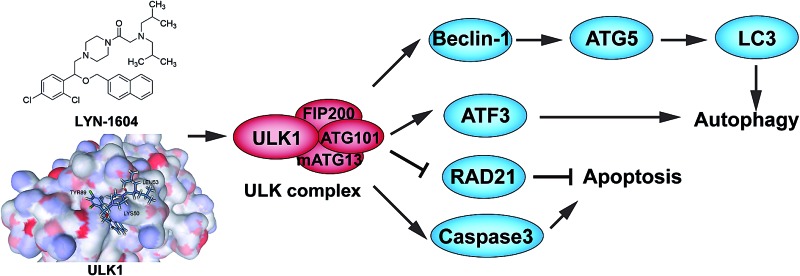
 ULK1 is identified as a target in TNBC; thus a small-molecule agonist is discovered by targeting ULK1-modulated cell death, associated with autophagy and apoptosis.
ULK1 is identified as a target in TNBC; thus a small-molecule agonist is discovered by targeting ULK1-modulated cell death, associated with autophagy and apoptosis.
Abstract
UNC-51-like kinase 1 (ULK1) is well-known to initiate autophagy, and the downregulation of ULK1 has been found in most breast cancer tissues. Thus, the activation of ULK1-modulated autophagy could be a promising strategy for breast cancer therapy. In this study, we found that ULK1 was remarkably downregulated in breast cancer tissue samples by The Cancer Genome Atlas (TCGA) analysis and tissue microarray (TMA) analysis, especially in triple negative breast cancer (TNBC). To design a ULK1 agonist, we integrated in silico screening and chemical synthesis to acquire a series of small molecule candidates. After rounds of kinase and anti-proliferative activity screening, we discovered the small molecule, LYN-1604, to be the best candidate for a ULK1 agonist. Additionally, we identified that three amino acid residues (LYS50, LEU53, and TYR89) were key to the activation site of LYN-1604 and ULK1 by site-directed mutagenesis and biochemical assays. Subsequently, we demonstrated that LYN-1604 could induce cell death, associated with autophagy by the ULK complex (ULK1-mATG13-FIP200-ATG101) in MDA-MB-231 cells. To further explore LYN-1604-induced autophagic mechanisms, we found some potential ULK1 interactors, such as ATF3, RAD21, and caspase3, by performing comparative microarray analysis. Intriguingly, we found that LYN-1604 induced cell death involved in ATF3, RAD21, and caspase3, accompanied by autophagy and apoptosis. Moreover, we demonstrated that LYN-1604 has potential for good therapeutic effects on TNBC by targeting ULK1-modulated cell death in vivo; thus making this ULK1 agonist a novel potential small-molecule drug candidate for future TNBC therapy.
Introduction
Macroautophagy (hereinafter referred as autophagy) is a highly conserved cellular self-digestion process by which cellular components are targeted by lysosomes for degradation.1 Autophagy is normally a cellular stress response and a quality control mechanism to maintain cell survival under stress conditions since it may provide energy and metabolic precursors, as well as get rid of some damaged proteins or organelles.2 Increasing evidence for cell death by autophagy is currently emerging.3,4 Although accurate mechanisms to distinguish the role of autophagy in cytoprotection or cell death still need to be addressed, there is evidence showing that both excessive activation of autophagy and the selective removal of autophagy substrates will lead to cell death.5–7
Defects in the autophagic machinery, such as those imposed by the heterozygous deletion of Beclin-1 or the homozygous loss of autophagy-related 5 (ATG5), predispose mice to malignant transformation, especially in breast cancer.8 UNC-51 like kinase 1 (ULK1), the mammalian homolog of ATG1, has been well-known as the autophagic initiator that may decide the subsequent cell fate.9 In the autophagic process, ULK1 is a component of the ULK complex (ULK1-mATG13-FIP200-ATG101) that is essential for autophagy induction in different types of cancer, such as breast cancer.10,11 Additionally, characterization of ULK1 expression in breast cancer tissues has identified that low expression of ULK1 is associated with operable breast cancer progression and is thus being regarded as an adverse prognostic marker of survival for patients.12 Besides inhibiting mTOR signaling, the negative regulators of ULK1 may result in the inhibition of breast cancer growth, suggesting the pivotal role of ULK1 in breast cancer.13 As abovementioned, activating ULK1-modulated autophagy will be a therapeutic strategy in breast cancer therapy.
Herein, we found that ULK1 was remarkably downregulated in breast cancer tissues (especially in TNBC) by using TCGA analysis and TMA analysis, suggesting that ULK1 may be a potential new drug target in TNBC. Subsequently, we integrated in silico screening, chemical synthesis, kinase and anti-proliferative activity screening, as well as site-directed mutagenesis, to eventually acquire a potent ULK1 agonist (LYN-1604). Moreover, we demonstrated that LYN-1604 induced cell death via the ULK complex involved in ATF3, RAD21 and caspase3 toward MDA-MB-231 cells. We found that LYN-1604 has therapeutic potential by targeting ULK1-modulated cell death associated with autophagy and apoptosis of TNBC in vivo, which together demonstrate that this ULK1 agonist could be utilized as a new anti-TNBC drug candidate.
Results and discussion
TCGA-guided identification of ULK1 is remarkably downregulated in breast cancer, especially TNBC
TNBC typically has a relatively poor outcome due to its inherently invasive and metastatic clinical behavior; therefore, the discovery of novel targets and relevant small-molecule drugs has been an emerging focus for TNBC therapy. To explore the function of ULK1 in TNBC, we investigated The Cancer Genome Atlas (TCGA) breast cancer gene expression difference between TNBC (n = 55) and non-TNBC tumors (n = 472). We found that ULK1 expression was downregulated in both non-TNBC breast cancer and TNBC. Interestingly, we demonstrated that ULK expression in TNBC tumors was significantly lower (p = 1.39 × 10–6) than in non-TNBC tumors (Fig. 1A). Consequently, we mapped all remarkably changed autophagic regulators that could be annotated by gene ontology (GO) into the core autophagic protein–protein interaction (PPI) network in TNBC. This network was composed of 203 nodes and 579 interactions. Among them, we found some low expressed autophagic positive regulators (e.g., ULK1, BECN1 and TBK1) that could be identified as hub proteins (e.g., ULK1 has 22 interactors that are more than the average number, 5.695), suggesting that ULK1 may be a hub protein or potential target in the autophagic network in TNBC (Fig. 1B). Moreover, we evaluated the expression of ULK1 in a tissue microarray (TMA) containing 40 samples of BRs with patient-matched adjacent tissues. Immunohistochemical analysis demonstrated decreased expression of ULK1 in breast cancer samples, compared with adjacent normal tissues (Fig. 1C and D). Notably, the expression of ULK1 in TNBC tissues is much lower than in non-TNBC ones, which displays a result consistent with TCGA analysis (p < 0.0001) (Fig. 1D). These results demonstrate that ULK1 is remarkably downregulated in TNBC.
Fig. 1. Identification of ULK1 is remarkably downregulated in TNBC. (A) ULK1 expression is significantly lower in TNBC tumors than in non-TNBC (p = 1.39 × 10–6). (B) In the core autophagic PPI network, ULK1, BECN1 and TBK1 are low expressed positive regulators of autophagy; ULK1 is a hub protein with 22 interactors. (C) Representative immunoreactivity intensities of ULK1 for negative (0), weakly positive (1), positive (2), strongly positive (3) staining. Scale bar = 50 μm. (D) ULK1 expression is downregulated in breast cancer tissues (TNBC: p < 0.0001, non-TNBC: p = 0.0478) compared with normal adjacent tissues (NAT), especially in TNBC (vs. non-TNBC: p = 0.0314).
More recently, the expression levels of ULK1 have been reported to be higher in hepatocellular carcinoma (HCC), nasopharyngeal carcinoma (NPC) and esophageal squamous cell carcinoma (ESCC) than those in adjacent tissues and are significantly associated with aggressive clinical features.14–16 In contrast, both mRNA and protein levels of ULK1 were found to be remarkably decreased in breast cancer tissues, and ULK1 expression is negatively correlated with tumor size, lymph node status and pathological stage, suggesting that decreased expression of ULK1 is associated with breast cancer progression.12 Thus, in our study, based upon TCGA analysis and TMA analysis, we found that ULK1 was downregulated in breast cancer tissue samples, which is especially remarkable in TNBC. In addition, we used a systematic biology network method to construct an autophagic PPI network in TNBC, which revealed ULK1 as a hub protein/potential target, not only at the protein expression level, but also at the PPI network level. Together, these results indicate that activating ULK1 may be a novel strategy for the current TNBC therapy.
Design, screening and synthesis of candidate ULK1 agonists
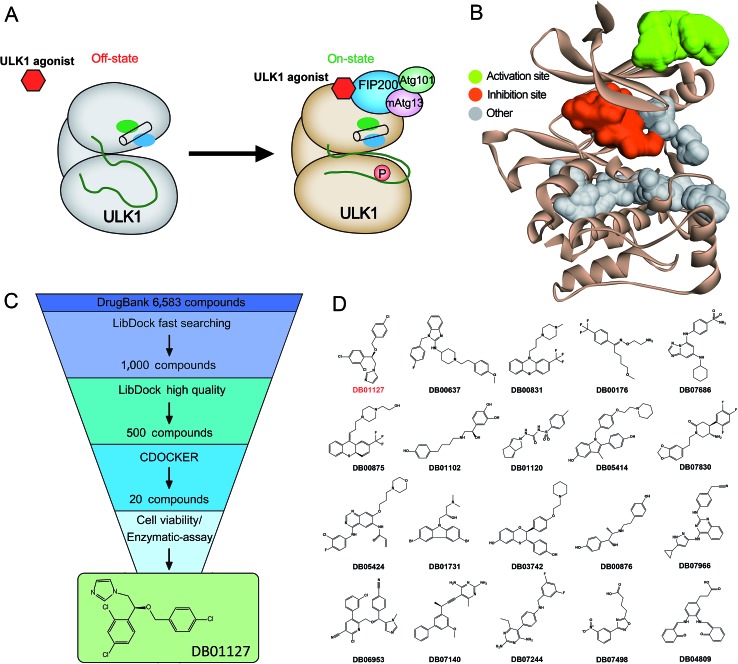
Based on the hypothesis that mATG13 and/or FIP200 bind to hydrophobic pockets of ULK1, thereby switching the conformation of the kinase domain to the on-state, we synthesized agonists to “lock in” their interaction (Fig. 2A). Firstly, we defined 9 candidate binding pockets of the kinase domain of ULK1 (PDB ID ; 4WNP) by Discovery Studio. As shown in Fig. 2B, we selected the nearest pocket (green) of the interaction site to perform the virtual screening of agonists. Three-step molecular docking was performed on FDA-approved drugs from Drugbank, searching for leading compounds (Fig. 2C). The top 500 hits were selected by the LibDock protocol in the first step. Subsequently, the top 20 (U-1–U-20) hits determined by the CDOCKER protocol were selected for in silico screening. As a result, one compound (DB01127) named U-11 (1-(2-((4-chlorophenyl)methoxy)-2-(2,4-dichlorophenyl)ethyl)-1H-imidazol) exhibited the best score (CDOCKER energy 17.6299; CDOCKER interaction energy 25.5461), suggesting that U-11 has priority as a candidate ULK1 agonist. Thus, we selected U-11 as a leading compound for structure modifications and SAR analysis.
Fig. 2. In silico high-throughput screening of candidate small-molecule compounds targeting ULK1. (A) Schematic drawing of the activation mechanism of ULK1 agonist. (B) Candidate binding pockets in the structure of the kinase domain of ULK1. The inhibitor pocket is shown in red and our virtual screening pocket is shown in green. (C) Virtual screening protocol for the identification of the novel ULK1 agonist. (D) The top twenty candidate small-molecule compounds targeting ULK1 from Drugbank.
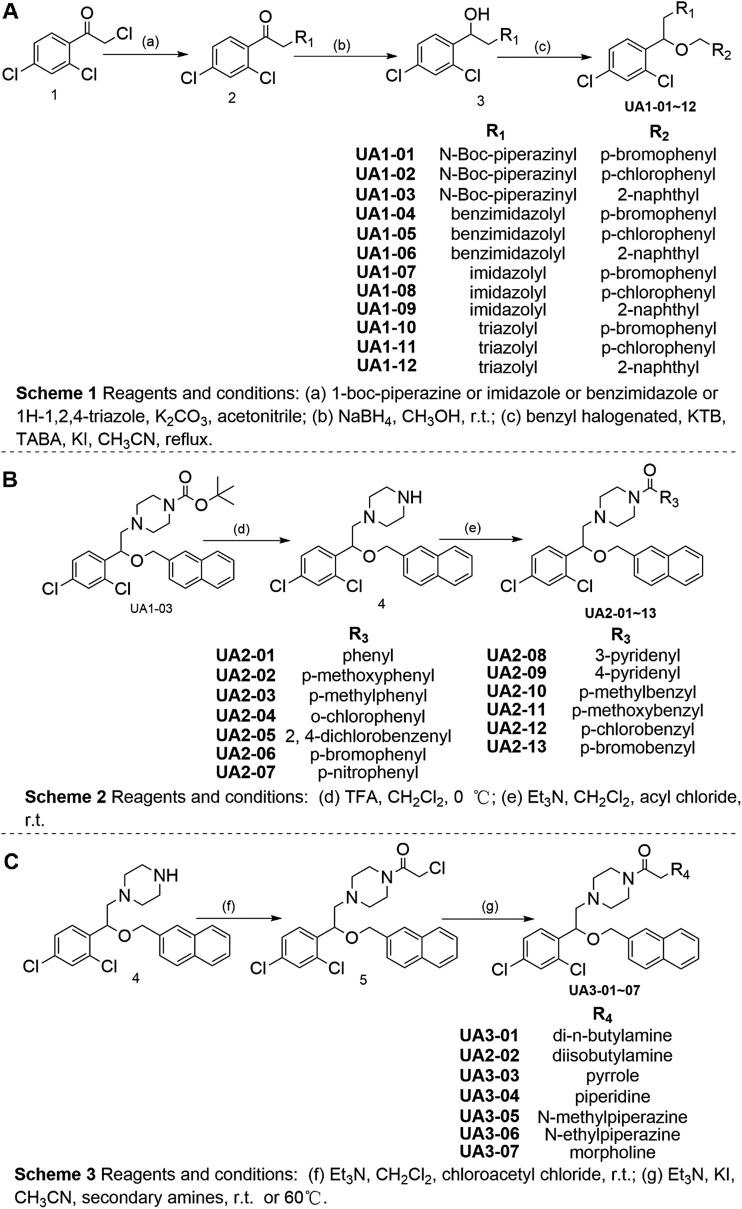
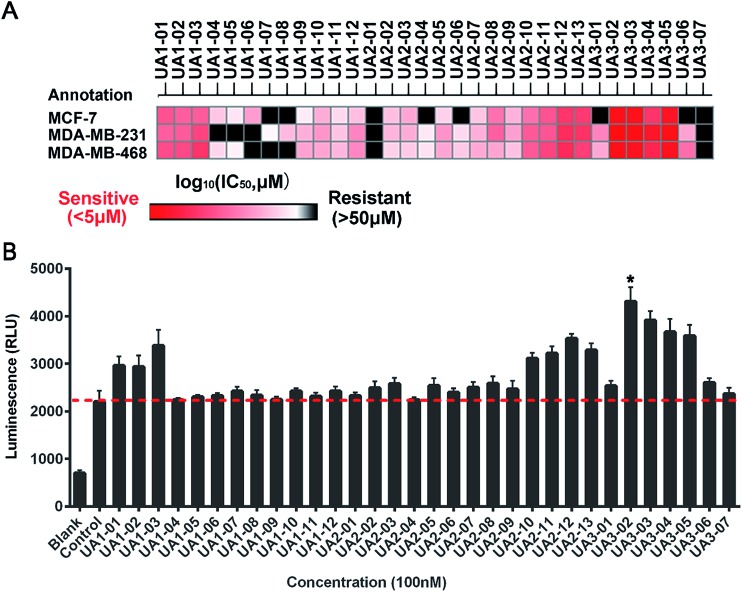
Next, a series of new UA1 derivatives, UA1-01–12, containing different terminal groups (R1 and R2 in Fig. 3 and Scheme 1) were synthesized. On the basis of molecular modeling, the nitrogen atoms of the piperazine ring represent hydrogen bond acceptors in an ideal orientation, which interact with the amino group of the LYS50 side chain and also provide suitable substitution vectors for improving activity. Therefore, the chemical library was enriched by different substituted phenyl or naphthyl-based hydrophobic ligands that were identified as favorable functional groups for biological potency in the first round of screening (Table S1†), and compound UA1-03 exhibited the best activity (enzymatic activity = 153.64% at 100 nM and IC50 = 20.94 μM toward MDA-MB-231 cells). Based on the preliminary results, UA2 analogs, UA2-01–13, containing specific 2-naphthyl and R3 (acylpiperazin-1-yl) were designed and prepared (Fig. 3 and Scheme 2). We found that the existence of methylene in UA2-10–13 played a key role in biological potency, which should be preserved in order to maintain the activity (Table S2†), and the mechanistic enzymatic potency of UA2-10–13 was outstanding (enzymatic activity = 141.37–160.29% at 100 nM). To explore the impact of R2, we used a methylene group to expand the hinge. Additionally, the α-chlorinated ketone group of the scaffold provided a straight forward entry for diversity generation from a list of available amines to afford UA3-01–07 (Fig. 3 and Scheme 3). Bulking up the N-substitution contributed to improving the activity (Table S3†). Compound UA3-02 (hereafter referred to as LYN-1604) showed the best potency, which is a 10-fold improvement over UA1–03. A preferred compound, LYN-1604, was finally found as a potential ULK1 agonist (enzymatic activity = 195.7% at 100 nM and IC50 = 1.66 μM against MDA-MB-231 cells) through three rounds of screening (Fig. 4A and B).
Fig. 3. Chemical synthesis of candidate ULK1 agonists.
Fig. 4. Biological evaluation of candidate ULK1 agonists toward human breast cancer cells. (A) Heat map of mean IC50 values of ULK1 agonists in breast cell lines. Cell viabilities were measured for each compound with various concentrations by MTT assay, the IC50 values were calculated by Prism 6.0. (B) The luminescence (RLU) of ULK1 agonists at 100 nM were determined by ADP-Glo ULK1 kinase assay. Blank: no ULK1 kinase (0% kinase activity), control: containing ULK1 kinase (100% kinase activity). Error bars represent the standard deviations of the results from three independent experiments.
Identification of LYN-1604 as a potent ULK1 agonist
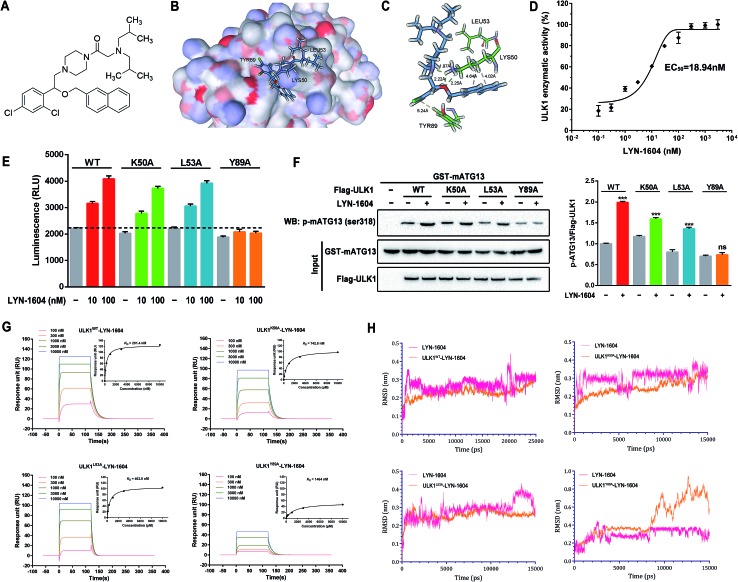
To confirm that LYN-1604 is a ULK1 agonist (Fig. 5A), the ligand structure with the most favorable binding free energies and reasonable orientations was selected as the optimal docked conformation. We found that LYN-1604 could form hydrogen bonds with LYS50 and form hydrophobic interaction with residues LEU53 and TYR89 (Fig. 5B and C). To validate the capacity of activation of ULK1 by LYN-1604, we used the ADP-Glo™ kinase assay to evaluate the EC50 value of LYN-1604. Intriguingly, LYN-1604 displayed a good effect on ULK1 activation with an EC50 (the concentration required to reach 50% of the maximal activation) of 18.94 nM, indicating that LYN-1604 is a potent ULK1 agonist (Fig. 5D). To further determine the binding mode of the activation site between ULK1 and LYN-1604, we generated three ULK1 mutants by site-directed mutagenesis. The lysine residue, leucine residue and tyrosine residue in the binding pocket were mutated to alanine (ULK1K50A, ULK1L53A, ULK1Y89A). ULK1 kinase activity tests indicated that both ULK1K50A and ULK1L53A mutants had less reduced kinase activity in the presence of LYN-1604, compared to the wild-type ULK1 protein (ULK1K50A: –11.93% at 10 nM, –8.73% at 100 nM; ULK1L53A: –3.22% at 10 nM, –4.06% at 100 nM). However, ULK1Y89A was not totally activated by LYN-1604 (Fig. 5E). The in vitro kinase assay revealed that the LYN-1604 increased the phosphorylation of mATG13 at ser318 in wild-type ULK1 transfected HEK-293T cells, indicating that LYN-1604 activates ULK1 in live cells. However, the ULK1K50A and ULK1L53A mutants only resulted in a negligible decrease of mATG13 phosphorylation in the presence of LYN-1604, whereas the ULK1Y89A mutant significantly decreased the phosphorylation of mATG13 (Fig. 5F). In addition, a known ULK1 inhibitor, SBI-0206965, was used to determine the kinase activity induced by LYN-1604. We found that SBI-0206965 (300 nM) could markedly reverse the LYN-1604-induced increase of ULK1 kinase activity (Fig. S1, ESI†). Subsequently, the binding properties of LYN-1604 to wild-type and mutant ULK1 were determined by surface plasmon resonance (SPR) analysis (Fig. 5G). We found that LYN-1604 was bound to wild-type ULK1 with a binding affinity in the nanomole range (K D = 291.4 nM), but the ULK1Y89A mutant protein caused a sharp decrease in binding affinity with lower response and K D than wild-type ULK1, ULK1K50A and ULK1L53A mutants. Moreover, we compared the binding conformation of LYN-1604 with wild-type and mutant ULK1 by molecular docking. Interestingly, LYN-1604 had a remarkable change in binding conformation with ULK1Y89A mutant, compared to wild-type ULK1 and the other two mutants (Fig. S2, ESI†). We then performed a 25 ns MD simulation on the LYN-1604/ULK1 complex. The low root-mean-square deviation (RMSD) fluctuations and the convergence of the energies, temperatures, and pressures of the system indicated that it is a well-behaved system. However, the LYN-1604/ULK1Y89A complex exhibited conformational change after 10 ns simulation, suggesting that TYR89 is a key amino acid residue of ULK1 for binding with LYN-1604 (Fig. 5H); thus, these results indicate that LYN-1604 is a potent ULK1 agonist.
Fig. 5. Identification of LYN-1604 as a potent ULK1 agonist, and its binding mode. (A) Chemical structure of ULK1 agonist LYN-1604. (B and C) The binding mode of LYN-1604 with ULK1. General (B) and detailed (C) views of the interactions between LYN-1604 and ULK1. A detailed view shows that LYN-1604 formed hydrophobic interactions with the binding pocket. (D) ULK1 kinase assay was performed with LYN-1604 to determine the level of ULK1 kinase activity. The luminescence change was normalized to the maximum response induced by LYN-1604 and quantified as 0–100% of ULK1 kinase activity. The EC50 was calculated from the fitted curve. (E) Mutations at LYS50, LEU53 and TYR89 in the binding pocket of ULK1 kinase were performed by site-directed mutagenesis. The wild-type and mutant ULK1 were expressed in insect cells and purified. ULK1 kinase activity was measured in the presence and absence of LYN-1604 using ADP-Glo assay. (F) In vitro kinase assay for the capability of wild type or mutant ULK1 purified from HEK-293T cells to phosphorylate purified mATG13. Phosphorylation of mATG13 at ser318 was detected by a specific antibody and equal inputs of enzymes and substrate are shown on the bottom. (G) Surface plasmon resonance measurements were carried out on a Biacore system to evaluate the binding affinities of wild type or mutant ULK1 to LYN-1604. (H) Molecular dynamics simulation of LYN-1604 binding to wild type or mutant ULK1. The binding conformation was stabilized after a 10 ns simulation, except in the LYN-1604/ULK1Y89A complex.
Recently, some small-molecule agents such as compound 6, SBI-0206965, MRT67307 and MRT68921 have been reported as inhibitors of ULK1; however, these inhibitors have not been widely used for cancer therapy.17–20 In addition, to our knowledge, no small-molecule agonist has been reported to activate ULK1 for potential cancer therapy. Thus, LYN-1604 is a ULK1 agonist with potent kinase activity for ULK1 activation and remarkable anti-proliferative effects against TNBC cell lines. Moreover, we used ACTP, a webserver for predicting potential targets and relevant pathways of autophagy-modulating compounds,21 to predict the potential autophagic target of LYN-1604. We found that ULK1 is the best predicted target of LYN-1604 and prone to activation patterns, which is in a good agreement with our aforementioned results. More interestingly, some ULK1 inhibitors such as compound 6 are designed based on the co-crystal structure of the inhibition pattern and its binding sites are determined as MET92, LYS46, ASP165 and ASN143.17 Unlike these, LYN-1604 was designed based upon the X-ray structure of ULK1, and we speculate that its activating binding sites are LYS50, LEU53 and TYR89. By mutation of these residues, we found TYR89 is a key amino acid residue of ULK1 for binding with LYN-1604. Thus, LYN-1604 as an ULK1 agonist will be a novel small molecule for TNBC therapy.
LYN-1604 induces cell death via the ULK complex in MDA-MB-231 cells
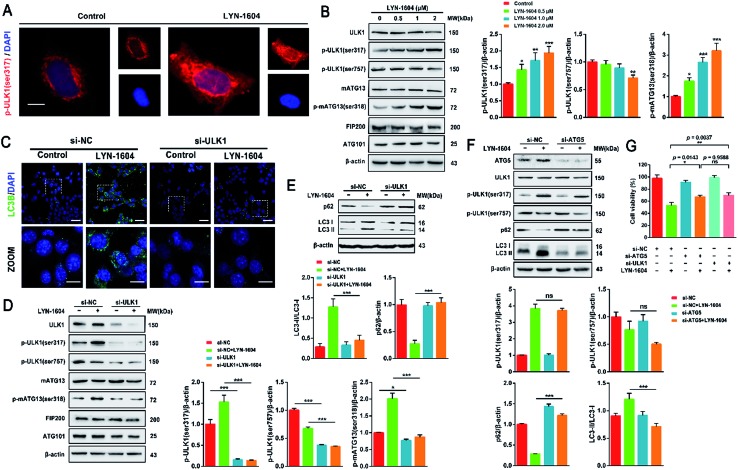
Next, we determined whether LYN-1604-induced cell death is associated with autophagy. The autophagic ratio was evaluated by MDC staining after treatment with LYN-1604. We found that LYN-1604-treated cells displayed a remarkable green fluorescence with MDC staining and the autophagy ratio was increased in a dose-dependent manner (Fig. 6A and B). We also found that LYN-1604 induced remarkable up-regulation of Beclin-1 and degradation of p62, as well as transformation of LC3-I to LC3-II (Fig. 6C). To assess whether LYN-1604-induced autophagy was a continual process, Bafilomycin A1 (BafA1) was used for the detection of p62 and LC3II/LC3I levels. Obvious aggregation of LC3 puncta was observed in the presence of BafA1, compared to LYN-1604-treated cells (Fig. 5D). We also found that BafA1 could increase the accumulation of p62 and LC3II, indicating that LYN-1604-induced autophagy is a continual process (Fig. 6E). To determine whether autophagy plays a role in LYN-1604-induced cell death, the autophagy inhibitor 3-methyladenine (3-MA), which inhibits autophagy by blocking autophagosome formation via inhibiting type III phosphatidylinositol 3-kinases (PI3KIII), was employed to block the induction of autophagy. We found that the cell viability was significantly increased following treatment with 3-MA (p < 0.001) (Fig. 5F). Subsequently, flow cytometry analysis with Annexin-V/PI demonstrated that 3-MA could suppress LYN-1604-induced cell death (Fig. 5G). These results indicate that LYN-1604 induces cell death in MDA-MB-231 cells.
Fig. 6. LYN-1604 induces cell death in MDA-MB-231 cells. (A) MDA-MB-231 cells were treated with 0.5, 1.0 and 2.0 μM LYN-1604, respectively, then stained with MDC and observed by fluorescence microscope. Scale bar: 50 μm (left panel), 20 μm (right panel). (B) MDA-MB-231 cells were treated with 0.5, 1.0 and 2.0 μM LYN-1604, respectively. Then, cells were collected and analyzed by flow cytometry, the MDC positive ratio was quantified. (C) MDA-MB-231 cells were treated with different concentrations of LYN-1604 for 24 h and then the expressions of Beclin-1, p62 and LC3 were detected by western blot analysis. (D) MDA-MB-231 cells were transfected with GFP/mRFP-LC3 plasmid, after co-incubation with 2.0 μM LYN-1604 in the presence or absence of 10 nM Bafilomycin A1 (BafA1), the GFP-LC3 puncta were observed by fluorescence microscope. Scale bar = 20 μm. (E) MDA-MB-231 cells were co-incubated with 2.0 μM LYN-1604 in the presence or absence of 10 nM Bafilomycin A1 (BafA1), then the expression levels of p62 and LC3 were detected. β-Actin was measured as the loading control. (F) MDA-MB-231 cells were treated with 2.0 μM LYN-1604, 3-MA (1 mM) was added 1 h before treatment of LYN-1604. After the above treatment, cell viabilities were detected by MTT assay; ***, p < 0.001 vs. LYN-1604-treated group. (G) MDA-MB-231 cells were treated with 2 μM LYN-1604 for the indicated times, apoptosis ratios were determined by flow cytometry analysis of Annexin-V/PI double staining. 3-MA (1 mM) was added 1 h before treatment of LYN-1604.
Considering that the ULK complex (ULK1-mATG13-FIP200-ATG101) is closely related to autophagosome formation, we detected the expressions of ULK1, mATG13, FIP200 and ATG101 in LYN-1604-treated cells. Firstly, we found that LYN-1604 significantly upregulated the activity of ULK1 in the cytoplasm as determined by an increasing fluorescence of anti-p-ULK1 (Fig. 7A). We also revealed that LYN-1604 treatment induced upregulation of p-ULK1 (ser317) and downregulation of p-ULK1 (ser757), as well as the increase in phosphorylation of its downstream substrate mATG13 at ser318 (Fig. 7B). In addition, we detected whether LYN-1604 activated AMPK. LYN-1604 decreased the phosphorylation of ACC and AMPK (Fig. S3, ESI†), indicating that LYN-1604 decreases AMPK activity, which may be due to the negative regulation of ULK1 on AMPK activity.22 To examine whether LYN-1604 induced autophagy by targeting ULK1, the specific small-interfering RNA was applied for silencing ULK1 expression. We found that the LC3B puncta almost disappeared in ULK1-siRNA treated cells, compared with the control-siRNA treated cells (Fig. 7C). Subsequently, we detected the expression of the ULK complex, LC3 and p62. The silencing of ULK1 resulted in a prominent inactivation of autophagy (Fig. 7D and E). Moreover, we knocked down another autophagy-related gene ATG5 by siRNA. Interestingly, we found that the silencing of ATG5 did not disrupt the activation of ULK1, but could block the degradation of p62 and the lipidation of LC3 (Fig. 7F). We determined the cell viability by the knocking down of ULK1 or ATG5, respectively. The cell viabilities were increased after silencing ULK1 or ATG5, compared to the negative control group (Fig. 7G). Simultaneously, the ULK1 inhibitor SBI-0206965 could also decrease LYN-1604-induced cell death (Fig. S4, ESI†). These results suggest that LYN-1604 can induce ATG5-dependent autophagy via the ULK complex.
Fig. 7. LYN-1604 induces ATG5-dependent autophagy via the ULK complex. (A) LYN-1604 can obviously up-regulate the expression of p-ULK1. The expression of p-ULK1 (Ser317) was detected by immunocytochemistry. Red: TRITC-anti-p-ULK1; blue: DAPI. Scale bar = 10 μm. (B) The expressions of the ULK complex were detected by western blot analysis. β-Actin was measured as loading control. (C) MDA-MB-231 cells were transfected with control or ULK1 siRNA, followed by treatment with 2.0 μM LYN-1604 for 24 h. Then, the expression of LC3B puncta was detected by immunocytochemistry. Green: anti-LC3B; blue: DAPI. Scale bar = 20 μm. (D and E) MDA-MB-231 cells were transfected with control or ULK1 siRNA, followed by treatment with 2.0 μM LYN-1604 for 24 h. Then, the expression levels of ULK complex (D), as well as p62 and LC3 (E) were determined by western blot analysis. β-Actin was measured as the loading control. (F) MDA-MB-231 cells were transfected with control or ATG5 siRNA, followed by treatment with 2.0 μM LYN-1604 for 24 h. Then, the expression levels of ATG5, ULK1, p-ULK1 (ser317, ser757), p62 and LC3 were determined by western blot analysis. β-Actin was measured as the loading control. (G) MDA-MB-231 cells were transfected with control, ATG5 or ULK1 siRNA, respectively. After treatment with 2.0 μM LYN-1604 for 24 h, cell viabilities were measured by MTT assay.
Notably, autophagy is regarded as a double-edged sword in the survival of cancer, and it is well known that ULK1-activated autophagy is mainly dependent on the ULK complex in different types of cancer cells.23,24 There are a number of small-molecule agents that have been reported to induce cell death, which are associated with autophagy in breast cancer cells. For instance, YM155 is a potent survivin inhibitor that can modulate autophagy and induce autophagy-dependent cell death in breast cancer cells.25 Ivermectin, a broad-spectrum anti-parasitic drug, has been found to induce cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer.26 However, these small molecules either have no specific targets or possess some other complex mechanisms that are not directly regulated by autophagy-related (ATG) proteins. Unlike the above-mentioned molecules, LYN-1604 can induce ATG5-dependent autophagy in TNBC cells by targeting the ULK complex, which may provide an autophagy-modulating agent for potential TNBC therapeutics.
Microarray-based analysis of potential ULK1 interactors in LYN-1604-induced MDA-MB-231 cell autophagy
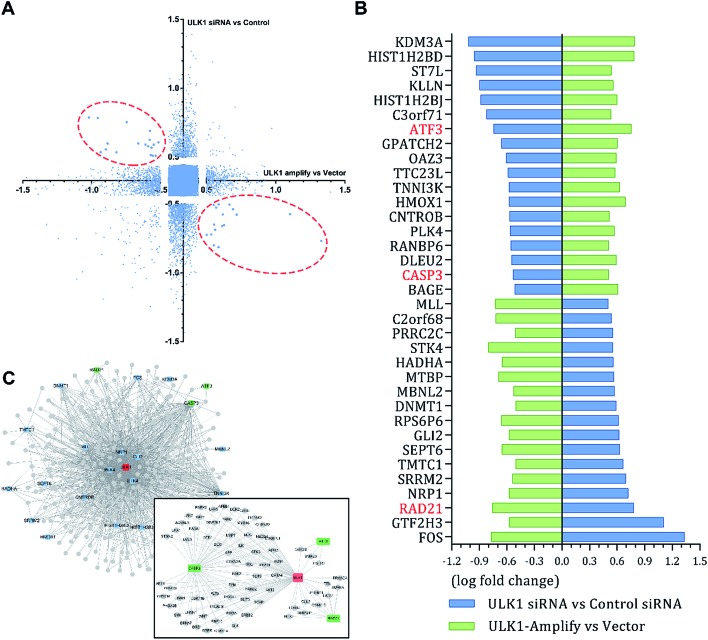
The different effects of ULK1 knockdown or amplification on gene expression by transcriptional analysis were investigated. We performed microarray analysis on MDA-MB-231 cells treated with ULK1 siRNA or transfected with pcDNA 3.1-ULK1. The analysis of significant differentially expressed genes (log fold change > 0.5) identified 491 down-regulated genes and 1001 up-regulated genes after ULK1 knockdown, while 712 genes were down-regulated and 1268 genes were up-regulated after ULK1 was amplified (Fig. 8A). There were 35 genes with opposite expression changes that were predicted to be closely involved in the ULK1-regulated network in breast cancer (Fig. 8B). Interestingly, 5 genes (ATF3, TTC23L, PLK4, CASP3 and RAD21) simultaneously showed the opposite expression changes after ULK1 knockdown or overexpression, while other genes showed expression changes to different extents, indicating that these genes may be directly interacting with ULK1. Moreover, the PPI network of ULK1 was modified to the microarray-based ULK1 network in the context of TNBC, which consisted of 23 possible ULK1-interacting genes. According to microarray-based analysis of the core ULK1-regulated autophagic subnetwork, we found that activating transcription factor 3 (ATF3), double-strand-break repair protein rad21 homolog (RAD21) and caspase3 had significant changes in their expressions and were closely correlated with ULK1 (Fig. 8C). Thus, it suggests the key role of ULK1 with its potential interactors in autophagy of TNBC cells. To validate the potential interactors and intricate mechanisms of ULK1, we used the ULK1 agonist LYN-1604, which is similar to the ULK1 overexpression plasmid, to activate ULK1-modulating cell death associated with autophagy.
Fig. 8. Cell-specific microarray-based analysis of core ULK1-regulated network in MDA-MB-231 cells. (A) Expression data for microarray analysis from two individual experiments. MDA-MB-231 cells were treated with ULK1 siRNA or transfected with pcDNA 3.1-ULK1. Significant differentially expressed genes with opposite expression change are circled by red dashed lines. (B) 35 opposite expression changed genes. (C) Microarray-based ULK1 subnetwork in breast cancer, 23 opposite expression changed genes (blue) were involved in this subnetwork. ATF3, RAD21 and caspase3 were verified to be regulated by ULK1.
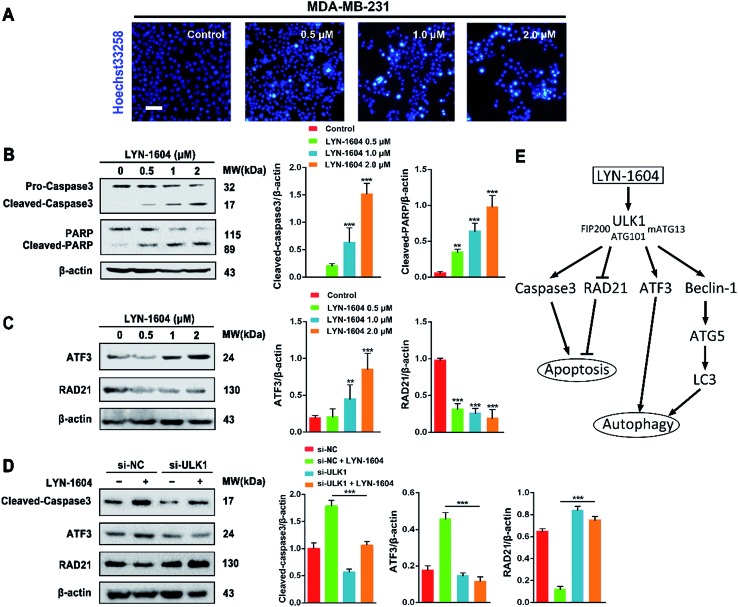
As caspase3 is known as the apoptotic executor, the morphological changes of MDA-MB-231 cells were observed with fluorescent dye. Interestingly, the apoptotic phenomenon was not obvious, which was determined by Hoechst 33258 staining (Fig. 9A). We checked the expressions of caspase3 and its cleavage substrate PARP, then found that they were slightly changed only with high concentration of LYN-1604 treatment, which suggests that LYN-1604 induces cell death, associated with autophagy and apoptosis (Fig. 9B). We also found that LYN-1604 obviously up-regulated ATF3 and down-regulated RAD21 (Fig. 9C). However, LYN-1604 had no effect on the expressions of cleaved-caspase3, ATF3 and RAD21, when ULK1 was knocked down by siRNA (Fig. 9D). These results indicate that caspase3, ATF3 and RAD21 are all involved in LYN-1604-induced cell death, which may be simultaneously regulated by ULK1 (Fig. 9E).
Fig. 9. LYN-1604 induces autophagy involved in ATF3, RAD21, and caspase3. (A) After treatment with LYN-1604, MDA-MB-231 cells were stained with Hoechst 33258. Morphological changes were observed under fluorescence microscope. Scale bar = 20 μm. (B) After LYN-1604 treatment, the expression levels of caspase3 and PARP were determined by western blot analysis. β-Actin was measured as loading control. (C) After LYN-1604 treatment, the expression levels of ATF3 and RAD21 were determined by western blot analysis. β-Actin was measured as loading control. (D) MDA-MB-231 cells were transfected with control or ULK1 siRNA, followed by treatment with 2.0 μM LYN-1604 for 24 h. Then, the expression levels of cleaved-caspase3, ATF3 and RAD21 were determined by western blot analysis. β-Actin was measured as loading control. (E) The schematic model of LYN-1604-induced cell death pathways, associated with autophagy and apoptosis.
ULK1 can be directly regulated by AMPK and mTORC1 via phosphorylation.27,28 Subsequently, ULK1 and its complex are activated, and modulate necessary downstream signals, including phosphorylation of Beclin-1 and activation of VPS34 lipid kinase.29,30 To explore the intricate mechanisms, we constructed the ULK1-regulated network by performing microarray analysis in TNBC cells. Intriguingly, we found several autophagic proteins such as ATF3, RAD21 and even the apoptotic protein caspase3 that could be regulated by ULK1, indicating that ULK1 plays dual roles in autophagy-initiation and apoptosis promotion with some potential new interactors in TNBC. Of note, ATF3 is an autophagy-related protein; therefore, activation of ATF3 can promote the process of autophagy.31 RAD21 is important and closely related to breast cancer cell growth and apoptosis; thus, inhibiting RAD21 can inhibit cancer cell proliferation.32,33 In our study, we show that LYN-1604 could modulate ATF3 and RAD21 through ULK1 in MDA-MB-231 cells, which is in good agreement with our above-mentioned microarray-based analysis of ULK1 in TNBC. In addition, LYN-1604 can also increase cleavage of caspase3 and induce apoptosis. We speculate that ULK1 probably regulates caspase3 activity to induce apoptosis, or ULK1 may be a substrate that could be cleaved by caspase3; however, the internal relations between ULK1 and caspase3 still need to be explored. Together, these findings provide some potential mechanisms for ULK1 agonists to induce cell death in TNBC therapy.
LYN-1604 inhibits the growth of xenograft TNBC by targeting ULK1-modulated cell death
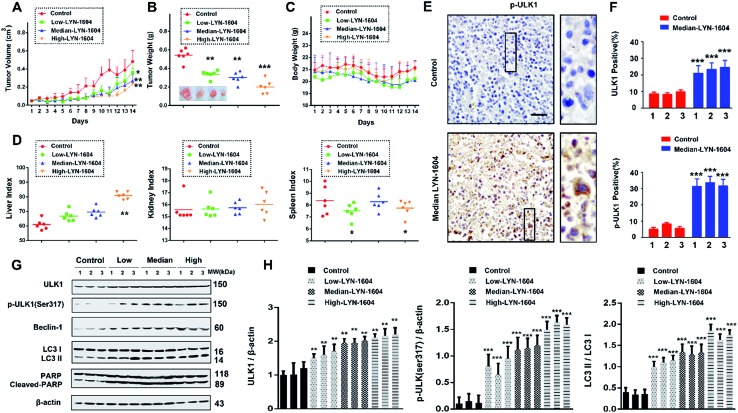
To evaluate the efficacy of LYN-1604 in vivo, we established a MDA-MB-231 xenograft model. Based on the results of tumor volume and weight, we found that LYN-1604 could significantly inhibit the growth of xenograft MDA-MB-231 cells (Fig. 10A and B). The body weights of mice were stable, with no obvious distinctions between LYN-1604-treated and control mice (Fig. 10C). By the end of the experiment, the liver and spleen weight indexes of mice were slightly increased in parts of the groups, while the kidney weight index was not affected in all dose groups (Fig. 10D). To confirm the mechanisms for the therapeutic efficacy of LYN-1604 in vivo, we examined the immunoreactivity of ULK1 and p-ULK1 in tumor tissues, and found both of them showed an increasing trend in LYN-1604-treated tumor tissues (Fig. 10E and F). Next, we carried out western blot analysis to clarify the mechanism of LYN-1604 in vivo. It revealed that LYN-1604 induced remarkable autophagy in vivo, which is consistent with the immunohistochemical analysis and the aforementioned in vitro results (Fig. 10G and H). These results suggest that LYN-1604 has good therapeutic potential by targeting ULK1-modulated cell death of TNBC in vivo, thus making a ULK1 agonist that can be utilized as a new drug for TNBC therapy. More recently, some agents have been broadly used for treatment of TNBC by modulating autophagy. Everolimus, an oral mTOR inhibitor, can effectively inhibit cell growth in different subtypes of TNBC cells through PI3K/Akt/mTOR cascade, which also demonstrates antitumor activity in a mouse xenograft model of basal-like breast cancer, rather than non-basal breast cancer.34 Chloroquine, an autophagy inhibitor that could prevent lysosome-fusion in the autophagy process, has been reported to inhibit xenograft tumor growth of TNBC, especially in combination with other anticancer drugs such as panobinostat, epirubicin and carboplatin.35–37 Beyond these small molecules, LYN-1604 can directly activate ULK1, the initiator of autophagy, thereby inducing cell death to inhibit the growth of TNBC in vitro and in vivo. More importantly, ULK1 is a promising, specific and potent therapeutic target of TNBC, thus enabling its targeted agonist LYN-1604 to be utilized as a small-molecule drug candidate for future TNBC therapy.
Fig. 10. LYN-1604 has therapeutic potential, targeting ULK-modulated cell death in vivo. Antitumor activities of LYN-1604 in representative tumors from mice after vehicle and LYN-1604 treatment: (A) tumor volume and (B) tumor weight are expressed as the mean ± SEM; ***, p < 0.001; **, p < 0.05 vs. control group (n = 6). Toxicity of LYN-1604: relative body weight (C), liver index, spleen index and kidney index (D) were evaluated at the end of treatment. (E) The expression of p-ULK1 (Ser317) in representative tumor sections of mice after vehicle and median dose. LYN-1604 treatment was determined by immunohistochemistry (×100 magnification); ***, p < 0.001 vs. control group. (F) Quantitative analysis of ULK1 and p-ULK1 positive ratios. (G) Western blot analysis of tumor tissues from vehicle or LYN-1604 treated mice for expression of ULK1, p-ULK1, LC3, Beclin-1 and PARP; β-actin was measured as loading control. (H) Quantification of immunoblot shown in (G).
Conclusion
Based on TCGA and TMA analyses, we found that ULK1 was more remarkably downregulated in TNBC tissues than other types of breast cancer, indicating that ULK1 may be a new target in TNBC. Combining in silico high-throughput screening, series synthesis, kinase and anti-proliferative activity screening, as well as site-directed mutagenesis, we discovered a potent ULK1 agonist (LYN-1604). Moreover, we demonstrated that LYN-1604 could activate the ULK1 and ULK complexes (ULK1-mATG13-FIP200-ATG101), and also trigger cell death via ATG5 involvement, as well as ATF3, RAD21 and caspase3. Additionally, we found that LYN-1604 has good therapeutic potential for TNBC by targeting ULK1-modulated cell death in vivo.
In summary, our integrative TCGA-based, microarray-based and experimental findings can together provide us a better understanding of how ULK1, a potential anti-TNBC target, can regulate cell death by the ULK complex (ULK1-mATG13-FIP200-ATG101) and other new potential interactors (ATF3, RAD21 and caspase3). Such results could help us in developing this ULK1 agonist (LYN-1604), with its remarkable anti-tumor activity and cell death mechanisms in vitro and in vivo, so that could be further exploited as a novel, small-molecule candidate drug for future TNBC therapeutics.
Experimental
TCGA analysis and network construction
The TCGA-based gene expression profile was measured using the Agilent G4502A_07_3 platform. Log2 lowess normalized (cy5/cy3) collapsed by gene symbol count was used as the gene level expression estimate in this study. A total of 527 breast cancer patients (55 TNBC and 472 non-TNBC) with gene expression datasets were studied. The core autophagic PPI network was downloaded from the ARN database.38 Only PPIs annotated as interactions between autophagy proteins were used. The PPI network was constructed using Cytoscape.39
Tissue microarray (TMA) analysis
The breast cancer tissue microarray (BR804a) was obtained from Cybrdi, Inc. Detection of ULK1 was performed by anti-ULK1 antibody (1:200) as previously described.26,40 The microarray contained breast cancer tumor tissues from 40 individuals plus matched adjacent normal tissues. The immunostaining intensity was indicated by four grades (0, negative; 1, weakly positive; 2, positive; 3, strongly positive) and the percentage of cell staining at each intensity level was graded as the proportion of stain-positive cells and divided into five grades 0 (<5%), 1 (5–25%), 2 (26–50%), 3 (51–75%), 4 (>75%). The final score was calculated as intensity score × the proportion of stain-positive cells.
Cell culture, antibodies and reagents
HEK-293T, MDA-MB-231, MDA-MB-468 and MCF-7 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in DMEM with 10% fetal bovine serum and incubated with 5% CO2.
Antibodies used in this study were as follows: ULK1 (8054, CST, MA, USA), ULK1 (ab128859, Abcam, UK), p-ULK1ser317 (12753, CST), p-ULK1ser757 (14202, CST), mAtg13 (13273, CST), p-mATG13ser318 (PAB19948, Abnova, Taiwan), ATG101 (13492, CST), FIP200 (12436, CST), AMPKα (5831, CST), p-AMPKαthr172 (2535, CST), ACC (3676, CST), p-ACCser79 (11818, CST), Beclin-1 (3495, CST), LC3B (3868, CST), p62 (5114, CST), ATG5 (12994, CST), PARP (9532, CST), caspase-3 (9665, CST), ATF3 (ab58668, Abcam), RAD21 (12673, CST), β-actin (66009-1-Ig, Proteintech, IL, USA).
MTT (M2128), Hoechst 33258 (14530), MDC (30432) and GST-tagged mATG13 (SRP5341) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Bafilomycin A1 (ab120497) was purchased from Abcam. The plasmid encoding myc-hULK1 (#31961) was purchased from Addgene and subcloned to pRK5-Flag. Flag-tagged ULK1 mutants (K50A, L53A and Y89A) were generated by site-directed mutagenesis with the Quikchange II site-directed mutagenesis kit (Stratagene).
Molecular docking and molecular dynamics (MD) simulations
The initial three dimensional geometric coordinates of the X-ray crystal structure of ULK1 (PDB code: 4WNP) was downloaded from the Protein Databank (PDB), and we constructed the screening library containing 6583 small molecule compounds from the latest version of Drugbank (; http://www.drugbank.ca/). The activators were constructed using the Accelrys Discovery Studio (version 3.5; Accelrys, SanDiego, CA, USA) molecular modeling software and were energy minimized with the CHARMm force field. Docking and molecular dynamics (MD) simulations were performed according to our previous reports.41,42 The LibDock and CDOCKER protocol were employed as docking approaches to conduct semi-flexible docking. MD simulations were carried out for LYN-1604-ULK1WT, LYN-1604-ULK1K50A, LYN-1604-ULK1L53A and LYN-1604-ULK1Y89A complexes using the GROMACS (version 4.5.5) software package.43 All MD simulation videos are supplied in the ESI.†
ADP-Glo™ kinase assay
ULK1 kinase activity assay was performed using ADP-Glo™ Kinase Assay + ULK1 Kinase Enzyme System (Promega, Madison, WI, USA) as previously described.44 ADP-Glo™ Kinase Assay is a luminescent kinase assay that measures ADP formed from a kinase reaction; ADP is converted into ATP, which is converted into light by Ultra-Glo™ Luciferase. The luminescent signal positively correlates with kinase activity. The ULK1 kinase enzyme, substrate, ATP and compound were diluted in kinase buffer (40 mM Tris pH 7.5; 20 mM MgCl2; 0.1 mg mL–1 BSA; 50 μM DTT). Then, 1 μL of compound or (5% DMSO), 2 μL of ULK1 kinase enzyme or purified wild-type and mutant ULK1 (K50A, L53A, Y89A) (10 ng), or 2 μL of MBP (0.1 μg μL–1)/ATP (10 μM) mix were added to the wells of a 384 well low volume plate. After incubation at room temperature for 60 minutes, 5 μL of ADP-Glo™ reagent was added per well. The plates were incubated at room temperature for 40 minutes and then 10 μL of kinase detection reagent was added. After incubation at room temperature for 30 minutes, the luminescence was recorded. The EC50 values were calculated using nonlinear regression with normalized dose–response fitting using Prism software (GraphPad Software, San Diego, CA, USA). To exclude the possibility that the increased kinase activity might be due to aggregation, 0.02% Triton-X was used as a detergent for eliminating the interference from non-specific compounds. The result is shown in Fig. S5.†
In vitro ULK1 kinase assay
The kinase assay of ULK1 was performed according to a previous report.45,46 To assay ULK1 kinase activity, flag-tagged ULK1WT, ULK1K50A, ULK1L53A or ULK1Y89A mutants expressed in HEK-293T cells were immunoprecipitated by anti-flag antibody, and then incubated with 200 ng of purified GST-tagged mATG13 at 37 °C for 20 minutes in a kinase reaction containing 20 mM HEPES (pH 7.5), 20 mM MgCl2, 20 mM NaF, 0.05 mM DTT, and 40 μM ATP at a final volume of 20 μL. The reaction was stopped by the addition of sample buffer, boiled and analyzed by western blot with p-mATG13 (ser318) antibody.
Surface plasmon resonance (SPR) analysis
The binding capacity of LYN-1604 with ULK1 and ULK1 mutations (K50A, L53A, Y89A) was assessed using a SPR analysis according to literature.47 The measurements were performed on a Biacore S51 device (GE Healthcare, Uppsala). For immobilization, PBS with 0.05% Tween 20 and pH 7.4, was used as a background buffer. The standard amine coupling procedure was applied. A series S sensor chip, CM5, was activated with a mixture of 0.1 M NHS (N-hydroxysuccinimide) and 0.4 M EDC (1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide-hydrochloride) at a flow rate of 10 μL min–1 for 10 min. Then, a 50 μg mL–1 solution of hCAII in 10 mM sodium acetate (pH 5.2) with 30 μM ethoxzolamide was injected for 2 min followed by an injection of 1 M ethanolamine hydrochloride (pH 8.5) for 7 min. For measuring the compound, PBS with 0.05% Tween 20 and 2% DMSO, pH 7.4, was used as the running buffer. The compound was injected at a flow rate of 30 μL min–1 for 120 s. Then, the dissociation was monitored for 360 s and the dissociation constant at equilibrium (K D) was evaluated with the Biacore S51 evaluation software (version: 1.2.1).
Cell viability assay
Cells were dispensed in 96-well plates at a density of 5 × 104 cells per mL. After 24 h of incubation, cells were treated with different concentrations of compounds for the indicated time periods. Cell viability was measured by the MTT assay.
Autophagy and apoptosis assays
For the autophagy assay, MDA-MB-231 cells were transfected with GFP/mRFP-LC3 (kindly provided by Prof. Canhua Huang, Sichuan University) or stained with MDC (0.05 mM) then observed under a fluorescence microscope. MDC positive ratio (here referring to autophagic ratio) was measured by FACScan flow cytometry (Becton Dickinson, Franklin Lakes, NJ). For apoptosis assay, MDA-MB-231 cells were stained with Hoechst 33258 in the dark at 37 °C for 30 min and then observed under a fluorescence microscope.
Immunofluorescence analysis
For immunofluorescence staining, nonspecific antibody binding was blocked by incubating with PBS containing 1.5% goat serum. The MDA-MB-231 cells were sequentially incubated, starting with p-ULK1 antibody (1:200) and LC3B antibody (1:200) diluted in PBS containing 1% BSA incubated overnight at 4 °C, followed by the addition of fluorescence-labeled secondary antibodies (TRITC, ab6718; FITC, ab6717) for 1 h at room temperature.
Western blot
Cells or tumor tissues were treated with LYN-1604 for the indicated times. Both adherent and floating cells were collected and western blot analysis was carried out. Briefly, the cell pellets were resuspended with lysis buffer consisting of 50 mM HEPES, pH 7.4; 1% Triton-X-100, 2 mM sodium orthovanadate, 100 mM sodium fluoride, 1 mM edetic acid, 1 mM PMSF, 10 mg L–1 aprotinin (Sigma, MO, USA) and 10 mg L–1 leupeptin (Sigma), and lysed at 4 °C for 1 h. After 12 000 rpm centrifugation for 15 min, the protein content of the supernatant was determined by the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of the total protein were separated by 10–15% SDS–PAGE and transferred to PVDF membranes, the membranes were soaked in blocking buffer (5% skimmed milk or BSA). Proteins were detected using primary antibodies, followed by HRP-conjugated secondary antibody and visualized using ECL as the HRP substrate. Quantification of immunoblot was performed by Quantity One 4.4.
siRNA transfection
Cells were transfected with ULK1, ATG5 and control siRNAs at 100 nM final concentration using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The transfected cells were used for subsequent experiments 48 h later.
Microarray profiling and significant analysis of microarray (SAM) analysis
The MDA-MB-231 cells were transfected with ULK1-overexpression plasmid (kindly provided by Prof. Canhua Huang, Sichuan University), empty plasmid, ULK1 siRNA or control siRNA using Lipofectamine 2000 reagent (Invitrogen), following the procedure recommended by the manufacturer. The transfected cells were used for microarray analysis, which was carried out following the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology). The significant analysis of the microarray (SAM) method was used to perform the unsupervised calculation. The proposed statistical technique is based on a t-test for finding significant genes in a set of microarray experiments. A hierarchical clustering of the differentially expressed genes was performed with the Cluster 3.0 (; http://bonsai.hgc.jp/∼mdehoon/software/cluster/software) software using the average linkage algorithm. The top scoring pair (TSP) algorithm was used to perform the supervised calculation.48 The basic principle of the k-TSP is to identify gene pairs that are oppositely expressed in two classes. All numerical analyses that are presented were performed using Matlab 7.0 (MathWorks Company, Natick, MA, USA).
Xenograft breast cancer model
The Institutional Animal Care and Treatment Committee of Sichuan University approved all studies herein. 24 female nude mice (BALB/c, 6–8 weeks, 20–22 g) were injected subcutaneously with MDA-MB-231 cells (5 × 106 cells per mouse). When the tumors reached 100 mm3 in volume (V = L × W 2/2), the mice were divided into four groups. Three groups were treated with different doses of LYN-1604 once a day by intragastric administration for 14 days (low dose, 25 mg kg–1; median dose, 50 mg kg–1; high dose, 100 mg kg–1), whereas the control group was treated with vehicle control. During the treatment, the tumor volumes and body weight were measured every day until the end of the study. At the end of treatment, all mice were sacrificed. The spleen, liver, kidney and tumor tissue were harvested, weighed, and photographed, then immediately frozen in liquid nitrogen or fixed in formalin.
Immunohistochemistry analysis (IHC)
Sections of the tumor were submerged in EDTA antigenic retrieval buffer (pH 8.0) or citrate buffer (pH 6.0), and microwaved for antigenic retrieval. The slides were then incubated with rabbit anti-p-ULK1 polyclonal antibody (1:400), or with anti-ULK1 antibody (1:400), for 30–40 min at 37 °C. Normal rabbit/mouse IgG was used as a negative control. The slides were then treated by HRP polymer conjugated secondary antibody for 30 min and developed with diamino-benzidine solution. Meyer's hematoxylin was used as a counterstain.
Statistical analysis
All the presented data and results were confirmed by at least three independent experiments. The data are expressed as means ± SEM and analyzed with GraphPad Prism 6.0 software. Statistical comparisons were made by One-way ANOVA and Student's t-test. P < 0.05 was considered statistically significant.
Other methods
Descriptions of chemical synthesis, see ESI.†
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Prof. Canhua Huang (Sichuan University) for providing the ULK1 and GFP-mRFP-LC3 plasmids in this study. This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81673455, 81602953, 81602627 and 81473091).
Footnotes
References
- Rabinowitz J. D., White E. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Li S. J., Guo M. R., Wu Y., Wang H. Y., Zhang K., Zhang X., Ouyang L., Liu J. Cell Prolif. 2015;48:119–139. doi: 10.1111/cpr.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. J., Lin M., Yu J. Y., Liu B., Bao J. K. Cancer Lett. 2011;300:105–114. doi: 10.1016/j.canlet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Wang S. Y., Yu Q. J., Zhang R. D., Liu B. Int. J. Biochem. Cell Biol. 2011;43:1263–1266. doi: 10.1016/j.biocel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Liu B., Wen X., Cheng Y. Cell Death Dis. 2013;4:e892. doi: 10.1038/cddis.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke B., Tian M., Li J., Liu B., He G. Med. Res. Rev. 2016;36:983–1035. doi: 10.1002/med.21398. [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Bravo-San Pedro J. M., Kroemer G. Mol. Cell. 2016;62:473–474. doi: 10.1016/j.molcel.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Chen Y., He J., Tian M., Zhang S. Y., Guo M. R., Kasimu R., Wang J. H., Ouyang L. Cell Prolif. 2014;47:494–505. doi: 10.1111/cpr.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S., Löffler A. S., Wesselborg S., Stork B. Cell Commun. Signaling. 2012;10:7. doi: 10.1186/1478-811X-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P. M., Puente C., Ganley I. G., Jiang X. J. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Deng R., Luo R. Z., Shen G. P., Cai M. Y., Du Z. M., Jiang S., Yang M. T., Fu J. H., Zhu X. F. Breast Cancer Res. Treat. 2012;134:549–560. doi: 10.1007/s10549-012-2080-y. [DOI] [PubMed] [Google Scholar]
- Jung C. H., Kim H., Ahn J., Jung S. K., Um M. Y., Son K. H., Kim T. W., Ha T. Y. J. Evidence-Based Complementary Altern. Med. 2013;2013:385219. doi: 10.1155/2013/385219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Li Y., Zhu Y. H., Wu X. Q., Tang J., Li Z., Feng G. K., Deng R., Li D. D., Luo R. Z., Zhang M. F., Qin W., Wang X., Jia W. H., Zhu X. F. Cancer Sci. 2011;102:1568–1575. doi: 10.1111/j.1349-7006.2011.01964.x. [DOI] [PubMed] [Google Scholar]
- Xu H. T., Yu H., Zhang X. Y., Shen X. Y., Zhang K. H., Sheng H. H., Dai S., Gao H. Int. J. Clin. Exp. Pathol. 2013;6:711–717. [PMC free article] [PubMed] [Google Scholar]
- Yun M., Bai H. Y., Zhang J. X., Rong J., Weng H. W., Zheng Z. S., Xu Y., Tong Z. T., Huang X. X., Liao Y. J., Mai S. J., Ye S., Xie D. PLoS One. 2015;10:e0117375. doi: 10.1371/journal.pone.0117375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Novotny C. J., Shokat K. M. ACS Chem. Biol. 2015;10:257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan D. F., Chun M. G., Vamos M., Zou H., Rong J., Miller C. J., Lou H. J., Raveendra-Panickar D., Yang C. C., Sheffler D. J., Teriete P., Asara J. M., Turk B. E., Cosford N. D., Shaw R. J. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petherick K. J., Conway O. J., Mpamhanga C., Osborne S. A., Kamal A., Saxty B., Ganley I. G. J. Biol. Chem. 2015;290:11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Shokat K. M. Bioorg. Med. Chem. 2015;23:5483–5488. doi: 10.1016/j.bmc.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Zhang L., Zhang S., Ouyang L., Cai H., Liu B. Oncotarget. 2016;7:10015–10022. doi: 10.18632/oncotarget.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A. S., Alers S., Dieterle A. M., Keppeler H., Franz-Wachtel M., Kundu M., Campbell D. G., Wesselborg S., Alessi D. R., Stork B. Autophagy. 2011;7:696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- Chen Y., Fu L. L., Wen X., Liu B., Huang J., Wang J. H., Wei Y. Q. Apoptosis. 2014;19:1177–1189. doi: 10.1007/s10495-014-0999-7. [DOI] [PubMed] [Google Scholar]
- Ganley I. G., Lam D. H., Wang J. R., Ding X. J., Chen S., Jiang X. J. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. M., Chang Y. C., Liu C. Y., Lee J. Y., Chan H. H., Kuo C. W., Lin K. Y., Tsai S. L., Chen S. H., Li C. F., Leung E., Kanwar J. R., Huang C. C., Chang J. Y., Cheung C. H. Br. J. Pharmacol. 2015;172:214–234. doi: 10.1111/bph.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Q., Chen H. N., Wang K., Yuan K., Lei Y., Li K., Lan J., Chen Y., Huang Z., Xie N., Zhang L., Xiang R., Nice E. C., Wei Y. Q., Huang C. Cancer Res. 2016;76:4457–4469. doi: 10.1158/0008-5472.CAN-15-2887. [DOI] [PubMed] [Google Scholar]
- Zhao M., Klionsky D. J. Cell Metab. 2011;13:119–120. doi: 10.1016/j.cmet.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan D., Kim J., Shaw R. J., Guan K. L. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko V. Y., Zhong Q. Nat. Cell Biol. 2013;15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., Guan K. L. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I., Sorbara M. T., Philpott D. J., Girardin S. E. Autophagy. 2012;8:1848–1850. doi: 10.4161/auto.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. A., Ouchi T. Cancer Res. 2005;65:10657–10662. doi: 10.1158/0008-5472.CAN-05-2087. [DOI] [PubMed] [Google Scholar]
- Atienza J. M., Roth R. B., Rosette C., Smylie K. J., Kammerer S., Rehbock J. Mol. Cancer Ther. 2005;4:361–368. doi: 10.1158/1535-7163.MCT-04-0241. [DOI] [PubMed] [Google Scholar]
- Yunokawa M., Koizumi F., Kitamura Y., Katanasaka Y., Okamoto N., Kodaira M., Yonemori K., Shimizu C., Ando M., Masutomi K., Yoshida T., Fujiwara Y., Tamura K. Cancer Sci. 2012;103:1665–1671. doi: 10.1111/j.1349-7006.2012.02359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R., Balusu R., Fiskus W., Mudunuru U., Venkannagari S., Chauhan L., Smith J. E., Hembruff S. L., Ha K., Atadja P., Bhalla K. N. Mol. Cancer Ther. 2012;11:973–983. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- Chittaranjan S., Bortnik S., Dragowska W. H., Xu J., Abeysundara N., Leung A., Go N. E., DeVorkin L., Weppler S. A., Gelmon K., Yapp D. T., Bally M. B., Gorski S. M. Clin. Cancer Res. 2014;20:3159–3173. doi: 10.1158/1078-0432.CCR-13-2060. [DOI] [PubMed] [Google Scholar]
- Liang D. H., Choi D. S., Ensor J. E., Kaipparettu B. A., Bass B. L., Chang J. C. Cancer Lett. 2016;376:249–258. doi: 10.1016/j.canlet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türei D., Földvári-Nagy L., Fazekas D., Módos D., Kubisch J., Kadlecsik T., Demeter A., Lenti K., Csermely P., Vellai T., Korcsmáros T. Autophagy. 2015;11:155–165. doi: 10.4161/15548627.2014.994346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T. Y., Wu C. N., Sie H. C., Cheng J. T., Lin Y. S., Liou H. H., Tseng Y. K., Shu C. W., Tsai K. W., Yen L. M., Tseng H. W., Tseng C. J., Ger L. P., Liu P. F. Oncotarget. 2016;7:85097–85108. doi: 10.18632/oncotarget.13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Zhang S., Zhang L., Tong X., Zhang J., Zhang Y., Ouyang L., Liu B., Huang J. Oncotarget. 2015;6:8071–8088. doi: 10.18632/oncotarget.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Fu L., Zhang C., Zhang L., Zhang Y., Ouyang L., He G., Huang J. Oncotarget. 2015;5:6762–6775. doi: 10.18632/oncotarget.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., Lindahl E. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzano D., Santaguida S., Musacchio A., Villa F. Chem. Biol. 2011;18:966–975. doi: 10.1016/j.chembiol.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Lin Y. C., Chen Y. H., Chen C. M., Pang L. Y., Chen H. A., Wu P. R., Lin M. Y., Jiang S. T., Tsai T. F., Chen R. H. Mol. Cell. 2016;61:84–97. doi: 10.1016/j.molcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., Hansen M., Shaw R. J. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari R., Rechlin C., Heine A., Bottegoni G., Rocchia W., Schwarz D., Bomke J., Gerber H. D., Klebe G., Cavalli A. J. Med. Chem. 2016;59:4245–4256. doi: 10.1021/acs.jmedchem.5b01643. [DOI] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.