Abstract
In recent years, the capability of synthetic biology to design large genetic circuits has dramatically increased due to rapid advances in DNA synthesis technology and development of tools for large-scale assembly of DNA fragments. Large genetic circuits require more components (parts), especially regulators such as transcription factors, sigma factors, and viral RNA polymerases to provide increased regulatory capability, and also devices such as sensors, receivers, and signaling molecules. All these parts may have a potential impact upon the host that needs to be considered when designing and fabricating circuits. DNA microarrays are a well-established technique for global monitoring of gene expression and therefore are an ideal tool for systematically assessing the impact of expressing parts of genetic circuits in host cells. Knowledge of part impact on the host enables the user to design circuits from libraries of parts taking into account their potential impact and also to possibly modify the host to better tolerate stresses induced by the engineered circuit. In this chapter, we present the complete methodology of performing microarrays from choice of array platform, experimental design, preparing samples for array hybridization, and associated data analysis including preprocessing, normalization, clustering, identifying significantly differentially expressed genes, and interpreting the data based on known biology. With these methodologies, we also include lists of bioinformatic resources and tools for performing data analysis. The aim of this chapter is to provide the reader with the information necessary to be able to systematically catalog the impact of genetic parts on the host and also to optimize the operation of fully engineered genetic circuits.
1. Introduction
The focus of synthetic biology has been the design and implementation of small-scale genetic circuits (Elowitz and Leibler, 2000; Ham et al., 2008; Tabor et al., 2009), including the transplantation and reconstruction of small metabolic pathways in suitable hosts (Lee et al., 2008; Steen et al., 2010). The focus on small systems reflected, in part, the laborious processes of DNA fragment construction and assembly required to optimize designed systems. The rapid expansion of DNA synthesis capacity (Czar et al., 2009; Tian et al., 2009) and the development of simple protocols for large-scale assembly of DNA fragments (Gibson et al., 2008, 2009) have broadened the potential focus of synthetic biology. However, larger synthetic circuits require more components (Voigt, 2006), and their reliable operation requires accurate assessments of the impact of each of these components on the host cell processes. When such circuits overburden the host, mutations will rapidly accumulate to relieve the stresses that are introduced. An accurate assessment of the impact of synthetic circuits on host physiology will enable intelligent choice of the circuits chosen for implementation.
The components of synthetic circuits and their impact on the host can be broadly classified into two categories: (1) Regulatory components comprised transcription factors, sigma factors, and viral RNA polymerases, which enable controlled expression of individual circuit components. Importantly, the DNA sequence specificities of the regulators may result in aberrant and possibly deleterious gene expression within the host. (2) Circuit devices comprised sensors, receivers, signaling molecules, enzymes, etc. These components receive information and then command the cell to perform a task, such as producing chemicals and fuels, secreting proteins, and sending out communication signals. Individual circuit components may be deleterious to the host when overexpressed. Additionally, certain combinations of components may be deleterious even when the individual components have no deleterious effect. Consequently, it is importantly to monitor and catalog the impact of individual and combinations of circuit components on the host in order to facilitate the design process, the choice of components for a particular circuit, and troubleshooting of large synthetic circuits. In addition, a good understanding of the impact of components may facilitate modification of the host to better tolerate the circuit. For example, high level expression of some proteins can result in the accumulation of unfolded products within the cytoplasm, triggering the cytoplasmic heat shock response. This can be relieved by overexpression of cytosolic chaperones.
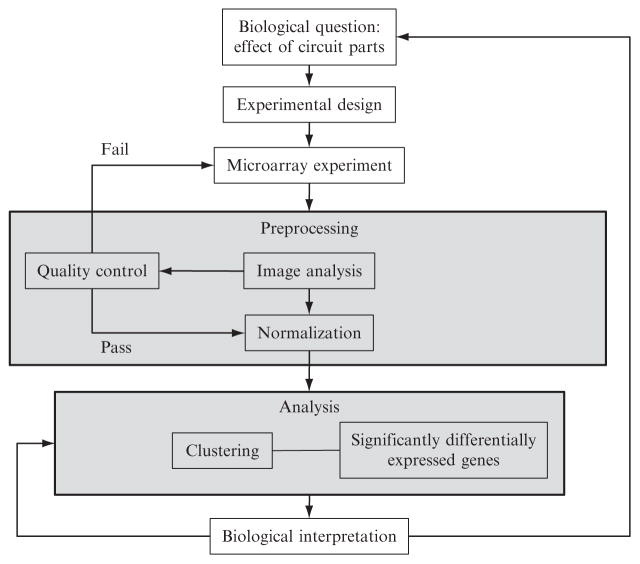
DNA microarrays provide an easy way to monitor changes in gene expression in the host (Rhodius et al., 2002). They can be used to pinpoint the effects of regulator parts of genetic circuits and provide a useful tool for identifying stress-response pathways that are upregulated in response to circuit devices. In this chapter, we will describe the process of performing microarray experiments and associated data analysis to monitor gene expression. The overall process is illustrated in Fig. 4.1 and involves the following steps: (1) Experimental setup: the biological question addressed, experimental design, and performing the microarray experiment(s); (2) Preprocessing: data quality control and normalization prior to analysis; and (3) Analysis: the statistical tools that identify significantly differentially expressed genes, clustering to identify coregulated genes or similar datasets, and functional annotation to identify common and/or enriched properties of the gene products in the final datasets. We discuss each step and indicate the utility of this technology for synthetic biology.
Figure 4.1.
Flowchart of the microarray process.
2. Different Microarray Platforms
The selection of microarray platforms is summarized in Table 4.1. Early microarrays used cDNA libraries, oligonucleotides, or PCR products fabricated by individual laboratories and printed onto polylysine- or epoxy-treated glass slides. Although inexpensive, the process is laborious, results can be inconsistent and usually limited to a single datapoint for each open reading frame (ORF). Commercially available platforms range from low density arrays with a single printed oligonucleotide probe per ORF, to various high-density platforms with multiple probes per ORF, in which the oligonucleotides are synthesized in situ. Additionally, tiled arrays have oligonucleotide probes that anneal to overlapping targets across the genome and some platforms contain probes for intergenic regions, both coding and noncoding strands. Many platforms duplicate their probes across the array surface to reduce hybridization artifacts, thereby improving data reliability. On most platforms, it is possible to perform competitive two-color hybridizations, in which both the reference sample and experimental sample are labeled with different dyes and hybridized to the same array, thereby increasing experimental efficiency. Affymetrix arrays are designed for one-color hybridizations; consequently, the reference and experimental sample are hybridized to two separate arrays. The advantage of one-color hybridizations is their flexibility in comparing samples: only one reference hybridization is required for multiple experimental hybridizations, and all samples can be directly compared with each other when calculating gene expression ratios.
Table 4.1.
Different microarray platforms
| Probea | Attachmentb | No. of features/arrayc | Hybridizationd | Supplier | Notese |
|---|---|---|---|---|---|
| 25 mers | Photolithography | 1.3 × 106~11 probes/ORF | One-color | Affymetrix (NimbleExpress), www.affymetrix.com | E. coli/custom: Requires specialist equipment |
| ≤60 mers | Photodeposition | 1 × 385K, 4 × 72K ≤22 probes/ORF | One- or two-color | NimbleGen, www.nimblegen.com | All sequenced bacteria/ custom-reusable × 3 |
| 35–40 mers | Electrochemical detritylation | 12K ~3 probes/ORF | Two-color | CombiMatrix, www.combimatrix.com | Custom-reusable × 3 |
| <60 mers | Ink jet in situ synthesis | 4 × 44K, 8 × 15K | Two-color | Oxford Gene Technology, www.ogt.co.uk | Select bacteria/custom |
| 60 mers | Ink jet in situ synthesis | 4 × 44K, 8 × 15K | Two-color | Agilent, www.agilent.com | No bacteria/custom |
| 50 mers | Self-spot | 1 probe/gene | Two-color | Ocimum, www.ocimumbio.com | Select bacteria/custom/ oligo sets |
| 70 mers | Self-spot | 1 probe/gene | Two-color | Operon, www.operon.com | Select bacteria/custom/ oligo sets |
| 65 mers | Self-spot | 1 probe/gene | Two-color | Sigma-Aldrich, www.sigmaaldrich.com | Select bacteria/custom/ oligo sets |
Probe length (nt): mer, oligonucleotide.
Method of probe synthesis and attachment to array surface. Self-spot, user self-spots oligos onto glass slides.
Number of features per array and number of probes per gene: K = 1000 (e.g., 72K = 72,000).
Sample hybridization: one-color, one sample is hybridized to the array (often labeled Cy3); two-color, competitive hybridization of two samples labeled Cy3 and Cy5.
Notes on whether arrays are predesigned for specific organisms or can be custom designed. Some arrays can be reused up to three times. Oligo sets are for self-spotting.
The choice of array platform depends upon the type of microarray experiment being performed. The advantage of commercial high-density arrays is that they provide multiple probes often in duplicate for each coding region, which increases data sensitivity and reliability. These probe sets are optimized for signal sensitivity and to reduce cross-hybridization with other homologous genomic sequences. Probes designed toward known ORFs are sufficient when the user is only concerned about the expression status of known annotated genes and transcribed regions. However, several studies demonstrate that there are large number of transcripts from nonannotated regions of the genome, such as intergenic regions and also antisense to known coding regions (Filiatrault et al., 2010; Guell et al., 2009; Selinger et al., 2000; Sharma et al., 2010). The functions of most of these transcripts are unknown, but likely include mRNAs of previously unannotated short coding regions (Hemm et al., 2008, 2010), sRNAs that regulate specific target mRNAs (Beisel and Storz, 2010), and regulatory antisense transcripts (Thomason and Storz, 2010). Consequently, high-density platforms that contain probes to both coding and noncoding strands and intergenic regions provide more comprehensive analysis of the transcriptome. It is also possible to custom design arrays to include specific probes of your choice, which is useful if you need to probe specific regions of the genome not covered in the commercial array sets. Finally, many commercial arrays are available in multiplex format in which the array surface is subdivided into sections. Each section contains a complete set of probes, enabling different samples to be separately hybridized to each section in the same experiment, thereby increasing the throughput of experiments.
Several studies have addressed the issue of data quality and reproducibility between commercial array platforms (Bammler et al., 2005; Irizarry et al., 2005; Larkin et al., 2005; Shi et al., 2006, 2008). Satisfyingly, these studies find that the results across platforms are remarkably consistent, and the observed fold change in gene expression levels correlates closely with qRT-PCR. However, it is important to standardize protocols for RNA labeling, hybridization, array processing, data acquisition, and normalization. If the array experiments are part of a consortium, agreeing upon a common array platform, strains and growth conditions will greatly facilitate the comparability of datasets produced in different laboratories. The final choice of array platform may also depend on the availability of additional resources in the host institute that are required for microarray experiments such as array hybridization stations and scanners. These are often quite expensive and limited to certain array platforms, and are often shared between multiple laboratories within institutes.
Deep sequencing (RNA-seq) is rapidly becoming a viable alternative for expression analysis (Cho et al., 2009; Filiatrault et al., 2010; Guell et al., 2009; Sharma et al., 2010; Sorek and Cossart, 2010; Wang et al., 2009). In this approach, purified RNA is converted to cDNA flanked with adaptamers that are then sequenced using high-throughput sequencers. The sequence reads are mapped back to the genome and the frequency of reads provides a digital read-out of RNA levels. The main advantage of this approach is that all transcripts are sequenced; consequently, the quantitative information is not limited to existing genomic sequences or restricted to annotations of ORFs. In addition, RNA-seq can provide nucleotide resolution information of transcript boundaries (5′ ends, operon structures, etc.), a greater dynamic range of expression values compared to microarrays and requires small amounts of RNA. However, microarrays currently are cheaper to perform, easier to high-throughput for large experiment sets, ideal for characterizing expression of known RNAs, and both the experimental and data handling issues have been optimized over several years.
3. Experimental Design
Microarray experiments can easily generate large lists of differentially expressed genes making it difficult to unravel the underlying biology. Consequently, careful experimental design is critical for interpretable data. Optimal datasets are where only a few cellular systems are disrupted by a designated perturbation (e.g., induced overexpression of candidate gene). Here, the response usually occurs within a short time frame of the perturbation and therefore can be monitored by following gene expression over a short time course (Nonaka et al., 2006; Rhodius et al., 2006). In contrast, cells exposed to long-term or steady-state differences (e.g., wild-type vs. constitutively expressing mutant strain) can generate complex responses as a result of a cascade of transcriptional effects, making data interpretation difficult.
The gene expression comparisons considered here will be the consequence to the host cell of expressing either a single part or device of a synthetic circuit or a combination of components. First, it is important to establish a standardized expression data set for all circuit parts. This enables cross-comparisons of the effects of different components on host gene expression and therefore can be used as a guide for selecting parts when designing and constructing a circuit. The aim is to catalog the short-term effects of induced overexpression of components within the host under a series of defined growth conditions. Here, gene expression is monitored after induction using a time course in which samples are removed at intervals for up to 1 doubling of culture growth (e.g., at 5, 10, 20, and 40 min after induction). Note that it is essential to standardize the defined growth conditions and the control wild-type host strain used in the transcriptome experiments to enable easy comparison of data between laboratories and scientific communities. It may also be necessary to examine the effects of components under the specific operating conditions of the proposed genetic circuits, especially if these conditions are dramatically different to the standardized conditions or involve long-term expression over several days. This may involve performing much longer time courses of part overexpression (e.g., at 8 h intervals for 48 h), or monitoring the effects of parts or even complete circuits under the specific growth conditions. This will enable fine-tuning of the circuit design and also provide information for whether it is necessary to modify the host in order to optimize circuit performance.
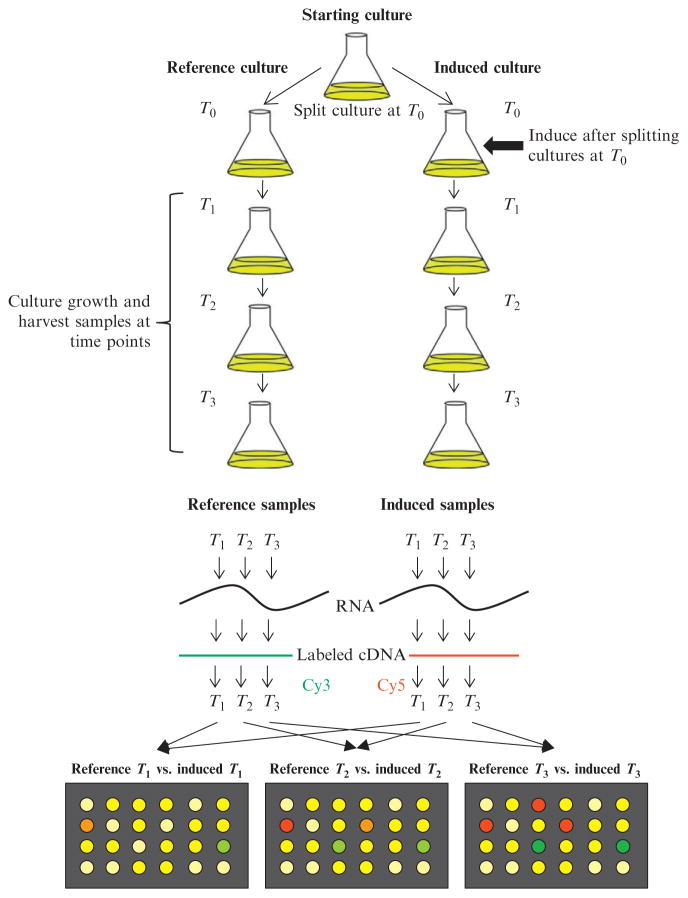
Time-course expression analysis is an effective method for yielding information about the succession of transcriptional changes induced by any component of a circuit. Typically, monitoring expression for 1 doubling after induction is sufficient for identifying direct and any indirect effects, as by that point, the induced protein will reach at least 50% of its maximum induced levels in the cell. Overexpression of a transcription factor part will likely result in a rapid transcriptional response, even if this is as a result of aberrant gene expression; however, overexpression of a device that may exert stresses upon the cell may take longer for transcriptional effects to become apparent as these will likely be indirect. Typically, RNA samples are harvested at select time intervals after induction and are compared on microarrays with samples harvested at equivalent time points from a noninduced control culture. This is best achieved by splitting the starting culture into two aliquots immediately prior to induction: inducing one aliquot and maintaining the other as a control (Fig. 4.2). Note that using completely separate starting cultures for any microarray comparison is inferior as there is increased biological variation in the separate cultures, and hence increased variability in the expression profiles. Harvesting samples at comparable time points from both the control and induced culture will correct for any growth state changes that may occur over the duration of the time course. Sometimes, overexpression of a protein may alter the growth rate of the induced culture. In this case, it is best to harvest samples at comparable culture densities to compensate. However, it is still important to be aware of growth rate differences when interpreting the final gene expression datasets as expression of the ribosomal operons are growth rate dependent and hence will likely be differentially expressed.
Figure 4.2.
Time-course microarray comparison for analyzing “part” effects. Example of a time-course comparison in which the original starting culture is split into two separate cultures ("Reference" and “Induced”) at T0. Immediately afterward, inducer (e.g., IPTG) is added to the “Induced” culture to overexpress the desired circuit part. Both cultures are maintained in identical growth conditions and samples from each culture harvested at the same time points after induction. From each sample, RNA is isolated and cDNA prepared. cDNA from the references samples is labeled with Cy3 and from the induced samples labeled with Cy5. For each time point, Cy3- (reference) and Cy5-labeled (induced) cDNAs are then mixed and hybridized to an array.
4. Experimental Variation
Experimental variation derives either from biological or technical issues. Variability in biology is more difficult to control and can come from issues with the biological sample, the growth conditions of the culture, and alteration in gene expression levels. We discuss each of these in turn and then conclude with the technical issues contributing to variability. Issues related to the biological sample itself are best described by its complexity, quantity, and quality.
Complexity relates to whether the samples are simple, that is, one organism under defined growth conditions (e.g., single homogenous cultures), or complex, that is, several organisms or across different growth states (e.g., mixed cultures, including biofilms and host-pathogen models). Sample complexity can dramatically increase biological variation due to poor reproducibility of growth conditions, variability of organism composition of mixed populations, and the isolation of total RNAs from multiple organisms that result in increased cross-hybridization and decreased signal specificity with target probes on microarray. Most biological sample effects are minimized when monitoring part effects under defined growth conditions; however, if the proposed operating conditions of the genetic circuit involve complex environments, then this will dramatically increase the biological variability and consequently the expression profiles measured under these conditions.
Sample quantity is a factor, especially from complex environments that are not standard cultures and therefore are difficult to grow in large enough quantities. Low sample yield reduces signal strength on microarrays, thereby increasing noise and signal variability.
Sample quality relates to the purity and integrity of the isolated RNA and labeled cDNA probes prior to hybridization on the microarray. For example, contaminating RNases reduces RNA quality: RNases vary with different host strains, growth conditions, and cellular stress levels.
Growth conditions are another important source of variation, even with defined homogenous cultures. Batch experiments relate to growing cultures in flasks and generally have low or moderate levels of variability. Samples compared from separate starting cultures have higher levels of variability than samples from the same culture. Consequently, it is generally better to split a culture and to induce gene expression in one culture and use the other as a comparative control (see Fig. 4.2). Experiments performed in chemo-stats tend to have low background variability; however, long chemostat runs can result in the accumulation of mutations, especially in stressed cultures, which can dramatically increase variability from run to run. Defined media, such as M9 (and others) decreases experiment to experiment variability, compared with complex media such as Lennox broth, where the composition of the broth can vary between lot numbers. Finally, the expression levels of weakly expressed genes are inherently more variable due to the sensitivity of probes on the array.
Technical variation comprises sample preparation and labeling, sample hybridization, and microarray slide variability (Yang et al., 2002). As discussed earlier, microarray variability has been shown to be small between most commercial arrays and largest with home-made arrays. Typical sources of variation include differences in array platform and probe design. This latter category includes probe length, specificity, cross-hybridization issues, and different probe sequences for the same target genes on different array platforms. For large-scale studies, these effects can be minimized by using the same array platform, and if possible, arrays manufactured from the same batch. Sample preparation, labeling, and hybridization will be discussed in the next section. With large-scale studies, confounding effects by extraneous factors can be minimized by orthogonalizing variable components, for example, by using equal numbers of arrays from different batches.
5. Sample Preparation
Sample preparation includes RNA harvesting, cDNA synthesis and labeling with fluorescent dyes, and sample hybridization on the microarray. Many array manufacturers recommend protocols for these steps, some of which are specific to the microarray platform (e.g., Affymetrix). In addition, there are many published protocols, for example, Beyhan and Yildiz (2007), Botwell and Sambrook (2003), and Rhodius and Wade (2009). Here, we will discuss important issues of sample preparation and present protocols successfully utilized by many laboratories for two-color hybridizations on most array platforms. Sample preparation involves harvesting the biological material, extracting and purifying the RNA, generating cDNA containing the modified nucleotide amino-allyl dUTP (aa-dUTP) and covalently linking the Cy3 or Cy5 fluorophores to the cDNA samples. Typically in two-color hybridizations to the same array, the reference sample is labeled with Cy3 (scans as red) and the experimental with Cy5 (scans as green). After labeling, both samples are mixed and hybridized to the same array. Reverse labeling in which the dyes are switched (reference is Cy5 and experimental Cy3) is often not necessary, as most dye-dependent biases are removed during data normalization (see later).
5.1. Materials
5 .1.1. Solutions
All solutions should be RNase free, either by treating with 0.1% DEPC or by purchasing as RNase free from suppliers such as Ambion.
Ethanol/phenol stop solution: H2O-saturated phenol (pH < 7.0) in ethanol (5%, v/v).
Lysozyme solution: 500 μg/mL lysozyme in 10 mM Tris (pH 8.0), 1 mM EDTA (pH 8.0). Prepare fresh just before use.
Nuclease-free water, Ambion #AM9938.
Sodium acetate, pH 5.5, 3 M (100 mL), Ambion #AM9740.
25× aa-dUTP/dNTP mix: 12.5 mM dATP/dGTP/dCTP, 5 mM dTTP, 7.5 mM aa-dUTP (Ambion, #8439). The 3:2 ratio of aa-dUTP:dTTP is optimized for Escherichia coli based on the (G + C) content of the template cDNA. Higher GC content organisms may require a lower aa-dUTP to dTTP ratio; lower GC content organisms may require a higher ratio.
1 M KPO4, pH 8.5: 9.5 mL 1 M K2HPO4, 0.5 mL KH2PO4. Prepare daily to ensure optimal pH.
Phosphate wash buffer: 5 mM KPO4, 80% EtOH. May be slightly cloudy. Prepare daily.
Phosphate elution buffer: 4 mM KPO4, pH 8.5. Prepare daily.
20× SSC (1 L), Ambion #AM9763.
SDS, 10% solution (100 mL), Ambion #AM9822.
5 .1.2. Supplies
All microfuge tubes and pipette tips should be RNase and DNase free.
TURBO DNA-free Kit, Ambion #AM1907. Contains TURBO DNase, 10× buffer, and DNase inactivation reagent.
Random octamer oligonucleotide, any oligo company.
SuperScript III Reverse Transcriptase, Invitrogen #18080-093. Includes 5× first-strand synthesis buffer.
MinElute PCR purification kit (50), QIAGEN #28004.
CyDye postlabeling reactive dye packs, GE Healthcare #RPN5661.
5.2. Sample harvesting
Careful isolation of RNA from biological samples is essential in order to accurately capture the RNA profile present at time of harvesting. Transcription profiles can rapidly change in response to the cellular stresses of harvesting (media change, centrifugation, temperature change, etc.). Also, RNA is extremely sensitive to degradation by RNases present both in the harvested cells and introduced during the purification procedure. Consequently, it is essential to use RNase-free materials and solutions throughout the purification and cDNA synthesis steps. It is also essential to “freeze” the RNA profile during sample harvesting. This is achieved by mixing the cells with a reagent designed to inactivate transcription and prevent RNA degradation; for example, RNAprotect (QIAGEN); a solution of 40% methanol, 62.5 mM HEPES, pH 6.5 at −45 °C (Pieterse et al., 2006); or 5% acid phenol in ethanol stop solution as outlined below:
Transfer 10 mL of culture (0.3 − 1 × 109 cells/mL) into a 15-mL conical tube containing 1.25 mL of ice-cold ethanol/phenol stop solution.
Harvest cells by centrifugation at 6700 × g for 2 min at 4 °C. Remove media by aspiration.
Rapidly freeze cell pellet in liquid nitrogen. Store at −80 °C until required.
5.3. Total RNA preparation
There are multiple RNA purification methods and kits available. The method of choice will be determined by the properties of your strain (growth conditions, ease of lysis, and level of RNases), the experimental design, and the quantity of available sample versus yield of RNA required for each array experiment. Consequently, it may be necessary to modify protocols to enhance lysis or to cope with high levels of endogenous RNases. It is also important to note that some kit protocols that have affinity purification steps may not give representative yields of small RNAs. Total RNA from bacterial cultures can be isolated using the hot phenol method outlined below, or by using several commercial reagents (e.g., TRIzol, Invitrogen), or kits (e.g., RNeasy, QIAGEN; RiboPure, Ambion). Also, there are protocols and kits available that (1) enrich mRNA from total RNA preparations by using oligos that bind to 16S and 23S rRNA, enabling these RNAs to be subtracted from the RNA prep (e.g., MICROBExpress, Ambion); (2) purify bacterial RNA from complex host-bacterial samples using a similar oligo subtraction method to remove contaminating rRNA and poly-A mRNAs from select eukaryote hosts (e.g., MICROBEnrich, Ambion); and (3) amplify RNA from low yield samples using a linear cDNA amplification step (Gao et al., 2007). We find that for E. coli cultures the hot phenol method outlined below yields the best quality RNA preps in terms of RNA integrity, size, purity, and yield.
Resuspend the cell pellet (from 10 mL of culture) in 800 μL lysozyme solution. Transfer lysate to 2-mL microfuge tube containing 80 μL of 10% SDS, mix by inversion, and incubate at 64 °C for 2 min.
Add 88 μL 1 M sodium acetate solution (pH 5.2) and mix by inversion.
To the lysate add an equal volume (~1 mL) of H2O-saturated phenol (pH < 7.0). Mix by inverting 10 times. Incubate in a 64 °C water bath for 6 min, continuing to mix the tube contents by inverting every 40–60 s.
Place tube on ice to chill for 2 min. Afterward, centrifuge at 16,000×g for 10 min at 4 °C.
Remove the upper aqueous phase into a fresh tube, taking care not to disturb the interface (this is a common point of RNase contamination of preps). Also, perform this step quickly, as the aqueous layer can rapidly become cloudy after centrifugation, making it difficult to separate the layers. If this happens, recentrifuge the sample.
Add to the solution an equal volume (~1 mL) of 1:1 mix of H2O-saturated phenol:chloroform. Invert the tube 6–10 times to mix and centrifuge at 16,000×g for 2 min.
Carefully remove the upper aqueous phase to a fresh microfuge tube. Repeat the H2O-saturated phenol:chloroform extractions until the interface is clear (usually ≥ 2–3 times). Some strains may require extensive phenol:chloroform extractions to completely remove contaminating RNases.
Divide the final extracted solution equally between two 1.5-mL micro-fuge tubes. Precipitate by adding 0.1 volume 3 M sodium acetate (pH 5.5) and 2.5 volumes 100% cold ethanol. Incubate at −80 °C for 30 min.
Recover the RNA by centrifugation at 16,000×g for 30 min at 4 °C.
Wash the RNA pellet with 1 mL 80% cold ethanol. Centrifuge at 16,000×g for 5 min at 4 °C. Carefully remove the ethanol solution by aspiration and dry the RNA pellet in a speed vacuum.
Redissolve and pool each pair of pellets in a final volume of 87 μL and place in a fresh 1.5-mL microfuge tube.
5.4. DNase treatment
To each RNA preparation (87 μL), add 10 μL 10× TURBO DNase buffer and 3 μL TURBO DNase. Incubate reaction at 37 °C for 30 min.
Add an additional 3 μL TURBO DNase and incubate a further 30 min at 37 °C.
Add 10 μL DNase inactivation reagent and incubate at room temp, mixing four times.
Centrifuge at 10,000×g for 1.5 min and transfer the supernatant containing the RNA to a fresh tube. Store at −20 °C until required.
5.5. Assessing RNA quality and yield
Determine RNA concentration by measuring the absorbance of a 1:100 dilution in H2O at 260 nm (concentration, c (μg/μL), in a 1-mL quartz cuvette with a 1 cm path length: c = A260 × f × 0.04 μg/μL, where f is the dilution factor). Typical yields are 70–300 μg RNA from 10 mL of culture, depending on strain of E. coli, growth conditions, and culture density upon harvesting.
Check purity by measuring the absorbance ratio of nucleic acid versus protein (A260/A280) of a 1:100 dilution in 10 mM Tris–HCl buffer, pH 7.5 (note that the absorbance ratio is sensitive to pH: as RNA is acidic, the ratio must be measured in a low salt neutral buffer). Good RNA preps free from protein contamination give values between 1.8 and 2.1.
If required, the integrity of the RNA can be analyzed on a denaturing formaldehyde 1% agarose gel (Ausubel et al., 1998). Upon visualizing the gel, the 23S and 16S ribosomal RNA should be easily observed. For good RNA, the 23S species should be twice as intense as the 16S with little or no smearing between or below these bands.
5.6. cDNA synthesis and RNA hydrolysis
cDNA is synthesized using random octamer primers and a dNTP mix containing aa-dUTP (aa-dUTP).
In 0.2-mL PCR tube, mix 15 μg total RNA with 16 μg random octamer primer to give a final volume of 35 μL. Incubate at 70 °C for 10 min and then chill on ice for 10 min.
cDNA synthesis reaction: Prepare a cocktail on ice containing for each reaction: 12 μL 5× first-strand synthesis buffer; 2.4 μL 25× aa-dUTP/ dNTP mix; 6 μL 0.1 M DTT, 2.3 μL SuperScript III RT; 2.3 μL H2O. Add 25 μL cocktail to annealed RNA sample and incubate at 50 °C for 3 h.
RNA is removed from the completed reverse transcription reaction by hydrolysis. To the sample, add 1.2 μL 0.5 M EDTA and 6 μL 1 N NaOH and incubate for 10 min at 65 °C.
Neutralize the reaction by adding 65 μL 1 M HEPES, pH 7, and mix well.
5 .7. Sample cleanup
Unincorporated aa-dUTP and competing free amines must be removed from the sample to enable successful coupling of the amino-allyl cDNA with the Cy3/Cy5 dyes. Consequently, Tris-based buffers cannot be used in the following cleanup steps. Each sample is cleaned using the QIAGEN MinElute PCR Purification Kit that has been modified, replacing the QIAGEN wash and elute buffers (PE and EB), which contain free amines, with phosphate-based wash and elute buffers (Beyhan and Yildiz, 2007; Hasseman, J., TIGR Aminoallyl Labeling of RNA for Microarrays & TIGR Microarray Labeled Probe Hybridization).
Remove the reverse transcription reactions from the PCR tube to a fresh 1.5-mL microfuge tube.
Add 500 μL QIAGEN Buffer PB to each sample.
Load samples on to MinElute Columns and centrifuge at ≥10,000×g for 1 min.
Discard the flow-through and add 750 μL phosphate wash buffer to each column and centrifuge at ≥ 10,000×g for 1 min.
Discard the flow-through and centrifuge again at ≥10,000×g for 1 min.
Add 10 μL phosphate elution buffer to the center of each column matrix, incubate for 1 min, and then elute using a fresh collection tube by centrifugation at ≥ 10,000 × g for 1 min. Samples can be stored at −20 °C if required.
5.8. Cy3/Cy5 coupling
The Cy dyes are shipped as a desiccate in sealed packs. Note that they are extremely sensitive to light and moisture, therefore each pack is opened and resuspended in DMSO immediately prior to use. Each pack is sufficient for approximately five reactions.
To each cDNA sample, add 1 μL 1 M sodium bicarbonate, pH 9.0 (note the bicarbonate becomes carbon dioxide with time; therefore, use fresh solution <1 month old).
Resuspend each fresh tube of Cy3 or Cy5 in 10 μL DMSO. Keep in dark until ready for use.
Add 2 μL of either Cy3 or Cy5 solution to each cDNA sample. Incubate for 2 h at room temperature in the dark.
Unincorporated Cy dyes are removed using the QIAGEN MinElute PCR purification kit following the procedure previously described in sample cleanup, steps 2–6. Note that the eluted samples should be colored red for Cy3 and blue for Cy5.
-
cDNA yield and labeling efficiency of the eluted samples can be calculated by using a nanodrop to measure their absorbance at 260, 550, and 650 nm against a water blank (Botwell and Sambrook, 2003). For each sample, calculateWhere
324.5 pg/pmol average MW of a dNTP
vol = sample volume (μL)
Optimal labeling values for hybridizations are incorporation of >150–200 pmol dye per sample and <20 nucleotides/dye molecule.
It is preferable to use the eluted samples the same day for hybridization, storing in the dark until required. For longer storage or if the hybridization volumes are small (<20 μL), combine pairs of Cy3 and Cy5 samples (2 μg each cDNA) to be hybridized together, dry down in a speedvac in the dark. Store at −20 °C.
5 .9. Sample hybridization
Hybridization protocols, sample volumes, and quantities vary depending on the microarray platform and hybridization chamber. For some array platforms, the hybridization sample is applied under a lifter-slip placed over the array on the glass slide. The slide is then placed in a small hybridization chamber and incubated between 42 and 65 °C (depending on the length of the array probes) for up to 16 h. In general, these hybridizations require large sample volumes (≥ 40 μL) and are prone to sample drying and uneven hybridization intensities over the array surface, resulting in poor or discarded data. Recently, several high-density array platforms (e.g., Nimblegen) use special hybridization systems (e.g., MAUI hybridization system) that dramatically improve hybridization efficiency. In these cases, mixers are applied over the array surface creating a sealed chamber, enabling the application of small sample volumes that are actively mixed during the hybridization process. This generates an even hybridization, minimizes sample evaporation, increases signal sensitivity, increases reproducibility, and shortens hybridization times. Below, we give a sample hybridization mix used for arrays with lifter-slips (volume = 50 μL): volumes can be scaled accordingly, maintaining the correct final concentration of SSC, HEPES, and SDS. Note water and all solutions must be filtered (e.g., with a 0.2 μm filter) to prevent small particles damaging the surface of the array.
For each hybridization, combine Cy3 and Cy5 sample pairs, using 2μg cDNA for each sample, in a 0.2-mL microfuge tube (if combined samples were dried down, resuspend in 10 μL H2O).
To each hybridization reaction, add 7.5 μL 20× SSC, 1.25 μL 1 M HEPES, pH 7.0, 1.25 μL 10% SDS and H2O to a final volume of 50 μL (3× SSC, 25 mM HEPES, 0.25% SDS final).
Incubate reaction at 99 °C for 2 min and then allow to cool at room temperature for 5 min. Lightly vortex sample to mix and spin down before applying to surface of microarray, following the hybridization instructions for your microarray and hybridization chamber.
5.10. Slide washing and scanning
Prior to scanning, hybridized slides are washed to remove any sample nonspecifically bound to the slide surface. As slide washing protocols will vary according to the manufacturer, we give a washing protocol commonly used for oligo and ORF PCR arrays printed onto polylysine-coated slides. Note that all wash stock solutions should be filtered before using. After washing, the Cy dyes are extremely unstable: Cy5 is rapidly degraded by ozone in minutes (Branham et al., 2007; Fare et al., 2003). Slides should be dried and scanned in a low ozone chamber; alternatively, some companies supply wash solutions that stabilize the dyes (e.g., Agilent).
Prepare the following wash solutions in glass slide dishes: two glass slide dishes each containing 500 mL Wash Solution I (897 mL Milli-Q-water, 100 mL 20× SSC, 3 mL 10% SDS). Place an empty slide rack in one of the dishes. If using oligo arrays, Wash Solution I should be preheated to 60 °C and poured into the slide dishes immediately prior to washing the slides. This is essential to remove nonspecific hybridization on oligo arrays; two glass slide dishes each containing 500 mL Wash Solution II (950 mL Milli-Q-water, 50 mL 20× SSC)and one glass slide dish containing 500 mL Wash Solution III (495 mL Milli-Q-water, 5 mL 20× SSC).
Carefully remove slide from the hybridization chamber; keeping the array level, submerge into the slide dish containing Wash Solution I with no slide rack.
Once submerged, using fine forceps carefully remove the cover slip or mixer-assembly following the manufacturer’s instructions, taking care not to scratch the surface of the array.
After removing the cover, place the array on the rack in the second slide dish containing Wash Solution I.
Repeat steps 2–4 for any other remaining slides. When finished, plunge the rack up and down 10–20 times.
Immediately transfer the slide rack to Wash Solution II, and plunge up and down for 60 s.
Drain the rack for 5 s, and then place in the second dish containing Wash Solution II and plunge up and down for 60 s.
Drain rack for 5 s, and then transfer to Wash Solution III and plunge up and down for 60 s.
Dry the arrays by centrifugation at 600 rpm for 2 min in a low ozone chamber.
Scan the arrays as soon as possible in a low ozone chamber to reduce degradation of Cy5.
There are several scanners and software available for processing slides and the generated image files; some are specific to certain slide platforms (e.g., Affymetrix). One of the most popular systems that handles several different slide platforms is the GenePix scanner and software from Molecular Devices and also SpotReader from Niles Scientific. Users should scan their slides following the manufacturer’s instructions. Finally, it is also possible to reuse slides from some manufacturers (e.g., Nimblegen) for up to three subsequent hybridizations without significant loss of hybridization signal by using a series of slide wash steps that remove the hybridized sample (stripping) but leaves the probes intact. Specific protocols are supplied by the manufacturer.
6. Microarray Preprocessing
For two-color arrays, each slide is excited at two wavelengths, 532 nm for the Cy3 (green)-labeled reference sample and 650 nm for Cy5 (red)-labeled experiment sample, to measure the fluorescence of the hybridized samples to each probe (feature) on the array. Note that during array scanning it is important to individually adjust the scanning voltages at each wavelength, which in turn controls the detected fluorescent intensities. This is required to (1) approximately balance the signal intensities from each channel and (2) to ensure an optimal signal dynamic range for the features on the array, reducing the number of saturated (overexcited) probes while maximizing the detection of weakly fluorescing probes. Scanning generates two high-resolution 16-bit tiff images (one for each channel) that contain the fluorescence intensities for each feature. Image files are also generated when scanning one-color Affymetrix arrays: here, two slides are scanned to generate separate image files for the reference and experimental samples. Preprocessing involves several steps that analyze the image files to generate a single expression ratio for each gene represented on the array. This involves
Image analysis and quality control. The fluorescent intensities of every feature are determined by overlaying a grid on each image file to map the location and identity of each feature, and to quantify their specific signal intensity and surrounding background signal. This generates separate data files for each channel that lists all features and their associated specific and background fluorescent intensities. From this preliminary analysis diagnostic reports can be generated on the quality of the hybridization. These measure the quality of grid alignment with the features, calculate average specific versus background intensity ratios for all features, the number of saturated features and features with signals above threshold, and assess the uniformity of the background and specific signals to determine if there was any bias in the hybridization across the array surface.
Probe set summarization. High-density arrays contain multiple probes for each gene. Summarization collates the fluorescent values of probe sets to generate a single intensity for each gene, and also discards or reduces the influence of any probes within the set that have unusual fluorescence values.
Within and between array normalization. Prior to calculating gene expression ratios between the reference and experiment data sets, it is important to correct for any systematic errors in fluorescence measurements and sample quantification. Within array normalization corrects for systematic errors between the two compared samples; between array normalization corrects for systematic errors across multiple arrays (experiments), enabling more accurate comparisons of expression ratios across multiple experiments. Normalization is discussed in more detail in the next section.
Most companies provide associated software for array preprocessing, and additional software is freely available that enables more advanced preprocessing and array normalizations (Table 4.2). Note that Bioconductor is an open source project that provides many high quality tools and documentation for microarray data analysis (Table 4.2). These tools run in the programming language, R, and whilst the learning curve is steep, they are highly recommended for the serious microarray analyst! Here, we focus on tools and procedures that are commonly available across freely available software. First, we present common diagnostic plots used to assess general features of the expression ratios and subsequent data normalization.
Table 4.2.
Free tools and resources
| Software | Functions | URL/Reference |
|---|---|---|
| R | Open source statistical computing environment | http://www.r-project.org/ |
| Bioconductor | Open source software for bioinformatics that runs in R. Extensive tools for preprocessing, normalization, cluster, statistical and gene set enrichment analysis | http://www.bioconductor.org/ |
| TM4 Microarray Software Suite | Four major software packages. Image analysis, normalization, clustering, statistical and gene set enrichment analysis | http://www.tm4.org/ (Saeed et al., 2006) |
| GenePattern | Genomic analysis platform. Clustering, statistical and gene set enrichment analysis | http://www.broadinstitute.org/cancer/software/genepattern/ (Reich et al., 2006) |
| limmaGUI | Linear Models for microarrays Graphical User Interface. Normalization, diagnostic plots, differential expression. Also available in Bioconductor (R/limma) | http://bioinf.wehi.edu.au/limmaGUI/ (Wettenhall and Smyth, 2004) |
| RMAExpress | Robust Multichip Average. Background adjustment, quantile normalization and probe summarization for Affymetrix Genechip data | http://rmaexpress.bmbolstad.com/ (Bolstad et al., 2003; Irizarry et al., 2003a,b) |
| SAM | Significance Analysis of Microarrays. Statistical technique for differential expression; gene set analysis. Available as Excel macro, Bioconductor (R/sam) and in TM4 suite (MeV) | http://www-stat.stanford.edu/~tibs/SAM/ (Storey, 2002; Taylor et al., 2005; Tusher et al., 2001) |
| Cyber-T | Statistics program for differential expression. Also available for Bioconductor (R/hdarray; R/bayesreg; R/bayesAnova) | http://cybert.microarray.ics.uci.edu/ (Baldi and Long, 2001) |
| Cluster 3.0 + Java TreeView | Clustering algorithms and visualization tools | http://bonsai.hgc.jp/~mdehoon/software/cluster/ |
6.1. Diagnostic plots of gene expression ratios
Plots of the gene expression ratios provide very useful information on the quality of array experiments and facilitate comparisons between experimental repeats and across experiment sets. Note that in all subsequent data analysis, the gene expression ratios are log transformed: log2 (R/G), where R = Cy5 intensities (experimental) and G = Cy3 intensities (reference). Generally, the Cy3 and Cy5 values are background subtracted, which is the default setting on most software programs. However, this can decrease the accuracy of determining expression ratios for weakly expressed genes with signals close to background. For this reason, it is common to filter datasets removing features that have signal intensities less than 2–3 standard deviations above the mean background.
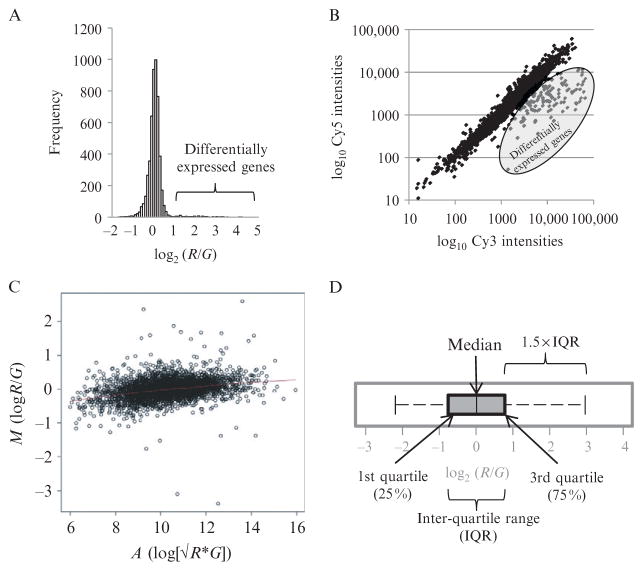
Histograms. The simplest gene expression plot is a histogram of log2 (R/G) expression ratios for all genes on the array (Fig. 4.3A). Most gene expression experiments only alter a small fraction of genes. Consequently, good histograms should contain a single symmetrical peak with no shoulders, and have a narrow distribution with only a small number of genes with large expression ratios in the distribution tails.
Scatter plots. These are plots of log Cy3 versus log Cy5 fluorescent intensities for every gene, and hence are more informative as they provide a visual overview of Cy3 and Cy5 signals for every gene (Fig. 4.3B). these plots should give a linear distribution with a good dynamic range and little scatter from the diagonal.
MA plots. MA plots enable visualization of variation of gene expression ratios (M = log2 [R/G]) as a function of average signal intensity (A = log2 √ [R × G]; Yang et al., 2002). Visually, MA plots are similar to scatter plots rotated clockwise by 45° (Fig. 4.3C). MA plots are good for detecting an artifact that arises when the labeling reaction is nonoptimal; this is apparent as a “skew” in the plot, such that low intensity ratios tend to be more negative (G > R) than high intensity ratios. This can be corrected by intensity-dependent normalization (see later).
Box plots. These are useful for comparing the spread and median of gene expression ratios either between sectors on an array or between arrays (Fig. 4.3D).
Figure 4.3.
Common diagnostic plots for analyzing microarray expression data. (A) Histogram of log expression ratios (log2 [R/G]) from one array. Histogram should be symmetrical with no shoulders. Tails represent differentially expressed genes. (B) Scatter plot of log Cy5 versus log Cy3 fluorescent intensities (background subtracted) of features on an array. Scatter plot should be linear and evenly distributed. Differentially expressed genes are indicated. (C) MA plot of log gene expression ratios (M = log2 [R/G]) versus average intensity (A = log2 √[R × G] for one array. Plot should be linear with good dynamic range (range of A values) (D) Box plot of log gene expression ratios (M = log2 [R/G]) for one array, illustrating the quartile distribution of M values.
6 .2. Data normalization
Normalization scales the red and green intensities within an array or across arrays by a common factor: log2 R/G → log2 R/(kG). Normalization attempts to correct systematic technical differences between the two channels (reference and experiment) or across experiments (multiple arrays). These include unequal RNA quantities, labeling efficiency, biases in measured expression levels, scanner settings, and array batch variations. It is also important that the normalization process maintains the original biological variation in the signal and that the assumptions of the normalization procedure are understood. Normalization requires common references between the two samples that remain unchanged. Possible approaches to normalization are:
Normalization to housekeeping genes. This is not considered to be an acceptable method as there is no evidence for a class of genes that has constant expression under different conditions.
Normalization to reference RNA. In these cases, each sample is spiked with RNA standards from another organism and accompanying target probes are present on the array. The expression values of each channel are then normalized to the standards. This is useful if there are large-scale changes in gene expression and hence the assumptions of global scaling are not valid (described next). However, the disadvantage of this approach is that errors are easily introduced from the accuracy of quantifying the references, application of references to the samples, and their signal intensity measurements from the array.
Normalization by global scaling. The most common form of normalization is by global scaling, which assumes that the total amount of mRNA remains constant under the various experimental conditions and only a small subset of genes change expression. Note, if this assumption is true, then the histogram of expression ratios remains “balanced”: that is, symmetrical with small tails. Simple forms of global scaling sum total mRNA intensities from each channel and scale one to the other. More advanced global normalizations discussed below correct for biases in expression data that are a function of signal intensity or have altered distributions of expression ratios that can be affected by array batches and labeling efficiencies.
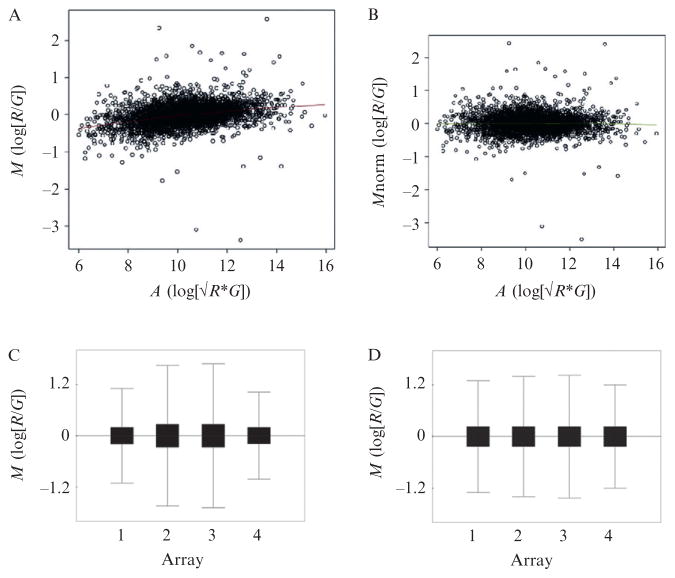
Intensity-dependent normalization (loess smoothing). Gene expression ratios visualized using MA plots often display a skew in the median intensities that is a function of signal intensity and is due to differences in dye stability, efficiency of dye incorporation, and scanner settings (Fig. 4.4A and B). This can be corrected using loess smoothing, in which a robust locally weighted regression curve (loess) is fitted to the data using overlapping windows of signal intensity (typically ~30% of the data). The data is then normalized to the curve such that the distribution of log gene expression ratios is centered on zero across the range of signal intensities (Yang et al., 2002).
Scale normalization. This adjusts the spread of gene expression ratios so that they have similar distributions between different arrays, and hence normalizes for differences in dye stability, dye incorporation, scanner settings, and array batch variations (Yang et al., 2002). Note that the assumption here is that the biological distribution of mRNA expression is similar across multiple experiments. This is applicable across replicates, but can reduce biological information if comparing different conditions that result in dramatically different variations in gene expression distributions; for example, time-course experiments. Simple scale normalization can be visualized using box-plots and involves regularizing the variance of log gene expression ratios across multiple experiments (Fig. 4.4C and D). Quantile normalization regularizes the distribution of probe intensities such that they are same across multiple arrays (Bolstad et al., 2003).
Figure 4.4.
MA and Box plots illustrating normalization of array data. (A) MA plot of prenormalized expression data from one array illustrating the loess curve. (B) MA plot of the same expression data in which the M values have been normalized to the loess curve (loess smoothing). (C) Box plots of four replicate microarray experiments, illustrating differences in distribution of M values. (D) Box plots of same four micro-array data in which the M values have been normalized by standard deviation regularization.
A variety of programs are freely available that perform array preprocessing tasks; some are listed in Table 4.2.
7. Clustering
Clustering is an exploratory data analysis process for datasets containing multiple array experiments. It is used to: discover patterns in the data; group “similar” patterns together either by clustering genes (rows) with similar expression profiles or by clustering arrays/experiments (columns) with similar profiles, or both genes and experiments; reduce the complexity of the data into several distinct patterns; and provide a method to order and organize the data (reviewed in Boutros and Okey, 2005; D’Haeseleer, 2005; Quackenbush, 2001). Consequently, clustering is extremely useful for data visualization, hypothesis generation, and selection of genes for further consideration. Clustering is an extremely useful tool for characterizing the transcriptional effects of circuit parts on the host. First, clustering expression experiments of different parts will identify parts that have similar effects on the host. This is useful for classifying parts into different categories based on their effects on the host, thereby aiding the design of large circuits with multiple parts. For example, the user may decide to select parts that affect different systems in the host to avoid “overstressing” any one particular system. Second, for a specific part, clustering genes across multiple experiments that measure the effect of the part under different growth conditions or across an induction time-course will identify genes with similar expression profiles and therefore aid detection of the cellular systems affected by the part across the different conditions. This is extremely useful, as it is well-documented that genes with similar expression profiles are involved in similar cellular processes and are often transcriptionally coregulated by regulators.
Most microarray data clustering is unsupervised; that is, no prior information (e.g., gene functional categories, operon structure, etc.) is used to guide the clustering. The goal of unsupervised clustering is to discover patterns from the data; however, even if the data is random such clustering assumes that there is still an underlying pattern. Consequently, clustering always works! Therefore it is important to cluster with caution, use different algorithms, and where possible, filter the data so that only genes with the most varied expression profiles are used. When clustering by array/experiment, the aim is to identify conditions that generate similar expression profiles; consequently, it is typical to use all the expression data. However, when clustering by genes to identify coregulated genes it is important to filter the dataset prior to clustering. This serves both to reduce the dataset to a meaningful size and, importantly, to remove genes with expression profiles that have little variance across the dataset, thereby reducing “noise” in the clusters. Commonly used filters are: (1) genes with expression ratios present in a given fraction of experiments; (2) number of genes with expression ratios above a certain threshold in a given fraction of experiments; (3) number of genes with variance or standard deviation of their expression ratios across the experiments exceeding a specified value. Finally, when preparing data for clustering, it is important to use normalized data that has been log transformed.
7.1. Measures of similarity between genes and distance between clusters
The basic principles of clustering will be discussed in the context of clustering only genes; however, these principles also apply to clustering by array/experiment. Clustering measures the similarity in expression profiles between genes and also the distance between clusters of genes with similar profiles. The aim of clustering algorithms is to identify clusters that maximize the similarity of gene expression profiles within clusters whilst maximizing the distance between clusters to obtain distinct clusters. It is also important to be able to identify outliers that contain expression profiles that do not easily fit into any particular cluster; however, only some algorithms do this.
There are two main measures of similarity between genes: correlation coefficients that are scale-invariant; and distance metrics that are scale-dependent. Scale-invariant measures identify similar patterns of “ups” and “downs” in gene expression ratios, while scale-dependent measures also consider the magnitude of the expression patterns. Commonly used correlations and distance metrics are listed in Table 4.3A. Euclidean distance is the most commonly used metric and considers both the patterns of ups and downs and also the magnitude of the expression ratios, thereby preserving more information about the data. There are three commonly used measures of distance between clusters listed in Table 4.3B. The average linkage is the least sensitive to outliers; however, complete distance often generates the most discrete clusters, while single linkage often performs very poorly.
Table 4.3.
Commonly used measures of similarity between genes and distance between clusters
| Measure | Comments |
|---|---|
| (A) Measures of similarity between genes | |
| Correlation (Pearson) | Identifies similar patterns of ups and downs |
| Uncentered correlation (cosine-angle) | Identifies similar patterns of ups and downs and also magnitude |
| Absolute correlation | Identifies similar patterns of ups or downs |
| City Block (Manhatten) distance | Corresponds to the sum of differences across dimensions. Yields diamond shaped clusters and is less sensitive to outliers |
| Euclidean distance | Corresponds to the geometric distance in multidimensional space. Identifies similar patterns of ups and downs and also the magnitude of the patterns. Yields spherical shaped clusters |
| (B) Measures of distance between clusters | |
| Single (minimum) linkage | Shortest distance between members of clusters. Yields elongated clusters and is sensitive to outliers |
| Compact (maximum) linkage | Largest (outside) distance between members of clusters. Yields compact clusters and is sensitive to outliers |
| Average (Mean) linkage | Average distance between members of clusters. Generates “in between” sized clusters and is insensitive to outliers |
7.2. Clustering algorithms
Clustering algorithms can be divided into two types: (1) hierarchical, in which genes with similar patterns are joined together by a dendrogram, for example, hierarchical clustering; and (2) partitioning, in which the data is divided into groups or clusters with similar patterns, for example, Self-Organizing Maps (SOMs) and k-means. Some freely available clustering software is listed in Table 4.2 and simple clustering algorithms outlined below.
Hierarchical clustering. This is an agglomerative process that starts with every gene considered as its own cluster (Eisen et al., 1998). The most similar pair of clusters are joined together to form a parent cluster, then the next most similar pair of clusters are joined together, and the process repeated until there is just one large cluster. During this process, all genes are scored using the similarity metrics, and the distance between clusters measured using the distance metrics. The merging of the clusters is illustrated using a dendrogram connecting each gene and the expression profiles illustrated using a heatmap (Fig. 4.5A). The dendrogram and associated heatmap provides a good overall visual guide of patterns in the data; however, there are several caveats. First, the order of the nodes in the dendrogram is arbitrary, placing genes adjacent to each other that may be not that similar. Some software programs flip the dendrogram nodes to optimize the ordering of genes based on their similarity (Fig. 4.5B). Second, as the agglomerative process of connecting gene expression profiles is rigid and relies on joining the most similar clusters first, any poor clusters generated early in the process affects the quality of clusters later on. Also, unrelated gene expression profiles are eventually joined. Third, it is difficult to define discrete clusters using hierarchical clustering.
Self-organizing maps (SOMs). SOMs is a partitioning algorithm that requires the user to input the desired number of clusters (centroids; Tamayo et al., 1999). The centroids take the form of a grid of dimensions x times dimensions y (e.g., 2 × 3) that is overlain on the data in n dimensional expression space. Next, an initial gene is chosen at random and the closest centroid moved toward that gene and the process repeated in turn for all genes, completing one cycle or iteration. Multiple iterations are performed until the program is terminated. As the initial order of gene selection is random, slightly different clusters are generated with each run; consequently, it is important to perform multiple runs in order to identify stable clusters. The disadvantage of SOMs is that all genes are forced into clusters; consequently, outliers can distort optimal clusters.
k-means. This is also a partitioning algorithm in which the user inputs the desired number of clusters (k) (Tavazoie et al., 1999). The algorithm then chooses k centroids at random that represent random gene expression profiles. Each gene is assigned to the closest centroid until all genes are assigned, then the centroids are reset to the average of their assigned genes (cluster). All genes are then reassigned to the new closest centroid and the process repeated for a defined number of iterations or until no more genes change cluster. Similar to SOMs, k-means generates different clusters in multiple runs depending on the initial centroid positions. Some software programs (e.g., k-means support in MEV; see Table 4.2) take advantage of the random initiation by deliberately rerunning the algorithm multiple times to identify genes that frequently cocluster (i.e., form “consensus” clusters). Here, a user-defined threshold is applied such that gene members are required to cocluster in X% of k-means runs. The advantage of this approach is that robust clusters are generated, giving the user a feel for the significance of clusters. Also, genes that do not commonly cocluster (i.e., outliers) remain unassigned and therefore do not distort existing clusters (see Fig. 4.5C).
Figures of Merit (FOM). The disadvantage of both SOMs and k-means is that the user is required to estimate the optimal number of clusters to describe the data. The FOM algorithm estimates the predictive power of a clustering algorithm and therefore can be used as a guide for determining the optimal number of clusters (Yeung et al., 2001). For example, FOM can be applied by running k-means repeatedly over a range of cluster numbers. This generates a curve of the predictive power of the clustering algorithm versus the number of clusters. The predictive power increases with the number of clusters; hence the curve can be used to select the optimal number of clusters for running k-means.
Figure 4.5.
Clustering algorithms. (A) Hierarchical clustering of top 1.5% most variable genes (59/4249 genes) across 45 experimental conditions (arrays). Color coded heat map indicates log2 (R/G) expression ratio of genes (red = R > G; green = G > R; black = no change). (B) Dendrograms linking four genes illustrating different arrangement of nodes. (C) k-means clustering of same dataset in (A) specifying seven (k) clusters, 10 k-means runs and requirement for genes to cocluster in at least eight runs. Note presence of 10 unassigned genes that did not meet cocluster threshold.
In summary, when clustering gene expression profiles, it is very important to use the most variable profiles for clustering and to “explore” the data using several different clustering algorithms in order to obtain robust clusters. The aim is to identify clusters that describe distinct expression patterns and to identify/remove outliers that can detract from the quality of the obtained clusters.
8. Differential Expression Analysis
Specialized statistical methods are required to identify significantly differentially expressed genes from microarray data (Allison et al., 2006; Dudoit et al., 2003). Use of these methods is essential to reliably identify genes that are perturbed by expression of parts in a host. Data from a single microarray experiment without further experimental validation is insufficient to reliably identify differentially expressed genes. This is because application of a fold cutoff does not take into account the uncertainty in measuring gene expression ratios introduced from biological sample variability and from technical issues. For example, expression ratios of weakly expressed genes are often inherently unreliable as their fluorescence measurements have low signal to noise ratios. Consequently, it is essential to perform replicate experiments from separate biological cultures (i.e., biological rather than technical replicates) to enable the construction of test statistics that incorporate variability estimates for each gene. This provides assessment of the statistical significance (e.g., p-value) in the differential expression values for each gene, enables a cutoff to be applied to identify significantly differentially expressed genes, and also provides an estimate of the type-I (false positive) and type-II (false negative) error rates at each applied cutoff. In addition, prior to statistical analysis, all microarray expression data should be normalized and log2 transformed.
Microarray datasets present unique problems for identifying significantly differentially expressed genes. Statistical analysis of expression measurements of a single gene in condition A versus condition B is relatively straightforward. Here, simple statistics such as the Student’s t-test can be used to derive p-values of whether the mean of the replicated log ratios differ from a null hypothesis of 0, in which there is no change in expression in condition A versus condition B. However, this approach is problematic when scaled up to large datasets such as microarrays that contain measurements of 1000s of genes with few experimental replicates. Here, additional methods are used including modified t-statistics and controls for the increased type-I error rate due to multiple testing.
Modified t-statistic. The t-statistic is derived from the difference in means divided by the sample variance. Due to the few replicates in microarray experiments, gene-specific variance estimates are imprecise, which can result in highly variable t-statistics when applied across 1000s of genes. Specialized microarray statistical methods such as SAM (Tusher et al., 2001), LIMMA (Smyth, 2004), and Cyber-T (Baldi and Long, 2001; see Table 4.2) calculate a modified t-statistic in which the estimated gene-specific variance is combined with a predicted variance derived from all genes on the microarray. This improves the estimate of variance for each gene, thereby increasing the power of the t-statistic. However, this approach assumes that the null distribution of the test statistics is the same across all transcripts and that all transcripts are independent, which is not necessarily true.
Controlling the type-I error rate. Applying a cutoff of p < 0.05 for replicates of a single gene experiment predicts 1 false positive from 20 independent trials. However, microarrays involve multiple statistical testing of 1000s of genes, which dramatically increases the type-I error (false positive) rate for a given α (p-value). For example, applying a cutoff of p < 0.05 for a typical microbial genome of 4500 genes would generate 4500 × 0.05 = 225 genes as false positives even if there is no significant differential gene expression. Consequently, specialized statistical methods are applied to microarray data to correct for multiple testing and thereby control for type-I errors. The Bonferroni correction is a simple method for controlling Family-Wise Error Rate (FWER; probability of making type-I errors) at level α, where p̃ ≤ α. Here, the p-value for each gene (pg) is adjusted: p̃ = Npg, where N = number of genes. However, the Bonferroni correction is a very stringent adjustment as it decreases FWER to 0, which can result in missing many true positives (i.e., false negatives, or type-II error rate), and also assumes independence amongst genes. There are multiple methods for controlling type-I error. The Ŝidák procedure, min P and max T (Westfall and Young, 1993) adjust p-values to control FWER (see also Dudoit et al., 2003). However, often it is more useful to have an estimate of the false detection rate (FDR): that is, for a given threshold, what fraction are false positives (Benjamini and Hochberg, 1995). Methods include controlling FDR below a certain level by adjusting p-values based on their ranking (Benjamini and Hochberg, 1995), and mixture-models that treat genes as either as differentially or not differentially expressed (Allison et al., 2002; Datta, 2005; Do et al., 2005; Pounds and Morris, 2003). One popular microarray statistical tool, SAM (Statistical Analysis of Microarrays; Tusher et al., 2001), provides a method for estimating FDR for a chosen cutoff value of test statistic (Storey, 2002). This is achieved by permuting the datasets to determine if the expression of any of the genes is significantly related to the response. Any test statistic of the permuted dataset exceeding the cutoff is counted as a “false positive.” The appealing feature of SAM is that the user chooses what cutoff to apply based on an FDR they are comfortable in dealing with in their significant data set. Recent versions of SAM have also incorporated a “Miss rate” table that also estimates the false negative rate of genes that do not make the cutoff (Taylor et al., 2005). In addition, SAM generates a q-value for each gene, which describes the lowest FDR rate at which the gene is called significant (Storey, 2002).
Most microarray statistical analysis is one-class where only one response variable is tested; for example, condition A versus condition B. The one-class problem tests whether the mean log expression ratios differ from the null hypothesis of 0. It is also possible to perform two-class comparisons; for example, condition A versus condition B (expt 1) compared against condition A versus condition C (expt 2), where the mean log ratios of expt 1 are compared for significant difference against the mean log ratios of expt 2. This is useful to compare if there are significant difference between different parts when compared to a common control. It is also possible to apply statistical analysis to time-course data to identify significant differentially expressed genes using either a one-class or two-class comparison based on the consistent increase or decrease in gene expression over time (e.g., SAM; Table 4.2).
A common question for statistical analysis of microarray data is: How many replicates are sufficient? Most algorithms require at least four biological replicates to obtain reasonable statistics. However, more replicates increase the statistical power: that is, maximizing the detection of true positives and minimizing FDR. Several approaches are available for estimating sample size (number of replicates) based on the observed variability of gene expression ratios in pilot experiments in order to achieve a desired statistical power (Lee and Whitmore, 2002; Li et al., 2005; Tibshirani, 2006; software SAM, R/size; Table 4.2).
Finally, it is common in many microarray experiments for statistical analysis to yield long lists of significant genes. Consequently, some users to employ both a statistical and a fold cutoff to reduce their candidate list and also employing the reasoning that genes with high-expression ratios are more likely to be directly regulated and therefore easier to biologically interpret. Note that, for published reports, it is necessary to describe the statistics and expected number of false positives within the significant dataset.
9. Data Analysis: Understanding the Perturbation
Biological interpretation of candidate gene lists identified through clustering and by significant differential expression is essential in order to identity the biological processes or systems perturbed by expression of parts. Expression of regulators may result in general aberrant ectopic gene expression due to recognition of miscellaneous sites throughout the genome. However, expression of circuit devices may target specific cellular processes that will likely be reflected in the expression patterns. Several approaches can be used to identify candidate cellular processes (see Table 4.4).
Table 4.4.
Resources for biological interpretation
| Resource | Function | URL |
|---|---|---|
| Metabolic pathways | ||
| KEGG | Kyoto Encyclopedia of Genes and Genomes database providing a metabolic pathway viewer for overlaying gene expression data | http://www.genome.jp/kegg/ |
| EcoCyc | Model organism database for E. coli providing a metabolic pathway viewer for overlaying gene expression data | http://biocyc.org/ecocyc/ |
| Functional categories | ||
| GO | Gene Ontology website provides gene functional classification and tools to analyze overrepresentation among lists of genes | http://www.geneontology.org/ |
| Protein–protein interactions | ||
| EcoCyc | Model organism database for E. coli providing experimentally verified protein–protein interaction networks | http://biocyc.org/ecocyc/ |
| Transcriptional networks and motif-finding algorithms | ||
| EcoCyc | Model organism database for E. coli providing experimentally verified transcription networks | http://biocyc.org/ecocyc/ |
| BioProspector | Motif-finding algorithm suited for two-block motifs | http://ai.stanford.edu/~xsliu/BioProspector/ |
| Consensus | Motif-finding algorithm | http://bifrost.wustl.edu/consensus/ |
| Gibbs Motif Sampler | Motif-finding algorithm | http://bayesweb.wadsworth.org/gibbs/gibbs.html |
| MEME | Motif-finding algorithm | http://meme.sdsc.edu/meme/intro.html |
Metabolic pathways. Expression data can be overlain on metabolic maps in order to identify genes in reactions or pathways that are differentially regulated.
Functional categories. A common query is whether particular functional groups are enriched within datasets. A useful classifier for this purpose is Gene Ontology (GO) that categorizes genes according to their associated biological processes, cellular components and molecular functions. A Fisher’s exact or chi-square test can be performed to determine if a particular GO term is overrepresented within a set of differentially expressed genes compared to the whole genome (additional tools are listed in Table 4.4). There are several limitations to this approach: an arbitrary cutoff is required to identify differentially expressed genes; many differentially expressed genes are required to provide robust statistics; nondifferentially expressed genes are not used; the level of gene expression is not incorporated. A better alternative is to ask whether the genes associated with a GO term are “differentially expressed” within the dataset. This is termed Gene Set Enrichment Analysis (GSEA) and can be tested using Wilcoxon rank sum test, modified Kolmogorov-Smirnov statistic (Subramanian et al., 2005) or using tools available in various software packages (see Table 4.4).
Protein interactions. High-throughput studies of protein–protein interactions have revealed that proteins involved in the same process often interact (Arifuzzaman et al., 2006; Butland et al., 2005). These interaction maps can be used to determine if any protein networks are over-represented within the expression data; however, these datasets may have many false positives as there is little overlap between them. Alternatively, the EcoCyc database has a collection of low throughput (experimentally verified) data (Table 4.4).
Transcriptional networks. Coregulated genes are often controlled by common transcription factors. Consequently, it is often useful to search for overrepresented motifs within the promoter regions of genes that cocluster or are differentially expressed (see algorithms listed in Table 4.4).
10. Closing Remarks
The key to successful microarray experiments are careful experiment design that enables the user to capture the direct effects of the introduced perturbation, in this case, the effect of expressing parts within a host. Systematic analysis of multiple parts requires the use of carefully defined growth conditions and control samples to enable cross-comparison of expression data, both within and between laboratories. Equally important is careful data analysis in order to maximize interpretation of the data. This requires knowledge of the assumptions behind data normalization, clustering, identification of significantly differentially expressed genes to select gene lists, and also biological interpretation to identify known cellular systems that are being modulated. The goal is to be able to identify and understand the cellular stress circuits that are being triggered by expression of parts of circuits. This enables the selection of parts that have orthogonal effects on the host to minimize the impact of the fully engineered circuit, and also to facilitate modifying the host to better tolerate the genetic circuit. Finally, it is important to accurately document the microarray experiment; both as a requirement for publication and also to enable others to repeat, modify, or critically evaluate your work. Sadly, this is a critical problem in the field; a recent study was unable to reproduce 10/18 published micro-array experiments (Ioannidis et al., 2009). Most journals require MIAME compliance (Minimum information about a microarray experiment) for publication of microarray data (Ball et al., 2004; Brazma et al., 2001). This requires uploading array data onto public repositories such as GEO (http://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/), and CIBEX (http://cibex.nig.ac.jp/). All these databases require detailed documentation of experimental design, samples and their preparation, hybridization, array design, and data measurement and analysis.
References
- Allison DB, Gadbury GL, Heo MS, Fernandez JR, Lee CK, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal. 2002;39:1–20. [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: From disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; Hoboken, NJ, USA: 1998. [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: Regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Ball CA, Brazma A, Causton H, Chervitz S, Edgar R, Hingamp P, Matese JC, Parkinson H, Quackenbush J, Ringwald M, Sansone SA, Sherlock G, et al. Submission of microarray data to public repositories. PLoS Biol. 2004;2:E317. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, Cunningham ML, Deng S, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- Beyhan S, Yildiz F. Bacterial gene expression analysis using microarrays. J Vis Exp. 2007;4:206. doi: 10.3791/206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Botwell D, Sambrook J. DNA Microarrays: A Molecular Cloning Manual. Cold Spring Harbor Press; New York: 2003. [Google Scholar]
- Boutros PC, Okey AB. Unsupervised pattern recognition: An introduction to the whys and wherefores of clustering microarray data. Brief Bioinform. 2005;6:331–343. doi: 10.1093/bib/6.4.331. [DOI] [PubMed] [Google Scholar]
- Branham WS, Melvin CD, Han T, Desai VG, Moland CL, Scully AT, Fuscoe JC. Elimination of laboratory ozone leads to a dramatic improvement in the reproducibility of microarray gene expression measurements. BMC Biotechnol. 2007;7:8. doi: 10.1186/1472-6750-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czar MJ, Anderson JC, Bader JS, Peccoud J. Gene synthesis demystified. Trends Biotechnol. 2009;27:63–72. doi: 10.1016/j.tibtech.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Datta S. Empirical Bayes screening of many p-values with applications to microarray studies. Bioinformatics. 2005;21:1987–1994. doi: 10.1093/bioinformatics/bti301. [DOI] [PubMed] [Google Scholar]
- D’Haeseleer P. How does gene expression clustering work? Nat Biotechnol. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- Do KA, Muller P, Tang F. A Bayesian mixture model for differential gene expression. J R Stat Soc C Appl Stat. 2005;54:627–644. [Google Scholar]
- Dudoit S, Shaffer JP, Boldrick JC. Multiple hypothesis testing in micro-array experiments. Stat Sci. 2003;18:71–103. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Fare TL, Coffey EM, Dai H, He YD, Kessler DA, Kilian KA, Koch JE, LeProust E, Marton MJ, Meyer MR, Stoughton RB, Tokiwa GY, et al. Effects of atmospheric ozone on microarray data quality. Anal Chem. 2003;75:4672–4675. doi: 10.1021/ac034241b. [DOI] [PubMed] [Google Scholar]
- Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, Khrebtukova I, Yang Y, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol. 2010;192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Yang ZK, Gentry TJ, Wu L, Schadt CW, Zhou J. Microarray-based analysis of microbial community RNAs by whole-community RNA amplification. Appl Environ Microbiol. 2007;73:563–571. doi: 10.1128/AEM.01771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA., 3rd Onestep assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, Rode M, Suyama M, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE. 2008;3:e2815. doi: 10.1371/journal.pone.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Paul BJ, Miranda-Rios J, Zhang A, Soltanzad N, Storz G. Small stress response proteins in Escherichia coli: Proteins missed by classical proteomic studies. J Bacteriol. 2010;192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, Falchi M, Furlanello C, Game L, Jurman G, Mangion J, Mehta T, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41:149–155. doi: 10.1038/ng.295. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Warren D, Spencer F, Kim IF, Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino G, Griffin C, Hilmer SC, et al. Multiple-laboratory comparison of microarray platforms. Nat Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat Methods. 2005;2:337–344. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- Lee ML, Whitmore GA. Power and sample size for DNA microarray studies. Stat Med. 2002;21:3543–3570. doi: 10.1002/sim.1335. [DOI] [PubMed] [Google Scholar]
- Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: From bugs to synthetic biology to fuels. Curr Opin Biotechnol. 2008;19:556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Li SS, Bigler J, Lampe JW, Potter JD, Feng Z. FDR-controlling testing procedures and sample size determination for microarrays. Stat Med. 2005;24:2267–2280. doi: 10.1002/sim.2119. [DOI] [PubMed] [Google Scholar]
- Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20:1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse B, Jellema RH, van der Werf MJ. Quenching of microbial samples for increased reliability of microarray data. J Microbiol Methods. 2006;64:207–216. doi: 10.1016/j.mimet.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Pounds S, Morris SW. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Wade JT. Technical considerations in using DNA microarrays to define regulons. Methods. 2009;47:63–72. doi: 10.1016/j.ymeth.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius V, Van Dyk TK, Gross C, LaRossa RA. Impact of genomic technologies on studies of bacterial gene expression. Annu Rev Microbiol. 2002;56:599–624. doi: 10.1146/annurev.micro.56.012302.160925. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigma(E) stress response in related genomes. PLoS Biol. 2006;4:43–59. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Perkins RG, Fang H, Tong W. Reproducible and reliable micro-array results through quality control: Good laboratory proficiency and appropriate data analysis practices are essential. Curr Opin Biotechnol. 2008;19:10–18. doi: 10.1016/j.copbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sorek R, Cossart P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc B Stat Methodol. 2002;64:479–498. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: Methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- Taylor J, Tibshirani R, Efron B. The “miss rate” for the analysis of gene expression data. Biostatistics. 2005;6:111–117. doi: 10.1093/biostatistics/kxh021. [DOI] [PubMed] [Google Scholar]
- Thomason MK, Storz G. Bacterial antisense RNAs: How many are there, and what are they doing? Annu Rev Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol Biosyst. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. A simple method for assessing sample sizes in microarray experiments. BMC Bioinform. 2006;7:106. doi: 10.1186/1471-2105-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment. Wiley; New York: 1993. [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]