ABSTRACT
Bocaparvoviruses are emerging pathogens of the Parvoviridae family. Human bocavirus 1 (HBoV1) causes severe respiratory infections and HBoV2 to HBoV4 cause gastrointestinal infections in young children. Recent reports of life-threatening cases, lack of direct treatment or vaccination, and a limited understanding of their disease mechanisms highlight the need to study these pathogens on a molecular and structural level for the development of therapeutics. Toward this end, the capsid structures of HBoV1, HBoV3, and HBoV4 were determined to a resolution of 2.8 to 3.0 Å by cryo-electron microscopy and three-dimensional image reconstruction. The bocaparvovirus capsids, which display different tissue tropisms, have features in common with other parvoviruses, such as depressions at the icosahedral 2-fold symmetry axis and surrounding the 5-fold symmetry axis, protrusions surrounding the 3-fold symmetry axis, and a channel at the 5-fold symmetry axis. However, unlike other parvoviruses, densities extending the 5-fold channel into the capsid interior are conserved among the bocaparvoviruses and are suggestive of a genus-specific function. Additionally, their major viral protein 3 contains loops with variable regions at their apexes conferring capsid surface topologies different from those of other parvoviruses. Structural comparisons at the strain (HBoV) and genus (bovine parvovirus and HBoV) levels identified differences in surface loops that are functionally important in host/tissue tropism, pathogenicity, and antigenicity in other parvoviruses and likely play similar roles in these viruses. This study thus provides a structural framework to characterize determinants of host/tissue tropism, pathogenicity, and antigenicity for the development of antiviral strategies to control human bocavirus infections.
IMPORTANCE Human bocaviruses are one of only a few members of the Parvoviridae family pathogenic to humans, especially young children and immunocompromised adults. There are currently no treatments or vaccines for these viruses or the related enteric bocaviruses. This study obtained the first high-resolution structures of three human bocaparvoviruses determined by cryo-reconstruction. HBoV1 infects the respiratory tract, and HBoV3 and HBoV4 infect the gastrointestinal tract, tissues that are likely targeted by the capsid. Comparison of these viruses provides information on conserved bocaparvovirus-specific features and variable regions resulting in unique surface topologies that can serve as guides to characterize HBoV determinants of tissue tropism and antigenicity in future experiments. Based on the comparison to other existing parvovirus capsid structures, this study suggests capsid regions that likely control successful infection, including determinants of receptor attachment, host cell trafficking, and antigenic reactivity. Overall, these observations could impact efforts to design antiviral strategies and vaccines for HBoVs.
KEYWORDS: capsid, cryo-EM, gastrointestinal infection, HBoV, human bocaviruses, parvovirus, respiratory infection
INTRODUCTION
Bovine parvovirus (BPV) and canine minute virus were the first bocaparvoviruses to be isolated in the 1960s from rectal samples of young cows and dogs, respectively, and were associated with symptoms such as acute bronchitis, diarrhea, interstitial pneumonia, and neonatal reproductive illnesses in their hosts (1–5). In 2005, the first human bocaparvovirus, HBoV1, was discovered in nasopharyngeal aspirates of children (<2 years of age) suffering from acute respiratory tract infections (6). Following HBoV1, three additional HBoV strains, namely, HBoV2, HBoV3, and HBoV4, were isolated from pediatric patients with acute gastroenteritis (7, 8). The HBoVs have also been detected in patients of all ages, albeit at lower frequencies (9, 10). Epidemiological studies reported the presence of episomal HBoV1 and HBoV3 DNA in infected patients, suggesting the ability of the viruses to cause persistent infections (11). Common patient symptoms include diarrhea, wheezing, nausea, vomiting, cough, dyspnea, pneumonia, pharyngitis, and bronchitis, similar to those caused by the animal bocaparvoviruses.
The lack of a stable cell line and animal model for replication has limited the study of the pathogenicity of human bocaviruses. Researchers initially debated the role of HBoV as a passenger or a pathogen in a diseased patient, as several studies detected bocavirus DNA with other coinfections in pediatric patients (12–17). However, recent diagnostic improvements and reports of HBoV1 as a sole pathogen in life-threatening diseases have confirmed the etiologic nature of HBoV1 infection (18–24). Additionally, according to a study conducted in 2006 and 2007, HBoV1 was the third most prevalent virus, after adenovirus and respiratory syncytial virus, detected in samples from young English and Thai children suffering from lower respiratory tract infections (25, 26). The report of HBoV-related respiratory and gastrointestinal tract infections worldwide (>17 countries), combined with a lack of direct treatment, drives the desire to understand the disease-causing mechanisms of these viruses (27). Moreover, HBoVs have also been detected in river water and urban sewage water samples, suggesting a possible transmission route (28, 29).
The parvovirus capsid plays a multifaceted role in infection and is reported to carry determinants of host/tissue cell recognition, endosomal trafficking, capsid assembly, genome packaging, and antibody recognition (30). Thus, the goal of this study was to determine the capsid structure of human bocaviruses with different tissue tropisms to delineate HBoV capsid regions that may serve as determinants of tropism and hence pathogenicity. The bocaparvoviruses are similar in genome organization, with three open reading frames (ORFs) (6). The right ORF encodes the overlapping capsid viral proteins (VPs) VP1 (∼74 kDa, 671 amino acids [aa] for HBoV1), VP2 (∼64 kDa, 581 aa for HBoV1), and VP3 (∼60 kDa, 542 aa for HBoV1). All of the VPs are assumed to be translated from the same transcript, with an expression ratio of 1:1:10, respectively (31, 32). However, it has been shown that VP3 alone is able to assemble capsids that are antigenically similar to wild-type capsids (33, 34). VP1, VP2, and VP3 share a C terminus, while the minor protein VP1 contains a unique N terminus (129 aa) (VP1u). VP1u contains a phospholipase A2 (PLA2) enzyme, which is required for modifying membranes and escaping from the endosomal/lysosomal pathway while trafficking to the nucleus for genome replication (35). For HBoV1, this PLA2 activity was also shown to be harmful to tight junctions found in human airway epithelial cells that normally function as barriers to pathogens and foreign entities by preventing infection and tissue injury (36, 37).
In this study, the structures of HBoV1, and HBoV3 and HBoV4, responsible for lower respiratory tract and gastrointestinal infections, respectively, were determined by cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction (cryo-reconstruction) to 2.9, 2.8, and 3.0 Å, respectively. All three capsids, assembled from VP3, display the surface features previously reported for other parvoviruses, including depressions at the icosahedral 2-fold axis and surrounding the 5-fold axis, protrusions that surround the icosahedral 3-fold axis, and a channel at the 5-fold axis. The VP structure also conserves the topology of other parvovirus capsids, including a βA strand, a core eight-stranded β barrel (βB-βI strand), and large loop insertions between these secondary structure elements. A unique α helix (αB) observed in the crystal structure of BPV between β strands βE and βF was conserved in the HBoVs. A comparison of these viruses identified differences in the specific variable loop regions between the secondary structure elements forming the surface of the capsids, particularly variable region I (VR-I) to VR-V and VR-VIIIB previously defined for autonomous parvoviruses (38). Compared to the crystal structure of BPV, differences were identified in all 10 reported VRs, VR-I to VR-IX (39). These VRs are important determinants of parvovirus infectivity, transduction efficiency, and antigenicity (39). Structural comparisons identified VR-III as a potential determinant of the tissue tropism of HBoVs and VR-VI to VR-VIII as well as VR-IX as potential determinants of bocaparvovirus host tropism. The data from these structures expand our knowledge of bocaparvovirus capsids and provide 3D platforms for probing of their capsid functional determinants, including tissue and host tropism and antigenic reactivity. The data can potentially guide efforts to design inhibitors and vaccines that control bocaparvovirus infection in humans and animals.
RESULTS AND DISCUSSION
The capsid structure of HBoVs.
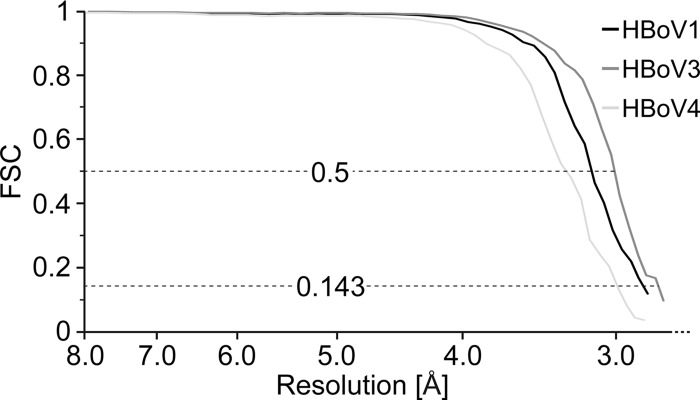
The 3D reconstructed structures of the HBoV1, HBoV3, and HBoV4 capsids were computed by using 31,064, 28,707, and 13,394 particle images, respectively (Table 1). The resolution of the 3D cryo-reconstructed HBoV1, HBoV3, and HBoV4 density maps was estimated by using a Fourier shell correlation (FSC) plot and is reported on the basis of threshold criteria of 0.143 and 0.5 (Fig. 1). For the 0.143 FSC threshold, the three HBoV structures were resolved to 2.9, 2.8, and 3.0 Å, respectively. While the HBoV3 and HBoV4 capsid structures are reported for the first time, the new map of the HBoV1 capsid structure represents a considerable improvement over the previously determined 7.9-Å resolution structure (33). The resolution of the HBoV1, HBoV3, and HBoV4 capsid structures reported here is among the highest that have been obtained by using cryo-reconstruction for virus capsids of this size.
TABLE 1.
Summary of data collection, image-processing parameters, and refinement statistics
| Parameter | HBoV1 | HBoV3 | HBoV4 |
|---|---|---|---|
| Total no. of micrographs | 3,519 | 1,719 | 1,494 |
| Defocus range (μm) | 0.79–4.43 | 0.82–3.91 | 0.90–4.26 |
| Electron dose (e−/Å2) | 63 | 63 | 60 |
| No. of frames/micrograph | 19 | 34 | 38 |
| Pixel size (Å/pixel) | 0.95 | 0.95 | 0.95 |
| Starting no. of particles | 51,756 | 31,896 | 14,882 |
| No. of particles used for final map | 31,064 | 28,707 | 13,394 |
| Inverse B factor used for final map (Å2) | 1/100 | 1/100 | 1/100 |
| Resolution of final map (Å) | 2.9 | 2.8 | 3.0 |
| PHENIX model refinement statistics | |||
| Residue range | 33–542 | 34–539 | 34–541 |
| Map CC | 0.819 | 0.837 | 0.810 |
| RMSD (Å) | |||
| Bonds | 0.01 | 0.01 | 0.01 |
| Angles | 1.46 | 1.15 | 1.33 |
| All-atom clash score | 20.36 | 16.57 | 23.39 |
| Ramachandran plot (%) | |||
| Favored | 85.7 | 82.1 | 80.6 |
| Allowed | 12.9 | 15.5 | 16.6 |
| Outliers | 1.4 | 2.4 | 2.8 |
| Rotamer outliers | 0.0 | 0.0 | 0.0 |
| No. of Cβ deviations | 0 | 0 | 0 |
FIG 1.
FSC plots for HBoV cryo-reconstructions. The estimated resolution of HBoV1 (black line) at FSC threshold values of 0.5 and 0.143 are 3.16 and 2.87 Å, respectively. The estimated resolution of HBoV3 (dark gray line) at FSC threshold values of 0.5 and 0.143 are 3.03 and 2.80 Å, respectively. The estimated resolution of HBoV4 (light gray line) at FSC threshold values of 0.5 and 0.143 are 3.31 and 3.02 Å, respectively.
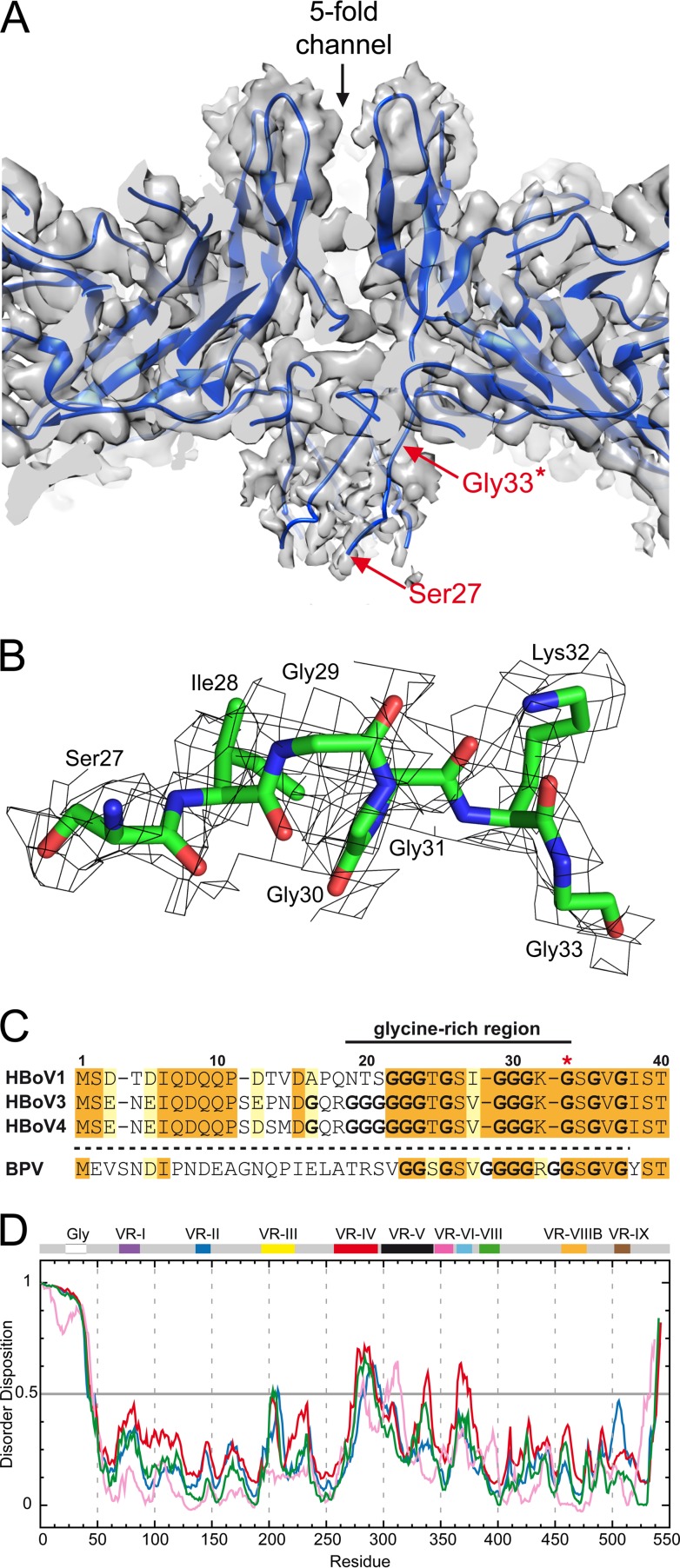
All three structures have characteristic features common to other available Parvovirinae subfamily structures but more closely resemble members of the Dependoparvovirus genus. These features include a shallow depression at each 2-fold symmetry axis, a trimeric protrusion that surrounds each 3-fold symmetry axis, and a channel at each 5-fold symmetry axis, which is surrounded by a wide, canyon-like region (Fig. 2A). However, HBoV3 and HBoV4 have more prominent 3-fold protrusions than HBoV1 that give their capsid surfaces a more “spiky” appearance (Fig. 2A). Comparison of the cross-sectional views of all three HBoV cryo-reconstructions shows the presence of a density (“basket”) beneath the 5-fold axis that extends the 5-fold channel into the capsid interior (Fig. 2B and 3A). This unique feature, reported previously for the crystal structure of BPV (39), for HBoV1 at 7.9-Å resolution (33), and low-resolution structures determined for the HBoV-Fab complexes (34), has not been observed in any other non-bocaparvovirus capsid structures determined to date.
FIG 2.
The capsid structure of HBoVs. (A) Capsid density maps of HBoV1, HBoV3, and HBoV4 contoured at a sigma (σ) threshold of 1.0. The reconstructed maps are colored according to radial distance from the particle center (blue to red), as indicated by the scale bar at the right. (B) Cross-sectional views of the HBoV structures as shown in panel A (contoured at 0.9 σ). Arrowheads point to the locations of the 5-fold (5f) symmetry axes and channel. Under each 5-fold channel, density forming a basket can be observed. These images were generated with UCSF-Chimera (68).
FIG 3.
The unique N termini of bocaparvoviruses. (A) Closeup of a cross-sectional view of the basket under the 5-fold channel in HBoV1. The ribbon diagram of the docked HBoV1 VP3 structure is shown within its semitransparent density. The first residue for which side chain density is ordered (glycine 33, also indicated by the asterisk in panel C) is labeled. The Cα chain was built into the basket density to residue 27. (B) Density for the basket under the 5-fold channel for HBoV1 (residue range, aa 27 to 33) at a sigma (σ) threshold of 0.6. The amino acids are shown in stick representation inside a gray mesh density map and colored according to amino acid type as follows: C, green; O, red; N, blue. (C) Sequence alignment of the VP3 N terminus of bocaviruses. Orange-shaded residues are identical amino acids among the HBoVs, while yellow-shaded residues are similar amino acids. (D) Disorder prediction of the HBoV1 (blue), HBoV3 (green), HBoV4 (red), and BPV (pink) VP3 proteins by the PONDR_fit application (45). Regions above 0.5 on the y axis are predicted to be disordered. The approximate regions corresponding to the VP3 glycine-rich region and VRs are indicated. Panel A was generated with UCSF-Chimera (68).
The high-resolution HBoV maps enabled the assignment of the VP3 amino acids with side chains starting from N-terminal residue 33 of HBoV1 or residue 34 of HBoV3 and HBoV4, respectively. These residues are situated after a glycine-rich region (Fig. 3B and C, indicated by an asterisk) and are part of the extended density inside the capsid under the 5-fold channel. When the reconstructed maps are rendered at lower σ density thresholds (0.7 to 0.8 sigma), this extended density could be interpreted as the main chain for residues 27 to 32 of HBoV1 and 28 to 33 of HBoV3 and HBoV4 (Fig. 3B). This observation supports a previous hypothesis that the N-terminal residues of the bocaparvoviruses lie inside the capsid, bundled under the 5-fold axis, and that the N termini of all of their VPs are similarly localized (34, 39).
For other parvoviruses, VP1u is postulated to become externalized through the 5-fold channel to enable their PLA2 function (40–44). However, how VP1u is extruded through the channel during infection requires further studies of bocaparvoviruses, especially given the unique bundling of the N-terminal residues under the 5-fold channel. The stretch of glycines at the N terminus of the HBoVs, also common to other parvoviruses, may provide the N terminus with enough flexibility for the externalization process. It will also be important, in future studies, to determine if capsids assembled from all three VPs, rather than VP3 alone, display a similar bundling of N termini under the 5-fold axis. However, the remaining N-terminal residues, 1 to 26 for HBoV1 and 1 to 27 for HBoV3/4, were disordered in these VP3-virus-like particle (VLP) structures. Indeed, such disorder of these residues is confirmed by results obtained with the PONDR_fit application, which predicts the N-terminal sequences to be disordered (Fig. 3D) (45).
The HBoV1, HBoV3, and HBoV4 VP3 models, where side chain assignments can be made, were built from N-terminal residues 33, 34, and 34 to the last C-terminal residues, 542, 539, and 541 (VP3 numbering), respectively. The amino acid side chain densities were well defined in the core secondary structure regions (e.g., βD and βG) and most of the surface loops (e.g., VR-IV) inserted between these regions (Fig. 4A to C). However, exceptions to the ordering of the side chain densities were observed for glutamic acid and aspartic acid residues. For a number of these types of residues, the density was less ordered for the terminal acidic groups past the Cβ or Cγ atom (e.g., Glu237 in Fig. 4B). This phenomenon has been previously described for other high-resolution cryo-EM maps and is caused by the high sensitivity of the negatively charged side chain to radiation damage (46). Furthermore, two short regions in HBoV1, aa 204 to 209 (GNAAGG) and aa 333 to 338 (GAAGFG), were not as well defined (not shown). This disorder likely reflects backbone flexibility conferred by the presence of multiple glycine residues in the sequences. The refinement statistics from the PHENIX program, used for further model refinement and evaluation of the models built into the reconstructed density maps for all three virus capsids, are shown in Table 1. These statistics show a high correlation coefficient (CC) between model and map for all three structures. The model geometries are consistent or better than for structures determined at comparable resolution, by either X-ray crystallography or cryo-reconstruction, for other parvovirus capsids, as well as those of other viruses (Table 1) (47).
FIG 4.
Representative regions of HBoV density maps and atomic models. (A) Example density for HBoV1 and HBoV3 at positions of amino acid differences. The resolution of the maps enabled the interpretation and modeling of these differences. (B) Example density for HBoV3 and HBoV4 at positions of amino acid differences. The resolution of the maps enabled the interpretation and modeling of these differences. The truncated density of the glutamic acid side chain (Glu237) is due to high sensitivity to radiation damage (46). (C) Densities of VR-IV of HBoV1, HBoV3, and HBoV4 with the modeled residues. HBoV4 possesses an insertion of 2 aa at the apex of the loop (see also Fig. 6). The amino acid residues are shown as a stick representation inside a gray mesh density map and colored according to atom type as follows: C, green; O, red; N, blue; S, yellow. This image was generated with PyMOL (73).
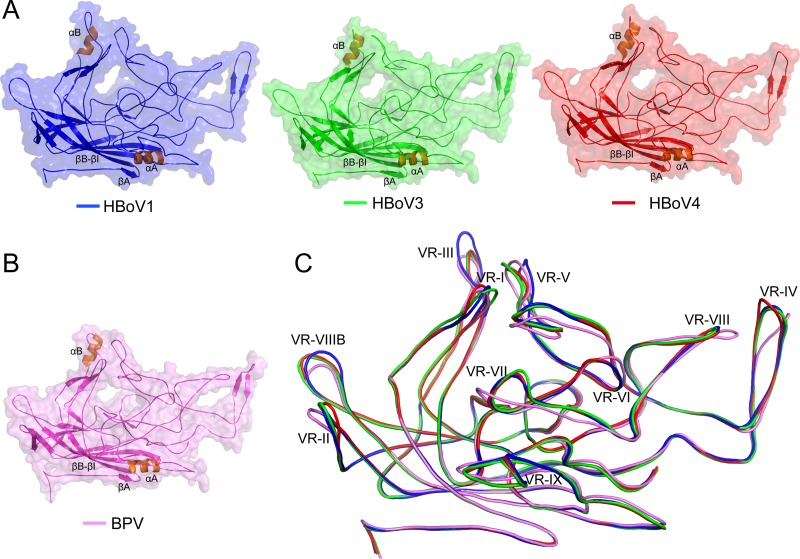
Overall, the VP3 structure of the HBoVs conserves the general topology of other parvoviruses: a βA strand, an eight-stranded β barrel formed by strands βB to βI, the α helix (αA), and large loops inserted between these secondary structure elements (Fig. 5A and 6). The loops, named after the β strands between which they are inserted, for example, the GH loop between βG and βH, form the surface features of the capsid. These features are similar to those reported for other parvoviruses (39). The bocavirus VPs contain a second α helix (αB) in the EF surface loop, which is not observed in most other parvoviruses. A similar helix was previously described in BPV (Fig. 5B), another bocaparvovirus (39). This observation suggests a genus level function for αB, as well as the density extension under the 5-fold axes.
FIG 5.
The bocaparvovirus VP3 structure. (A) Ribbon diagrams of the HBoV VP3 structures within a transparent surface representation. The conserved β-barrel core motif (βB-βI), βA, and the αA and αB helices (orange) are indicated. The loops inserted between these secondary structure elements also contain β-strand regions, as indicated (see also Fig. 6). (B) Ribbon diagram of the BPV VP3 structure displayed as in panel A. (C) Structural superposition of HBoV1 (blue), HBoV3 (green), HBoV4 (red), and BPV (pink) VP3 shown as coil diagrams with the positions of VR-I to VR-X indicated. This image was generated with PyMOL (73).
FIG 6.
Structure-based sequence alignment of HBoV1, HBoV3, HBoV4, and BPV. Secondary structure elements, β strands and α helices, are indicated by blue arrows and red cylinders, respectively. Orange highlighting indicates sequence identity among the HBoVs and BPV. VR locations are indicated. Amino acid numbering, based on the HBoV1 sequence, is shown above the alignment. Amino acids whose Cα atoms are >2 Å apart when each virus VP3 Cα atom is superposed onto HBoV1 are shown offset below the aligned residues.
Comparison of the HBoVs identified strain level differences in the VRs that confer their unique surface topology.
The overall VP3 sequence identity range among HBoV1, HBoV3, and HBoV4 is ∼78 to 91%, with HBoV3 and HBoV4 being the most similar at 91%. The structural alignment resulted in Cα root mean square deviations (RMSDs) of 0.88 Å (498 residues aligned and six gaps), 0.97 Å (480 residues aligned and eight gaps), and 0.86 Å (484 residues aligned and four gaps) for pairwise comparisons of HBoV1 and HBoV3, HBoV1 and HBoV4, and HBoV3 and HBoV4, respectively. Thus, while only minor, structurally, the main chains of HBoV1 and HBoV4 differ the most. The β barrel and αA helix secondary structural elements that form the core of the capsid and the wall of the 2-fold depression, respectively, are conserved in sequence and structure in all three HBoVs (Fig. 5C and 6). In contrast, the surface loops demonstrate variability between the viruses in the superposed structures (Table 2; Fig. 5C to 7). The largest differences are localized to VR-I, VR-II, VR-III, VR-IV, VR-V, and VR-VIIIB. Among the human bocaviruses, HBoV1 is more divergent at VR-III, VR-V, and VR-VIIIB and HBoV4 is more divergent at VR-II and VR-IV (Fig. 5C and 7). These differences cluster on the capsid surface at the 2-fold, 2/5-fold wall (between the depressions at the 2- and 5-fold axes), the 3-fold protrusions, the 5-fold channel, and the floor of the 5-fold depression to create local capsid surface variations between the viruses (Fig. 2A and 8).
TABLE 2.
Range of Cα distances for the aligned bocaparvoviruses in the VRs
| Comparison | Range of Cα distances (Ä) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VR-I | VR-II | VR-III | VR-IV | VR-V | VR-VI | VR-VII | VR-VRIII | VR-VIIIB | VR-IX | |
| HBoV1 vs HBoV3 | 0.6–5.2 | 0.1–2.2 | 0.2–4.9 | 0.2–1.8 | 0.5–6.6 | 0.2–1.1 | 0.7–2.9 | 0.4–1.3 | 0.3–4.7 | 0.4–2.5 |
| HBoV1 vs HBoV4 | 0.1–4.4 | 0.4–3.5 | 0.5–4.3 | 0.3–5.6 | 0.5–5.5 | 0.4–1.2 | 0.8–1.8 | 0.3–1.9 | 0.5–3.9 | 0.6–1.9 |
| HBoV1 vs BPV | 0.3–4.0 | 0.8–4.4 | 0.3–6.8 | 0.5–9.5 | 0.7–6.4 | 0.4–3.3 | 1.1–8.7 | 1.0–5.2 | 0.7–4.1 | 1.2–9.9 |
| HBoV3 vs HBoV4 | 0.6–6.1 | 0.3–2.1 | 0.1–1.2 | 0.2–3.9 | 0.4–2.8 | 0.2–1.1 | 0.5–2.0 | 0.4–2.3 | 0.3–1.6 | 0.4–1.7 |
| HBoV3 vs BPV | 0.5–4.6 | 0.4–5.7 | 0.4–5.3 | 0.2–9.8 | 0.6–4.5 | 0.4–2.7 | 1.7–11.1 | 0.6–6.3 | 0.4–4.9 | 0.7–9.9 |
| HBoV4 vs BPV | 0.2–3.0 | 0.4–7.3 | 0.5–5.6 | 0.5–10.7 | 0.9–4.4 | 0.9–3.1 | 0.5–10.1 | 1.0–4.2 | 0.4–4.9 | 1.0–10.1 |
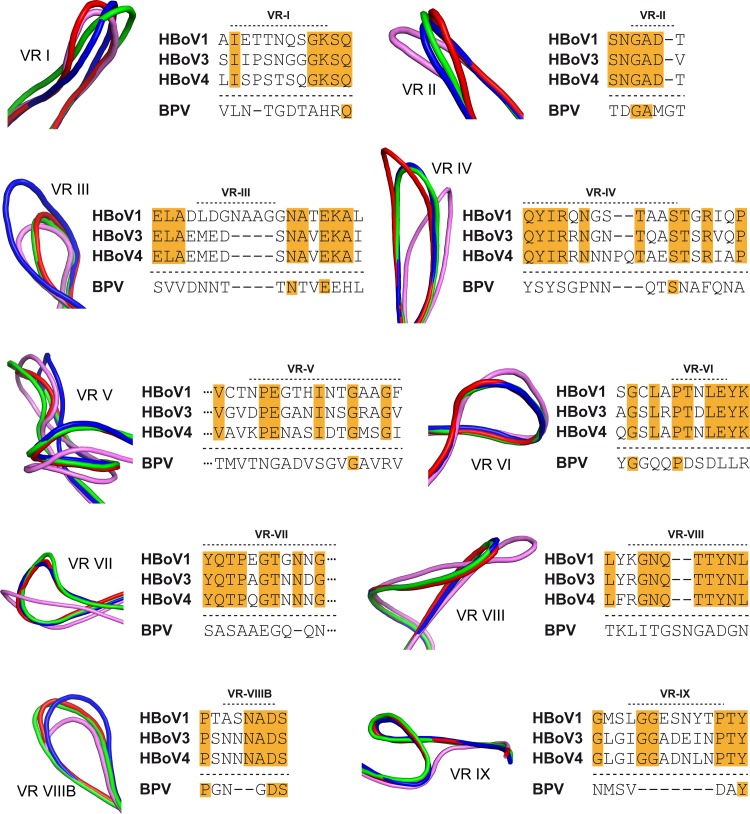
FIG 7.
The bocaparvovirus VP3 VRs. Shown are closeup views of the VRs when HBoV1 (blue), HBoV3 (green), HBoV4 (red), and BPV (pink) are compared. Next to each VR, a sequence alignment of the loops of the different bocaviruses is shown. Orange highlighting indicates identical residues among the HBoVs. The VR structure images were generated with PyMOL (73).
FIG 8.
Surface representations of HBoV capsids. (A, B) 3D capsid surface representations generated with PyMol (37) (A) of the HBoV capsid models viewed down the 2-fold axis and Roadmap projections generated with the RIVEM program (74) (B). For the roadmap image, amino acid residues that are exposed on the capsid exterior are visible and are indicated by the three-letter code and their number. The viral asymmetric unit, with the 5-fold axis (pentagon), the 2-fold axis (ellipse), and the two 3-fold axes (triangles), is shown. In both panels, the VRs are colored as indicated in the legend at the bottom.
The amino acids in VR-I (BC loop) differs significantly among the HBoVs, resulting in a conformational difference in the VPs of the viruses (Fig. 5C and 7). This VR, along with VR-III, VR-VII, and VR-IX, forms the 2/5-fold wall (Fig. 8), a capsid region shown to be important for antigenic reactivity and infectivity in other parvoviruses and strain-specific antibody recognition for HBoV1 (30, 34, 48). Five DE loops, containing VR-II at their apex, form the channel at the 5-fold symmetry axis of the capsid (Fig. 5 and 8) and are proposed as the portals for VP1u externalization and genomic DNA packaging and uncoating for the parvoviruses. The critical nature of this function is reflected in the conserved sequence and structure of VR-II among the HBoVs, with the minor flexibility observed likely required to enable the externalization/extrusion functions of the 5-fold channel (Fig. 7). Consistent with its conservation, VR-II is part of the footprint for a cross-reactive antibody that recognizes HBoV1, HBoV2, and HBoV4 (34). In contrast, VR-III is highly divergent in HBoV1 compared to HBoV3 and HBoV4 because of an insertion of 4 aa at the apex of the loop (within the EF loop) (Fig. 5 to 7). Residues within VR-III are part of an HBoV1-specific antigenic region (34). Furthermore, regions analogous to VR-III serve as determinants of tissue tropism, pathogenicity, transduction efficiency, and antibody recognition in other parvoviruses (30, 48). The structural diversity of HBoV1 at VR-III, a respiratory tract-infecting strain, compared to HBoV3 and HBoV4, which predominantly infect the gastrointestinal tract, suggests a possible role for this VR in dictating the tissue tropism of HBoVs. VR-IV, which forms part of the 3-fold protrusions, is similar in HBoV1 and HBoV3 but differs in HBoV4. The latter virus contains a larger loop because of a 2-aa insertion and adopts a different conformation that contributes to the “spikier” appearance of the protrusions surrounding its 3-fold axis (Fig. 2A, 5C, and 7). This loop region is a determinant of antibody recognition for other parvoviruses such as adeno-associated virus (AAV), human parvovirus B19, and Aleutian mink disease parvovirus (30, 49–51). The loop that corresponds to VR-V displays significant differences at both the sequence and structure levels among all three viruses (Fig. 7). VR-V, a component of the protrusions surrounding the 3-fold symmetry axis, is part of an HBoV1-specific antibody footprint and has been shown to be important for transduction efficiency and antigenicity in other parvoviruses (34, 52–54). VR-VI, VR-VII, and VR-VIII are structurally similar among the HBoVs and show only minor amino acid differences (Fig. 7). These loops are located on the side of the 3-fold protrusions (Fig. 8). These regions have also been associated with a role in parvovirus infectivity and antigenic reactivity, including HBoV1 (30, 34, 48). For VR-VIIIB, which is the HI loop that lines the floor of the depression surrounding the 5-fold channel, the HBoV1 conformation is the most structurally divergent (Fig. 7 and 8). However, this region forms part of the footprint of an antibody generated against HBoV1 that cross-reacts with HBoV2 and HBoV4 (34). Thus, its role as part of this cross-reactive epitope remains to be determined. This region of the parvovirus capsid has been reported to form antigenic epitopes for other parvoviruses and plays a role in genome packaging for the AAVs (49, 53–55). Finally, VR-IX, located at the icosahedral 2-fold axis of the capsid, is structurally identical in the HBoVs despite sequence differences (Fig. 7). This region is implicated in receptor attachment and transduction efficiency in the protoparvovirus minute virus of mice and in the dependoparvovirus AAV, respectively (56, 57).
Genus level structural differences exaggerate diversity in the VRs.
BPV shares ∼46% sequence identity with the HBoVs. Superposition of the BPV crystal structure onto the HBoV crystal structure resulted in Cα RMSDs ranging between 1.28 and 1.37 Å with conservation of the core secondary structure elements, including αB (Fig. 5). However, the conformations of the surface loops differed significantly (Fig. 5C and 7). This observation is consistent with the differences seen in the amino acid sequences within the surface loops (Fig. 6 and 7). Interestingly, VR-III, one of the most divergent VRs of the HBoVs, as discussed above, is structurally more similar between HBoV3 and -4 and BPV because of an insertion of 4 aa in HBoV1. The former three viruses show predominant tropism for the gastrointestinal tract, while HBoV1 infects the respiratory tract, again pointing to the potential role of this capsid region as a tissue tropism determinant for bocaparvoviruses. The most striking difference between the HBoVs and BPV is observed in VR-IX, which is absent in BPV compared to the HBoVs, which have a 7-aa insertion (Fig. 7). This VR is structurally similar in all three HBoVs and suggests a potential role as a host determinant for the bocaparvoviruses. BPV also adopts unique loop conformations compared to the HBoVs in VR-VI to VR-VIII, which may also contribute to host tropism. As mentioned above, all of the HBoVs (Fig. 2B) and BPV display a density extension under the 5-fold axis that extends the 5-fold channel into the capsid interior (39). Once the role of this genus level feature is understood, its function may become a potential target for the control of bocaparvovirus infections.
Utilization of the HBoV structure information for HBoV-based gene therapy vectors.
Similar to recombinant AAVs, efforts to develop HBoV capsids as vectors for potential gene therapy applications are under way (58). The utilization of HBoV vectors is advantageous, especially for the treatment of respiratory genetic diseases by using HBoV1 vectors and the treatment of gastrointestinal diseases by using HBoV2 to HBoV4 vectors by taking advantage of their natural tissue tropism. Another advantage of the HBoVs is the higher genome packaging capacity of ∼5.3 kb, which is larger than that of AAVs (∼4.7 kb). In the last decade, the capsids of many AAV serotypes have been engineered for improved vector efficacy on the basis of their structural information. These modifications include the mutation of capsid surface-exposed serines, threonines, tyrosines, and lysines to prevent phosphorylation and subsequent degradation following cellular entry (59, 60). Furthermore, peptides have been inserted into various surface loops to improve or alter the tropism of these vectors (61) and mutations are introduced into the capsid surface to generate AAV variants that escape preexisting neutralizing antibodies (62). The high-resolution HBoV structures presented in this report will allow similar modifications of HBoV vectors to improve their application in human gene therapy.
Summary.
This study produced the highest-resolution structures of HBoVs determined to date. Their VP and capsid structures exhibit the conserved features reported for other parvoviruses but contain an extended 5-fold axis region and an additional α helix (αB) that are unique, suggestive of genus level functions. Comparative structural analysis of these viruses suggests a previously identified VR, VR-III, as a determinant of tissue tropism, while comparison to the previously reported BPV structure identified VR-VI to VR-VIII and VR-IX as possible host range determinants. Other regions showing structural variability localize to previously identified functional regions of parvovirus capsids, including those controlling tissue and host tropism, pathogenicity, and antigenicity. This study thus provides a basis for further functional mapping of the bocaparvoviruses to generate data to help develop strategies to control virus infection.
MATERIALS AND METHODS
HBoV VLP expression and purification.
Protocols for the expression of HBoV VLPs with the baculovirus-Spodoptera frugiperda (Sf9) cell system, along with methods for purification and quality control of protein purity and capsid integrity, have been previously reported (34). Briefly, recombinant baculoviruses expressing VP3 of HBoVs were generated and used to infect Sf9 cells at a multiplicity of infection of 5. The infected cells were harvested 3 days postinfection, and the VLPs were purified from lysed cells and the medium supernatant by sucrose cushion and sucrose density gradient ultracentrifugation. The HBoV samples were dialyzed into 1× phosphate-buffered saline, concentrated, and analyzed by SDS-PAGE and negative-stain electron microscopy (EM) (Spirit; FEI Co.) to verify VLP purity and integrity, respectively.
Cryo-EM and data collection.
Three-microliter aliquots of HBoV1, HBoV3, and HBoV4 VLPs (∼0.2 to 1 mg/ml) were applied to C-flat holey carbon grids (Protochips, Inc.) following glow discharge treatment to increase their hydrophilicity and vitrified with a Vitrobot Mark IV (FEI Co.). These grids were screened for suitability for cryo-data collection with a 16-megapixel charge-coupled device camera (Gatan, Inc.) in a Tecnai G2 F20-TWIN transmission electron microscope operated at 200 kV under low-dose conditions (∼20 e−/Å2) prior to data collection on a Titan Krios electron microscope (FEI Co.) operated at 300 kV with a DE20 (Direct Electron) direct electron detector. The data collection parameters are summarized in Table 1. Frame alignment of the micrographs was performed with the DE_process_frames software (Direct Electron). For the alignment of each data set, the individual parameters of the data collection and the corresponding dark and bright reference images were used without radiation dose damage compensation (63). Data collection for HBoV2 was also attempted in this study. However, problems with aggregation and preference for the carbon surface rather than holes in the grids inhibited these efforts. Strategies to overcome these problems are under way.
Structure determination by 3D reconstruction.
The RobEM subroutine in the AUTO3DEM software suite (64) was used to extract individual particles from the recorded micrographs, followed by normalization with AUTO3DEM. Estimations of the defocus values for the micrographs were made with the ctffind4 subroutine (65) in AUTO3DEM to enable correction of the microscope-related contrast transfer functions (CTFs) in AUTO3DEM. Low-resolution (30-Å) cryo-reconstructed initial model maps were generated from the images of 100 particles by an ab initio random model method while imposing icosahedral symmetry (66). This initial model was used to determine the orientations and origins of each particle, followed by multiple refinement steps (particle recentering, solvent flattening, and individual particle CTF refinement) for the entire set of particle images with AUTO3DEM. The gold standard protocol was utilized throughout the structure determination process. The resolutions of the reconstructed maps are reported on the basis of FSC threshold criteria of 0.143 and 0.5. The numbers of particles used to compute the final density maps are listed in Table 1. An inverse temperature factor of 1/100 Å2 was applied to each of the final reconstructed density maps to sharpen the high-resolution features following manual screening of a range of 1/100 to 1/500 Å2 and inspection of the sharpened maps. A noise suppression factor was applied in AUTO3DEM to avoid amplification of noise in the density maps.
Model building and structure refinement.
A 3D homology model of HBoV1 VP3 was generated from the protein sequence (NCBI accession number: AKG93794.1) in UCSF-CHIMERA with the crystal structure of BPV (Research Collaboratory for Structural Bioinformatics [RCSB] PDB code 4QC8) as the template (67). An icosahedral capsid model based on 60 copies of the VP3 model was generated with VIPERdb2 (47). The 60-mer capsid model was docked into the HBoV1 cryo-reconstructed density map by rigid-body rotations and translations by using the “fit in map” subroutine in UCSF-Chimera (68), which uses a CC calculation to assess the quality of the fit between the map generated from the model and the cryo-reconstructed map. The pixel size of the cryo-reconstructed map was adjusted to obtain the best fit with the 60-mer model, and the cryo-reconstructed map was filtered to the new pixel size by using the e2proc3d.py subroutine in EMAN2 to enable interactive model adjustment in the Coot program (Table 1) (69, 70). The high resolution of the HBoV1 density map, at 2.9 Å, enabled the interpretation of individual amino acid positions. The reference VP3 monomer model was adjusted into the density map guided by the amino acid sequence by using interactive model building and the real-space-refine options available in Coot (69). An approach similar to that described for HBoV1 was used to build the VP3 and capsid models for HBoV3 (accession no. YP_002808457.1) and HBoV4 (NCBI accession no. YP_002916063.1). After the VP3 residues were built into the density maps of HBoV1, HBoV3, and HBoV4, the models were refined with PHENIX (http://www.phenix-online.org/documentation/reference/real_space_refine.html) (71). The real-space refinement subroutine was run with the default settings for five macrocycles. Subsequently, the newly generated model was inspected with Coot. Side chain modifications were made, if necessary, followed by real-space refinement. A 60-mer of the resulting model was further refined by using the rigid-body and B-factor refinement options in PHENIX. The final statistics for each refined HBoV model are listed in Table 1.
Strain and genus level structure comparisons.
The Cα atoms of the VP3 atomic coordinates of HBoV1, HBoV3, and HBoV4 were superposed with the secondary structure matching (SSM) program in Coot (72). For a genus level comparison, the HBoV1 VP3 Cα atom coordinates were superposed onto those of the BPV VP3 crystal structure (RCSB PDB code 4QC8) with the same program. The SSM subroutine generated RMSDs for the superposed structures and calculated the distances between the aligned Cα positions that were used to identify regions of variability between the aligned structures. VRs were identified as previously defined for autonomously replicating viruses (as regions with two or more consecutive residues with an RMSD of >2.0 Å between the superposed structures) (38). Cartoon representations of the VP3 structures and side chain density images were generated with the PyMOL program (73). Surface representations were generated in the UCSF-Chimera program (68).
Accession number(s).
The final cryo-reconstructed maps and atomic models of HBoV1, HBoV3, and HBoV4 have been deposited in the EMDB (accession numbers EMD-8598, EMD-8604, and EMD-8605), as well as the PDB (ID codes 5URF, 5US7, and 5US9), respectively.
ACKNOWLEDGMENTS
We thank the University of Florida (UF) Interdisciplinary Center for Biotechnology Research (ICBR) EM lab for providing negative-stain EM and cryo-EM screening services.
Data collection at Florida State University was made possible by NIH grants S10 OD018142-01 (purchase of a direct electron camera for the Titan-Krios at FSU [P. I. Taylor]), S10 RR025080-01 (purchase of a FEI Titan Krios for 3D EM [P. I. Taylor]), and U24 GM116788 (the Southeastern Consortium for Microscopy of MacroMolecular Machines [P. I. Taylor]). Financial support was provided by the UF Division of Sponsored Research and the College of Medicine to establish EM facilities at the UF ICBR and the research efforts at the University of Florida, as well as the Sigrid Jusélius Foundation and the Research Funds of the University of Helsinki, Helsinki Finland. This work was also partially support by NIH grant GM-033050 (T.S.B.).
REFERENCES
- 1.Abinanti FR, Warfield MS. 1961. Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virology 14:288–289. doi: 10.1016/0042-6822(61)90206-9. [DOI] [PubMed] [Google Scholar]
- 2.Binn LN, Lazar EC, Eddy GA, Kajima M. 1970. Recovery and characterization of a minute virus of canines. Infect Immun 1:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmichael LE, Schlafer DH, Hashimoto A. 1994. Minute virus of canines (MVC, canine parvovirus type-1): pathogenicity for pups and seroprevalence estimate. J Vet Diagn Invest 6:165–174. doi: 10.1177/104063879400600206. [DOI] [PubMed] [Google Scholar]
- 4.Carmichael LE, Schlafer DH, Hashimoto A. 1991. Pathogenicity of minute virus of canines (MVC) for the canine fetus. Cornell Vet 81:151–171. [PubMed] [Google Scholar]
- 5.Jordan EK, Sever JL. 1994. Fetal damage caused by parvoviral infections. Reprod Toxicol 8:161–189. doi: 10.1016/0890-6238(94)90023-X. [DOI] [PubMed] [Google Scholar]
- 6.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog 5:e1000391. doi: 10.1371/journal.ppat.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis 201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnott A, Vong S, Rith S, Naughtin M, Ly S, Guillard B, Deubel V, Buchy P. 2013. Human bocavirus amongst an all-ages population hospitalised with acute lower respiratory infections in Cambodia. Influenza Other Respir Viruses 7:201–210. doi: 10.1111/j.1750-2659.2012.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo L, Wang Y, Zhou H, Wu C, Song J, Li J, Paranhos-Baccala G, Vernet G, Wang J, Hung T. 2012. Differential seroprevalence of human bocavirus species 1-4 in Beijing, China. PLoS One 7:e39644. doi: 10.1371/journal.pone.0039644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapoor A, Hornig M, Asokan A, Williams B, Henriquez JA, Lipkin WI. 2011. Bocavirus episome in infected human tissue contains non-identical termini. PLoS One 6:e21362. doi: 10.1371/journal.pone.0021362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissbrich B, Neske F, Schubert J, Tollmann F, Blath K, Blessing K, Kreth HW. 2006. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect Dis 6:109. doi: 10.1186/1471-2334-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schildgen O, Muller A, Allander T, Mackay IM, Volz S, Kupfer B, Simon A. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev 21:291–304, table of contents. doi: 10.1128/CMR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smuts H, Workman L, Zar HJ. 2008. Role of human metapneumovirus, human coronavirus NL63 and human bocavirus in infants and young children with acute wheezing. J Med Virol 80:906–912. doi: 10.1002/jmv.21135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streiter M, Malecki M, Prokop A, Schildgen V, Lusebrink J, Guggemos A, Wisskirchen M, Weiss M, Cremer R, Brockmann M, Schildgen O. 2011. Does human bocavirus infection depend on helper viruses? A challenging case report. Virol J 8:417. doi: 10.1186/1743-422X-8-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi EH, Lee HJ, Kim SJ, Eun BW, Kim NH, Lee JA, Lee JH, Song EK, Kim SH, Park JY, Sung JY. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000–2005. Clin Infect Dis 43:585–592. doi: 10.1086/506350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canducci F, Debiaggi M, Sampaolo M, Marinozzi MC, Berre S, Terulla C, Gargantini G, Cambieri P, Romero E, Clementi M. 2008. Two-year prospective study of single infections and co-infections by respiratory syncytial virus and viruses identified recently in infants with acute respiratory disease. J Med Virol 80:716–723. doi: 10.1002/jmv.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edner N, Castillo-Rodas P, Falk L, Hedman K, Söderlund-Venermo M, Allander T. 2012. Life-threatening respiratory tract disease with human bocavirus-1 infection in a 4-year-old child. J Clin Microbiol 50:531–532. doi: 10.1128/JCM.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Körner RW, Söderlund-Venermo M, van Koningsbruggen-Rietschel S, Kaiser R, Malecki M, Schildgen O. 2011. Severe human bocavirus infection, Germany. Emerg Infect Dis 17:2303–2305. doi: 10.3201/eid1712.110574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ursic T, Steyer A, Kopriva S, Kalan G, Krivec U, Petrovec M. 2011. Human bocavirus as the cause of a life-threatening infection. J Clin Microbiol 49:1179–1181. doi: 10.1128/JCM.02362-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vries JJ, Bredius RG, van Rheenen PF, Smiers FJ, Scholvinck EH, Vossen AC, Claas EC, Niesters HG. 2009. Human bocavirus in an immunocompromised child presenting with severe diarrhea. J Clin Microbiol 47:1241–1243. doi: 10.1128/JCM.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ergul AB, Altug U, Aydin K, Guven AS, Altuner Torun Y. 1 January 2017. Acute necrotizing encephalopathy causing human bocavirus. Neuroradiol J doi: 10.1177/1971400916687586:1971400916687586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moesker FM, van Kampen JJ, van der Eijk AA, van Rossum AM, de Hoog M, Schutten M, Smits SL, Bodewes R, Osterhaus AD, Fraaij PL. 2015. Human bocavirus infection as a cause of severe acute respiratory tract infection in children. Clin Microbiol Infect 21:964.e1-8. doi: 10.1016/j.cmi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Söderlund-Venermo M, Lahtinen A, Jartti T, Hedman L, Kemppainen K, Lehtinen P, Allander T, Ruuskanen O, Hedman K. 2009. Clinical assessment and improved diagnosis of bocavirus-induced wheezing in children, Finland. Emerg Infect Dis 15:1423–1430. doi: 10.3201/eid1509.090204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning A, Russell V, Eastick K, Leadbetter GH, Hallam N, Templeton K, Simmonds P. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J Infect Dis 194:1283–1290. doi: 10.1086/508219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fry AM, Lu X, Chittaganpitch M, Peret T, Fischer J, Dowell SF, Anderson LJ, Erdman D, Olsen SJ. 2007. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis 195:1038–1045. doi: 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jartti T, Hedman K, Jartti L, Ruuskanen O, Allander T, Söderlund-Venermo M. 2012. Human bocavirus—the first 5 years. Rev Med Virol 22:46–64. doi: 10.1002/rmv.720. [DOI] [PubMed] [Google Scholar]
- 28.Hamza IA, Jurzik L, Wilhelm M, Uberla K. 2009. Detection and quantification of human bocavirus in river water. J Gen Virol 90:2634–2637. doi: 10.1099/vir.0.013557-0. [DOI] [PubMed] [Google Scholar]
- 29.Iaconelli M, Divizia M, Della Libera S, Di Bonito P, La Rosa G. 2016. Frequent detection and genetic diversity of human bocavirus in urban sewage samples. Food Environ Virol 8:289–295. doi: 10.1007/s12560-016-9251-7. [DOI] [PubMed] [Google Scholar]
- 30.Halder S, Ng R, Agbandje-McKenna M. 2012. Parvoviruses: structure and infection. Future Virol 7:253–278. doi: 10.2217/fvl.12.12. [DOI] [Google Scholar]
- 31.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou W, Cheng F, Shen W, Engelhardt JF, Yan Z, Qiu J. 2016. Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J Virol 90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurda BL, Parent KN, Bladek H, Sinkovits RS, DiMattia MA, Rence C, Castro A, McKenna R, Olson N, Brown K, Baker TS, Agbandje-McKenna M. 2010. Human bocavirus capsid structure: insights into the structural repertoire of the parvoviridae. J Virol 84:5880–5889. doi: 10.1128/JVI.02719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kailasan S, Garrison J, Ilyas M, Chipman P, McKenna R, Kantola K, Söderlund-Venermo M, Kucinskaite-Kodze I, Zvirbliene A, Agbandje-McKenna M. 2016. Mapping antigenic epitopes on the human bocavirus capsid. J Virol 90:4670–4680. doi: 10.1128/JVI.02998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zádori Z, Szelei J, Lacoste MC, Li Y, Gariepy S, Raymond P, Allaire M, Nabi IR, Tijssen P. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell 1:291–302. doi: 10.1016/S1534-5807(01)00031-4. [DOI] [PubMed] [Google Scholar]
- 36.Chiu CC, Shi YF, Yang JJ, Hsiao YC, Tzang BS, Hsu TC. 2014. Effects of human parvovirus B19 and bocavirus VP1 unique region on tight junction of human airway epithelial A549 cells. PLoS One 9:e107970. doi: 10.1371/journal.pone.0107970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol 79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kailasan S, Halder S, Gurda B, Bladek H, Chipman PR, McKenna R, Brown K, Agbandje-McKenna M. 2015. Structure of an enteric pathogen, bovine parvovirus. J Virol 89:2603–2614. doi: 10.1128/JVI.03157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stahnke S, Lux K, Uhrig S, Kreppel F, Hosel M, Coutelle O, Ogris M, Hallek M, Buning H. 2011. Intrinsic phospholipase A2 activity of adeno-associated virus is involved in endosomal escape of incoming particles. Virology 409:77–83. doi: 10.1016/j.virol.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 41.Bleker S, Sonntag F, Kleinschmidt JA. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol 79:2528–2540. doi: 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorsch S, Liebisch G, Kaufmann B, von Landenberg P, Hoffmann JH, Drobnik W, Modrow S. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J Virol 76:2014–2018. doi: 10.1128/JVI.76.4.2014-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farr GA, Zhang LG, Tattersall P. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci U S A 102:17148–17153. doi: 10.1073/pnas.0508477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mani B, Baltzer C, Valle N, Almendral JM, Kempf C, Ros C. 2006. Low pH-dependent endosomal processing of the incoming parvovirus minute virus of mice virion leads to externalization of the VP1 N-terminal sequence (N-VP1), N-VP2 cleavage, and uncoating of the full-length genome. J Virol 80:1015–1024. doi: 10.1128/JVI.80.2.1015-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. 2010. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. 2014. Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci U S A 111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL III, Reddy VS. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res 37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 49.Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74:9281–9293. doi: 10.1128/JVI.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chipman PR, Agbandje-McKenna M, Kajigaya S, Brown KE, Young NS, Baker TS, Rossmann MG. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci U S A 93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bloom ME, Martin DA, Oie KL, Huhtanen ME, Costello F, Wolfinbarger JB, Hayes SF, Agbandje-McKenna M. 1997. Expression of Aleutian mink disease parvovirus capsid proteins in defined segments: localization of immunoreactive sites and neutralizing epitopes to specific regions. J Virol 71:705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pulicherla N, Shen S, Yadav S, Debbink K, Govindasamy L, Agbandje-McKenna M, Asokan A. 2011. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol Ther 19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen WJ, Muzyczka N, Olson NH, Sinkovits RS, Chiorini JA, Zolotutkhin S, Kozyreva OG, Samulski RJ, Baker TS, Parrish CR, Agbandje-McKenna M. 2013. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng YS, Gurda BL, Chipman P, McKenna R, Afione S, Chiorini JA, Muzyczka N, Olson NH, Baker TS, Kleinschmidt J, Agbandje-McKenna M. 2015. Adeno-associated virus serotype 1 (AAV1)- and AAV5-antibody complex structures reveal evolutionary commonalities in parvovirus antigenic reactivity. J Virol 89:1794–1808. doi: 10.1128/JVI.02710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiPrimio N, Asokan A, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J Virol 82:5178–5189. doi: 10.1128/JVI.02721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.López-Bueno A, Rubio MP, Bryant N, McKenna R, Agbandje-McKenna M, Almendral JM. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J Virol 80:1563–1573. doi: 10.1128/JVI.80.3.1563-1573.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol 80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan Z, Keiser NW, Song Y, Deng X, Cheng F, Qiu J, Engelhardt JF. 2013. A novel chimeric adenoassociated virus 2/human bocavirus 1 parvovirus vector efficiently transduces human airway epithelia. Mol Ther 21:2181–2194. doi: 10.1038/mt.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong L, Li B, Jayandharan G, Mah CS, Govindasamy L, Agbandje-McKenna M, Herzog RW, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. 2008. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology 381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong L, Li B, Mah CS, Govindasamy L, Agbandje-McKenna M, Cooper M, Herzog RW, Zolotukhin I, Warrington KH Jr, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. 2008. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc Natl Acad Sci U S A 105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Münch RC, Janicki H, Volker I, Rasbach A, Hallek M, Buning H, Buchholz CJ. 2013. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol Ther 21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. 2012. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol 86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spear JM, Noble AJ, Xie Q, Sousa DR, Chapman MS, Stagg SM. 2015. The influence of frame alignment with dose compensation on the quality of single particle reconstructions. J Struct Biol 192:196–203. doi: 10.1016/j.jsb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol 157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. 2012. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol 179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 69.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. 2013. New tool: phenix. real_space_refine. Comput Crystallogr Newsl 4:43–44. [Google Scholar]
- 72.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 73.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 74.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]