Abstract
Par-6 is a scaffold protein that organizes other proteins into a complex required to initiate and maintain cell polarity. Cdc42-GTP binds the CRIB module of Par-6 and alters the binding affinity of the adjoining PDZ domain. Allosteric regulation of the Par-6 PDZ domain was first demonstrated using a peptide identified in a screen of typical carboxyl terminal ligands. Crumbs, a membrane protein that localizes a conserved polarity complex, was subsequently identified as a functional partner for Par-6 that likely interacts with the PDZ domain. Here we show by NMR that Par-6 binds a Crumbs carboxyl terminal peptide and report the crystal structure of the PDZ-peptide complex. The Crumbs peptide binds Par-6 more tightly than the previously studied carboxyl peptide ligand and interacts with the CRIB-PDZ module in a Cdc42-dependent manner. The Crumbs:Par-6 crystal structure reveals specific PDZ-peptide contacts that contribute to its higher affinity and Cdc42-enhanced binding. Comparisons with existing structures suggest that multiple C-terminal Par-6 ligands respond to a common conformational switch that transmits the allosteric effects of GTPase binding.
Graphical Abstract

Epithelial apical-basolateral polarization is primarily governed by the action of the Crumbs, Par, and Scribble complexes, which assemble at the plasma membrane to organize the proper distribution and orientation of the cytoskeleton and other cellular components1–3. PSD-95/Dlg/ZO-1 (PDZ) domains mediate many of the interactions within and between these conserved polarity proteins. PDZ domains are small (~10 kDa) protein-protein interaction modules that typically bind a short sequence at the C-terminus of another protein4. Partition defective 6 (Par-6/Pard-6) is a central component of the apical Par complex that contains a single PDZ domain. The Par-6 PDZ domain is unusual in two respects. First, it can bind an internal (non-C-terminal) sequence from Stardust/protein associated with Lin7-1 (Pals1)5, 6, a component of the Crumbs complex. Second, it combines with an adjoining Cdc42/Rac Interaction Binding (CRIB) domain to form a novel GTPase-activated molecular switch that modulates binding affinity for a peptide with the sequence VKESLV, originally identified from a library of likely C-terminal PDZ ligands7.
Binding of VKESLV to the Par-6 CRIB-PDZ module is regulated by the monomeric GTPase Cdc427. Upon Cdc42-GTP binding, a portion of the flexible CRIB motif folds into a stable β-strand (‘β0’) that pairs with the β1 strand of the PDZ domain and forms a continuous nine-stranded intermolecular β-sheet with the GTPase8. GTPase binding to the CRIB increases the affinity for VKESLV in the distal PDZ ligand binding cleft ~10-fold7. In contrast, CRIB-PDZ binding to the internal sequence ligand from Pals1 is unaffected by Cdc427. Our previous structural studies showed that allosteric Cdc42 activation of Par-6 binding to VKESLV involves rearrangement of two adjacent sidechains – Leu 164 and Lys 165, the ‘dipeptide switch’ – in the β2–3 loop9, and that interconversion between the low and high affinity PDZ conformations requires partial unfolding of the domain10.
Par-6 activation by Cdc42-GTP alters localization of the Par complex and other proteins in both epithelial cell monolayers and migrating cells7–9, 11–19, presumably by promoting its interaction with a C-terminal ligand. Crb3, one of three human orthologs of the Crumbs protein, was the first C-terminal Par-6 PDZ ligand to be identified in the context of cell polarity20. However, the Cdc42-dependence of this interaction has not been measured and the VKESLV peptide does not correspond to any known Par-6 physiologically relevant binding partner. Thus, the CRIB-PDZ dipeptide switch has not yet been shown to regulate formation of a Par-6 complex known to function in cell polarization.
Herein we report the X-ray crystal structure of the Par-6 PDZ:Crb complex, and show that VKESLV and Crb peptides exhibit similar binding modes by NMR. Crumbs binds Par-6 with significantly higher affinity than VKESLV, consistent with its role as a functional link between the mammalian Par-6 and Crb3 proteins. Importantly, when Cdc42-GTP is bound to the CRIB-PDZ module the binding energy increases by the same margin for both Crb and VKESLV. We conclude that allosteric activation of C-terminal ligand binding by Cdc42 promotes the association of the Par and Crumbs complexes and may alter other Par-6 interactions as a consequence.
Materials and Methods
Protein expression and purification
D. melanogaster Par-6 constructs were produced as previously described9. All protein expression utilized the pBH4 vector and BL21 (DE3) E. coli as a host strain grown in minimal 15N-labeled media. The pBH4 vector contains an Ampicillin-resistance gene and an IPTG-inducible T7 promoter sequence, and expresses proteins as a fusion with a Tobacco Etch Virus (TEV) cleavage sequence and hexahistidine affinity tag (6His) N-terminal to the protein sequence. Following purification with Ni-NTA beads, treatment with TEV protease allows removal of the 6His tag in final purification steps. Synthesized peptides were ordered either from the internal MCW Protein and Nucleic Acid Facility or commercially and purified with reverse-phase HPLC. Final purified proteins and peptide were measured at >99% purity by SDS-PAGE and MALDI-TOF spectroscopy.
X-ray Crystallography
The Par-6 PDZ domain (residues 156–255)–Crb3 C-terminal peptide complex was prepared by mixing Par-6 PDZ domain at 10 mg/mL with a 5-fold molar excess of Crb3 C-terminal peptide (sequence: LPPEERLI). Hanging drop vapor diffusion crystallization experiments were carried out at 16 °C by mixing equal volumes of the protein with well solution containing 100 mM HEPES (pH 7.1) and 26% PEG 6,000. The resulting crystals were flash frozen after passing through a cryoprotection solution of the well solution plus an additional 10% glycerol. X-ray diffraction data were gathered for a single crystal at 100 K using beamline 8.2.2 of the Advanced Light Source. Data reduction was carried out using HKL200021.
Molecular replacement was used to evaluate the initial phases using the Par-6 PDZ domain:VKESLV complex (PDB entry: 1RZX) as the search model. Phenix.AutoMR22 solved the initial phases and automatically built in the majority of the residues for the binary complex. Models were completed through iterative rounds of manual model building in Coot and refinement with Phenix.refine. Geometry of the final structure was validated using Molprobity23. Ramachandran statistics for the binary complex were 98 and 2% for the favored and additionally allowed regions of the Ramachandran plot, respectively. Data collection and refinement statistics for the final model (PDB ID 5I7Z) are listed in Table 1. Residues 156, 157, 254, and 255 were not visible in the electron density for PDZ156–255. No density was observed for the N-terminal leucine of the peptide ligand. The proline residues were placed into interpretable electron density but exhibited significantly higher B-factors (~55 Å2) than to the C-terminal peptide residues (~30 Å2).
Table 1.
X-ray data collection and refinement statistics.
| Par-6–Crb | |
|---|---|
| Data collection | |
| Space group | P 63 |
| Cell dimensions | |
| a, b, c (Å) | 65.1, 65.1, 52.6 |
| α,β,γ (°) | 90, 90, 120 |
| Resolution (Å) | 50–1.80 (1.86–1.80) |
| Rmerge | 0.073 (0.418) |
| I/σI | 43.4 (1.9) |
| Completeness (%) | 95.6 (70.6) |
| Redundancy | 12.6 (6.1) |
| Refinement | |
| Resolution (Å) | 38.4–1.80 (1.88–1.80) |
| No. reflections | 10,639 |
| Rwork/Rfree | 0.197/0.218 |
| No. atoms | 859 |
| Protein | 792 |
| Ligand/ion | 7 |
| Water | 60 |
| B-factors | |
| Protein | 37.0 |
| Ligand/ion | 40.3 |
| Water | 34.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.321 |
Values in parentheses are for highest-resolution shell.
NMR spectroscopy
As described previously9, NMR experiments were performed at 25 °C on a Bruker Avance II 600 MHz or a Bruker Avance III 500 MHz spectrometer equipped with a triple resonance z-axis gradient CryoProbe. All NMR samples contained 0.1–0.25 mM 15N-labeled protein, and final samples were prepared as 90% H2O/10% D2O in a buffer containing 20mM sodium phosphate, 20 mM sodium acetate, 50 mM sodium chloride and 0.05% sodium azide (pH 5.5). Titrations were executed using a sequence of 1H-15N HSQC experiments following incremental additions of an unlabeled peptide. NMR data were processed using the NMRPipe24 and XEASY25 packages. Par-6 peak assignments were determined in previous work for PDZ156–255 9, CRIB-PDZ130–255 7, and CRIB-PDZQ144C/L164C 9.
Fluorescence polarization
All fluorescence experiments were executed using a Photon Technology International spectrofluorometer as previously described9. Using the buffer conditions described above, a series of N-terminally Rhodamine B-labeled peptide samples at 150 nM were prepared with increasing concentrations of protein. Fluorescence polarization values were recorded using 560 and 585 nm wavelengths for excitation and emission, respectively. Binding curves were nonlinearly fitted to an equation describing 1:1 binding stoichiometry, as previously described26. The reported uncertainties in fitted Kd values represent estimates from the nonlinear regression algorithm, and agree closely with the standard deviation of replicate experiments.
Isothermal titration calorimetry
ITC experiments were performed on a Microcal VP-ITC microcalorimeter (Thermo) at 25 °C. Measurements were collected by titration of peptide (2 mM) into a 0.2 mM Par-6 protein solution. Peptide and protein buffer conditions were 20 mM sodium phosphate, 20 mM sodium acetate, 50 mM sodium chloride, and 0.05% sodium azide (pH 5.5). Prior to beginning each experiment, pH of peptide and protein solutions were verified to be within 0.02 pH units of 5.5 at 25 °C, and each solution was degassed for 5–10 minutes at 10 °C prior to start of experiments. C-values were ~12 and ~105 for VKESLV and GEERLI, respectively. Identical calorimetry experiments were performed without protein present for each of VKESLV and GEERLI to account for the heat of dilution. Data were fitted according to a one-site binding model using the ORIGIN 5 software (Thermo).
Results
Par-6 PDZ binds a Crb C-terminal peptide
Members of the Crumbs protein family share a conserved C-terminal sequence (Fig. 1A) that was implicated in the interaction of human Crb3 with the Par-6 PDZ domain6. Most of the Crb family proteins end with the amino acid sequence –PEERLI, which shares only the −1 and −3 positions in common with the VKESLV peptide that was used as a C-terminal PDZ ligand in previous studies of Par-6. To confirm that the Crb C-terminus can interact directly with Par-6, we collected 2D 1H-15N HSQC spectra of the PDZ domain (PDZ156–255) of D. melanogaster Par-6 in the presence of increasing amounts of a peptide containing the conserved Crb C-terminus (LPPEERLI) (Fig. 1B). Significant shifts were observed for many amino acids in and near the PDZ ligand binding cleft consistent with the specific interaction reported for Crb and Par-6. We solved the X-ray crystal structure of LPPEERLI bound to PDZ156–255 to 1.8 Å resolution as summarized in Table 1. Residues 156, 157, 254, and 255 were not visible in the electron density for PDZ156–255. No density was observed for the N-terminal leucine of the LPPEERLI peptide ligand. The proline residues were placed into interpretable electron density but exhibited significantly higher B-factors (~55 Å2) than the C-terminal peptide residues (~30 Å2). The Crb peptide occupies the PDZ binding cleft in a manner typical of other PDZ-ligand complexes (Fig. 1C) and forms antiparallel β-sheet hydrogen bonds with the β2 strand of the PDZ domain.
Figure 1. The conserved Crb C-terminus binds the Par6 PDZ domain.
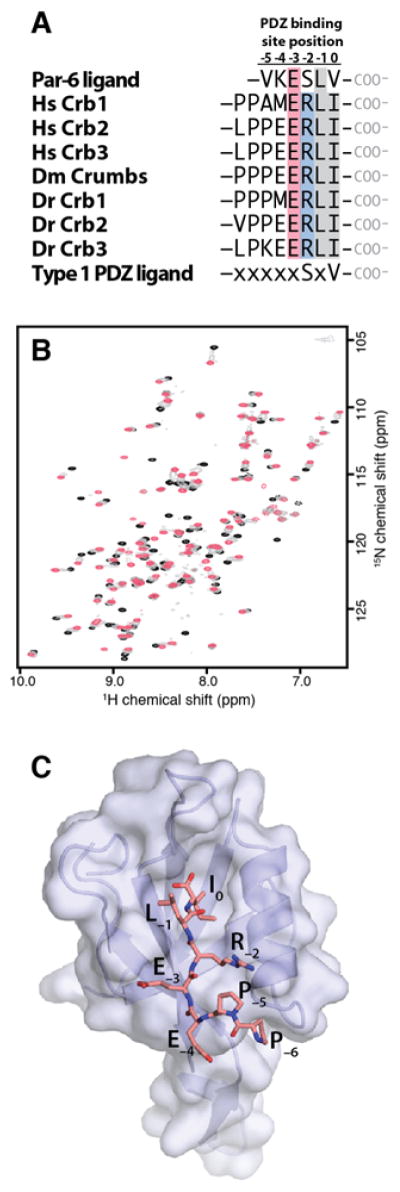
A. C-terminal sequences from human, fly and zebrafish proteins in the Crumbs family. Conserved hydrophobic (gray) and charged (red/blue) residues are highlighted. B. 1H-15N HSQC spectra of PDZ156–255 in the presence of 0 (black contours), 0.5 (gray) and 1 (red) molar equivalent of LPPEERLI. C. X-ray crystal structure of PDZ156–255 :LPPEERLI complex.
NMR comparison of Crb and VKESLV binding to Par-6 CRIB-PDZ
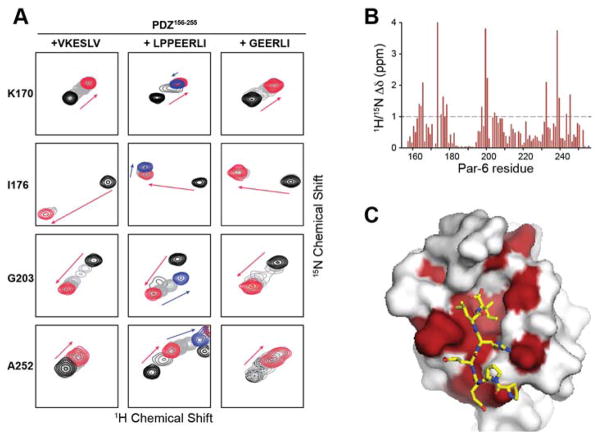
We previously solved NMR structures of the Par-6 PDZ156–255 9, the CRIB-PDZ module (CRIB-PDZ130–255)7 and a disulfide-stabilized CRIB-PDZ (CRIB-PDZQ144C/L164C)9. CRIB-PDZQ144C/L164C mimics Cdc42 activation by linking the CRIB domain to the β1–2 loop of the PDZ and constraining the dipeptide switch in the high affinity configuration. In previous NMR studies of CRIB-PDZ130–255, the VKESLV peptide exhibited simple two-state binding similar to the linear peak shifts observed upon VKESLV binding to PDZ156–255 (Fig. 2A). In contrast, at least seventeen HSQC peaks in the NMR titration of PDZ156–255 with LPPEERLI shift in different directions at low and high peptide concentrations (Fig. 2A). Most peaks exhibit fast or intermediate exchange titration behavior, with more pronounced broadening approaching slow exchange for peaks with larger shift differences. The same pattern of non-linear peak shifting was observed in titrations with CRIB-PDZ130–255 (Fig. S1), but the affected residues do not define a clear secondary binding site when mapped to the PDZ156–255 structure (Fig. S2). We speculated that the di-proline motif of LPPEERLI might be a contributing factor and synthesized a shorter peptide with the sequence GEERLI. Simple two-site binding with linear peak shifts was observed in NMR titrations with the GEERLI peptide (Fig. 2A and Fig. S3), so it was used for all subsequent studies of Crb peptide binding. To ensure that the truncated GEERLI peptide bound the Par-6 PDZ similarly to LPPEERLI, we mapped GEERLI-induced 1H/15N chemical shift perturbations (Fig 2B) to the surface of the LPPEERLI:PDZ156–255 crystal structure (Fig 2C). Residues that experience large (> 1 ppm) combined chemical shift perturbations upon GEERLI binding are located in or adjacent to the binding site occupied by LPPEERLI in the structure of the complex.
Figure 2. Detection of a proline-dependent alternative binding mode.
A. Representative peaks from HSQC titrations of Par-6 PDZ156–255 with VKESLV (left) shift in a linear pattern consistent with saturable binding (red arrows and contours) at a single site in the fast exchange regime on the chemical shift time scale. Binding of 1 molar equivalent of LPPEERLI (middle) causes similar perturbations (red arrows and contours) to the same residues but ~17 peaks, including residues G167, I176, E190, F196, R199, G203, G204, I219, N222, E225, M237, M238, I247, T248, V249, K250, and A252, move in a different direction in the presence of excess peptide (blue arrows and contours) consistent with binding at a second, low affinity site. An equivalent titration with a modified Crumbs peptide lacking the proline residues (GEERLI) (right) displays saturable binding (red arrows and contours) with no evidence of an alternative binding mode. B. Par-6 PDZ156–255 1H/15N chemical shift perturbations induced by GEERLI binding. C. Significant shift perturbations (combined 1H/15N shift > 1 ppm) are highlighted on the surface of the Crb:PDZ156–255 crystal structure. LPPEERLI peptide is shown in yellow sticks.
Comparison of Crb and VKESLV binding affinities
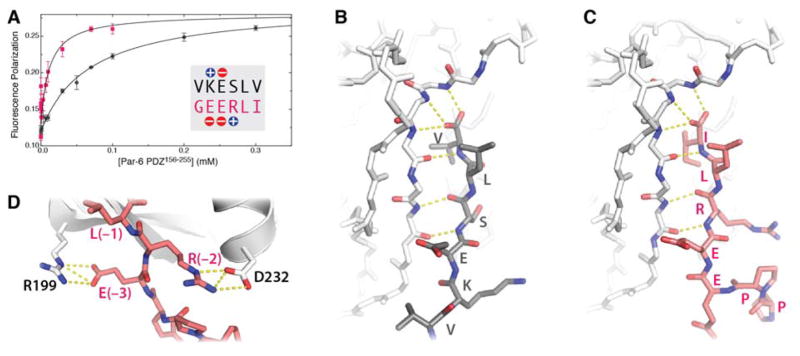
We speculated that GEERLI might bind the Par-6 PDZ domain with higher affinity than the VKESLV peptide, since it represents a functional link between the Crumbs and Par polarity complexes27. We compared GEERLI and VKESLV binding to PDZ156–255 by fluorescence polarization using rhodamine-labeled peptides (Fig. 3A). VKESLV bound with relatively low affinity (Kd = 72 ± 5 μM) consistent with previous results9. In contrast, GEERLI bound PDZ156–255 with six-fold higher affinity (Kd = 12 ± 3 μM).
Figure 3. Crb binds Par-6 with higher affinity than the VKESLV peptide ligand.
A. Fluorescence polarization measurements of rhodamine-labeled VKESLV (black) or GEERLI (red) peptides yielded Kd values of 72 ± 5 and 12 ± 3 μM, respectively. B. Intermolecular β-sheet hydrogen bonds in the VKESLV:PDZ156–255 complex (PDB ID 1RZX). C. An equivalent set of PDZ-ligand hydrogen bonds is observed in the Crumbs:PDZ156–255 crystal structure. D. Both C-terminal peptide ligands contain a glutamic acid at the −3 position that interacts with R199 of Par-6. The arginine side chain of GEERLI makes an additional electrostatic contact with Asp 232 at the start of the PDZ156–255 α-helix.
We compared the structures of PDZ156–255 bound to each peptide to identify specific contacts that might enhance the binding of the Crb peptide relative to VKESLV. As shown in Figures 3B and C, each peptide makes virtually identical main chain contacts with the β2 strand of the PDZ, forming six intermolecular hydrogen bonds typical of other PDZ ligands. Both peptides make a favorable electrostatic contact between a glutamic acid side chain at the −3 and the R199 side chain of Par-6, as shown for GEERLI in Figure 3D. An arginine at the −2 position of GEERLI contacts D232 in a similar manner. The salt bridge or hydrogen bonds arising from this interaction would likely contribute the ~1 kcal/mol to ΔGbind that corresponds to the six-fold higher affinity of GEERLI, since the serine at this position in VKESLV does not make an equivalent contact with the PDZ.
We reasoned that the additional electrostatic contacts or hydrogen bonds contributed by R(−2) of the Crb peptide should enhance the enthalpic contributions to the free energy of binding relative to VKESLV. Attempts to measure VKESLV binding to PDZ156–255 or CRIB-PDZ130–255 by isothermal titration calorimetry (ITC) failed due to the relatively low affinity of the interaction. However, the disulfide-stabilized CRIB-PDZQ144C/L164C protein permitted a direct comparison of VKESLV (Fig. 4A) and GEERLI (Fig. 4B) binding by ITC, which yielded Kd values of 17 ± 1 and 1.9 ± 0.1 μM, respectively. As summarized in Table 2, the Crb peptide binds with a two-fold more favorable change in enthalpy compared to VKESLV, consistent with the additional contacts observed in the crystal structures. Interestingly, the entropic contribution to GEERLI binding is five-fold less than VKESLV, leading to an overall difference in free energy of binding (ΔΔG) of ~1.3 kcal/mol.
Figure 4. Comparison of FP and ITC assays for Par-6 PDZ ligand binding.
A. ITC of VKESLV binding to CRIB-PDZQ144C/L164C. B. ITC of GEERLI binding to CRIB-PDZQ144C/L164C. Fitted parameters for FP and ITC data presented in Table 2. C. FP of VKESLV (black circles) and GEERLI (red circles) to CRIB-PDZQ144C/L164C.
Table 2.
Isothermal titration calorimetry results for CRIB-PDZQ144C/L164C binding
| VKESLV | GEERLI | |
|---|---|---|
| N | 1.09 | 1.05 |
| ΔH (cal/mol) | −3400 ± 40 | −7210 ± 30 |
| ΔS (cal/(mol*T) | 10.3 | 2.0 |
| Kd (μM) | 17 ± 1 | 1.9 ± 0.1 |
Because FP measurements of peptide biding rely on an N-terminally linked rhodamine B to generate the fluorescence signal, nonspecific PDZ-dye interactions could influence the measured affinity. We measured binding of VKESLV and GEERLI to CRIB-PDZQ144C/L164C by FP (Fig. 4C) and obtained Kd values of 13 ± 1 and 2.5 ± 0.4 μM, respectively, in reasonably close agreement with the values from ITC, providing additional validation of the FP assay for Par-6 PDZ ligand binding.
Cdc42 regulates Crumbs binding
The hallmark of VKESLV binding to Par-6 is a 9-fold increase of binding affinity upon allosteric activation of the Par-6 CRIB-PDZ130–255 protein associated with the binding of Cdc42-GTP to the CRIB module 7, 9. We subsequently showed that Cdc42-GTP accomplishes this activation by flipping a ‘dipeptide switch’ comprised of the L164 and K165 sidechains in the β1–2 loop of the PDZ domain. In the low affinity conformation, represented by the NMR structure of PDZ156–255 9, the hydrophobic L164 sidechain is buried in the PDZ core and K165 extends into solution (Fig. 5A). When Cdc42-GTP binds Par-6, it stabilizes a CRIB-PDZ interface that flips the dipeptide switch, placing K165 in the PDZ core near the carboxylate binding pocket (Fig. 5B). The high affinity configuration of the L-K dipeptide switch is also attained upon binding of the C-terminal ligand VKESLV (Fig. 5C). We inspected the crystal structure of the Crb: PDZ156–255 complex and found that dipeptide switch in the ‘K165 in’ high affinity arrangement (Fig. 5D).
Figure 5. Crumbs binding to Par-6 is allosterically regulated by Cdc42.
A. NMR structure of PDZ156–255 (PDB ID 2LC7). The side chains of L164 (white sticks) and K165 (blue sticks) are oriented into and away from the PDZ core, respectively, corresponding to the low affinity configuration of the dipeptide switch. B. X-ray crystal structure of Cdc42:CRIB-PDZ130–255 (PDB ID 1NF3) shown in teal. The side chains of C164 (tan) and K165 (blue) are in the high-affinity dipeptide switch configuration with the lysine side chain projecting into the carboxylate binding pocket. C. X-ray crystal structure of the PDZ156–255:VKESLV complex (PDB ID 1RZX). The VKESLV peptide is displayed in gray. D. X-ray crystal structure of the PDZ156–255:Crb complex. The GEERLI peptide is displayed in pink. E. FP measurements of GEERLI binding to CRIB-PDZ130–255 (black circles) and Cdc42:CRIB-PDZ130–255 (black squares).
Based on the close structural similarities exhibited by the L164/K165 dipeptide switch in different complexes, we reasoned that binding of the Crb peptide to the CRIB-PDZ130–255 module should also be allosterically regulated. We measured GEERLI binding to CRIB-PDZ130–255 in the presence or absence of Cdc42 loaded with GMPPNP (a hydrolysis-resistant GTP analog) (Fig. 5E). Based on the change in Kd from 7.2 to 1.2 μM, it is apparent that Cdc42 association with the CRIB-PDZ130–255 module exerts an equivalent allosteric enhancement on the binding of both C-terminal peptide ligands, as summarized in Table 3. These results show that, while the Crb C-terminal peptide binds with higher affinity to Par-6 than does VKESLV, it remains sensitive to the activation state of the PDZ domain.
Table 3.
Par-6 PDZ:peptide binding affinities from fluorescence polarization.
| VKESLV | GEERLI | |||
|---|---|---|---|---|
|
| ||||
| Construct | Kd (μM) | ΔΔGactivation(kcal/mol) | Kd (μM) | ΔΔGactivation (kcal/mol) |
| PDZ156–255 | 72 ± 5 | −0.2 | 12 ± 3 | −0.3 |
| CRIB-PDZ130–255 | 54 ± 5 | – | 7.3 ± 0.8 | – |
| CRIB-PDZQ144C/L164C | 13 ± 1 | 0.8 | 2.5 ± 0.4 | 0.6 |
| Cdc42:CRIB-PDZ130–255 | 6 ± 2 | 1.3 | 1.2 ± 0.1 | 1.1 |
Discussion
The network of polarity complexes is assembled from of an array of stable and dynamic protein-protein interactions. The Par complex is linked to Crumbs and Pals1, which are both members of the Crumbs complex, by mutually exclusive interactions with the Par-6 PDZ domain. Our previous work showed that an internal sequence in Pals1 binds the Par-6 CRIB-PDZ module with a Kd of 6 μM28. In this work we demonstrated that the Par-6:Crumbs interaction is modulated by Cdc42-GTP binding to the CRIB-PDZ module and shifts the Kd from 7 to 1 μM. The Crb:PDZ structure presented here also reveals specific contacts that likely contribute significantly to the enhanced binding affinity relative to the previously studied VKESLV peptide ligand.
A hallmark of Par-6 function is its ability to segregate to either the apical or leading edge of a cell upon activation by Cdc42-GTP29, 30. One implication of the comparison of Pals1 and Crumbs peptide binding affinities is that Cdc42 activation (by exchange of GTP for GDP) may alter the function or localization of Par-6 by favoring the interaction with Crumbs over the interaction with Pals1. Interestingly, the C-terminal residues of Crumbs can bind to other binding partners, as revealed by crystal structures of the conserved ERLI motif bound to the PDZ domain of Pals131 and the FERM domain of moesin32. In the case of Pals1, a membrane-associated guanylate kinase (MAGUK) protein, the typical PDZ interaction is augmented by contributions from the adjoining SH3 and GK domains, which together form a functional unit that binds the Crb C-terminal domain with very high affinity (Kd ~ 0.1 μM)31. Given the complex network of protein-protein interactions within and between the three major polarity complexes, additional studies will be needed to fully define the role of Cdc42-regulated PDZ binding in the function of Par-6.
In previous work, we observed conformational exchange dynamics in the Par-6 PDZ10 that are quenched when the high-affinity conformation is stabilized by C-terminal ligand binding33. We also showed that the disulfide-linked CRIB-PDZQ144C/L164C mutant, which mimics Cdc42 binding, is ~2.5 kcal/mol more stable than CRIB-PDZ130–255 33. Thus, a portion of the Cdc42-GTP:Par-6 binding energy (~10 kcal/mol; Kd ~ 50 nM8) is required to stabilize intramolecular contacts at the CRIB-PDZ interface. Our current results show that when Cdc42-GTP binds the CRIB-PDZ module, the binding energies of GEERLI and VKESLV increase by similar margins (ΔΔGactivation ≈ 1.3 kcal/mol, Table 3). This suggests that the allosteric switch of the Par-6 CRIB-PDZ module is tuned to transfer a specific portion of the Cdc42-GTP:Par-6 binding energy to flip the dipeptide switch to the high-affinity configuration for any C-terminal ligand.
The accumulating structural data illustrates how the conformational plasticity of the Par-6 PDZ domain supports interactions with multiple ligand types, including the internal sequence ligand Pals128. Given the generality of the dipeptide switch mechanism for C-terminal ligands of its PDZ domain, Par-6 likely interacts with other proteins in a Cdc42-regulated manner. For example, the Crb3 protein is expressed only in epithelial cells16, 34 yet Par-6 activation by Cdc42-GTP is required during T lymphocyte chemotaxis17, 19, 35–37. The modular nature of Par-6 allostery as illustrated by its interaction with Crumbs may enable the regulated assembly and localization of different polarity complexes depending on the tissue type and cellular context, many details of which remain to be discovered.
Supplementary Material
ABBREVIATIONS
- PDZ
PSD-95/Dlg/ZO-1
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- ITC
isothermal titration calorimetry
- FP
fluorescence polarization
Footnotes
Supporting Information Available. 1H-15N HSQC titrations of Par-6 CRIB-PDZ with Crb peptides LPPEERLI or GEERLI. Surface mapping of residues with nonlinear shift patterns upon LPPEERLI binding to Par-6.
References
- 1.Wang Q, Margolis B. Apical junctional complexes and cell polarity. Kidney international. 2007;72:1448–1458. doi: 10.1038/sj.ki.5002579. [DOI] [PubMed] [Google Scholar]
- 2.Bilder D. PDZ domain polarity complexes. Current biology : CB. 2003;13:R661–662. doi: 10.1016/s0960-9822(03)00599-2. [DOI] [PubMed] [Google Scholar]
- 3.Ngok SP, Lin WH, Anastasiadis PZ. Establishment of epithelial polarity--GEF who’s minding the GAP? J Cell Sci. 2014;127:3205–3215. doi: 10.1242/jcs.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris BZ, Lau FW, Fujii N, Guy RK, Lim WA. Role of electrostatic interactions in PDZ domain ligand recognition. Biochemistry. 2003;42:2797–2805. doi: 10.1021/bi027061p. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Hurd TW, Margolis B. Tight junction protein Par6 interacts with an evolutionarily conserved region in the amino terminus of PALS1/stardust. J Biol Chem. 2004;279:30715–30721. doi: 10.1074/jbc.M401930200. [DOI] [PubMed] [Google Scholar]
- 6.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 7.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Molecular cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 8.Garrard SM, Capaldo CT, Gao L, Rosen MK, Macara IG, Tomchick DR. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. Embo J. 2003;22:1125–1133. doi: 10.1093/emboj/cdg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitney DS, Peterson FC, Volkman BF. A conformational switch in the CRIB-PDZ module of Par-6. Structure. 2011;19:1711–1722. doi: 10.1016/j.str.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitney DS, Peterson FC, Kovrigin EL, Volkman BF. Allosteric Activation of the Par-6 PDZ via a Partial Unfolding Transition. Journal of the American Chemical Society. 2013 doi: 10.1021/ja400092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. Journal of cell science. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruewer M, Hopkins AM, Hobert ME, Nusrat A, Madara JL. RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am J Physiol Cell Physiol. 2004;287:C327–335. doi: 10.1152/ajpcell.00087.2004. [DOI] [PubMed] [Google Scholar]
- 14.Gerard A, Mertens AE, van der Kammen RA, Collard JG. The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. The Journal of cell biology. 2007;176:863–875. doi: 10.1083/jcb.200608161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroschewski R, Hall A, Mellman I. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nature cell biology. 1999;1:8–13. doi: 10.1038/8977. [DOI] [PubMed] [Google Scholar]
- 16.Mertens AE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16:308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, Muthuswamy SK. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer research. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prehoda KE. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb Perspect Biol. 2009;1:a001388. doi: 10.1101/cshperspect.a001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Medina E, Arsanto JP, Le Bivic A. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W, WCC Processing of X-ray diffraction data collected in oscillation mode. 1997 doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic acids research. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The Program XEASY for Computer-Supported NMR Spectral Analysis of Biological Macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 26.Tyler RC, Peterson FC, Volkman BF. Distal interactions within the par3-VE-cadherin complex. Biochemistry. 2010;49:951–957. doi: 10.1021/bi9017335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 28.Penkert RR, DiVittorio HM, Prehoda KE. Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nature structural & molecular biology. 2004;11:1122–1127. doi: 10.1038/nsmb839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 30.Etienne-Manneville S, Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol. 2003;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Wei Z, Yan Y, Wan Q, Du Q, Zhang M. Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17444–17449. doi: 10.1073/pnas.1416515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei Z, Li Y, Ye F, Zhang M. Structural basis for the phosphorylation-regulated interaction between the cytoplasmic tail of cell polarity protein crumbs and the actin-binding protein moesin. The Journal of biological chemistry. 2015;290:11384–11392. doi: 10.1074/jbc.M115.643791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney DS, Peterson FC, Kovrigin EL, Volkman BF. Allosteric activation of the Par-6 PDZ via a partial unfolding transition. Journal of the American Chemical Society. 2013;135:9377–9383. doi: 10.1021/ja400092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochimica et biophysica acta. 2008;1778:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. Journal of cell science. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 36.Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 37.Walker SJ, Brugge JS. SePARating polarity and proliferation in ErbB2 oncogenesis. Nat Cell Biol. 2006;8:1220–1222. doi: 10.1038/ncb1106-1220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.