Abstract
Current methods for antibody structure prediction rely on sequence homology to known structures. Although this strategy often yields accurate predictions, models can be stereo-chemically strained. Here, we present a fully automated algorithm, called AbPredict, that disregards sequence homology, and instead uses a Monte Carlo search for low-energy conformations built from backbone segments and rigid-body orientations that appear in antibody molecular structures. We find cases where AbPredict selects accurate loop templates with sequence identity as low as 10%, whereas the template of highest sequence identity diverges substantially from the query’s conformation. Accordingly, in several cases reported in the recent Antibody Modeling Assessment benchmark, AbPredict models were more accurate than those from any participant, and the models’ stereo-chemical quality was consistently high. Furthermore, in two blind cases provided to us by crystallographers prior to structure determination, the method achieved <1.5 Ångstrom overall backbone accuracy. Accurate modeling of unstrained antibody structures will enable design and engineering of improved binders for biomedical research directly from sequence.
Keywords: combinatorial-backbone modeling, loop prediction, protein structure prediction, rosetta, AbDesign
Introduction
Antibodies are the main soluble component of the mammalian immune system.1 Most antibodies comprise two chains, light and heavy, each of which is generated by random recombination of variable (V) and junctional (J) genomic segments; the heavy chain is further diversified by recombination of diversity (D) segments. Each antibody variable domain comprises three hypervariable complementarity-determining regions (CDRs), which encode binding specificity, and a conserved framework, which provides structural stability. During affinity maturation, antibodies are further diversified through a random process of somatic hypermutation, which introduces mutations primarily at the CDRs. The antibody repertoire and affinity maturation process are powerful natural mechanisms for producing a versatile and nearly unlimited capacity for molecular recognition. Owing to their versatility in molecular recognition and their compatibility with the mammalian immune system antibodies are ubiquitous in biomedical research and biotechnology.
Structure modeling is an important tool for antibody design and optimization. Several methods were therefore advanced to model structures directly from sequence.2 Current methods typically use a combination of sequence homology to existing antibody structures and energy-based conformation sampling. For example, RosettaAntibody3 parses the variable domain (Fv) amino acid sequence into its constituent CDRs and frameworks, and for each segment searches the Protein Data Bank (PDB) for the highest sequence-identity templates. These segments are combined to generate a starting model, and the conformationally variable CDR H3 is predicted ab initio. Finally, the resulting model is relaxed by side-chain packing and minimization. The MAPS method, by contrast, segments the antibody according to its germline gene segments [V gene and (D)J segments] instead of along the lines of the CDRs.4 Similar to homology-modeling based methods, the segments with the highest sequence identity to the target are then combined to create the final model. Other methods use expert rules to model H3,5 or a combination of sequence identity and stereo-chemical quality to select models.6 For recent reviews of antibody structure modeling see Refs. 7,8.
To provide an unbiased analysis of the strengths and weaknesses of modeling algorithms, two community-wide blinded Antibody Modeling Assessments (AMA-I and AMA-II) were conducted.7,8 The assessors of AMA-II determined that CDRs 1 and 2 were generally well predicted by existing methods, but CDR H3 and the rigid-body orientation (RBO) between the variable light and heavy chains were usually predicted poorly: on average, among the best predicted structures from each participating group in AMA-II 35% were tilted >3° relative to the experimental structure, and 75% had H3 carbonyl root mean square deviation (rmsd) >1.5 Å. The assessors further noted that some models exhibited substantial stereo-chemical strain, including steric overlaps and backbone distortions. Modeling inaccuracy, stereo-chemical strain, and the use of expert rules and manual intervention limit the usefulness of modeling in design and simulation by non-experts.
Recently, we proposed an antibody-design strategy, called AbDesign, that uses combinatorial backbone modeling.9 Early antibody-engineering studies suggested that CDRs could be grafted independently of one another and regardless of the framework.2 We found, however, that combining CDRs and frameworks arbitrarily resulted in models with high stereo-chemical strain, including voids and steric overlaps between the CDRs and the framework. Instead, AbDesign starts by segmenting all >1,300 antibodies in the PDB into two regions in both the light and heavy chains: a region that roughly corresponds to CDR3 and another that encompasses much of the V gene, comprising the framework and CDRs 1 and 2. The design calculations arbitrarily combine these backbone fragments and subsequently optimize the amino acid sequence for high stability and ligand affinity. The AbDesign strategy is therefore inspired by the natural antibody-generation process by genomic recombination.10 We found that designs produced using this strategy had superior stereo-chemical quality compared to those produced by grafting the CDRs independently of one another on a single framework. Here, we develop a method called AbPredict to model the structures of antibodies directly from their sequences by using a conformation-sampling approach that extends AbDesign. For every target sequence, AbPredict searches for low-energy combinations of backbone fragments from natural antibody structures. Unlike current prediction algorithms, all of which rely on sequence homology,3–6,11–14 AbPredict does not use expert rules or sequence homology to select conformations. We find that this approach results in models that are as accurate as, and in several cases more accurate, than prediction models submitted to AMA-II, and that stereo-chemical strain is uniformly low.
Materials and Methods
Torsion and rigid-body orientation databases
Conformation database construction followed AbDesign,9 which used the SabDab database15 to select antibodies from the PDB.16 We chose a template κ antibody (PDB entry: 2BRR) as a reference for all conformation and rigid-body calculations of κ light-chain antibodies, and a λ antibody (PDB entry: 2G75) for λ light-chain antibodies. To avoid bias in comparing to AMA-II participants, all antibodies deposited in the PDB after February 2014 were excluded from the conformation databases. By contrast, conformation databases including recent structures were used for the two blind prediction cases. The conformation databases contained the following number of entries for κ antibodies: 961 Vh; 962 H3; 1,169 Vl; and 1,223 L3. For the λ antibodies the databases contained 1,028 Vh, 1,039 H3, 221 Vl, and 232 L3.
The previous implementation of antibody modeling in AbDesign only allowed limited rigid-body minimization. Since this degree of freedom is important for prediction accuracy we extended modeling to include it. In accordance with the general strategy employed in AbDesign of sampling degrees of freedom according to their observed values in actual molecular structures, we constructed a rigid-body orientation (RBO) database that encodes the spatial relationships between light and heavy chains in antibody molecular structures. Visual inspection of a structural superimposition of light chains and heavy chains from diverse antibodies showed that the C-terminal disulfide-linked cysteines in each chain (Kabat numbering: L88 and H92) were highly conserved in structure, recommending the spatial relationship between these cysteines as a natural axis about which to define the RBO. Briefly, for each antibody structure we recorded in a database the Rosetta rotation-translation matrix (implemented as the Rosetta Jump object17) relating the three backbone heavy atoms of these two cysteines. During structure-prediction simulations, this database is read into memory and entries from it can be efficiently imposed to change the RBO in the modeled antibody, thereby sampling RBOs that are observed in natural antibody structures. The RBO recording and imposition routines were implemented in the RosettaScripts18 movers RBOut and RBIn, respectively. Furthermore, since κ and λ light chains exhibited different RBO distributions, we constructed a separate RBO database for each class.
Simulated-annealing Monte Carlo sampling of conformation degrees of freedom
The protocol (Fig. 1) is available in Supporting Information.
Rosetta source code and availability
All runs used git version 6f79e55bdcb86e92269495-b363ecd745536914c5 of the Rosetta biomolecular modeling software, which is available to academics at http://www.rosettacommons.org. The torsion databases and the RBO databases are distributed with the Rosetta release.
Running AbPredict and analyzing output
A Python wrapper manages cluster job runs on a load-sharing facility (LSF), and a suite of analysis tools generates energy landscapes against the RBO or the rmsd of backbone conformations. Both the python wrapper and the suite of analysis tools are available at https://github.com/chnorn/AbPredict.git together with the setup needed to reproduce the benchmark results. The MolProbity score for each model was calculated using a standalone tool provided with MolProbity (git entry: 920791-day5a3e1672be808be385606c0c524c10da0).
Data acquisition and analysis of benchmark results
All structures from AMA-II were obtained from http://www.3dabmod.com. Each participant provided three unranked models. As in AMA-II, we measured the RBO prediction accuracy as the difference in torsion angle between the light and heavy chains (tilt) as described in Ref. 19. For each metric (L1—3, H1—3, H3 stem, tilt, and total rmsd) reported in Figure 4 we compared the most accurate model of the three predictions deposited by each participant in AMA-II. Similarly, three models were selected from AbPredict for comparison. The AbPredict models were selected without manual intervention by clustering the 5% lowest-energy models using a radius of 1.0 Å across the CDRs with a hierarchical agglomerative clustering method,20 followed by selection of the lowest-energy models from each of the three most populated clusters.
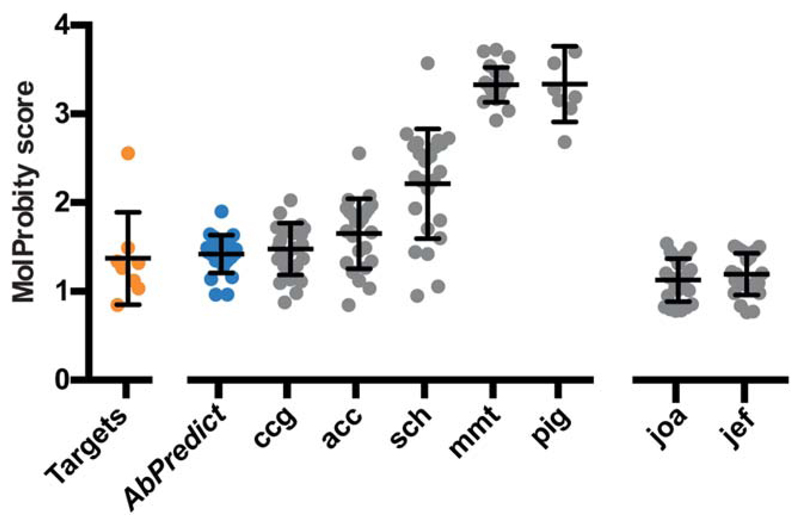
Figure 4.
AbPredict models are stereo-chemically relaxed. Methods: joa,5 jef,3 ccg,6 acc,13 sch,12 mmt,11 pig,14 Jef and joa use MolProbity scores to select models, whereas the other methods, including AbPredict (blue), do not. The targets category (orange) represents the MolProbity scores of the experimental benchmark target structures deposited in the PDB. Error bars represent the standard deviation from the mean.
Results
AbPredict: combinatorial backbone modeling in antibodies
Our previous implementation of combinatorial backbone design (AbDesign)9 starts with a pre-computation step, in which all antibodies in the PDB are segmented into their four constituent conformation fragments (corresponding roughly to Vl, Vh, and CDRs L3 and H3). During modeling, fragments are randomly combined, and Rosetta combinatorial sequence optimization is applied. In structure prediction we are also interested in modeling the RBO between the light and heavy chains, a degree of freedom that was not modeled in AbDesign. In keeping with the AbDesign strategy of sampling conformation degrees of freedom from known structures, we derive the rotation and translation degrees of freedom from all experimentally determined antibody structures and record them in a database for efficient lookup during modeling (Fig. 1).
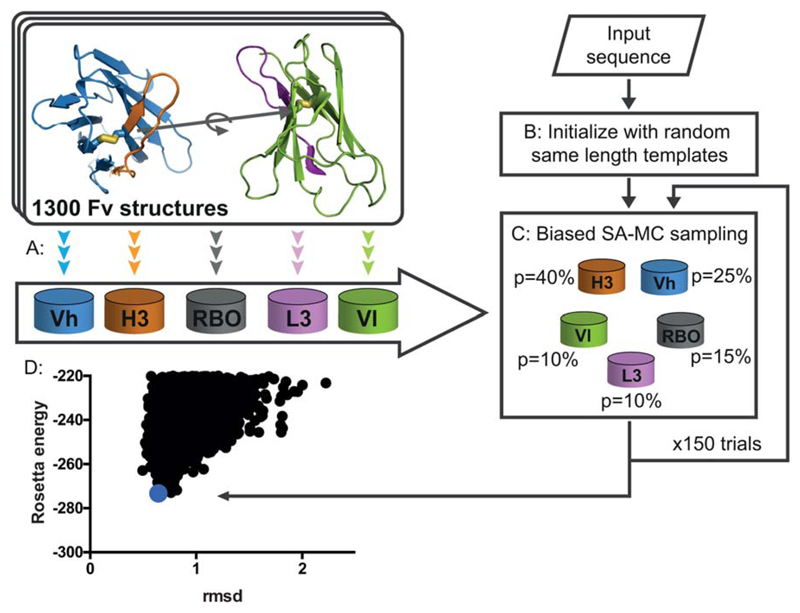
Figure 1.
AbPredict: A method for combinatorial backbone modeling of antibodies. (A) Database preparation: The antibody variable domain (Fv) is segmented into regions comprising CDRs L3 and H3 (magenta and orange, respectively), and regions comprising much of the V segments, Vh (blue) and Vl (green), including CDRs 1 and 2.9 In a pre-computation step the backbone torsion angles of each segment as well as the RBO between light and heavy chains (gray) are stored in five databases for efficient lookup during modeling simulations. (B & C) Schematic description of a structure prediction trajectory: starting from the input sequence an arbitrary combination of backbone and rigid-body degrees of freedom is chosen from the databases and 150 simulated-annealing Monte Carlo (SA-MC) steps are conducted to identify low-energy conformations. (D) 3,000 independent trajectories are run to produce an energy landscape, where every point is the lowest-energy structure from an individual modeling trajectory and carbonyl root mean square deviation (rmsd) is measured to the experimentally determined structure.
Before AbPredict starts we determine whether the light chain belongs to the λ or κ class, and further determine the sequence length of each backbone conformation fragment (Vl, Vh, L3, and H3) (Fig. 1). During modeling, all backbone-sampling moves are restricted to conformations within the same sequence-length class. Unlike current prediction methods3–6,11–14, sequence homology to the target is not used to select or bias conformations and RBOs. At the start of the simulation, we select a random combination of backbone segments and a RBO, and thread the target sequence on this arbitrary backbone. We then conduct a simulated-annealing Monte Carlo search comprising 150 steps (Fig. 1), in each of which a move (conformation change in Vl, Vh, L3, H3, or the RBO) is randomly sampled from the pre-computed databases (see movie in Supporting Information). To reduce stereo-chemical strain, residues within 6 Å of the region affected by the move are optimized using combinatorial side-chain packing,21 and the segment’s backbone and side chains and the RBO are simultaneously minimized subject to harmonic backbone-coordinate restraints.9 Following minimization, the new conformation is accepted if its energy passes the Metropolis criterion, and the temperature is gradually reduced to 0 during the simulation, saving at each step both the last-accepted and the lowest-energy model recorded up to that step; each trajectory outputs the lowest-energy model sampled during the trajectory. For each target sequence, 3,000 independent trajectories are computed to produce an energy landscape. The 5% top-energy models are then clustered according to carbonyl rmsd, and the lowest-energy representatives of the three most populated clusters are selected as the predicted structures. Each trajectory takes, on average, 1.5 hours on an Intel Xeon 2.4 GHz CPU. The protocol is fully automated requiring no manual intervention and no sequence alignment to template antibodies, and is therefore accessible to non-experts.
AbPredict discretizes antibody conformation space
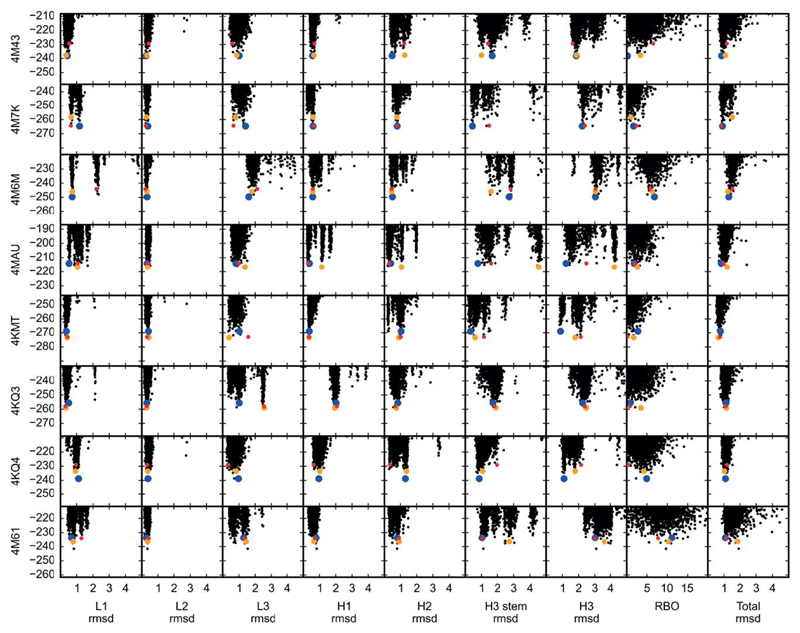
Several antibody structure-prediction methods optimize homology models using energy minimization3,11–13. Although minimization eliminates stereo-chemical strain, resulting structures could fill voids, where waters typically bind, or introduce non-native contacts due to force-field inaccuracies. Combinatorial backbone modeling, by contrast, relies on a discrete set of backbone conformations that are actually observed in natural antibodies. Therefore, one advantage of AbPredict is that it reduces the risk of modeling non-native contacts. Furthermore, AbPredict energy landscapes show multiple distinct funnels rather than a continuum of different conformations as observed in unconstrained modeling (Fig. 2).22,23 This discretization simplifies discrimination between near-native and far-from-native conformations; in effect, low-energy conformations comprise a small set of distinct alternatives rather than a continuum.
Figure 2.
Combinatorial-backbone modeling in AbPredict explores the energy landscape discretely. Unlike ab initio methods,3,11–13 combinatorial backbone modeling as implemented in AbPredict only explores combinations of conformations, which are observed in natural structures; since these conformations fall into clusters,2 the resulting energy landscape in AbPredict is more discrete than seen in ab initio methods. The rigid-body orientation between light and heavy chains (RBO) is a more continuous degree of freedom, and accordingly the energy landscape for RBO is less discrete than that of the backbone conformations. Carbonyl rmsd or RBO deviation from the experimental antibody structure is measured on the x axis, and energy (in Rosetta energy units) on the y axis. The lowest-energy model of the most, the second most, and the third most populated clusters are automatically selected (see Methods) and are marked by blue, yellow, and red, respectively.
Strain-free, atomic-accuracy modeling in a benchmark
We benchmarked AbPredict’s performance on targets from AMA-II.8 From the original AMA-II benchmark of 11 antibodies, we eliminated two structures with poor crystallographic resolution (>2.5 Å; PDB entries: 4M6O and 4KUZ) that additionally form crystal-packing interactions through their CDR H3s. Additionally, we excluded an antibody with an unusually long CDR L3 (PDB entry: 4MA3), as there is only one structure in the PDB with this L3 length. We note that exclusion of these three structures does not bias the comparison between AbPredict and the participants in AMA-II as no participant successfully predicted the H3 of any of these structures (H3 rmsd >2.2 Å), and the 4MA3 L3 (rmsd >3.3 Å).
To remove bias in assessing prediction accuracy, we eliminated from our conformation databases all PDB entries deposited after the start of the benchmark study (February 2014), including the entries from the benchmark. Furthermore, to ensure fair comparison between other groups’ predictions and AbPredict’s we obtained the models submitted to AMA-II and applied to them the same assessment metrics as to AbPredict’s. Following the AMA-II assessors we analyzed the carbonyl rmsd for each of the six CDRs, the H3 stem positions (Kabat numbering H94 and H100A), which capture the portion of H3 that is either kinked or extended,24 the RBO between the light and heavy chains, the complete paratope comprising the six CDRs taken as a whole, and the models’ stereo-chemical quality according to their MolProbity score, which measures van der Waals clashes and backbone and side-chain conformation outliers.25,26 The three AbPredict models for each of the AMA-II entries are available in the online supplement.
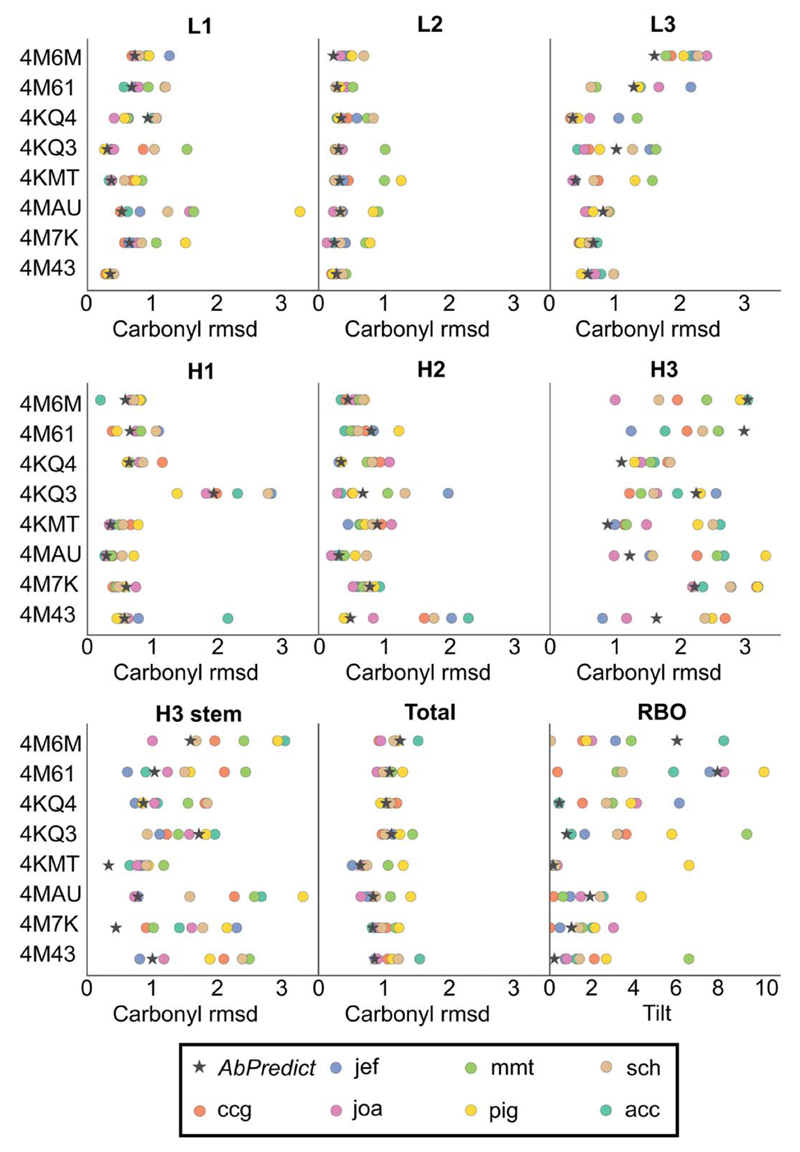
All the methods surveyed in AMA-II perform well in predicting the conformationally restricted CDRs L1, L2, and H2 (average rmsd <1.0 Å), and AbPredict is similarly accurate (Fig. 3). H1 is also conformationally restricted, and except in one case (PDB entry: 4KQ3), for which no correct predictions were made in AMA-II, AbPredict performs as well as the other methods (rmsd <1.0 Å). CDR 3 is conformationally more diverse, but despite this potential source of uncertainty AbPredict achieves accurate predictions for L3, which in the worst case (PDB entry: 4M6M) shows 1.6 Å rmsd to the correct conformation. Furthermore, AbPredict achieves the most accurate predictions for the H3 stem positions. In principle, the solvent-exposed tip of H3 is where a knowledge-based method such as AbPredict might be inferior to others. Nevertheless, performance is on par with the other methods’, and three of the eight entries exhibit rmsd <1.5 Å with average rmsd for the lowest-energy models measuring 1.9 Å. The RBO between light and heavy chains is predicted with an average error of 2.2 ± 2.9°, similar to the average error of other methods (2.7 ± 2.5°). We note that accuracy in RBO correlates with H3 prediction accuracy: when two antibodies (PDB entries: 4M61 and 4M6M) with poorly predicted H3 conformations are eliminated, the models’ average error improves to 0.7°.
Figure 3.
Prediction accuracy in AbPredict relative to AMA-II participants. Methods: joa,5 jef,3 ccg,6 acc,13 sch,12 mmt,11 pig.14 Carbonyl rmsd is measured in Ångstrom and tilt in degrees.
The AMA-II assessors noted that models from several groups suffered from stereo-chemical strain, as judged by MolProbity.27 Unlike some other methods surveyed in AMA-II,3,5 AbPredict selects models solely on their computed energy and not their MolProbity scores. Nevertheless, we find that predicted structures have uniformly low MolProbity scores, with an average composite score of 1.4 and a worst score of 1.9 (PDB entry: 4MAU), corresponding to the quality one may expect from crystal structures with resolution 1.9 Å.25,26 By contrast, those AMA-II methods that did not use MolProbity in selection, submitted models with MolProbity average scores of 1.5–3.3 (Fig. 4). We conclude that in all of the structural and stereo-chemical parameters used to assess antibody structure-prediction algorithms, AbPredict performs as well as, and in some cases (L1, H1, H2, and H3 stem), better than other methods. Our analysis suggests that this fully automated algorithm captures some aspects of expert-knowledge implemented in supervised methods.
Search for lowest-energy structure improves accuracy compared to homology modeling
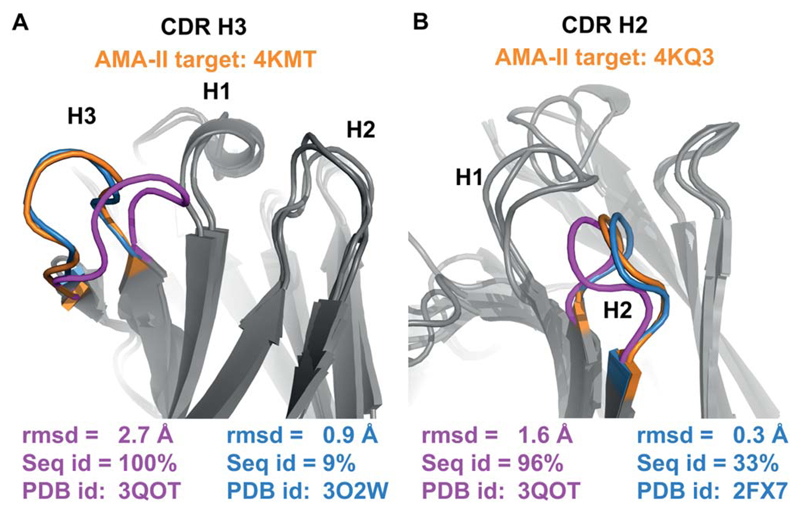
The strategy adopted by all participants in the AMA-II benchmark relies on homology modeling, with some groups employing homology modeling followed by ab initio prediction of CDR H33,11–13. AbPredict, by contrast, searches for the lowest-energy conformation of the target sequence, ignoring sequence homology as a selection criterion. For several AMA-II entries AbPredict shows higher accuracy than any other method (Fig. 3). To investigate the underlying reason for higher accuracy we analyzed the highest sequence-identity template (which is presumably the default for homology-based modeling), AbPredict’s chosen template, and the template of lowest rmsd to the experimentally determined structure (Fig. 5). For instance, AMA-II entry 4KMT shows 100% sequence identity to PDB entry 3QOT in H3, but the H3 backbone conformations exhibit 2.7 Å rmsd [Fig. 5(A)]. Modeling the H3 backbone conformation from 3QOT on the background of the 4KMT structure reveals significant backbone side-chain clashes between the modeled H3 and the 4KMT Vh. In this case, AbPredict achieves higher prediction accuracy than any participant in AMA-II by selecting 3O2W as template, even though 3O2W’s H3 only shares one identical amino acid with 4KMT’s out of 11; the rmsd for 3O2W relative to 4KMT is only 0.9 Å [Fig. 5(A)]. Of the eight AMA-II benchmark targets we find four other examples, where the highest sequence-identity template shows H3 rmsd >1.5 Å and where a template with lower rmsd exists (PDB entries: 4M43, 4MAU, 4M6M, and 4KQ4). In these cases, we find that the average H3 rmsd can decrease from 2.8 Å to 1.1 Å if the lowest-rmsd templates were selected, and AbPredict selects templates with 1.6 Å average rmsd. Our observation that low sequence identity structures could provide superior templates for modeling extends to segments other than H3 as shown for H2 [Fig. 5(B)].
Figure 5.
Examples of accurate low-energy predictions that use templates of low sequence identity. High sequence identity in loop regions does not guarantee high structural similarity, and even identical (A) or highly similar (B) sequences diverge in structure. In these examples AbPredict selects templates that have lower rmsd to the target, despite their low sequence identity. Orange: experimental structure; magenta: highest sequence-identity template; blue: AbPredict model.
High-accuracy, fully blind predictions
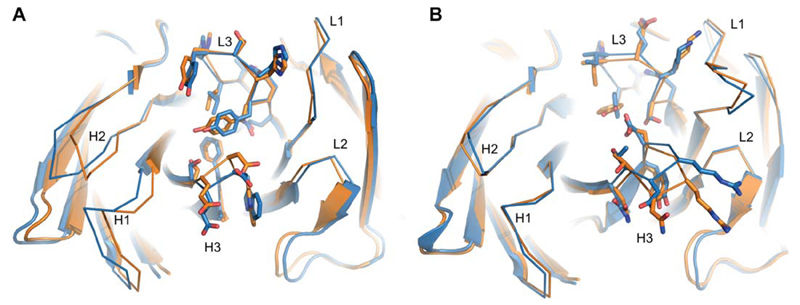
Encouraged by AbPredict’s performance on the AMA-II benchmark, we obtained from other research groups the sequences of two antibodies targeting unrelated proteins that were in the process of structure determination; we treated these sequences as blind-prediction cases. In these cases, we used the complete conformation databases based on known antibody structures available at the beginning of 2015 (prior to these antibodies’ structure determination), and predicted the structures using the same strategy as above. For Antibody A all predictions outside CDR H3 have rmsd <1.5 Å to the correct structure (Fig. 6). Antibody A has an H3 of length 13 residues, which is as long as the longest H3 in the AMA-II benchmark. Nevertheless, the lowest-energy model achieves 1.6 Å rmsd in H3; the deviation between the predicted and observed rigid-body angle is 1.7°, and the rmsd over the entire paratope ranges from 0.8 to 1.1 Å in all three predictions. In antibody B, all CDRs of the lowest-energy model are below 1.5 Å and the rigid-body angle is 1.3° off-target. Throughout much of the paratope, we observe that even side-chain conformations are predicted accurately (Fig. 6). These fully-blind results reinforce the benchmark results above, that AbPredict can be used in an unsupervised and fully automated way to obtain high-accuracy model structures.
Figure 6.
High-accuracy blind predictions. The lowest rmsd of three predicted structures (orange) for two antibodies that were experimentally elucidated (blue) after modeling. Models and experimental structures overlap with (A) 1.2 Å carbonyl rmsd and (B) 1.0 Å carbonyl rmsd. Selected, high-accuracy side-chain conformations on L3 and H3 are shown as sticks. Sequences and solved structures were kindly provided by Aliza Katz and Ron Diskin (A), and Iris Grossman and Deborah Fass (B) prior to publication.
Discussion
Existing antibody structure-prediction algorithms use sequence homology to select templates for threading. This report demonstrates, however, that high sequence similarity could misguide modeling, particularly in the conformationally heterogeneous CDRs H2 and H3. An alternative to homology modeling is ab initio structure prediction, where the conformation is predicted from scratch based on a search for low-energy conformations.3 Ab initio modeling, however, is complicated in loop regions owing to the large conformation space open to regions lacking secondary structure and due to energy-function inaccuracies, and often results in flat energy landscapes and poor discrimination between near and far-from-native conformations.22,23 Combinatorial backbone modeling is similar to homology modeling in using template structures from the PDB, but it only uses energy to select models. Since combinatorial backbone modeling chooses fragments from a limited set of conformations that are actually observed in nature it does not force non-native contacts due to energy-function inaccuracies, and yields discretized energy landscapes that improve discrimination between native and non-native backbone conformations.
Conclusion
AbPredict selects models strictly according to energy and does not use sequence alignments or expert rules. This simplifies automation, opening the way for adoption by non-experts. The resulting models’ high accuracy and low stereo-chemical strain suggest that they can be used as templates for design calculations.28 Combinatorial backbone modeling, moreover, can be extended to other modular folds of high conformational diversity, such as TIM barrels and β-propellers.
Supplementary Material
Additional Supporting Information may be found in the online version of this article.
Acknowledgments
The authors thank Liam Longo, Lior Zimmerman, and Adi Goldenzweig for testing AbPredict and Shira Warszawski for valuable discussions on antibody loop flexibility. They also thank Aliza Katz, Ron Diskin, Iris Grossman, and Deborah Fass for providing the blind-prediction cases. C.N. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds. S.J.F. is a Martha S Sagon Career Development Chair.
Grant sponsors: Starter’s Grant from the European Research Council (to Fleishman laboratory), the Human Frontier Science Program, the Israel Science Foundation (ISF) through an individual grant, the ISF-UGC framework, and the Center for Research Excellence in Structural Cell Biology, the Israel Chief Scientist’s Office, the Marie Curie Reintegration Career Development Award, and a charitable donation from Sam Switzer and family.
Abbreviations
- LSF
load-sharing facility
- PDB
Protein Data Bank
- RBO
rigid-body orientation
References
- 1.Janeway CA, Travers P, Walport M, Shlomchik M. Immunobiology: The immune system in health and disease. New York: Garland Science; 2001. [Google Scholar]
- 2.Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 3.Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3.0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins. 2014;82:1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pantazes RJ, Maranas CD. MAPs: a database of modular antibody parts for predicting tertiary structures and designing affinity matured antibodies. BMC Bioinform. 2013;14:168. doi: 10.1186/1471-2105-14-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirai H, Ikeda K, Yamashita K, Tsuchiya Y, Sarmiento J, Liang S, et al. High-resolution modeling of antibody structures by a combination of bioinformatics, expert knowledge, and molecular simulations. Proteins. 2014;82:1624–1635. doi: 10.1002/prot.24591. [DOI] [PubMed] [Google Scholar]
- 6.Maier JKX, Labute P. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014;82:1599–1610. doi: 10.1002/prot.24576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almagro JC, Beavers MP, Hernandez-Guzman F, Maier J, Shaulsky J, Butenhof K, et al. Antibody modeling assessment. Proteins. 2011;79:3050–3066. doi: 10.1002/prot.23130. [DOI] [PubMed] [Google Scholar]
- 8.Almagro JC, Teplyakov A, Luo J, Sweet RW, Kodangattil S, Hernandez-Guzman F, et al. Second antibody modeling assessment (AMA-II) Proteins. 2014;82:1553–1562. doi: 10.1002/prot.24567. [DOI] [PubMed] [Google Scholar]
- 9.Lapidoth GD, Baran D, Pszolla GM, Norn C, Alon A, Tyka MD, et al. AbDesign: an algorithm for combinatorial backbone design guided by natural conformations and sequences. Proteins. 2015;83:1385–1406. doi: 10.1002/prot.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khersonsky O, Fleishman SJ. Why reinvent the wheel? Building new proteins based on ready-made parts. Protein Sci. 2016:1–9. doi: 10.1002/pro.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berrondo M, Kaufmann S, Berrondo M. Automated Aufbau of antibody structures from given sequences using Macromoltek’s SmrtMolAntibody. Proteins. 2014;82:1636–1645. doi: 10.1002/prot.24595. [DOI] [PubMed] [Google Scholar]
- 12.Zhu K, Day T, Warshaviak D, Murrett C, Friesner R, Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins. 2014;82:1646–1655. doi: 10.1002/prot.24551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasnacht M, Butenhof K, Goupil-Lamy A, Hernandez-Guzman F, Huang H, Yan L. Automated antibody structure prediction using Accelrys tools: results and best practices. Proteins. 2014;82:1583–1598. doi: 10.1002/prot.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcatili P, Olimpieri PP, Chailyan A, Tramontano A. Antibody structural modeling with prediction of immunoglobulin structure (PIGS) Nat Protoc. 2014;9:2771–2783. doi: 10.1038/nprot.2014.189. [DOI] [PubMed] [Google Scholar]
- 15.Dunbar J, Krawczyk K, Leem J, Baker T, Fuchs A, Georges G, et al. SAbDab: the structural antibody database. Nucleic Acids Res. 2014;42:1140–1146. doi: 10.1093/nar/gkt1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, et al. The Protein Data Bank. Eur J Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 18.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abhinandan KR, Martin ACR. Analysis and prediction of VH/VL packing in antibodies. Protein Eng Des Sel. 2010;23:689–697. doi: 10.1093/protein/gzq043. [DOI] [PubMed] [Google Scholar]
- 20.Alexander N, Woetzel N, Meiler J. Bcl::Cluster: a method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. 2011 IEEE 1st Int Conf Comput Adv Bio Med Sci IEEE; 2011. pp. 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.London N, Schueler-Furman O. Funnel hunting in a rough terrain: learning and discriminating native energy funnels. Structure. 2008;16:269–279. doi: 10.1016/j.str.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Tyka MD, Jung K, Baker D. Efficient sampling of protein conformational space using fast loop building and batch minimization on highly parallel computers. J Comput Chem. 2012;33:2483–2491. doi: 10.1002/jcc.23069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirai H, Kidera A, Nakamura H. Structural classification of CDR-H3 in antibodies. FEBS Lett. 1996;399:1–8. doi: 10.1016/s0014-5793(96)01252-5. [DOI] [PubMed] [Google Scholar]
- 25.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teplyakov A, Luo J, Obmolova G, Malia TJ, Sweet R, Stanfield RL, et al. Antibody modeling assessment II. Structures and models. Proteins. 2014;82:1563–1582. doi: 10.1002/prot.24554. [DOI] [PubMed] [Google Scholar]
- 28.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.