations. Our findings widen the mutation spectrum of known HM-genes and provided additional genetic evidence thatSLC39A5 and LEPREL1may be associated with high myopia in Chinese population. Nevertheless, given the paucity of our data on the pathogenicity of these genes, further studies are needed to better understand the potential roles of these genes in the development of high myopia.
Electronic supplementary material
Acknowledgements
We thank the patients for their participation in this study. This study was supported by the National Key Basic Research Program (2013CB967502 to Z.J.), National Natural Science Foundation of China (81371059, 81522014, 81500741), Zhejiang Provincial Natural Science Foundation of China (LR13H120001, LY12H12003), Zhejiang Provincial Key Research and Development Program (2015C03029), NHFPC Grant-in-Aid for Medical and Health Science (201472911), Wenzhou Science and Technology Innovation Team Project (C20150004), National Key Clinical Specialty (Ophthalmology), MOST Projects (2012YQ12008004), the project of Zhejiang Provincial Top Key Discipline and Key Construction Discipline of Medicine, Wenzhou Science and Technology Foundation (Y20140358).
Author Contributions
Conceived and designed the experiments: Z.J. Performed the experiments: C.F., X.H., X.C. Analyzed the data: C.F. Contributed reagents/materials/analysis tools: X.H., R.W., F.L. Wrote the paper: C.F., Z.J.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Chun-Yun Feng and Xiao-Qiong Huang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-01285-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buch H, et al. Prevalence and causes of visual impairment and blindness among 9980 Scandinavian adults: the Copenhagen City Eye Study. Ophthalmology. 2004;111:53–61. doi: 10.1016/j.ophtha.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Xu L, et al. Causes of blindness and visual impairment in urban and rural areas in Beijing: the Beijing Eye Study. Ophthalmology. 2006;113:1134.e1131–1111. doi: 10.1016/j.ophtha.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 3.Percival SP. Redefinition of high myopia: the relationship of axial length measurement to myopic pathology and its relevance to cataract surgery. Developments in ophthalmology. 1987;14:42–46. doi: 10.1159/000414364. [DOI] [PubMed] [Google Scholar]
- 4.Young, T. L., Metlapally, R. & Shay, A. E. Complex trait genetics of refractive error. Archives of ophthalmology (Chicago, Ill.: 1960) 125, 38–48, doi:10.1001/archopht.125.1.38, 125, 38–48 (2007). [DOI] [PubMed]
- 5.Benhamou N, Massin P, Haouchine B, Erginay A, Gaudric A. Macular retinoschisis in highly myopic eyes. American journal of ophthalmology. 2002;133:794–800. doi: 10.1016/S0002-9394(02)01394-6. [DOI] [PubMed] [Google Scholar]
- 6.Baba T, et al. Prevalence and characteristics of foveal retinal detachment without macular hole in high myopia. American journal of ophthalmology. 2003;135:338–342. doi: 10.1016/S0002-9394(02)01937-2. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi, K. et al. Long-term pattern of progression of myopic maculopathy: a natural history study. Ophthalmology117, 1595–1611, 1611-611.e1591–1594, doi:10.1016/j.ophtha.2009.11.003 (Long-term pattern of progression of myopic maculopathy: a natural history study, Ophthalmology, 2010). [DOI] [PubMed]
- 8.Ripandelli G, et al. Macular vitreoretinal interface abnormalities in highly myopic eyes with posterior staphyloma: 5-year follow-up. Retina (Philadelphia, Pa.) 2012;32:1531–1538. doi: 10.1097/IAE.0b013e318255062c. [DOI] [PubMed] [Google Scholar]
- 9.Ma F, Dai J, Sun X. Progress in understanding the association between high myopia and primary open-angle glaucoma. Clinical & experimental ophthalmology. 2014;42:190–197. doi: 10.1111/ceo.12158. [DOI] [PubMed] [Google Scholar]
- 10.Williams KM, et al. Increasing Prevalence of Myopia in Europe and the Impact of Education. Ophthalmology. 2015;122:1489–1497. doi: 10.1016/j.ophtha.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kempen, J. H. et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Archives of ophthalmology (Chicago, Ill.: 1960) 122, 495–505, doi:10.1001/archopht.122.4.495 (2004). [DOI] [PubMed]
- 12.Vitale S, Sperduto RD, Ferris FL., 3rd Increased prevalence of myopia in the United States between 1971-1972 and 1999-2004. Archives of ophthalmology (Chicago, Ill.: 1960) 2009;127:1632–1639. doi: 10.1001/archophthalmol.2009.303. [DOI] [PubMed] [Google Scholar]
- 13.Woo WW, et al. Refractive errors in medical students in Singapore. Singapore medical journal. 2004;45:470–474. [PubMed] [Google Scholar]
- 14.Sawada, A., Tomidokoro, A., Araie, M., Iwase, A. & Yamamoto, T. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology115, 363–370.e363–370.e3, doi:10.1016/j.ophtha.2007.03.075 (2008). [DOI] [PubMed]
- 15.He M, Zheng Y, Xiang F. Prevalence of myopia in urban and rural children in mainland China. Optometry and vision science: official publication of the American Academy of Optometry. 2009;86:40–44. doi: 10.1097/OPX.0b013e3181940719. [DOI] [PubMed] [Google Scholar]
- 16.Wojciechowski R. Nature and nurture: the complex genetics of myopia and refractive error. Clinical genetics. 2011;79:301–320. doi: 10.1111/j.1399-0004.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan IG, Ohno-Matsui K, Saw SM. Myopia. Lancet (London, England) 2012;379:1739–1748. doi: 10.1016/S0140-6736(12)60272-4. [DOI] [PubMed] [Google Scholar]
- 18.Hammond CJ, Snieder H, Gilbert CE, Spector TD. Genes and environment in refractive error: the twin eye study. Investigative ophthalmology & visual science. 2001;42:1232–1236. [PubMed] [Google Scholar]
- 19.Feldkamper M, Schaeffel F. Interactions of genes and environment in myopia. Developments in ophthalmology. 2003;37:34–49. doi: 10.1159/000072037. [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi, H. et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS genetics5, e1000660, doi:10.1371/journal.pgen.1000660 (2009). [DOI] [PMC free article] [PubMed]
- 21.Solouki AM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nature genetics. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hysi PG, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nature genetics. 2010;42:902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YJ, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–375. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Human molecular genetics. 2011;20:2861–2868. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, et al. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. American journal of human genetics. 2011;88:805–813. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Q, et al. Childhood gene-environment interactions and age-dependent effects of genetic variants associated with refractive error and myopia: The CREAM Consortium. Scientific reports. 2016;6:25853. doi: 10.1038/srep25853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS genetics. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran-Viet KN, et al. Study of a US cohort supports the role of ZNF644 and high-grade myopia susceptibility. Molecular vision. 2012;18:937–944. [PMC free article] [PubMed] [Google Scholar]
- 29.Tran-Viet KN, et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. American journal of human genetics. 2013;92:820–826. doi: 10.1016/j.ajhg.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, et al. SLC39A5 mutations interfering with the BMP/TGF-beta pathway in non-syndromic high myopia. Journal of medical genetics. 2014;51:518–525. doi: 10.1136/jmedgenet-2014-102351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mordechai S, et al. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. American journal of human genetics. 2011;89:438–445. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H, et al. Homozygous loss-of-function mutation of the LEPREL1 gene causes severe non-syndromic high myopia with early-onset cataract. Clinical genetics. 2014;86:575–579. doi: 10.1111/cge.12309. [DOI] [PubMed] [Google Scholar]
- 33.Aldahmesh MA, et al. Mutations in LRPAP1 are associated with severe myopia in humans. American journal of human genetics. 2013;93:313–320. doi: 10.1016/j.ajhg.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang D, et al. Detection of mutations in LRPAP1, CTSH, LEPREL1, ZNF644, SLC39A5, and SCO2 in 298 families with early-onset high myopia by exome sequencing. Investigative ophthalmology & visual science. 2015;56:339–345. doi: 10.1167/iovs.14-14850. [DOI] [PubMed] [Google Scholar]
- 35.Kent, W. J. BLAT–the BLAST-like alignment tool. Genome research12, 656–664, doi:10.1101/gr.229202. Article published online before March 2002 (2002). [DOI] [PMC free article] [PubMed]
- 36.Kent, W. J. et al. The human genome browser at UCSC. Genome research12, 996–1006, doi:10.1101/gr.229102. Article published online before print in May 2002 (2002). [DOI] [PMC free article] [PubMed]
- 37.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine: official journal of the American College of Medical Genetics. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 41.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England) 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic acids research. 2009;37:e67–e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic acids research. 2011;39:e118–e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic acids research. 2015;43:D257–260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg M. The Phyre2 web portal for protein modeling, prediction and analysis. J. The Phyre2 web portal for protein modeling, prediction and analysis. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.5 (2015).
- 50.Sun W, et al. Exome Sequencing on 298 Probands With Early-Onset High Myopia: Approximately One-Fourth Show Potential Pathogenic Mutations in RetNet Genes. Investigative ophthalmology & visual science. 2015;56:8365–8372. doi: 10.1167/iovs.15-17555. [DOI] [PubMed] [Google Scholar]
- 51.Wang F, Kim BE, Petris MJ, Eide DJ. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. The Journal of biological chemistry. 2004;279:51433–51441. doi: 10.1074/jbc.M408361200. [DOI] [PubMed] [Google Scholar]
- 52.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 53.Capellini TD, Dunn MP, Passamaneck YJ, Selleri L, Di Gregorio A. Conservation of notochord gene expression across chordates: insights from the Leprecan gene family. Genesis (New York, N.Y.: 2000) 2008;46:683–696. doi: 10.1002/dvg.20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hudson DM, et al. Post-translationally abnormal collagens of prolyl 3-hydroxylase-2 null mice offer a pathobiological mechanism for the high myopia linked to human LEPREL1 mutations. The Journal of biological chemistry. 2015;290:8613–8622. doi: 10.1074/jbc.M114.634915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willnow TE, et al. RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. The EMBO journal. 1996;15:2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 56.Willnow, T. E., Armstrong, S. A., Hammer, R. E. & Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo, Herz, J. Functional expression of low density lipoprotein receptor-related protein is controlled by receptor-associated protein in vivo. Proceedings of the National Academy of Sciences of the United States of America92, 4537–4541 (1995). [DOI] [PMC free article] [PubMed]
- 57.McBrien NA. Regulation of scleral metabolism in myopia and the role of transforming growth factor-beta. Experimental eye research. 2013;114:128–140. doi: 10.1016/j.exer.2013.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


