Abstract
The composition and activity of microorganisms in the gut, the microbiome, is emerging as an important factor to consider with regard to the treatment of many diseases. Dysbiosis of the normal community has been implicated in inflammatory bowel disease, Crohn’s disease, diabetes and, most notoriously, Clostridium difficile infection. In Canada, the leading treatment strategy for recalcitrant C. difficile infection is to receive faecal material which by nature is filled with microorganisms and their metabolites, from a healthy individual, known as a faecal microbiota transplantation. This influx of bacteria into the gut helps to restore the microbiota to a healthy state, preventing C. difficile from causing further disease. Much of what is known with respect to the microbiota and faecal microbiota transplantation comes from animal studies simulating the human disease. Although these models allow researchers to perform studies that would be difficult in humans, they do not always recapitulate the human microbiome. This makes the translation of these results to humans somewhat questionable. The purpose of this review is to analyse these animal models and discuss the advantages and the disadvantages of them in relation to human translation. By understanding some of the limitation of animal models, we will be better able to design and perform experiments of most relevance to human applications.
Keywords: Faecal microbiota transplantation, microbiome, animal models
Introduction
It is widely known that faecal microbiota transplantations (FMTs) have been very effective in treating recurrent Clostridium difficile infection with reported success rates of up to 90%.1 Currently, in Canada, it is one of the best treatment options for C. difficile infection and it is being performed on a routine basis. Due to this success, it has been postulated that FMTs may provide relief from other conditions, including some that do not yet have a clear microbial component. Recent studies have shown that malnutrition, obesity, diabetes and other conditions have a dysfunctional microbiome.2 These findings have been supported by animal studies where the human condition has been invoked, despite the intestinal systems of humans and animals differing significantly.3 For example, humans are unique in that the number of recoverable bacteria from the stomach is considerably less than those of other hosts including rodents, ruminants and other mammals.4 The reason for this discrepancy is unclear. If animal models are to provide important insight into a condition afflicting humans, the findings from their use must translate to, and be replicated in, a human. The purpose of this review is to discuss what animal models have taught us in regard to the human microbiome and to recommend ways to alleviate some shortcomings.
Animal models for human microbiome studies
Animal models can be of great benefit to science, but many studies pertaining to the microbiome have disregarded the basic physiology of the host. The gastrointestinal (GI) tract varies greatly between species, and although there are some basic structural similarities, the differences in dietary requirements, frequency of intake, need for storage, body size and shape cannot be lightly discounted.
The stomachs of rodents are divided into glandular and non-glandular sections. The non-glandular part of the stomach is lined by keratinized stratified squamous epithelium and is used in the storage and digestion of food. In humans, pigs, dogs and monkeys, the stomach is entirely glandular and is lined with cardiac, gastric and pyloric mucosa. Differences in gut motility, transit time, capacity, intestine length and pH are also apparent between species.5 Immunological structures such as gut-associated lymphoid tissue and Peyer’s patches also vary between species with respect to their development, abundance and cellular composition.6 Therefore, in basic physiology alone, much needs to be considered when aiming to use animal models to advance the understanding of human conditions.
Besides the physiological differences, the GI tract of mammals differs in secretions. Mucins are heavily glycosylated proteins that are the major component of the mucus layer in the gut and protect the epithelial cells. The mucous layer extends through the entire intestinal tract and is an important component in bacterial persistence and metabolism. Different mammals have considerably different mucins. These alter bacterial composition despite similar microbial species being present or administered. For example, Lactobacillus rhamnosus GG adheres to mucins that have carbohydrate moieties containing β-galactoside at the non-reducing end of the compound.7 Thus, presumably, a model that contains these types of mucins would respond better to probiotic treatment than those models without, and the conclusions drawn might not have human relevance.
Disturbed mucus layer integrity is invariably part of the pathology of disorders of the GI tract, such as Crohn’s disease and ulcerative colitis. The genes that encode mucins are not well conserved between species and need to be considered when using animal models.8 Similarly, cholic acid is the major bile acid in humans, whereas hyocholic acid is the major bile acid in pigs. These are important for the breakdown of ingested lipids and as such would have a major impact on the diversity and composition of the microbiome. One would postulate that with this variability in the GI tract between mammals, there would be great diversity between the microbiomes, but this is only true to some extent. In our own studies with a guinea pig model of placental insufficiency, the prototypical decrease in relative abundance of Bacteroidetes accompanied by an increase in Firmicutes (B/F) was not observed (unpublished data). Similar conclusions were drawn from studies of obesity and metabolic syndrome.9,10 However, a metagenomic comparison between 33 mammalian species that included humans and many different digestive systems with a range of diets found that the microbiome composition and genetically encoded abilities were largely driven by diet (carnivore, omnivore or herbivore) not species. Many of the genes encoding the metabolic enzymes distinguished the faecal microbiomes between carnivorous and herbivores. This suggests that the carnivorous microbiome has been specialized to degrade proteins as an energy source, whereas herbivorous communities have a significant genetic component focussed on synthesizing amino acids.11 This makes it important even when studying humans, as some have a high protein diet and others high carbohydrate.
Gnotobiotic animal models have certainly impacted microbiome research. By creating germ-free or defined microbiota variants, researchers have observed the effect of a particular microorganism type on the host. But, how do physiologically challenged germ-free mice eating the same daily chow relate in any way to humans? Many conclusions have been drawn from mouse studies for obesity, autism and other conditions of societal and physiological interest,12–14 without proving their relevance in humans. Indeed, even without a proven link because of flawed statistical analyses,15 the lay media report the conclusions with vigour, and patients then seek treatments because of them, yet, too often the cause-and-effect is unproven in human subjects. There are no gnotobiotic humans on this microbial planet, so while the guise of this mechanistic research is appealing to granting agencies and some scientists, the relevance is too often frail. This is even more so if the microbiome differences between species are clearly dissimilar.
Consumption of faeces in non-humans: what can we learn?
In nature, many animals commonly supplement their indigenous bacteria by eating their own or another species’ faeces. Some social insects (e.g. termites) and rodents require coprophagy to extract specific nutrients out of their food. Guinea pigs consume their own faeces to allow the bacteria in their stomach to break down many of the nutrients, as this extraction typically occurs in the lower GI tract where absorption is low. This trait is especially important during periods of famine when a food source is scarce. Certain young herbivores eat their mother’s faeces (allocoprophagy) in order to obtain nutrients quicker. Not surprisingly, coprophagy has also been associated with disease outbreaks among rabbits, dogs and many other animals,16,17 since pathogenic microbes are also constituents of the faeces. Although not widely associated with humans,18 other primates such as gorillas and chimpanzees eat their own faeces to a limited degree, but it is unknown what benefit this provides.17 As these traits are not shared by humans, animal studies must carefully evaluate the effect on outcomes.
It is intuitive to think that any factor that will broadly kill microorganisms, like antibiotics, will directly alter the diversity and abundance of the gut microbiome. A recent review by Ianiro et al.19 delves into the plethora of changes that occur in the microbiome depending on the antibiotic used (for review, see Ianiro et al.19). Many of these compounds will also have indirect effects on the host, such as changes in weight, which has been utilized since the 1940s by farmers to increase the weight of livestock.20 By manipulating the microbiome, other secondary effects may arise. Many cattle and other animals are also fed faecal matter from poultry as a source of cheap protein, yet the transfer of microbes has been ignored. Faeces can be high in unwanted nutrients, such as copper, but generally fresh livestock manure seems to contain few toxic products unless it is allowed to putrefy. To maintain safety, the proposed Food and Drug Administration (FDA) recommends removal of pathogens prior to feeding faeces to livestock.21 This might be easy for some unwanted organisms, but knowing what constitutes a bacterial pathogen and what does not, and then trying to remove it without antibiotic use is all but impossible. In the near future, as FMTs in humans become more commonplace, guidelines are going to be needed to ensure that sampling is performed correctly and there is no transfer of ‘unwanted material’. It would be wiser to remove the faecal component from the process altogether and use bioreactors to grow only the bacteria without the production of faeces. This would help alleviate much of the variability between different FMTs as well as help the general public cope with consuming faecal samples. Unfortunately, guidelines currently in place make these experiments impossible to test in a human study despite the fact that using a real faecal sample is okay.
FMT
FMT is a relatively new treatment for refractory C. difficile infection, and despite its widespread use, no adequate meta-analysis has been published to prove its efficacy. The success of the therapy has been documented through numerous case reports, case series and a randomized controlled trial.1 To date, FMTs offer resolution rates of 90%,1 although long-term follow-up data are still sparse. Essentially, FMTs provide a very high dose of bacteria to the gut (Figure 1(a)). The mechanism of action of FMTs remains unknown, and its success may be due to a variety of factors including the microbiota and/or the metabolites present in the donors sample. Bacterial diversity of the microbiome is considered a hallmark of intestinal health. FMTs may provide a source to increase diversity which may reduce the susceptibility to infection by C. difficile,22 although an increase in diversity is not always associated with a decrease in susceptibility.21 There has also been an association between the presence of specific bacterial types and protection against disease. For instance, Clostridium scindens is able to directly inhibit C. difficile through the production of secondary bile acids, deoxycholate and lithocholate.23 These metabolites are found in the stool at concentrations that can inhibit the growth of C. difficile in vitro; however, the susceptibility of different C. difficile strains to secondary bile acids varies.24 FMTs do not constitute a probiotic, since the donor material is not reproducible and defined,25 yet the transfer of these organisms in their active and interlinked metabolic state is much more effective than a dried or food probiotic whose strains do not colonize the GI tract for long. Although it appears to be a more recent advancement in healthcare practice, ancient writings in Chinese history dating back over 1700 years ago have described the consumption of human faeces for many ailments.26 This demonstrates that ancient populations were modulating their microbiome for health benefits, although our understanding of the consequences to human microbial evolution is unclear. An advantage of FMTs is that they are permitted quite freely in most countries. Given the side effects or pharmaceutical agents, their poor efficacy in many cases and the lack of new agents in the pipeline, it is no surprise that FMTs are being applied to attempt to treat a variety of diseases including metabolic syndrome, non-alcoholic fatty liver disease, ulcerative colitis, Crohn’s disease and even those associated with the brain, because of the gut-brain vagus nerve link.27 This is human experimentation at its core, raising the question of what other way could studies by-pass the use of scantly relevant animal models?
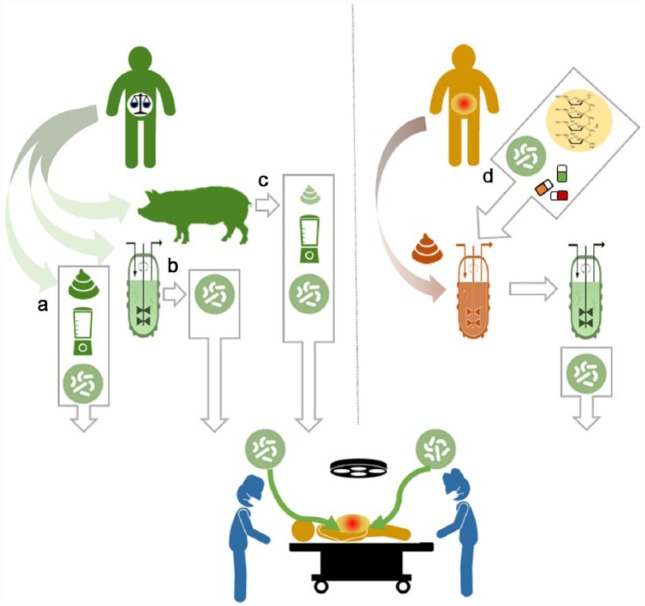
Figure 1.
Various potential sources of material could be used for microbiome transplant in the future beyond direct faecal transplant (a). These include (b) synthetic stool, essentially a defined collection of ‘cornerstone’ bacteria from the microbiome of a healthy person thought to be the minimal requirement to positively change the microbial constitution and support each other’s growth. These maybe grown together or combined from individual stocks, though typically like synergistic growth. (c) Animals such as disease-free pigs given a humanized microbiome and specialized diet. (d) Microbiome reconditioning of faecal material taken from a person with an ‘ailing’ microbiome. While diversity is typically reduced, not all of the cornerstone bacterial types have being deleted, but rather at much lower levels. Faecal specimens are grown in chemostat culture and pulsed with compounds (prebiotic, probiotics, antibiotics and other selective dietary ingredients) to ‘revive’ the minor beneficial components and to deplete less desirable microbial types.
To date, there have been no serious adverse events reported after using FMTs, but due to the experimental nature of the therapy, it is not yet recommended as the first-line treatment for C. difficile–associated diarrhoea by the Canadian Association of Gastroenterology.28 Our willingness to continue the experiment suggests that humans are prepared to be guinea pigs rather than wait for the results of guinea pig experiments.
Animal versus human models of FMTs
Animal models are widely used throughout many aspects of research with the understanding that they provide many benefits, but this does not always reflect the human situation. The microbiota of a mouse differs substantially from a human. As more is discovered with regard to the microbiome of certain conditions, many researchers have begun to humanize the microbiota of their mice prior to experimentation;29 however, this has not become standard practice. Initially, mice had been used extensively as models of C. difficile infection and FMTs;30–33 however, human trials are now highly abundant.
FMTs are also being considered in many other diseases, such as obesity and other metabolic disorders.34 It is widely known when a germ-free mouse receives an FMT from an obese donor versus a lean donor, the mouse with the obese-donor FMT will have increased rates of adiposity.35 Ridaura et al.36 noticed an interesting phenomenon where if mice that received an FMT for either a lean donor or an obese donor were caged together, the obese-donor mice were protected from this increased adiposity. The researchers believed this was due to coprophagy, showing the power FMTs have. However, making this finding a bit more convoluted, a study showed that when two groups of mice were fed a high-fat diet but one received faeces from a lean mouse, mice that received the FMT had an increase in weight.34 When trying to relate this to humans, a clinical study in humans found there was no change in weight observed after FMT with patients continuing to eat their regular diet.37
Similarly, another study investigating the use of FMTs to treat a mouse model of ulcerative colitis found that responses were not only host-specific, but strain-specific as well.38 BALB/c mice were responsive to FMTs as a treatment for ulcerative colitis; however, C57BL/6J mice did not show any improvement in symptom severity.38 The continual use of animal models to study the effects of FMT appears to be redundant and lacks applicability to humans. Fortunately, this is one field where human trials have become more standard practice and animal studies are not required.39–41 However, this is not to say that animal studies are not useful. For instance, Vétizou et al.42 demonstrated in a mouse model of metastatic melanoma that the success of the immunotherapy relied on the commensal microbiome. If germ-free or treated with antibiotics, the mice did not respond to cytotoxic T lymphocyte antigen-4 (CTLA-4) blockade and the tumour grew significantly larger. As a human cannot be germ-free, these mechanisms would never have been found in human studies.
Issues with FMT donor samples
One of the biggest concerns faced by FMT clinics is identifying good donors. In clinical practice, related family members and anonymous donors are common for FMTs. Some patients prefer using a close relative or spouse as their donor since they are familiar with that person, their health and their eating habits. Other patients do not have any relatives that qualify to be a donor so they must use an anonymous one instead. Prior to being considered a donor, subjects need to be screened for high-risk activities, personal and family history of disease, as well as different transmissible agents and pathogenic organisms (including but not limited to: C. difficile; parasites; norovirus; rotavirus; sapovirus; enterovirus; parechovirus; adenovirus; Epstein–Barr virus (EBV); Hepatitis A, B and C; HIV; and human T-cell lymphotropic virus (HTLV)).43 There are currently no regulations on how often donors must be screened for these pathogens,1 but most clinics screen every 6 months or after international travel. Screening a related donor for every patient can be quite costly so there is a shift to use anonymous donors because one person can be screened and used for multiple FMTs to different recipients. It has been estimated that the cost of screening a donor to be US$885, which includes the cost of a clinical assessment, stool test and serum test.44 Interim testing costs approximately US$135 per subject.44 Differences in the microbiota between successful and unsuccessful donors have not been identified. This has led to the concept of preparing humanized artificial FMT samples grown in the laboratory. Propagating human faecal material can be achieved in a defined mixed culture or chemostat (Figure 1(b)). This synthetic stool contains what current research believes are the essential a minimal number of bacterial strains (i.e. not the whole microbiome).45,46 Although this approach has been successful in a few subjects, the concept has faced a regulatory impasse.47 There are minor concerns that like re-cultivated probiotic strains, phenotypic characteristics important to confer health benefits to the host may be lost due to multiple propagations.
To address some regulatory concerns, researchers are being guided to perform more experiments in animal models. This seems ironic given the issues with animal model relevance to humans, and the fact that animals are not considered by regulators as a suitable source of FMT.48 The argument is being made that using animals could provide a large, continuous source of non-infectious material at low cost, eliminating the need to comprehensively screen human donors on every occasion. A humanized porcine microbiome has been proposed (Figure 1(c)), as the pig intestine can be colonized with human faecal microbiomes.49 Piglets delivered germ-free could be colonized with healthy human faeces and then maintained on an optimal ‘human’ diet. A single colony of totally disease-free pigs exists, originally from the Auckland Islands, left by mariners some two centuries ago as a source of food in case of shipwrecking. These pigs are already being used for their islets to replace the blood glucose regulatory system for type I diabetes, but would be an ideal starting point for examining the feasibility of creating ‘high grade’ FMT material.50 Understandably, researchers are interested in testing this concept, but could the donor microbiome be more affected by interactions inside a pig than if they are grown in a chemostat? Is it the microbe–microbe and metabolic linkages that are the key to successful FMT or is it what the organisms do with the host? We would suggest that the host interactions are best left to once the microbes enter the humans from the chemostat, rather than transplanting them from an animal to a human. Time will tell which option works best and which is deemed acceptable to regulators.
Conclusion
Animal models continue to be a mainstay in medical research. For application and expansion of FMTs, the necessity for animals is questionable. The process has been effective without this intermediary step, and questions are being raised about the relevance of tests in mice, which share little in common with humans. Researchers and regulators need to carefully consider the end goal, and if an animal or in vitro system is able to truly answer a specific question. If the key mechanisms for FMT success are which microbes are present, how they interact with each other in a given nutrient-rich environment, then propagating them in chemostats may prove to be efficient, effective and reproducible and most importantly safe for human use.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The W. Garfield Weston Foundation provides funding to J.P.B. No other authors received any specific grant from any funding agency in the public, commercial or not-for-profit sectors for this research.
References
- 1. Kassam Z, Lee C, Yuan Y, et al. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 2013; 108: 500–508. [DOI] [PubMed] [Google Scholar]
- 2. Lozupone C, Stombaugh J, Gordon J, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tannock GW. The intestinal microflora: potentially fertile ground for microbial physiologists. Adv Microb Physiol 2000; 42: 25–46. [DOI] [PubMed] [Google Scholar]
- 4. Tannock GW. Normal microflora: an introduction to microbes inhabiting the human body. London: Chapman and Hall, 1995. [Google Scholar]
- 5. Kararli T. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos 1995; 16: 351–380. [DOI] [PubMed] [Google Scholar]
- 6. Jung C, Hugot J, Barreau F. Peyer’s patches: the immune sensors of the intestine. Int J Inflam 2010; 2010: 823710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishiyama K, Ueno S, Sugiyama M, et al. Lactobacillus rhamnosus GG SpaC pilin subunit binds to the carbohydrate moieties of intestinal glycoconjugates. Anim Sci J 2015; 87: 809–815. [DOI] [PubMed] [Google Scholar]
- 8. Gum J, Hicks J, Lagace R, et al. Molecular cloning of rat intestinal mucin: lack of conservation between mammalian species. J Biol Chem 1991; 266: 22733–22738. [PubMed] [Google Scholar]
- 9. Ley R, Turnbaugh P, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- 10. Ley R, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science 2008; 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muegge B, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011; 332: 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heo J, Seo M, Park J, et al. Gut microbiota modulated by probiotics and Garcinia cambogia extract correlate with weight gain and adipocyte sizes in high fat-fed mice. Sci Rep 2016; 6: 33566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsiao E, McBride S, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayer E, Knight R, Mazmanian S, et al. Gut microbes and the brain; paradigm shift in neuroscience. J Neurosci 2014; 34: 15490–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gloor G, Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol 2016; 62: 692–703. [DOI] [PubMed] [Google Scholar]
- 16. Akers J, Schildkraut D. Regurgitation/reingestion and coprophagy in captive gorillas. Zoo Biol 1985; 4: 99–109. [Google Scholar]
- 17. Soave O, Brand CD. Coprophagy in animals: a review. Cornell Vet 1991; 81: 357–364. [PubMed] [Google Scholar]
- 18. Lewin R. Merde: excursions in scientific, cultural, and socio-historical coprology. New York: Random House, 1999. [Google Scholar]
- 19. Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut 2017; 65: 1906–1915. [DOI] [PubMed] [Google Scholar]
- 20. Cromwell GL. Why and how antibiotics are used in swine production. Anim Biotechnol 2002; 13: 7–27. [DOI] [PubMed] [Google Scholar]
- 21. Food and Drug Administration (FDA). Guidance for industry: ensuring safety of animal feed maintained and fed on-farm. Rockville, MD: FDA, 2016. [Google Scholar]
- 22. Chang J, Antonopoulos D, Kalra A, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197: 435–438. [DOI] [PubMed] [Google Scholar]
- 23. Buffie C, Bucci V, Stein R, et al. Precision microbiome restoration of bile-acid mediated resistance to Clostridium difficile. Nature 2015; 517: 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weingarden A, Dosa P, DeWinter E, et al. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS ONE 2016; 11: e0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill C, Guarner F, Reid G, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014; 11: 506–514. [DOI] [PubMed] [Google Scholar]
- 26. Zhang F, Luo W, Shi Y. Should we standardize the 1700-year-old fecal microbiota transplantation? Am J Gastroenterol 2012; 107: 1755–1756. [DOI] [PubMed] [Google Scholar]
- 27. Montiel-Castro A, González-Cervantes R, Bravo-Ruiseco G, et al. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front Integr Neurosci 2013; 7: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moayyedi P, Marshall J, Yuan Y, et al. Canadian Association of Gastroenterology position statement: fecal microbiota transplant therapy. Can J Gastroenterol 2014; 28: 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Collins J, Auchtung J, Schaefer L, et al. Humanized mice as a model of recurrent Clostridium difficile disease. Microbiome 2015; 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seekatz A, Theriot C, Molloy C, et al. Fecal microbiota transplantation eliminates Clostridium difficile in a model of relapsing disease. Infect Immun 2015; 83: 3838–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reeves A, Theriot C, Bergin I. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes 2011; 2: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X, Katchar K, Goldsmith J, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterol 2008; 135: 1984–1992. [DOI] [PubMed] [Google Scholar]
- 33. Lawley T, Clare S, Walker A, et al. Targeted restoration of the intestinal microbiota with a simple defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 2012; 8: e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kulecka M, Paziewska A, Zeber-Lubecka N, et al. Prolonged transfer of feces from the lean mice modulates gut microbiota in obese mice. Nutr Metab 2016; 13: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Turnbaugh P, Ley R, Mahowald M, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 36. Ridaura V, Faith J, Rey F, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterol 2012; 143: 913–916. [DOI] [PubMed] [Google Scholar]
- 38. Tien Z, Liu J, Liao M, et al. Beneficial effects of fecal microbiota transplantation on ulcerative colitis in mice. Dig Dis Sci 2016; 61: 2262–2271. [DOI] [PubMed] [Google Scholar]
- 39. Kunde S, Pham A, Bonczyk S, et al. Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr 2013; 56: 597–601. [DOI] [PubMed] [Google Scholar]
- 40. Cui B, Li P, Xu L, et al. Step-up fecal microbiota transplantation strategy: a pilot study for steroid-dependent ulcerative colitis. J Transl Med 2015; 13: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wei Y, Zhu W, Gong J, et al. Fecal microbiota transplantation improves the quality of life in patients with inflammatory bowel disease. Gastroenterol Res Pract 2015; 2015: 517597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015; 350: 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rossen N. Fecal microbiota transplantation as novel therapy in gastroenterology: a systematic review. World J Gastroenterol 2015; 21: 5359–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kazerouni A, Burgess J, Burns L, et al. Optimal screening and donor management in a public stool bank. Microbiome 2015; 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen-Vercoe E. Bringing the gut microbiota into focus through microbial culture: recent progress and future perspective. Curr Opin Microbiol 2013; 16: 625–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petrof E, Gloor G, Vanner S, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 2013; 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petrof E, Claud E, Gloor G, et al. Microbial ecosystems therapeutics: a new paradigm in medicine? Benef Microbes 2013; 4: 53–65. [DOI] [PubMed] [Google Scholar]
- 48. Kostic A, Howitt M, Garrett W. Exploring host-microbiota interactions in animal models and humans. Genes Dev 2013; 27: 701–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Q, Widmer G, Tzipori S. A pig model of the human gastrointestinal tract. Gut Microbes 2013; 4: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wynard S, Nathu D, Garkavenko O, et al. Microbiological safety of the first clinical pig islet xenotransplantation trial in New Zealand. Xenotransplantation 2014; 21: 309–323. [DOI] [PubMed] [Google Scholar]