Abstract
Amphioxus or lancelets have been regarded as a key animal in understanding the origin of vertebrates. However, the evolutionary history within this lineage remains unexplored. As the amphioxus lineage has likely been separated from other chordates for a very long time and displays a marked left-right asymmetry, its evolutionary history is potentially helpful in better understanding chordate and vertebrate origins. We studied the phylogenetic relationships within the extant amphioxus lineage based on mitochondrial genomes incorporating new Asymmetron and Epigonichthys populations, and based on previously reported nuclear transcriptomes. The resulting tree patterns are consistent, showing the Asymmetron clade diverging first, followed by the Epigonichthys and Branchiostoma clades splitting. Divergence time estimates based on nuclear transcriptomes with vertebrate calibrations support a shallow diversification of the extant amphioxus lineage in the Tertiary. These estimates fit well with the closure of seaways between oceans by continental drift, ocean currents, and present geographical distributions, and suggest a long cryptic history from the origin of amphioxus to its most recent diversification. Deduced character polarities based on phylogenetic analyses suggest that the common ancestor of the extant amphioxus existed in a tiny epibenthic state with larva-like appearance of extant amphioxus, likely with ciliate epidermis.
Introduction
Among chordates, the deepest branching chordate subphylum Cephalochordata displays a unique asymmetrical development1. Cephalochordata is a small group divided into three genera, Asymmetron, Branchiostoma, and Epigonichthys 2–6, and comprises about 30 species at present7. All extant species are generally inactive suspension feeders living in sand substrata (e.g. Branchiostoma 8). Curiously, however, all species display streamlined appearances similar to actively swimming fish, and do not develop a mucus layer or cuticles on the skin unlike in many other sand dwellers. These incongruous features of amphioxus provide clues for understanding the origin of the chordate lineage. To reconstruct an accurate image of the last common ancestor (LCA) of the chordate lineage, it is essential to understand the LCA of the amphioxus lineage by elucidating character polarities from varying features found in living amphioxus species based on reliable molecular phylogenetic analyses.
A recent genome-based phylogenetic study suggests that the Cephalochordata appeared in the Precambrian, soon after the split of bilaterian animals into deuterostomes and protostomes9. The branching order of the three extant amphioxus genera was also suggested based on the mitochondrial genomic (mitogenomic) sequences of Asymmetron and Epigonichthys specimens from the Maldives, Bermuda, and western Japan3, 5; with the genus Asymmetron diverging first followed by Epigonichthys and Branchiostoma. This phylogenetic framework is important for understanding the origin of chordates as the amphioxus clades that develop gonads only on the right side (Asymmetron, Epigonichthys) are not monophyletic. However, considering the wide distribution of amphioxus genera, each possibly including cryptic species4, there is a need to expand the phylogenetic dataset utilised in reconstruction of the evolutionary history of the amphioxus lineage.
Divergence times for the amphioxus lineage have also been previously proposed3–5, 10. Some of these studies applied deep reference points such as 652 Ma and 891 Ma5, but these are inappropriate for mitogenomes due to substitution saturation11, 12, while other studies have depended exclusively on outgroup reference points and have estimated divergence of Asymmetron in the Mesozoic or Paleozoic (120–360 Ma) with a very wide upper confidence limit that reaches to ~25 Ma in the Cenozoic5, 10. These studies accordingly suggest very slow evolution within the amphioxus lineage. However, a whole genome study on Branchiostoma belcheri found that in this species gene turnover was active and also that the amino acid substitution rate was comparable to that of vertebrates with rapid substitutions13. Thus, although the evolutionary rate of the amphioxus lineage has been regarded as being slow, a re-evaluation of this theory is clearly needed due to uncertainty.
Northwest Pacific coastal waters surrounding China, Taiwan, and western Japan harbour six or more amphioxus species, including representatives of all three genera2, 14–17; Asymmetron lucayanum Andrews, 1893, A. inferum Nishikawa, 2004, Epigonichthys maldivensis (Forster Cooper, 1903), E. cultellus Peters, 1877, Branchiostoma belcheri (Gray, 1847), and B. japonicum (Willey, 1897). Asymmetron and Epigonichthys are characterised by gonads that develop only on the right side of the body. Asymmetron lucayanum is currently thought to be distributed circumtropically in the Indian, Pacific, and Atlantic Oceans7, but based on analyses of sequences of cytochrome c oxidase subunit I (coxI), it has been suggested that this taxon contains at least three cryptic species4. Around Kuroshima Island, Okinawa, Japan, A. lucayanum and E. maldivensis have been reported as occurring sympatrically, and the Kuroshima A. lucayanum population has been suggested to contain two cryptic species3, 4. In Taiwan, A. lucayanum and E. maldivensis are sympatric in most habitats as seen at Kuroshima Island14, 16. Epigonichthys cultellus has been reported from the northern South China Sea16, 18–20. Although all of these amphioxus species with dextral gonads are distributed in tropical or subtropical waters, undescribed Epigonichthys-like amphioxus specimens were collected in this study from temperate Kyushu Island, Japan.
Whole mitochondrial DNA sequences allow comparisons among a wide range of populations and can serve as a useful tool for understanding phylogenetic relationships within the amphioxus lineage3, 5. In this study we performed mitogenomic analyses by utilising new data acquired from Epigonichthys and Asymmetron specimens from Taiwan, from one of the Epigonichthys-like specimens from Kyushu, Japan, and from Asymmetron species from the Bahamas, West Atlantic (the type locality of A. lucayanum 21), and combined our acquired data with all publically available amphioxus mitogenomic sequences. We also analysed a nuclear transcriptome-based phylogeny complied from publically available data that included Branchiostoma, Asymmetron, and vertebrate species. Furthermore, we estimated divergence times with an expanded ingroup dataset of nuclear transcriptomes with carefully selected calibration points, and then verified if the resulting estimates corresponded to any reliably dated or well-known isolating events.
This study found that contrary to previous studies3–5, 10 the extant amphioxus lineage has experienced rather recent diversification events in the Tertiary. Our divergence estimates are consistent with the genetic proximity between species observed within the Asymmetron clade as we found that nucleotide substitution rates in amphioxus genomes were comparable to mammalian rates. In this phylogenetic framework, we suggest that Asymmetron lucayanum should be divided into two species, Asymmetron pelagicum (Günter, 1889) (Pacific and West Atlantic clades in ref. 4) and Asymmetron orientale Parker, 1904 (Indian and West Pacific clade in ref. 4). Character distributions in the three genera, when based on the present phylogenetic analyses, suggest that the amphioxus crown lineage survived in a tiny epibenthos form for a long period from its origin to the onset of the most recent diversification.
Results
Phylogeny within the amphioxus lineage
Phylogenetic analyses based on the mitogenomes’ nucleotide (nt) sequences resulted in trees topologically identical between Bayesian inference (BI) and maximum likelihood (ML) methods (Fig. 1). The phylogeny confirmed that the Asymmetron clade first diverged from the ancestral Epigonichthys + Branchiostoma clade (Fig. 1), as proposed in previous studies5, 17, 22. Within the Asymmetron clade, Asymmetron inferum, which is found near whale falls2, was sister to the other groups. The Indian Ocean Asymmetron clade (specimen from the Maldives)4 then separated from a clade comprising specimens from Taiwan, Bermuda4, the Bahamas, and Okinawa3 (Fig. 1).
Figure 1.
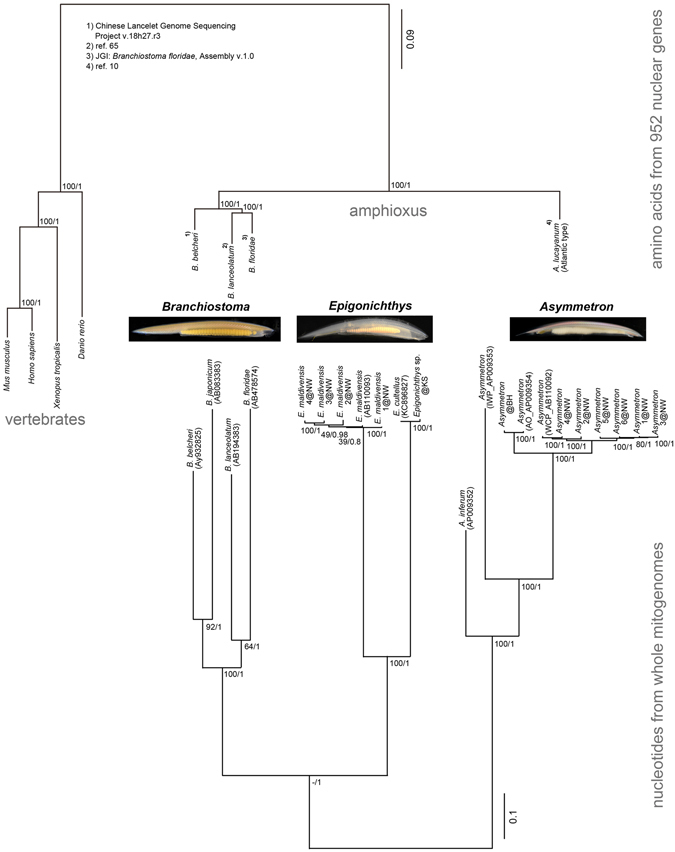
Phylogenetic trees of three extant amphioxus genera based on mitogenomes and nuclear transcriptomes. Bayesian and maximum likelihood methods gave the same topology for both trees based on amino acid sequences of nuclear transcriptomes (top) and on nucleotides of whole mitogenomes (bottom). Values at nodes are bootstrap values (left side) and posterior probabilities (right side). The top tree includes four vertebrate species and shows the monophyly of B. lanceolatum and B. floridae as shown in the bottom tree. The Asymmetron sequence in the top tree is derived from Atlantic type4. Numerals and initials in Epigonichthys and Asymmetron clades denote specimen numbers and types4/locations. IWP, Indo-West Pacific type; AO, Atlantic type; WCP, West-Central Pacific type; BH, Bahamas; NW, Nanwan Bay; KS, Kyushu.
When we compared amino acid (aa) sequences, the branching pattern of the Branchiostoma clade was inconsistent between BI and ML analyses. The most probable tree resulting from ML was almost identical to that of nt, whereas the BI tree showed that B. floridae was sister to all other Branchiostoma species with low posterior probability (also see ref. 17).
Phylogenetic analyses based on aa sequences of 952 protein-coding genes deduced from nuclear transcriptomes of amphioxus and vertebrate species generally supported the mitogenomic analyses, although these analyses lacked Epigonichthys data, and the branching order within Branchiostoma showed a monophyletic relationship between B. lanceolatum and B. floridae in both BI and ML methods (Fig. 1).
Inference of divergence times
In estimates of divergence times within a single lineage, calibration time points should be ideally related to the ingroup history rather than inferred based on outgroup calibrations23. Unfortunately, however, there is no reliable calibration point within the amphioxus lineage, and we thus calculated pairwise nucleotide substitutions per unit nucleotide length (p-distance). Our results found that substitution rates in mitochondria and nuclear genes between amphioxus species are comparable to those between humans and mice, whose divergence is dated to 81 ± 10 Ma24 (Fig. 2). In the comparison between amphioxus species, the nucleotide substitution rate in mitochondrial genes was three times as high as that in nuclear genes, as seen in humans and mice. In higher vertebrate taxa and vertebrate-amphioxus comparisons, the substitution per unit length did not increase in mitochondrial genes unlike as seen in nuclear gene analyses, suggesting substitution saturation of mitochondrial genes (Fig. 2). In amino acid substitution analyses, saturation in mitochondrial genes between deep branching taxa was weaker than as seen in nucleotide substitution analyses. A comparison including mammalian taxa noticed faster substitutions in mitochondrial genes than in nuclear genes (Fig. 2). These substitution results have two possible meanings; either diversification of extant amphioxus started as far back in time as the human-mouse divergence (81 ± 10 Ma), or alternately that substitutions in the amphioxus lineage are slower than those in the vertebrate lineage, as has been previously suggested10.
Figure 2.
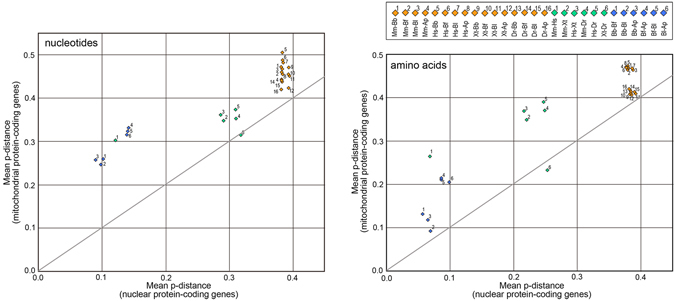
p-distances of mitochondrial and nuclear protein-coding genes between amphioxus and vertebrate taxa. Pairwise p-distances of nucleotide and amino acid sequences of 13 mitochondrial and 952 nuclear genes were calculated for all combinations of four amphioxus and four vertebrate species. The pairwise distances were calculated using MEGA-CC71 and plotted by mean values for all corresponding species pairs.
We first estimated divergence times based on nuclear transcriptome data with four reference points; the amphioxus-vertebrates (550.0 ± 16.0 Ma25), ray-finned fish-tetrapods (419.0 ± 1.4 Ma25), amphibians-amniotes (340.0 ± 5.0 Ma24), and primates-rodents (81.0 ± 10.0 Ma24) splits. Fossil evidence for the earliest vertebrate, Haikouichthys ercaicunensis 26, is more reliable than disputable amphioxus-related fossils such as Haikouella spp.27 and Pikaia gracilens 28 as the reference point for the split of the amphioxus lineage from other chordates. In fact, as the latter fossils are the same age as or younger than the former, the choice of fossil actually does not affect the calibration of this reference point. Calculations showed two convergence regions of estimates depending on seed values; one converged at >100 Ma for the divergence of Asymmetron- (Branchiostoma + Epigonichthys) as in previous studies3, 5, 10, while the other converged at 46.0 (32.4–56.6) Ma for the same divergence. As the latter result gave a significantly higher mean Bayesian posterior probability summarizing three runs of each result [older divergence (lnL = −137011) vs. younger divergence (lnL = −137008) p < 3.3 × 10−12, Welch’s t-test after Bonferroni correction], we accepted the latter result. This estimate dated the divergence times between extant amphioxus clades as two- to four times younger than previous estimates (Fig. 3A)5, 10. When calibration points are set at deep nodes in mitogenome-based estimates, saturation of nucleotide substitution can lead to over-estimation of divergence timing11. We thus inferred divergence times in this study by using mitogenomes with reference points obtained from the nuclear transcriptome-based estimates of the Asymmetron-(Branchiostoma + Epigonichthys) (46.0 ± 5.5 Ma), B. belcheri-(B. lanceolatum + B. floridae) (28.2 ± 5.5 Ma), and B. lanceolatum-B. floridae (22.6 ± 2.3 Ma) splits. These analyses estimated the timing of the splits for A. inferum diverging from the remainder of Asymmetron at 16.4 Ma, for the divergence of the Indian Asymmetron population from the Pacific + Atlantic population at 12.4 Ma, and for divergence of the Atlantic and Pacific populations at 3.1 Ma (Fig. 3B). Our estimates based on the mitogenomes also showed splits into the three current extant genera during a short period, and more recent diversifications at the species-level within Asymmetron (Fig. 3B) than previously has been assumed.
Figure 3.
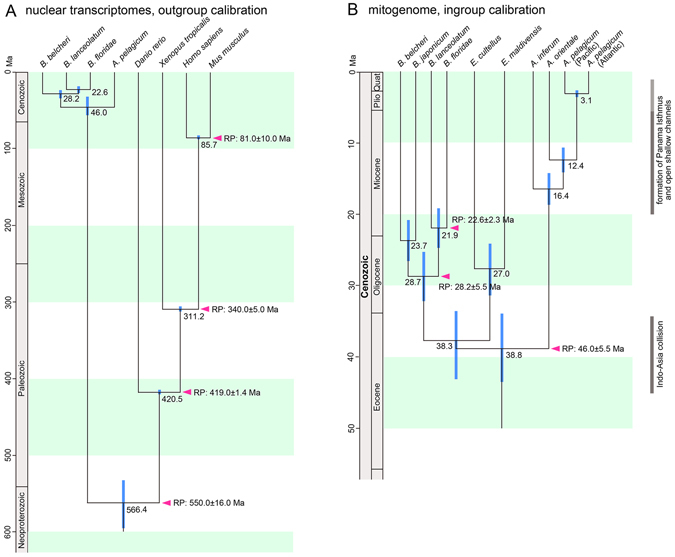
Divergence time estimates in amphioxus lineage. (A) Estimate based on nuclear transcriptome comparison calibrated with vertebrate fossil records26. Numerals at nodes denote estimated split time in Ma. (B) Estimate based on mitogenomic comparison with ingroup calibration points derived from (A). Timings of formation of the Isthmus of Panama36 and of Arabia-Eurasia38 and Indo-Asia collisions72, 73 are also shown. Blue bars denote 95% confidence interval.
The mitogenome-based phylogenetic analyses in this study showed that the Atlantic and Pacific Asymmetron populations were more closely related compared to the more distant Indian population (Fig. 1), as seen in a previous study4. We thus searched for isolation events that could explain this pattern, and based on our estimated divergence timings, we focused on the formation of the Isthmus of Panama and the closure of the Neo-Tethys Sea. We then again estimated divergence times within the amphioxus lineage based on the mitogenomic data utilizing these two geological reference points separately. We verified the congruence between estimated and given times, and then finally conducted an estimate with both reference points. With both reference points, the initial split of the amphioxus crown lineage into the Asymmetron and Branchiostoma + Epigonichthys clades was 42.2 (49.6–35.1) Ma, followed by the split of Brachiostoma and Epigonichthys at 35.6 (42.0–29.7) Ma (Supplementary Fig. S2). These estimates are consistent with our estimates obtained based on calibration points derived from nuclear transcriptome sequences (Fig. 3B).
Mitogenomic properties and taxonomic identity of sympatric Asymmetron and Epigonichthys specimens from Taiwan
One species each of Asymmetron and Epigonichthys are sympatric in tropical Nanwan Bay at the southern tip of Taiwan, where they are found in fine sand substratum containing a considerable amount of mud at 10–15 m depths, as reported previously14. The annual seawater temperature (monthly means) ranges between 23–29 °C29, and there is an extensive coral reef ecosystem at this location30. We collected Asymmetron individuals measured approximately 10–15 mm in body length, and Epigonichthys individuals ranging from 10 to 25 mm from Nanwan Bay. Some individuals of both species were developing gonads in March and May 2014 and in May 2015.
Previously, based on cox1 sequence analyses, two different clades of A. lucayanum were confirmed to exist sympatrically in Okinawa, Japan; these have been previously designated as ‘West-Central Pacific’ (herein Pacific type) and ‘Indo-West Pacific’ (herein Indian type) clades based on specimen origin4. We therefore compared sequences of coxI including all sequences in ref. 4 and found 21 unique haplotypes that include two previously reported4 within the Taiwanese population. All neighbour-joining, ML and BI trees based on coxI sequences showed the same branching pattern of Indian-(Pacific + Atlantic) (Fig. 4), which was also recovered in the whole mitogenomic sequence analyses (Fig. 1). In the network tree, all haplotypes from the Taiwanese population (n = 25) belonged to the Pacific type and was divided into two clusters; one contained haplotypes from the West Pacific while the other contained mainly haplotypes from Hawai’i (Fig. 4).
Figure 4.
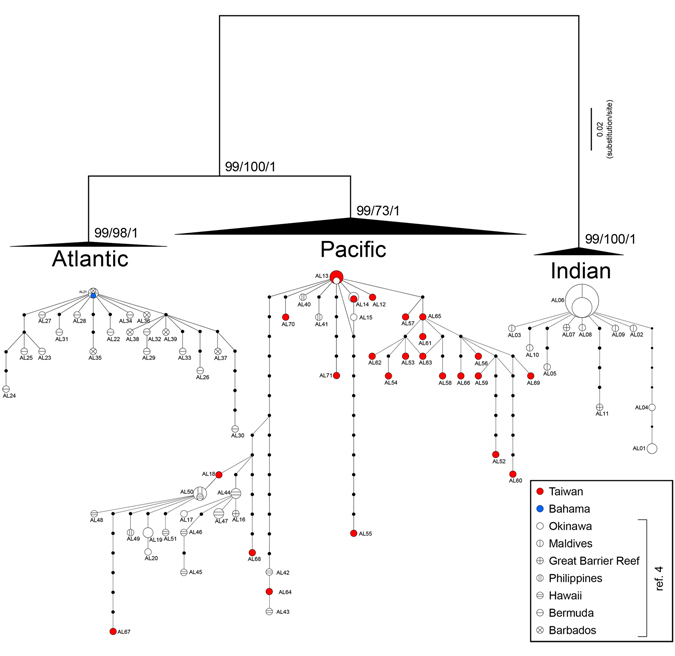
Genetic relationships of Asymmetron populations. Neighbour-joining, maximum likelihood, and Bayesian methods produced trees with the same topology for cytochrome c oxidase subunit I sequences (top) and show a close relationship between Pacific and Atlantic haplotype groups. Parsimony haplotype networks based on the same gene (bottom) show that haplotypes of all Taiwanese specimens are within the Pacific group. Sizes of circles indicate haplotype frequency and dots represent missing haplotypes.
As all Asymmetron specimens from Nanwan Bay were included within the Pacific type, we sequenced the full-length of mitogenomes for six randomly selected Asymmetron specimens from Nanwan Bay: AL52 (1@NW), AL13 (2@NW), AL57 (3@NW), AL67 (4@NW), AL70 (5@NW), and AL71 (6@NW), as well as for one Asymmetron specimen from the Bahamas: AL21 (@BH). The gene orders were all identical to those previously reported for A. lucayanum 3, 5, but the lengths of nucleotide sequences slightly varied, from 15,095 to 15,097 bp (Accession Numbers AP015017-015023), due to indel sites in 12S rRNA, 16S rRNA, tRNA-Ile genes, and/or the control region.
For Epigonichthys specimens, mitogenomes obtained from four specimens from Nanwan Bay clustered as a clade with a mitogenome of E. maldivensis from Okinawa, Japan (Fig. 1), and the gene order was identical to that of E. maldivensis 3. However, the lengths of nucleotide sequences slightly varied from 14,967 to 14,970 bp (AP015025-015028) due to indels as seen in the Asymmetron mitogenomes. A previous study on the complete mitogenome of an Epigonichthys specimen from the South China Sea has clarified its difference at the species level from E. maldivensis from Okinawa, Japan17, and therefore we examined the numbers of myomeres, gonads, and dorsal and preanal finboxes of Epigonichthys specimens from Nanwan Bay and the South China Sea (Table 1). These data reconfirmed clear morphological differences between the specimens from Nanwan Bay and the South China Sea (Fig. 5). Previously, based on morphometric data, the specimens from the South China Sea were assigned to E. cultellus 17 while specimens from Nanwan Bay were assigned to E. maldivensis 14, 16. Our results showed that whole mitogenomic sequences of Epigonichthys from Nanwan Bay are most similar to and had gene orders identical to that of E. maldivensis from Okinawa and reconfirm that the Epigonichthys species found sympatrically with Asymmetron species in Nanwan Bay is E. maldivensis.
Table 1.
Metrical characteristics of Epigonichthys species from China-Taiwan-Japan waters.
| Specimens | Body length (mm) | No. of myomeres | No. of dorsal finboxes | No. of preanal finboxes | No. of gonads |
|---|---|---|---|---|---|
| Em@Taiwan #1 | 15.2 | 66 | 320 | 34 | 25 |
| Em@Taiwan #2 | 29.0 | 67 | 294 | 32 | 23 |
| Em@Taiwan #3 | 23.0 | 67 | 308 | 25 | NA |
| Em@Taiwan #4 | 24.0 | 67 | 288 | 32 | NA |
| Em@Taiwan #5 | 26.0 | 66 | 312 | 36 | NA |
| Em@Taiwan #6 | 15.4 | 68 | 293 | 29 | NA |
| Em@Taiwan #7 | 14.1 | 69 | 303 | 25? | NA |
| Em@Taiwan #8 | 13.3 | 58 | 292 | 20 | NA |
| mean ± s.d. | 20.00 ± 5.77 | 66.0 ± 3.2 | 301.3 ± 10.6 | 29.7 ± 5.1 (n = 7) | |
| Ec@China #1 | 15.6 | 51 | 222 | NA | NA |
| Ec@China #2 | 14.1 | 47 | 228 | NA | NA |
| E sp@Kyushu #1 | 30.0 | 46 | 202 | NA | 11 |
| E sp@Kyushu #2 | NA | 50+ | 230 | 20 | 15 |
Figure 5.

Left and right lateral views of dextral gonad amphioxus species. (A) Live Asymmetron pelagicum from Nanwan Bay under incident light. (B) Live A. pelagicum without gonads from Nanwan Bay under transmitted light. (C) Live Epigonichthys maldivensis from Nanwan Bay under incident light. (D) Live E. maldivensis without gonads from Nanwan Bay under transmitted light. (E) Fixed E. cultellus from the South China Sea under incident light. (F) Lateral view of live E. cultellus from northern Kyushu under incident light. Scale bars 2 mm.
Identity of Epigonichthys-like specimens from temperate waters in Kyushu, Japan
In Japan, a single Epigonichthys specimen was previously recorded from the coastal waters of Shirahama, Wakayama (33°42′N 135°18′E), representing the most northern record of the genus Epigonichthys, and this specimen was assigned to E. maldivensis based on morphometric data31. In this study we collected two specimens morphologically distinct from E. maldivensis (Fig. 5) from northern Kyushu (Fukuoka, 33°47′40.3″N 130°24′25.1″E; Ariake Sea, 32°32′09″N 130°19′10″E) during field collections between 2001 to 2014. The collection sites are located at almost the same latitude as Shirahama, and similarly influenced by the Kuroshio Current. The morphometric data of the Kyushu specimens (Table 1) suggested an affinity to E. australis (Raffe, 1912)7 or E. cultellus, and the complete mitogenome was 14,985 bp in length (AP015024) and almost identical to that of E. cultellus from the South China Sea [99.73% (40/14,985 substitution sites) similarity] with identical gene order17. We thus assigned these Epigonichthys-like specimens from Kyushu to E. cultellus, although their body sizes were 1.5 times larger than the mean size of specimens from the South China Sea17.
Discussion
Generic phylogeny and evolutionary history
Our analyses of mitogenomic sequence data reconfirm the previously observed divergence pattern of the cephalochordate lineage, in which Asymmetron diverged first, followed by Epigonichthys and Branchiostoma (Fig. 1). Species within the Asymmetron clade are less divergent compared with those in the Branchiostoma + Epigonichthys clade. In particular, the close relationship between the Pacific and Atlantic Asymmetron populations is remarkable. To explain the genetic divergence within Asymmetron, it has been suggested that Asymmetron originated in the eastern Tethys Sea during the breakup of Pangaea in the Mesozoic, then separated into Indo-West Pacific and Atlantic populations, and finally the Atlantic population expanded into the West Pacific from the Atlantic to become sympatric with the Indo-West Pacific population as found in the Kuroshima population4. However, this scenario is not without weak points. We found no Indian haplotype of coxI in Taiwanese specimens (n = 25). Furthermore, whole mitogenomic sequence comparisons separate the Indian group from the Pacific + Atlantic group but not the Atlantic group from the Indian + West Pacific group. If gene flow is directly ongoing between Indian and Japanese populations as suggested previously4 or via an intermediate population on the eastern coast of Australia (Indian haplotypes of coxI have been reported from this region4), we would expect to find the Indian haplotype in the Taiwanese population as currents from the northern South China Sea seasonally join the Kuroshio Current via the Luzon Strait, onto which Nanwan Bay opens32. Additionally, the Kuroshio Current passes through this strait seasonally32. However, we did not find any Indian haplotype in Nanwan Bay, and this absence is consistent with proposed Cenozoic currents that flowed from the Pacific to the Indian Ocean with no opposite direction flow except at high latitudes33. Additionally, in the Cenozoic there was no northward current along the eastern coast of Australia34. We thus speculate that the Indian coxI haplotype found in the Okinawan population may be due to recent human introduction, as has been seen with the introduction of other marine species via the discharge of ballast water35.
The mitogenomic analyses suggest that the Asymmetron clade after the divergence of A. inferum was present in the Indian Ocean and migrated westward, passing through the Neo-Tethys into the Atlantic, and then into the Pacific through a seaway between North and South America. However, we could not detect any evidence of direct migration from the Indian Ocean towards the eastern coast of Eurasia. The genetic proximity between the Pacific and Atlantic populations suggests that gene flow between these two groups was not terminated until the formation of the Isthmus of Panama. The formation of the isthmus has been suggested to have been a long and complex process, but many marine taxa living in shallow waters have divergence times between Caribbean and Pacific groups between 1.03 to 4.35 Ma36, 37. The formation of the isthmus and the closure of the Neo-Tethys, estimated as 14 Ma38, appear to have caused species diversification within Asymmetron as our divergence timing estimates corresponded well to these geological events.
The absence of Epigonichthys from the Atlantic is interesting. When we take into account the fact that E. maldivensis is sympatric with the Pacific population of Asymmetron and that some Epigonichthys species are distributed in temperate regions such as the Bass Strait in southern Australia (annual water temperatures 12–22 °C39) and northern Kyushu, Japan (14–28 °C), it seems unlikely that Epigonichthys was once distributed in the Atlantic and has since gone extinct. The diversity of extant Epigonichthys species in Oceania7 suggests that the origin of the Epigonichthys lineage may be in this region. On the other hand, the Branchiostoma lineage shares a common ancestor with the Epigonichthys lineage but is distributed worldwide. Differences in present distributions between the Epigonichthys and Branchiostoma clades may be attributable to large egg numbers in Branchiostoma. Larger numbers of larvae with long pelagic life like amphioxus40 could increase chances for wider distributions via ocean currents41.
Character polarity in amphioxus lineage
There is a large time gap between the origin of the cephalochordate lineage, which likely occurred in the Precambrian, and the diversification into extant taxa in the Cenozoic, and therefore it is difficult to ascertain the form of the LCA of the amphioxus crown lineage. However, the character polarity of major traits deduced from the phylogenetic tree may provide insights into the LCA of extant amphioxus taxa. The most conspicuous feature of the amphioxus lineage is left-right asymmetrical development. Gonad development is also asymmetrical, with Asymmetron and Epigonichthys sharing dextral gonads while Branchiostoma spp. develop gonads on both sides (Fig. 6). Dextral gonads are thus shared by the two major clades, which supports the evolution from ancestral unilateral to derived bilateral gonads3. This polarity suggests that the asymmetrical development of amphioxus is tightly related to the origin of this lineage as there are no comparable outgroups, and this idea has also been supported by a comparative developmental study42. In amphioxus, the most pronounced left-right asymmetry occurs in the larval body1 with some variations between genera; Asymmetron larvae display less-pronounced left-right asymmetry in the location of the primary gill openings (future left gill openings) and anus compared to Branchiostoma larvae43.
Figure 6.
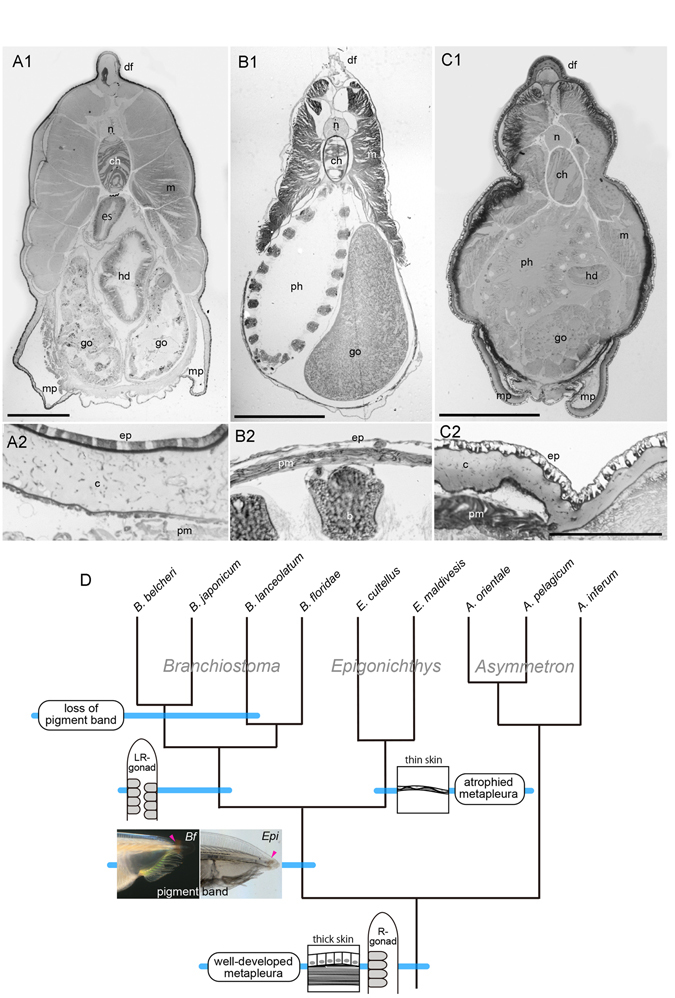
Transverse sections of three amphioxus genera and parsimoniously expected character polarity. (A1) Transverse section at posterior pharyngeal region of Branchiostoma japonicum. (A2) Thick epidermal epithelium and collagen layer in B. japonicum. (B1) Transverse section at pharyngeal region of Epigonichthys maldivensis. (B2) Squamosal epidermal epithelium and very thin collagen layer in E. maldivensis. (C1) Transverse section at posterior pharyngeal region of Asymmetron pelagicum. (C2) Cuboidal epidermal epithelium and well-developed collagen layer. (D) Metapleura, thick skin, and dextral gonads are ancestral characters in amphioxus lineage. Genus Epigonichthys displays most derived features. Branching pattern in Branchiostoma clade follows nucleotide-based trees. b, branchial bar; Bf, Branchiostoma floridae; c, collagen layer; ch, notochord; df, dorsal fin; ep, epidermis; Epi, Epigonichthys; es, esophagus; go, gonad; hd, hepatic diverticulum; m, myomeric muscle; mp, metapleuron; ph, pharynx; pm, pterygial muscle. Scale bars 0.5 mm for A1, B1, C1, and 0.2 mm for C2 applicable to A2, B2.
Molecular studies have demonstrated that amphioxus larval asymmetry is controlled by the Nodal-Pitx signaling unit in early development44, especially in oropharyngeal development, in which the asymmetry is most pronounced45. Nodal signaling also controls apoptosis of germ cells in sea urchins46. These studies suggest that Nodal signaling and its downstream gene regulatory networks may have been key players in the evolution of the amphioxus lineage, and subtle differences in these molecular functions may explain the variations in the developmental left-right asymmetry including gonad development in the amphioxus lineage.
Branchiostoma and Epigonichthys are larger than Asymmetron species in body size, and have similar appearances, sharing preanal finboxes, caudal myomeres, and head patterns with similar buccal cirri and six myomeres anterior to the velum. Further, B. floridae shares a pigment band at the base of the rostrum with Epigonichthys species (Fig. 6). These features support the monophyly of these two genera as recovered in the phylogenetic analyses. In contrast, the skin of Epigonichthys is specialised thin squamous epithelium with a thin subepidermal collagenous layer, contrary to the cuboidal epithelium with a thick collagenous layer in Asymmetron and Branchiostoma (Fig. 6). Other unique conspicuous features of Epigonichthys are a dorso-ventrally expanded flat body with tall dorsal finboxes (Fig. 5), and atrophied metapleura (longitudinal folds that develop on both sides of the belly in the other two genera), that are particularly noticeable when gonads are developing (Fig. 6), as well as a slight leftward shift of the buccal opening (the amphioxus mouth is located at the end of buccal cavity separating it from the pharynx and is called the ‘velum’) (Fig. 5). These features suggest that Epigonichthys is the most derived group among extant amphioxus lineages.
All extant amphioxus clades do not develop a mucus or cuticle layer on the epidermis, and have a naked simple epithelium despite inhabiting sandy substrata (Fig. 6). These unique features are thus likely an ancestral character in the lineage. Asymmetron and Branchiostoma embryos and larvae move with monocilia that develop throughout the epidermis similar to cnidarian planula larvae even after they develop locomotive myomeres43, 47, and the cilia disappear during metamorphosis when larval length reaches approximately five mm in B. japonicum 40. Although Branchiostoma larvae are regarded as pelagic, they frequently slide by ciliary movement on the bottom when maintained in culture tanks48, suggesting that this small ciliated stage is not necessarily pelagic. The naked epidermal surface of extant amphioxus taxa may be a retained feature of their ciliated ancestral form as deduced from the fact that most ciliate larvae found across animal phyla do not develop a thick mucus layer on the epidermis as the mucus negatively influences locomotion and feeding. It should be noted that some animal larvae develop epidermal mucus cells to secret mucus strands for collecting food particles or locomotion control49, 50.
We estimated the diversification into the three extant clades occurred in the Eocene (38.8–46.0 Ma with 33.9–56.6 Ma 95% confidence interval). These estimates are more recent than compared to previous estimates3–5, 10. Additionally, they are consistent with the genetic proximity between species observed within the Asymmetron clade, and also with the recent finding that amphioxus displays a rapid evolutionary rate comparable to that of vertebrates13. The estimates for species splits within the Asymmetron clade likely correspond with geological events that occurred onward from the Miocene, such as the closure of the Neo-Tethys38 and the formation of the Isthmus of Panama36, 37. Given the Precambrian origin of the amphioxus lineage, the amphioxus lineage survived many mass extinction events before its most recent diversification. Taking into account this and the character polarities mentioned above, one possible scenario for the long evolutionary history until diversification is that soon after separation from the main chordate lineage, the amphioxus ancestor acquired its modern developmental pattern with metamorphosis, and diversified as sandy bottom dwellers as seen in extant amphioxus clades. All but one ancestral group went extinct, and the surviving species then diversified into the extant clades. In this case, the cephalochordate LCA might have grown to the size of modern amphioxus. For example, the Cambrian fossils Haikouella 27, 51 and Pikaia 28, which may have had affinity to the amphioxus lineage, had body sizes comparable to or larger than those of modern amphioxus. However, the fact that many animal groups rapidly increased their body sizes by the Middle Cambrian52 suggests another possibility; that the LCA of chordates was comparatively tiny and that the common ancestor of the extant amphioxus lineage retained this tiny size until the most recent diversification. This idea seems more likely when considering the character polarity found in extant species, especially the naked simple epithelial epidermis without mucus coat that is found in all amphioxus clades. This idea is also consistent with the small-sized ancestral state expected from the polarity of the body size, in which the Asymmetron clade has smaller sizes than the Branchiostoma clade, and the Epigonichthys clade displays a variety of sizes.
We hypothesise, therefore, that the LCA of extant amphioxus had a few-mm long ciliated body with left-right asymmetry, and that diversification into extant clades may have been triggered by increasing body size and the acquisition of metamorphosis. The LCA thus may have retained the larva-like anatomy of extant amphioxus with unilateral gonads developing near the mid-ventral region (anatomically derived from the right coelom). Given the small body size of the proposed LCA and a likely limited number of eggs [egg sizes of 120–140 μm do not vary among extant taxa, suggesting an ancestral character (for A. pelagicum, ref. 43)], the proposed tiny LCA might have inhabited shoals on the surface of fine sandy seafloors.
Mitogenomic systematics and population identities
The genus Asymmetron currently comprises two species, A. lucayanum and A. inferum. The former species is distributed circumtropically and proposed to contain at least three cryptic species4. Our phylogenetic analyses reconfirm the existence of these three clades; Indian, Pacific and Atlantic Ocean groups. Although these clades have been proposed to be three species4, it is reasonable to group the Pacific and Atlantic clades into a single species based on a lack of sequence divergence (~7%; interspecific differences in Epigonichthys and Branchiostoma are ~25% in nt comparison), and consider the Indian Ocean clade as a different species (~23% compared with the Pacific or Atlantic clade in nt). As the Pacific + Atlantic Asymmetron clade included haplotypes from Hawai’i (Fig. 4 and ref. 4), we propose that Asymmetron pelagicum Günther, 1889 described from Hawai’i should be used as the correct binomial name for the Pacific + Atlantic clade, replacing the junior synonym Asymmetron lucayanum Andrew, 1893, described from the Bahamas. If, in the future, researchers decide the Atlantic clade should be split from A. pelagicum, then the name A. lucayanum should be utilised for this species-group. Similarly, specimens from the Maldives in the Indian Ocean were described as Asymmetron orientale Parker, 1904, and we propose this binomial should be used for the Indian clade of the A. lucayanum complex. The number of Asymmetron species in the world is, therefore, at least three, including the anaerobic sulfidophilic A. inferum 2. The Asymmetron specimens from Nanwan Bay at the southern tip of Taiwan are assigned to A. pelagicum (=former A. lucayanum) based on the present mitogenomic analyses and morphometric data.
The genus Epigonichthys has been reported from the eastern coast of African continent to Hawai’i, and is not present in the Atlantic7. This genus currently comprises six species, most of which are distributed in Oceania7, but taxonomically many questions remain. Previous studies on Epigonichthys from Taiwan have assigned specimens to E. maldivensis based on morphometric data14, 16. Our mitogenomic sequence analyses showed a close similarity of Taiwanese Epigonichthys to E. maldivensis from Okinawa, supporting the assignment of Taiwanese Epigonichthys specimens to E. maldivensis for now, although molecular examination of specimens from the Maldives are needed to confirm this.
On the other hand, the Epigonichthys specimens from northern Kyushu, Japan, are morphologically distinct from E. maldivensis specimens from Taiwan. The mitogenomic sequence of this rare amphioxus was almost identical to that of E. cultellus from the South China Sea17, and we have identified these specimens to this species. Although the very low numbers of specimens of this amphioxus in Japan raises the question of whether the species breeds at the collecting sites or not, this study confirms that two species of Epigonichthys, E. maldivensis and E. cultellus, are present in Japanese coastal waters. To further understand the Epigonichthys clade, studies on Oceanian populations that lack modern analytical data are needed.
Methods
Animal sampling and animal care
Amphioxus (n = 83) with dextral gonads were collected from three sites (21°57′12.8″N 120°46′06.2″E, 21°57′11.1″N 120°46′08.5″E, 21°57′11.5″N 120°46′06.7″E) in Nanwan Bay, Taiwan, by SCUBA diving with collecting bags in 2014 and 2015 (permission No. 1032900823 issued by Kenting National Park Headquarters, Taiwan) (Supplementary Fig. S1). Two specimens were collected from Branchiostoma habitats in northern Kyushu, Japan, one in Hakata Bay (33°47′40.3″N 130°24′25.1″E), Fukuoka, in 2012, and the other in the Ariake Sea (32°32′09″N 130°19′10″E), Kumamoto, in 2008 (no permission required for amphioxus collection). DNA from a specimen collected in Bimini Lagoon, the Bahamas (25°43′22.7″N 79°17′38.0″W) (courtesy of Dr. Jr-Kai Yu) was also utilised (Supplementary Fig. S1). Specimens from Nanwan Bay were maintained in the laboratory and individuals damaged during culture were fixed with 4% paraformaldehyde in 0.1 M 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (pH 7.5) and then stored in 75% ethanol or directly fixed with 75% ethanol. The specimens from Fukuoka were fixed with the same fixatives in the field. All fixed specimens were stored at −20 °C until use. Branchiostoma japonicum specimens for histological observations were collected from a laboratory colony53, and Epigonichthys cultellus specimens used for morphometrical data were collected from off Guandong, China17. All animals in the present study were maintained in accordance with guidelines established by Hiroshima University for the care and use of experimental animals. Our experimental protocols were approved by the Hiroshima University Animal Research Committee (Approval number: G14-2.). Animals were fixed at the laboratory under anaesthesia with 0.02% amino benzoic acid ethyl ester in millipore-filtered seawater.
DNA extraction, amplification, and sequencing of short DNA fragments
DNA was extracted individually from 29 Asymmetron specimens from Nanwan Bay and 5 Epigonichthys specimens from Nanwan Bay and Hakata Bay by using NucleoSpin Tissue (MACHEREY-NAGEL, Germany). For the Nanwan Bay Asymmetron specimens, PCR was performed initially to amplify a fragment of the cytochrome c oxidase subunit I (coxI) gene with the primer set AmphL109/AmphH13254. The PCR was carried out with genomic DNA as template (95 °C for 3 min, 95 °C for 30 sec x35, 55 °C for 30 sec, 72 °C for 30 sec). The amplicons were directly sequenced as described previously54. Amplification of two DNA fragments that cover the full length of the mitogenomic DNA excluding the cox1 region was performed by PCR (95 °C for 3 min and 95 °C for 30 sec x35, 68 °C for 15 min) with primer sets shown in Supplementary Table S1. The sizes of amplicons were confirmed by gel electrophoresis, and the DNA fragments in the gel blocks were purified using FastGene Gel/PCR Extraction Kit (Nippon Genetics, Japan).
Whole mitogenomic DNA sequencing
Whole mitogenomic DNA was sequenced using two long DNA fragments with Ion Torrent PGM™ (Thermo Fisher Scientific, MA). The two fragments had adapters with different barcode sequences attached (Ion Xpress Barcode Adapters) that were mixed individually and used as templates for constructing an amplicon library with NEBNext Fast DNA Fragmentation & Library Prep Set (New England Biolabs, MA) for Ion Torrent. The libraries constructed were quantified by using KAPA Library Quantification Kits (KAPA Biosystems, MA) for Ion Torrent and pooled into a single tube. The pooled library sample was further amplified by an emulsion PCR with Ion PGM Template OT2 200 Kit (Thermo Fisher Scientific, MA). The product was finally sequenced on an Ion PGM sequencer (Thermo Fisher Scientific, MA) with Ion 318 Chip Kit v. 2 and Ion PGM Sequencing 200 Kit v. 2.
Phylogenetic analyses
Haplotypes of partial segments of cytochrome c oxidase I (coxI) from 26 specimens of Asymmetron species sequenced in this study, and 51 unique haplotypes of the same coxI segments from 80 specimens were identified4. A neighbour-joining (NJ) tree was constructed under the p-distance55 within MEGA 5.2 based on the sequences of coxI. Phylogenetic trees based on maximum likelihood (ML) with T92 + G models56 (ML model selected after model selection analysis) and Bayesian inference (BI) were also constructed by the method mentioned below. Bootstrap values were calculated with 1,000 pseudoreplicates. A minimum spanning network was also constructed using TCS 1.2157. The maximum number of steps to connect haplotypes parsimoniously was calculated with a 99% limit.
From 12 full-length mitogenomic sequences obtained in this study (7 Asymmetron, 1 E. cultellus, and 4 E. maldivensis individuals) and 10 mitogenomes from GenBank (Supplementary Table S2), we prepared two subsets of data: amino acid (aa) sequences of 13 protein genes, and all nucleotide (nt) sequences of the mitogenome excluding the control region. Both datasets were aligned separately for each gene by using clustalW58 within MEGA 5.259 with default settings. For protein coding genes, we first aligned aa and then aligned the nt correspondingly to their codons. Poorly aligned regions and gap sites were deleted by using gBlocks v. 0.91b60 while specifying the type of sequence.
We also constructed an aa sequence dataset from 952 orthologous protein coding nuclear genes of Branchiostoma belcheri, B. lanceolatum, B. floridae, Asymmetron lucayanum, and four vertebrate species (Danio rerio 61, Xenopus tropicalis 62, Mus musculus 63, and Homo sapiens 64). The aa sequences of four vertebrates (GRCz10, JGI 4.2, GRCm38.p4, and GRCh38.p7) were retrieved from Ensembl Release 85 (July 2016) and those of B. belcheri (v.18h27.r3) and B. floridae (v.1.0) were retrieved from each portal site of genome sequencing project (http://genome.bucm.edu.cn/lancelet/ and http://genome.jgi.doe.gov/Brafl1/Brafl1.home.html). The aa sequences of B. lanceolatum were deduced from a published transcriptome65 by using TransDecoder 3.0.0 (https://transdecoder.github.io/). For A. lucayanum, raw RNA-seq reads10 were retrieved from the DDBJ Sequence Read Archive (DRA) and transcriptome data was assembled by using Trinity v. 2.1.1 after quality control of the reads10. The transcriptome data of A. lucayanum and B. lanceolatum were then translated into aa sequences. Orthologous 952 gene sets were also constructed by using Proteinortho (v. 5.12) and the PhyloTreePruner pipeline10.
Phylogenetic trees were constructed by ML and Bayesian (BI) methods based on the nuclear transcriptome and whole mitogeninic sequences. To select the optimum substitution models for each gene, Aminosan and Kakusan466 were used based on the Akaike information criterion (AIC). ML analyses were conducted using RAxML v. 8.1.2467, evaluated by bootstrap values of 1,000 pseudoreplicates. BI analyses were conducted using MrBayes 5D68 with two independent runs of four Markov chain Monte Carlo (MCMC) chains. Analyses were run for ten million generations, and trees were sampled every 1,000 generations. Convergence among runs was verified by examining the likelihood plots using Tracer 1.669. The first 50% of trees were discarded as burn-in and the remaining trees were summarized with posterior probabilities at the nodes.
Inference of divergence time
We estimated divergence times based either on nuclear transcriptomes with outgroup reference points or mitogenomes with ingroup reference points. First we used aa sequences derived from nuclear transcriptomes with calibration points at the cephalochordate-vertebrate (550.0 ± 16.0 Ma)25, osteichthyan-tetrapode (419.0 ± 1.4 Ma)25, anamniote-amniote (340.0 ± 5.0 Ma)24 splits, and rodent-primate split (81.0 ± 10.0 Ma)24. For the mitogenomic sequences, we excluded 22 tRNAs and applied ingroup calibration points obtained from the first estimate based on the nuclear transcriptomes; Branchiostoma-Asymmetron (46.0 ± 5.5 Ma), B. belcheri-(B. lanceolatum + B. floridae) (28.2 ± 5.5 Ma), and B. lanceolatum-B. floridae (22.6 ± 2.3 Ma). Divergence times were estimated by using BEAST 1.8.4 with the random local clock model that assumes rate changes across branches70. The substitution model used in the analysis was selected in Kakusan4 under AIC according to each codon position. For the mitogenome-based estimation, we also applied geological calibration points to the split between the Pacific and Atlantic populations of A. pelagicum (=former A. lucayanum) (2.06 Ma: 1.03–4.35 Ma = 95% credible interval (CI))36, 37 and to the split between the Indian and Atlantic populations (14 Ma: 6.16–21.84 Ma = 95% CI)38. Tree topology was fixed to the ML tree and prior distributions of the time of the most recent common ancestors of these species were constrained by normal distributions to cover the 95% CIs arbitrarily. All other model parameters were set to default priors. For MCMC analysis, we performed a run of 10 million generations, sampling every 1,000th generation and removing the initial 10% of samples as burn-in. Convergence of the chains was confirmed using Tracer v. 1.669.
Morphometric analyses
Live or fixed specimens were photographed under microscope or digital camera (D800 Nikon, Japan) with a close-up lens. Digital photographs were visually optimized by using Photoshop CS6 (Adobe, CA), and body length, as well as the numbers of dorsal finboxes, preanal finboxes, gonads, and myomeres were measured or counted from images.
Histological sectioning
Small specimens with gonads of Asymmetron pelagicum (=former A. lucayanum), Branchiostoma japonicum, and Epigonichthys maldivensis were cut into anterior and posterior halves and fixed with 75% ethanol or 4% paraformaldehyde in 0.1 M MOPS buffer (pH 7.5) with 0.5 M NaCl at 4 °C overnight. After washing with Millipore filtered seawater (MFSW), they were stained with 1% tannic acid in MFSW for 2 hours, washed again with MFSW, and then fixed with 1% osmium tetroxide in MFSW at 4 °C for 2 hours. The postfixed specimens were dehydrated through a graded ethanol series and embedded in hydrophilic Epon. The pharyngeal region of embedded specimens was sectioned with glass knives at 1 μm and stained with 0.1% toluidine blue containing 1% sodium borate at 60 °C.
Electronic supplementary material
Acknowledgements
We thank Jr-Kai Yu of Academia Sinica for his coordination of field collection in Nanwan Bay, Taiwan, and for the photograph of Branchiostoma floridae, and Ching-Yi Lin, Teng-Sung Chen, Ming-Jay Ho, and Tzu-Kai Huang of the same institute for their help in SCUBA diving to collect amphioxus specimens in the field; Michiyasu Yoshikuni and other staff of the Fishery Research Laboratory of Kyushu University, Hiroshi Wada of Tsukuba University, and Hideyuki Shimasaki of Kumamoto University for collecting Epigonichthys specimens in Hakata Bay and the Ariake Sea; Guang Li of Xiamen University for his help in the observation of E. cultellus specimens; Kun-Lung Li, Luok Wen Yong, Angela Hsu, and Jr-Kai Yu of Academia Sinica for their cooperation in sequencing mitochondrial genomes; and Kanako Onizuka and Chie Iwamoto of the National Institute of Genetics for helping with next generation sequencing. The authors have no external support or funding to report.
Author Contributions
K.Y. and T.I. conceived and designed the experiments. T.I., M.N., A.R.M., Y.W., D.G.S. and K.Y. performed the experiments. All authors analyzed the data. K.Y., T.I., M.N. and Y.H. contributed reagents/materials/analysis tools. K.Y., T.I., D.G.S., J.D.R. and M.N. wrote the paper. All authors read and edited the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-00786-5
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kowalevsky A. Entwicklungsgeschichte des Amphioxus lanceolatus. Mém L’Acad Imp Sci St-Pétersboug. 1867;11:1–17. [Google Scholar]
- 2.Nishikawa T. A new deep-water lancelet (Cephalochordata) from off Cape Nomamisaki, SW Japan, with a proposal of the revised system recovering the genus Asymmetron. Zool Sci. 2004;21:1131–1136. doi: 10.2108/zsj.21.1131. [DOI] [PubMed] [Google Scholar]
- 3.Nohara M, Nishida M, Miya M, Nishikawa T. Evolution of the mitochondrial genome in cephalochordata as inferred from complete nucleotide sequences from two Epigonichthys species. J Mol Evol. 2005;60:526–37. doi: 10.1007/s00239-004-0238-x. [DOI] [PubMed] [Google Scholar]
- 4.Kon T, Nohara M, Nishida M, Sterrer W, Nishikawa T. Hidden ancient diversification in the circumtropical lancelet Asymmetron lucayanum complex. Mar Biol. 2006;149:875–883. doi: 10.1007/s00227-006-0271-y. [DOI] [Google Scholar]
- 5.Kon T, et al. Phylogenetic position of a whale-fall lancelet (Cephalochordata) inferred from whole mitochondrial genome sequences. BMC Evol Biol. 2007;7:127. doi: 10.1186/1471-2148-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 7.Poss SG, Boschung HT. Lancelet (Chephatochordata: Branchiosotmidae): how many species are valid? Isr J Zool. 1996;42:S13–S66. [Google Scholar]
- 8.Ueda H, Sakaki K. Effects of turbation of the Japanese common lancelet Branchiostoma japonicum (Cephalochordata) on sediment condition: laboratory observation. Plankt Benthos Res. 2007;2:155–160. doi: 10.3800/pbr.2.155. [DOI] [Google Scholar]
- 9.Erwin DH, et al. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science. 2011;334:1091–1097. doi: 10.1126/science.1206375. [DOI] [PubMed] [Google Scholar]
- 10.Yue J-X, Yu J-K, Putnam NH, Holland LZ. The transcriptome of an amphioxus, Asymmetron lucayanum, from the Bahamas: a window into chordate evolution. Genome Biol Evol. 2014;6:2681–2696. doi: 10.1093/gbe/evu212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igawa T, Kurabayashi A, Usuki C, Fujii T, Sumida M. Complete mitochondrial genomes of three neobatrachian anurans: A case study of divergence time estimation using different data and calibration settings. Gene. 2008;407:116–129. doi: 10.1016/j.gene.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Peng R, Kuro-O M, Zeng X. Exploring patterns and extent of bias in estimating divergence time from mitochondrial DNA sequence data in a particular lineage: A case study of salamanders (Order Caudata) Mol Biol Evol. 2011;28:2521–2535. doi: 10.1093/molbev/msr072. [DOI] [PubMed] [Google Scholar]
- 13.Huang S, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishikawa T, et al. First find of Epigonichthys maldivensis (Cooper) and rediscovery of E. lucayanus (Andrews) from Nanwan Bay, southern Taiwan (Cephalochordata) Benthos Res. 1997;52:103–109. doi: 10.5179/benthos1996.52.2_103. [DOI] [Google Scholar]
- 15.Zhang Q-J, Zhong J, Fang S-H, Wang Y-Q. Branchiostoma japonicum and B. belcheri are distinct lancelets (Cephalochordata) in Xiamen waters in China. Zool Sci. 2006;23:573–579. doi: 10.2108/zsj.23.573. [DOI] [PubMed] [Google Scholar]
- 16.Lin H-C, Chen J-P, Chan BKK, Shao K-T. The interplay of sediment characteristics, depth, water temperature, and ocean currents shaping the biogeography of lancelets (Subphylum Cephalochordata) in the NW Pacific waters. Mar Ecol. 2015;36:780–793. doi: 10.1111/maec.12183. [DOI] [Google Scholar]
- 17.Li W-Y, Fang S-H, Wang Y-Q. Complete mitochondrial genome of Epigonichthys cultellus (Cephalochordata: Branchiostomatidae) Zool Sci. 2014;31:766–72. doi: 10.2108/zs130163. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Lü X. Discovery and distribution of Asymmetron cultellus in southern Fujian seas. Fujian Fish. 1990;1:1–2. [Google Scholar]
- 19.Jiang S, Lin S. Reappear of Asymmetron cultellum in Shantou seas. J Ocean Taiwan Strait. 1990;9:61. [Google Scholar]
- 20.Zhang X. Discovery of genus Asymmetron in China Seas and the geographic distribution of Branchiostoma belcheri (Gray) Acta Zool Sin. 1962;14:525–528. [Google Scholar]
- 21.Andrews E. An undescribed acraniate: Asymmetron lucayanum. Stud Biol Lab Johns Hopkins Univ. 1893;5:213–247. [Google Scholar]
- 22.Zhong J, et al. Complete mitochondrial genomes defining two distinct lancelet species in the West Pacific Ocean. Mar Biol Res. 2009;5:278–285. doi: 10.1080/17451000802430817. [DOI] [Google Scholar]
- 23.Ho SYW, Phillips MJ, Drummond AJ, Cooper A. Accuracy of rate estimation using relaxed-clock models with a critical focus on the early metazoan radiation. Mol Biol Evol. 2005;22:1355–1363. doi: 10.1093/molbev/msi125. [DOI] [PubMed] [Google Scholar]
- 24.Benton, M. J. et al. Constraints on the timescale of animal evolutionary history. Palaeontol Electron 1–107 (2015).
- 25.Benton, M., Donoghue, P. C. J. & Asher, R. J. In The Timetree of Life 35–86 (2009).
- 26.Shu D-G, et al. Lower Cambrian vertebrates from south China. Nature. 1999;402:42–46. doi: 10.1038/46965. [DOI] [Google Scholar]
- 27.Shu D, et al. A new species of yunnanozoan with implications for deuterostome evolution. Science. 2003;299:1380–1384. doi: 10.1126/science.1079846. [DOI] [PubMed] [Google Scholar]
- 28.Conway-Morris S, Caron JB. Pikaia gracilens Walcott, a stem-group chordate from the Middle Cambrian of British Columbia. Biol Rev. 2012;87:480–512. doi: 10.1111/j.1469-185X.2012.00220.x. [DOI] [PubMed] [Google Scholar]
- 29.Shikina S, et al. Germ cell development in the scleractinian coral Euphyllia ancora (Cnidaria, Anthozoa) PLoS One. 2012;7:e41569. doi: 10.1371/journal.pone.0041569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CA, Yang YW, Wei NV, Tsai WS, Fang LS. Symbiont diversity in scleractinian corals from tropical reefs and subtropical non-reef communities in Taiwan. Coral Reefs. 2005;24:11–22. doi: 10.1007/s00338-004-0389-7. [DOI] [Google Scholar]
- 31.Nishikawa T. Records of two lancelet species, Asymmetron maldivense and A. lucayanum, from the western North Pacific. Publ Seto Mar Biol Lab. 1980;25:167–173. [Google Scholar]
- 32.Hwang J-S, Dahms H-U, Tseng L-C, Chen Q-C. Intrusions of the Kuroshio Current in the northern South China Sea affect copepod assemblages of the Luzon Strait. J Exp Mar Bio Ecol. 2007;352:12–27. doi: 10.1016/j.jembe.2007.06.034. [DOI] [Google Scholar]
- 33.Barron EJ, Peterson WH. The Cenozoic ocean circulation based on ocean General Circulation Model results. Palaeogeogr Palaeoclimatol Palaeoecol. 1991;83:1–28. doi: 10.1016/0031-0182(91)90073-Z. [DOI] [Google Scholar]
- 34.Ridgway, K. & Hill, K. In A Marine Climate Change Impacts and Adaptation Report Card for Australia 2009 (eds Poloczanska, E. S., Hobday, A. J. & Richardson, A. J.) 1–16 (NCCARF, 2009).
- 35.Floerl O, Inglis GJ, Hayden BJ. A risk-based predictive tool to prevent accidental introductions of nonindigenous marine species. Environ Manage. 2005;35:765–778. doi: 10.1007/s00267-004-0193-8. [DOI] [PubMed] [Google Scholar]
- 36.Bacon CD, et al. Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc Natl Acad Sci. 2015;112:6110–6115. doi: 10.1073/pnas.1423853112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lessios HA. The Great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Ann Rev Ecol Evol Syst. 2008;39:63–91. doi: 10.1146/annurev.ecolsys.38.091206.095815. [DOI] [Google Scholar]
- 38.Hamon N, Sepulchre P, Lefebvre V, Ramstein G. The role of eastern tethys seaway closure in the middle miocene climatic transition (ca. 14 Ma) Clim Past. 2013;9:2687–2702. doi: 10.5194/cp-9-2687-2013. [DOI] [Google Scholar]
- 39.Locarnini, R. A. et al. In NOAA Atlas NESDIS 73 (ed. Mishonov, A.) 1, 1–40 (NOAA, 2013).
- 40.Urata M, Yamaguchi N, Henmi Y, Yasui K. Larval development of the oriental lancelet, Branchiostoma belcheri, in laboratory mass culture. Zool Sci. 2007;24:787–797. doi: 10.2108/zsj.24.787. [DOI] [PubMed] [Google Scholar]
- 41.Shanks AL. Pelagic larval duration and dispersal distance revisited. Biol Bull. 2009;216:373–85. doi: 10.1086/BBLv216n3p373. [DOI] [PubMed] [Google Scholar]
- 42.Morov AR, Ukizintambara T, Sabirov RM, Yasui K. Acquisition of the dorsal structures in chordate amphioxus. Open Biol. 2016;6:160062. doi: 10.1098/rsob.160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holland ND, Holland LZ. Laboratory spawning and development of the Bahama lancelet, Asymmetron lucayanum (Cephalochordata): Fertilization through feeding larvae. Biol Bull. 2010;219:132–141. doi: 10.1086/BBLv219n2p132. [DOI] [PubMed] [Google Scholar]
- 44.Soukup V, et al. The Nodal signaling pathway controls left-right asymmetric development in amphioxus. EvoDevo. 2015;6:5. doi: 10.1186/2041-9139-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaji T, Reimer JD, Morov AR, Kuratani S, Yasui K. Amphioxus mouth after dorso-ventral inversion. Zool Lett. 2016;2:2. doi: 10.1186/s40851-016-0038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y-J, Su Y-H. Opposing Nodal and BMP signals segulate left–right asymmetry in the sea urchin larva. PLoS Biol. 2012;10:e1001402. doi: 10.1371/journal.pbio.1001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaji T, Hoshino Y, Henmi Y, Yasui K. Longitudinal observation of Japanese lancelet, Branchiostoma japonicum, metamorphosis. Dataset Pap Biol. 2013;2013:1–6. doi: 10.7167/2013/839671. [DOI] [Google Scholar]
- 48.Yasui K, Urata M, Yamaguchi N, Ueda H, Henmi Y. Laboratory culture of the oriental lancelet Branchiostoma belcheri. Zool Sci. 2007;24:514–520. doi: 10.2108/zsj.24.514. [DOI] [PubMed] [Google Scholar]
- 49.Strathmann RR, Grünbaum D. Good eaters, poor swimmers: compromises in larval form. Integr Comp Biol. 2006;46:312–322. doi: 10.1093/icb/icj031. [DOI] [PubMed] [Google Scholar]
- 50.Larsson AI, et al. Embryogenesis and larval biology of the cold-water coral Lophelia pertusa. PLoS One. 2014;9:e102222. doi: 10.1371/journal.pone.0102222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen J, Huang D, Li C. An early Cambrian craniate-like chordate. Nature. 1999;402:518–522. doi: 10.1038/990080. [DOI] [Google Scholar]
- 52.Heim NA, Knope M. Cope's rule in the evolution of marine animals. Science. 2015;347:867–870. doi: 10.1126/science.1260065. [DOI] [PubMed] [Google Scholar]
- 53.Yasui K, Igawa T, Kaji T, Henmi Y. Stable aquaculture of the Japanese lancelet Branchiostoma japonicum for 7 years. J Exp Zool B Mol Dev Evol. 2013;320B:538–547. doi: 10.1002/jez.b.22540. [DOI] [PubMed] [Google Scholar]
- 54.Igawa T, et al. Inbreeding Ratio and Genetic Relationships among Strains of the Western Clawed Frog, Xenopus tropicalis. PLoS One. 2015;10:e0133963. doi: 10.1371/journal.pone.0133963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nei, M. & Kumar, S. Molecular Evolution and Phylogenetics. (Oxford Univ, 2000).
- 56.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;9:678–87. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 57.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 58.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2002;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 61.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hellsten U, et al. The genome of the western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 64.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 65.Oulion S, Bertrand S, Belgacem MR, Le Petillon Y, Escriva H. Sequencing and analysis of the mediterranean amphioxus (Branchiostoma lanceolatum) transcriptome. PLoS One. 2012;7:e36554. doi: 10.1371/journal.pone.0036554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanabe AS. KAKUSAN: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol Ecol Notes. 2007;7:962–964. doi: 10.1111/j.1471-8286.2007.01807.x. [DOI] [Google Scholar]
- 67.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanabe, A. S. MrBayes5D. (2008).
- 69.Rambaut, A. & Drummond, A. J. Tracer 1.5 (2007).
- 70.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aitchison JC, Ali JR, Davis AM. When and where did India and Asia collide? J Geophys Res. 2007;112:B05423. doi: 10.1029/2006JB004706. [DOI] [Google Scholar]
- 73.Ali JR, Aitchison JC. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166-35 Ma) Earth-Science Rev. 2008;88:145–166. doi: 10.1016/j.earscirev.2008.01.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


