Abstract
The lymphotropic Herpesvirus saimiri (HVS) causes acute leukemia, T-cell lymphoma, and death in New World monkeys. HVS encodes seven small RNAs (HSURs) of unknown function. The HSURs acquire host Sm proteins and assemble Sm cores similar to those found on the spliceosomal small nuclear RNPs (snRNPs). Here we show that, like host snRNPs, HSURs use the SMN (survival of motor neurons) complex to assemble Sm cores. The HSURs bind the SMN complex directly and with very high affinity, similar to or higher than that of host snRNAs, and can outcompete host snRNAs for SMN-dependent assembly into RNPs. These observations highlight the general utility of the SMN complex for RNP assembly and suggest that infectious agents that engage the SMN complex may burden SMN-dependent pathways, possibly leading to a deleterious reduction in available SMN complex for essential host functions.
The small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U5, and U4/U6 are major components of the spliceosome. Each snRNP is comprised of one U snRNA (U1, U2, U5, or U4/U6), seven common Sm proteins, and a set of proteins that are specific to the individual snRNAs (32, 33, 55). The Sm proteins B/B′, D1, D2, D3, E, F, and G are common to all spliceosomal snRNPs and are arranged into a seven-membered ring (25, 51) on a consensus sequence (PuAU4-6GPu) known as the Sm site of the U snRNA (6, 44). The process of bringing these proteins and RNA components together (snRNP assembly) occurs in the cytoplasm and is mediated by the SMN (survival of motor neurons) protein complex (7, 16, 30, 31, 36, 38, 49, 50). SMN is the protein product of the spinal muscular atrophy (SMA) disease gene (28). SMA is a severe neurodegenerative disease that is characterized by degeneration of motor neurons in the spinal cord (10, 13, 22). More than 98% of SMA patients carry deletions or loss-of-function mutations in the SMN1 gene and produce reduced levels of the protein that correlate with the phenotypic severity of the disease (12, 28, 29). SMN, as an oligomeric protein, is part of a large multiprotein complex that contains Gemin2 (31), the DEAD box RNA helicase Gemin3 (8), Gemin4 (9), Gemin5 (21), Gemin6 (46), and Gemin7 (3). Although the function of the SMN complex in snRNP assembly is best characterized, it most likely functions in the assembly and metabolism of various other RNPs, including snoRNPs, miRNPs, and the machineries that carry out transcription and pre-mRNA splicing (7, 17, 24, 35, 39, 40, 45, 47-49).
To function in the assembly of the snRNP Sm core, the SMN complex must bring together both protein and RNA components. Several components of the SMN complex bind directly to the Sm proteins, including the binding of SMN to the RG-rich C-terminal domains of the Sm proteins B, D1, and D3 (3, 7-9, 17, 21, 31, 46, 47). This interaction is enhanced by the symmetric dimethylarginine modification of specific arginines by the 20S methylosome that contains an arginine methyltransferase (JBP1/PRMT5) (18-20, 37). The SMN complex also binds directly and with sequence specificity to the Sm site-containing U snRNAs (56, 57). These and other studies suggest that through the specific recognition of its RNA targets, the SMN complex acts as a specificity factor and a surveillance machine to ensure that Sm cores are only assembled on the correct RNAs (50, 56).
Herpesvirus saimiri (HVS) encodes seven small RNAs (75 to 143 nucleotides), named HSURs (2, 26, 27, 41, 54). HVS strain A11, the prototype gamma 2 herpesvirus, causes acute leukemias and T-cell lymphomas in some New World primates (15). This virus family includes the human herpesvirus type 8, which is more commonly known as Kaposi's sarcoma-associated herpesvirus (15). Although HSURs are the most abundant viral gene products expressed in latently infected, transformed T cells (41), their function remains unknown since they are not essential for viral replication or transformation of T cells in vitro (14, 41, 42). The HSURs contain a canonical Sm sequence (AUUUUUG), and their predicted secondary structures are reminiscent of the spliceosomal U snRNAs (2, 26, 27, 54). Further studies revealed that similar to host U snRNAs, HSURs are transcribed by RNA polymerase II, acquire a trimethyl guanosine cap, and associate with Sm proteins (26). In transformed T cells, there are about 20,000 copies of HSUR1 and HSUR4 per cell, whereas only about 2,000 copies of each of the other five HSURs can be detected per cell (11). Individual HSURs can be expressed by transient transfection in HeLa cells and assemble Sm cores in the absence of other viral genes (27).
Because the SMN complex binds directly to Sm site-containing snRNAs and mediates the assembly of Sm cores on them (56, 57), we wanted to determine whether it plays a similar role in the assembly of Sm cores on the HVS snRNAs or whether HSURs have an alternative route to acquire Sm cores. Here, we show that the SMN complex binds directly to HSURs with an affinity similar to, or higher than, that of the host snRNAs. Furthermore, we show that the SMN complex is both necessary and sufficient for Sm core assembly on these viral RNAs. Importantly, the HSURs can effectively outcompete host snRNAs for SMN-dependent snRNP assembly. These findings reinforce the central role of the SMN complex as an assembly machine for RNPs. It seems plausible that infectious agents that sequester the SMN complex may lead to a reduction in the amount of SMN complex available for essential host functions and thus cause cytopathology. If such a burden on the SMN complex were to occur, it could be particularly deleterious to cells already compromised in their levels of SMN, such as those found in SMA patients.
MATERIALS AND METHODS
Plasmids.
Plasmids for in vitro transcription of HSURs 1 to 5 were kindly provided by Joan A. Steitz; HSUR EL-1, EL-3 and EL-5 vectors have the coding regions of HSUR1, HSUR3, and HSUR5, respectively, cloned into pSP64, and HSUR EL-4 has the coding region of HSUR4 cloned into pGem-3Z. The plasmid HSUR4ΔSm has a substitution mutation in the Sm site (CTCGAG) and was constructed by PCR according to the method of Imai et al. (23). For transient-transfection experiments, plasmids that contain HSUR genes were used. Murthy et al. (42) described the numbering of the HVS 11 genome to begin at +1, which is the leftmost L DNA nucleotide adjacent to the H DNA repeat unit. pT7.4 (kindly provided by Ronald C. Desrosiers) contains HVS L-DNA sequences from +21 to approximately +7400, which encompasses the genes for HSURs 1 to 5, cloned into vector pBR322 (42). For in situ hybridization of HSUR5, the gene for HSUR5 (27) was cloned from pT7.4 and inserted into pGem-3Z.
Labeling of RNAs.
In vitro transcription and [32P]UTP labeling of RNAs were carried out as described previously (57). [32P]UTP-labeled RNAs were purified by electrophoresis on 7 M urea-6% polyacrylamide gels and precipitated with ethanol. RNAs were resuspended in deionized distilled water.
Preparation of HeLa cell cytoplasmic extracts.
HeLa cell cytoplasmic extracts competent for snRNP assembly were prepared as described previously (56).
Purification and analysis of native SMN complex.
The SMN complex was purified from Flag-Gemin2 HeLa Tet-ON cells as described previously (56). The parental HeLa cell line served as a negative control. For purification of SMN complex under low-salt conditions, SMN complex or control bound to anti-Flag beads (Sigma) was washed extensively with RSB-100 (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2.5 mM MgCl2) containing 0.02% NP-40. For complex purification under more stringent conditions, three additional washes were performed for 15 min each at 4°C with 10 bead volumes of RSB-500 containing 0.02% NP-40. The bound proteins were either equilibrated with 10 bead volumes of RSB-100 containing 0.01% NP-40 for binding experiments or eluted for 1 h at 4°C with 3× Flag peptides (Sigma) at a final concentration of 0.5 mg/ml for in vitro snRNP assembly or analysis by silver staining or Western blotting. Proteins were resolved on NuPAGE Novex Bis-Tris precast gradient 4 to 12% minigels (Invitrogen). The following mouse monoclonal antibodies were used for Western blot: 2B1 (anti-SMN), 2E17 (anti-Gemin2), 12H12 (anti-Gemin3), 17D10 (anti-Gemin4), 10G11 (anti-Gemin5), Y12 (anti-Sm), 1F12 (Y14), 4F4 (hnRNP C), and 3C2 (hnRNP K). A rabbit polyclonal antibody was used to detect Gemin6.
In vitro binding of RNAs.
In vitro binding and competition experiments were performed as previously described (56). The bound RNAs were isolated and analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels.
Equilibrium binding experiments.
Equilibrium binding assays were carried out by using a nitrocellulose filter attached to a multiwell vacuum manifold as described previously (56).
Assay for in vitro assembly of snRNPs.
In vitro Sm core assembly and electrophoretic mobility shift assays were carried out as described previously (50). For the anti-Sm monoclonal antibody (Y12) supershift experiment, 3 μg of purified Y12 antibody was incubated with the completed in vitro assembly reaction for 5 min on ice prior to the addition of loading buffer. For in vitro assembly competition experiments, nonradioactive competitor RNA was added to the assembly reaction at the same time as 32P-labeled RNA, and reactions were carried out for 1 h at 30°C. Assembly reaction products were quantitated by phosphorimager analysis.
Immunodepletion of the SMN complex.
Cytoplasmic extracts (250 μl) from HeLa S3 cells were incubated with 25 μl of GammaBind G Sepharose beads (Amersham) conjugated to 4 μg of either purified anti-SMN (2B1) monoclonal antibody or control antibody (SP2/0). After 1 h at 4°C, the supernatants were transferred to a new tube of conjugated antibody and again incubated for 1 h at 4°C. This procedure was repeated four times and, after the final incubation, glycerol was added, and the supernatants were stored in aliquots at −80°C. Western blots were performed to verify the immunodepletion of the SMN complex in the extracts.
S5 cell culture and snRNP assembly.
Chicken DT40 cells that have a deletion in the endogenous SMN gene and stably carry a tetracycline-repressible SMN cDNA (S5 cells) were maintained as previously described under normal-SMN (10 ng of tetracycline/ml) or low-SMN (18 ng of tetracycline/ml) conditions (53). After 72 h in the specified media, the cells were harvested, and cytoplasmic extracts were prepared and tested for in vitro snRNP assembly. The extracts were assayed by Western blotting with anti-SMN (2B1), anti-Sm (Y12), and anti-Y14 (1F12) mouse monoclonal antibodies to confirm the specific reduction of SMN in vivo.
Transient-transfection and immunoprecipitation of HSURs.
Flag-Gemin2 cells grown in the presence of doxycycline (5 μg/ml) were transiently transfected with 1 μg of pT7.4 or empty vector by using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. At 48 h posttransfection, cells were harvested by scraping into ice-cold phosphate-buffered saline, washed twice, and pelleted. Cell pellets were resuspended in 300 μl of RSB-100 buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2.5 mM MgCl2) containing 0.01% NP-40 plus protease inhibitors and lysed by sonication. After centrifugation for 15 min at 10,000 rpm (17,000 × g) at 4°C, supernatants were collected, 10% of the total (30 μl) was treated with TRIzol reagent (Invitrogen) to isolate total RNA, and the remainder was equally divided and subjected to immunoprecipitation for 1 h at 4°C with either anti-Flag beads (Sigma) or anti-Sm (Y12) monoclonal antibody conjugated to protein A-Sepharose CL-4B (Amersham). After extensive washing, the beads were treated with 20 U of DNase I (Ambion) for 15 min at 37°C, followed by proteinase K, and the RNAs were purified by phenol-chloroform extraction, followed by ethanol precipitation. RNA pellets were resuspended in 30 μl of nuclease-free water, and 1 μl was added to each reverse transcription (RT) reaction by using the ThermoScript RT-PCR System (Invitrogen). The following DNA oligonucleotides were used for RT and amplification: U1-RT, CAGGGGAAAGCGCGAACGCAGTCC; U1-PCR, GATACACCTGGCAGGGGAGATACCA; HSUR1-RT, TGGTACCGGTCATCATATTTAC; HSUR1-PCR, GACACTACATATTTATTTATTTATTTCTT; HSUR2-RT, CAGCGCTGGTTTTTAAATATGTAG; HSUR2-PCR, GACACTACATATTTATTGTTTATTTATACC; HSUR3-RT, TGGCACTGGTTTGGACCTAA;HSUR3-PCR, GAAGACTTGCTATAGGAGATTAACAACC; HSUR4-RT, TGGCACTGGTTTGGACTACCCCAGA; HSUR4-PCR, GGCCCACAGCCAGAGAGTTACTCT; HSUR5-RT, CGGCTCTGGTTGTTAGTAACACAC; and HSUR5-PCR, GAACACTACATATTTATTTTTCGCTC. RT reactions were carried out at 50°C for 1 h. Subsequently, 2 μl of each RT reaction was used for 30 cycles of PCR. One-half of each PCR product was analyzed by electrophoresis on 1.5% agarose gels and visualized by ethidium bromide staining under UV light.
In situ hybridization and indirect immunofluorescence.
In situ hybridization of RNAs and indirect immunofluorescence of proteins were performed as previously described (49). HeLa PV cells grown in six-well plates were transfected with 0.4 μg of the HSUR5 gene with its endogenous promoter and terminator elements (27) cloned into pGem-3Z or empty vector alone with Effectene transfection reagent (Qiagen) according to the manufacturer's protocol, and cells were fixed at 48 h posttransfection. The following 2′-O-methylated probes were used for in situ hybridization: HSUR5 (complementary to nucleotides 21 to 42), CUCAGUUACAGCUUUGCGAGCG 4-Biotin-U (visualized with antibiotin secondary antibody conjugated to Texas red); and U1 snRNA, UGCCAGGUAAGUAU-fluorescein isothiocyanate (FITC) (34). For indirect immunofluorescence of proteins, cells were incubated with anti-SMN (2B1) and anti-Sm (Y12) mouse monoclonal antibodies, followed by secondary antibody conjugated to FITC.
RESULTS
HSURs associate directly with the SMN complex in vitro and in vivo.
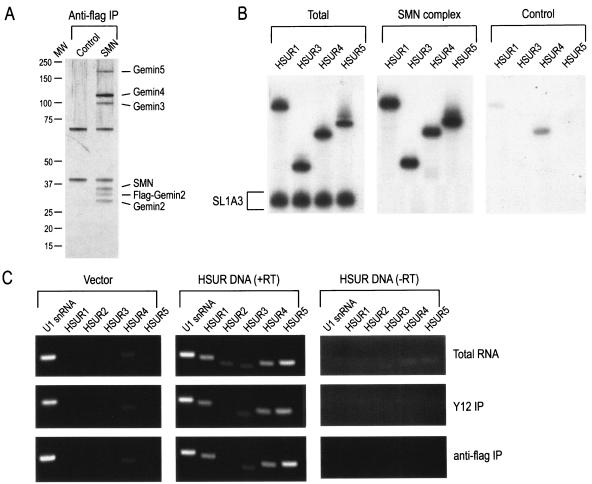
To determine whether the SMN complex interacts directly with HSURs, native SMN complexes were purified under stringent conditions (500 mM NaCl) from a Flag-Gemin2 stable HeLa cell line, which is expressing tetracycline-inducible Flag-Gemin2 (3, 46). Under these conditions, anti-Flag antibodies immunopurified complexes that contain all of the known core components of the SMN complex, including SMN and Gemin2 to Gemin7, but no detectable substrates, such as Sm proteins (Fig. 1A) (3, 46, 50). For direct binding to the SMN complex, 32P-labeled HSURs 1, 3, 4, and 5 were mixed with labeled SL1A3 RNA (a mutant of stem-loop 1 of U1 snRNA that does not efficiently bind to the SMN complex [57]) as a negative control and incubated with purified SMN complexes. As shown in Fig. 1B, HSURs 1, 3, 4, and 5 bound specifically and efficiently to the SMN complex. Based on the percentage of RNAs bound, it appears that HSURs bind to the SMN complex with at least equal or likely greater efficiency than U snRNAs. Thus, the SMN complex binds directly and specifically to HSURs in vitro and this interaction occurs in the absence of Sm proteins.
FIG. 1.
The SMN complex binds directly to HSURs in vitro and associates with HSURs in vivo. (A) Native SMN complexes (SMN) were purified under high-salt conditions from stable cell lines expressing Flag-Gemin2 (as described in Materials and Methods) and were analyzed by electrophoresis on 4 to 12% gradient polyacrylamide gels and by silver staining. Immunoprecipitation with anti-Flag antibody from the parental HeLa cell line was used as a control (Control). Gemin6 and Gemin7 are not shown. The total amount of SMN complex shown in this gel was used for direct RNA-binding experiments. (B) [32P]UTP-labeled HSUR1, HSUR3, HSUR4, or HSUR5 was mixed with SL1A3 RNA (a mutant of SL1 of U1 snRNA that does not efficiently bind to the SMN complex) and incubated for 1 h at 4°C with Flag-purified SMN complex (SMN complex) or nonspecific proteins purified from HeLa cells (Control). Bound RNAs were washed, isolated, and analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels and autoradiography. Total represents 20% of input. (C) HeLa cells stably expressing Flag-Gemin2 were transiently transfected with empty vector (Vector) or with a 7.4-kb fragment of genomic HVS DNA that contains the genes for HSURs 1 to 5 (HSUR DNA). After 48 h, cell extracts were made and subjected to immunoprecipitation with either anti-Sm (Y12) or anti-Flag monoclonal antibodies, and RNAs were purified by phenol-chloroform extraction and ethanol precipitation. Immunoprecipitated RNAs were reverse transcribed (+RT), and the cDNAs were amplified by using primers for U1 snRNA and HSURs 1 to 5. RT in the absence of transcriptase was performed as a negative control (−RT). cDNAs were run on 1.5% agarose gels, stained with ethidium bromide, and visualized by UV light. Total RNA represents 10% of input.
To determine whether the HSURs interact with the SMN complex in vivo, Flag-Gemin2-expressing cells were transfected with a plasmid encoding the first 7.4 kb of the HVS strain 11 genome (pT7.4) that contains all seven HSURs (42). Immunoprecipitations from total cell extract were performed with either anti-Sm antibody (Y12) to detect Sm core assembly on HSURs or anti-Flag antibody to coimmunoprecipitate RNAs that interact with the SMN complex in vivo. The isolated RNAs were purified and detected by RT-PCR. Agarose gel analysis shows RT-PCR products for U1 snRNA, HSUR1, HSUR3, HSUR4, and HSUR5, whereas cDNA for HSUR2 was only visible with increased exposure (Fig. 1C). HSUR2 is immunoprecipitated at low efficiency either because it has a lower copy number than other HSURs when expressed in cells (26) and/or it does not amplify well in the RT-PCR because of its sequence or structure. Both explanations are consistent with the decreased amount of HSUR2 expressed in the cells, as assessed in the total RNA fraction, prior to immunoprecipitation. RT-PCR of HSUR4 consistently produced a faint background band that, when cloned, was revealed to be primer-dimer artifact. These data suggest that the SMN complex associates with HSURs in vivo.
The SMN complex mediates the assembly of Sm cores on HSURs.
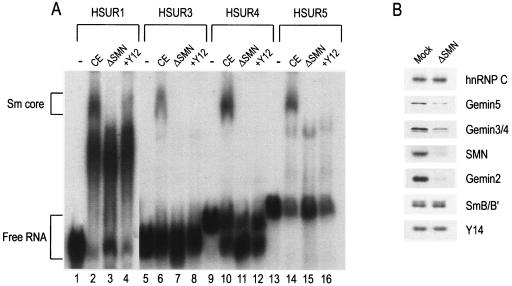
To assay Sm core assembly in vitro, 32P-labeled HSURs 1, 3, 4, and 5 were incubated with HeLa cytoplasmic extracts and the assembly reaction products were analyzed by electrophoresis on native polyacrylamide gels. Figure 2A shows that all four HSURs tested assemble Sm cores (lanes 2, 6, 10, and 14). To confirm further that the slowly migrating RNA-protein band is indeed an assembled HSUR-Sm core complex, the completed assembly reaction products were incubated with Y12 antibodies prior to gel loading. The addition of Y12 supershifted the Sm core band to a protein-antibody complex that remained in the well of the native gel (Fig. 2A, wells not shown, lanes 4, 8, 12, and 16). For HSUR1, the large band that migrates slightly faster than the Sm core band and does not change upon addition of Y12 antibody most likely consists of HSUR1 complexed to the HuR protein that has been shown to bind to a consensus sequence (AUUUA) at the 5′ end of HSURs 1, 2, and 5 (43) (Fig. 2A, lanes 2 to 4). Of the three RNAs, HSUR1 contains the most copies of this motif. Importantly, to examine the requirement of the SMN complex in Sm core assembly on HSURs, cytoplasmic extracts were immunodepleted of the SMN complex prior to the assembly reaction. Lanes 3, 7, 11, and 15 of Fig. 2A show that immunodepletion of the SMN complex inhibited the Sm core assembly, despite the abundance of Sm proteins in the immunodepleted extract (Fig. 2B). These results suggest that the SMN complex is necessary for the assembly of Sm cores on HSURs.
FIG. 2.
HSURs assemble SMN-dependent Sm cores in vitro. (A) [32P]UTP-labeled HSUR1, HSUR3, HSUR4, and HSUR5 were incubated with buffer (−), HeLa mock-depleted extracts (CE), or SMN complex-depleted HeLa extracts (ΔSMN) for 1 h at 30°C. Anti-Sm monoclonal antibody (Y12) was added to supershift Sm cores assembled in CE (+Y12). The assembly reaction products were analyzed by electrophoresis on 6% native polyacrylamide gels and autoradiography. Sm cores and free RNAs are each indicated by brackets. (B) The HeLa cytoplasmic extracts used in Fig. 2A were immunodepleted by using control (SP2/0) antibody (Mock) or anti-SMN (2B1) monoclonal antibody (ΔSMN). Subsequently, proteins were resolved on 4 to 12% gradient polyacrylamide gels and immunodepletion of SMN complex components was confirmed by Western blotting.
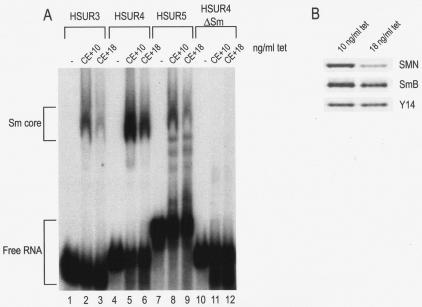
In addition to immunodepletion of SMN in vitro, we used a chicken pre-B cell line (DT-40) to specifically reduce the level of SMN protein in vivo since chicken SMN can functionally complement human SMN in vivo (53) and can assemble Sm cores on U snRNAs (data not shown). Previously, we developed a cell line (S5 cells) from which the endogenous chicken SMN gene has been deleted and that stably carries a tetracycline-repressible SMN cDNA. Cell culture in the presence of 10 ng of tetracycline/ml produces wild-type levels of SMN, whereas 18 ng of tetracycline/ml results in SMN levels that are ∼20% of the wild-type level (Fig. 3B [53]). These extracts were then incubated with 32P-labeled HSURs 3, 4, and 5, along with an HSUR4 mutant that has a substitution in the Sm site (HSUR4ΔSm), and the assembly products were analyzed on native gels. As seen in Fig. 3A, reduction of SMN caused a decrease in the Sm core assembly on HSURs 3, 4, and 5, despite the presence of equivalent amounts of Sm proteins (Fig. 3B), compared to the wild type. As expected, the HSUR4ΔSm mutant did not form an Sm core in either extract. In a similar experiment, reduction of SMN by RNAi in HeLa cells also reduced Sm core assembly on HSURs (data not shown). These findings demonstrate that the SMN complex of host cells is strictly required for Sm core assembly on HSURs in vivo.
FIG. 3.
Reduction of SMN in vivo leads to decreased Sm core assembly on HSURs. (A) HSUR3, HSUR4, HSUR5, and HSUR4ΔSm were transcribed in the presence of [32P]UTP and incubated with buffer (−), cytoplasmic extract derived from S5 cells grown under wild-type SMN conditions (CE + 10 ng of tetracycline [tet]/ml) or from S5 cells grown under low SMN conditions (CE + 18 ng of tetracycline/ml) as described in Materials and Methods. After 1 h of incubation at 30°C, the assembly reaction products were analyzed by electrophoresis on 6% native polyacrylamide gels and autoradiography. Sm cores and free RNAs are each indicated by brackets. (B) The S5 cell extracts used in Fig. 3A were run on 4 to 12% gradient polyacrylamide gels and Western blotted to confirm the in vivo reduction of SMN.
The SMN complex is sufficient for Sm core assembly on HSURs.
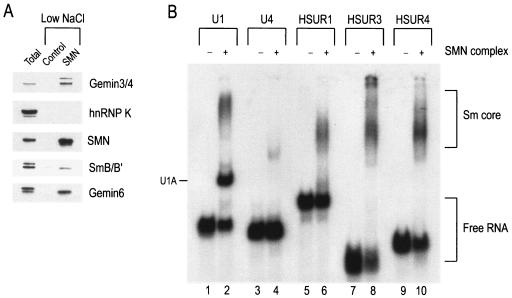
We have previously reported that high-salt-purified SMN complex that consists only of SMN and the Gemins (as illustrated in Fig. 1A) is necessary and sufficient for Sm core assembly only when Sm proteins are added back to the complex (50). Alternatively, we can purify native SMN complexes under low-salt conditions (100 mM NaCl) so that the SMN complex core components and its cellular substrates, such as the Sm proteins, remain associated. This SMN complex preparation is sufficient for assembly of Sm cores on U snRNAs (46, 50). Figure 4A shows a Western blot of several proteins associated with SMN after complex purification at low-salt conditions, including the Gemins and SmB/B′. Native SMN complexes purified at 100 mM NaCl were incubated with 32P-labeled U1 snRNA, U4 snRNA, or HSURs 1, 3, or 4, and assembly reaction products were analyzed on native gels. As seen in Fig. 4B, purified SMN-Sm protein complex assembles Sm cores on both the U snRNAs and HSURs 1, 3, and 4. Taken together, these results demonstrate that the SMN complex is both necessary and sufficient for Sm core assembly on HSURs.
FIG. 4.
SMN complex purified with Sm proteins is sufficient for Sm core assembly on HSURs. (A) Native SMN complexes (SMN) or nonspecific proteins (Control) were purified from Flag-Gemin2 cells or the parental HeLa cells, respectively, under low-salt conditions as described in Materials and Methods. Flag-purified proteins were eluted with 3× Flag peptides, resolved by electrophoresis on 4 to 12% gradient polyacrylamide gels, and Western blotted for components of the SMN complex. (B) [32P]UTP-labeled U1 snRNA, U4 snRNA, HSUR1, HSUR3, and HSUR4 were incubated with buffer (−) or with Flag-purified SMN complex (+) isolated under low-salt conditions for 1 h at 30°C. Assembly reaction products were analyzed by electrophoresis on 6% native polyacrylamide gels and autoradiography. Sm cores and free RNAs are each indicated by brackets.
HSURs bind to the SMN complex with high affinity.
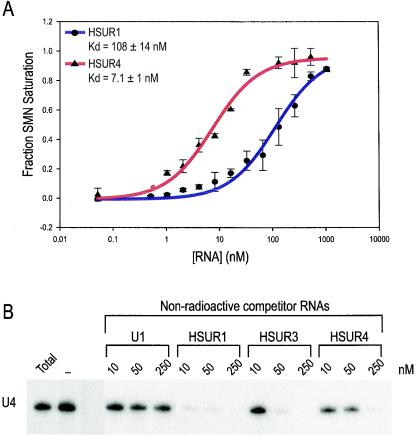
Recently, we determined the apparent equilibrium dissociation constant for U4 snRNA binding to the SMN complex to be 17 ± 2.8 nM and, through competitive binding experiments, we concluded that the SMN complex most likely has at least two independent high-affinity RNA binding sites—one for U1 and the other for U4 snRNA (56). To determine the affinity of the SMN complex for the HSURs, we performed equilibrium binding experiments similar to those used for U4 snRNA. Native SMN complexes were purified at high salt and incubated under equilibrium conditions with increasing amounts of either 32P-labeled HSUR1 or HSUR4. As illustrated in Fig. 5A, the fraction of SMN complex saturated with RNA was quantitated and plotted. For both HSUR1 and HSUR4, the data are consistent with the prediction of a single high-affinity binding site, with apparent equilibrium dissociation constants [Kd (apparent)] of 108 ± 14 nM for HSUR1 and 7.1 ± 1 nM for HSUR4 (Fig. 5A).
FIG. 5.
HSURs bind to the SMN complex with high affinity. (A) A nitrocellulose filter-binding assay was used to determine the affinities of HSUR1 and HSUR4 to the SMN complex. Flag-purified SMN complexes were incubated with various amounts of HSUR1 or HSUR4. A plot of the fraction of SMN complex saturation as a function of RNA concentration is shown. Error bars represent the standard deviations from at least four independent experiments for each RNA. (B) U4 snRNA was transcribed in the presence of [32P]UTP, and 10,000 cpm was mixed with increasing concentrations of nonradioactive U1 snRNA, HSUR1, HSUR3, and HSUR4 (10, 50, or 250 nM) or no competitor (−) and immediately incubated with purified SMN complexes for 1 h at 4°C. Bound RNAs were isolated and analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels. Total represents 10% of input.
We next investigated whether HSURs and host snRNAs share a common binding site on the SMN complex. Competition binding experiments were carried out by incubating high-salt-purified SMN complex with 32P-labeled U4 snRNA and increasing amounts (10, 50, and 250 nM) of nonradioactive U1 snRNA, HSUR1, HSUR3, and HSUR4. The bound RNAs were purified and analyzed by electrophoresis. Figure 5B shows that nonradioactive HSUR1, HSUR3, and HSUR4 effectively compete with labeled U4 at 10, 50, and 250 nM, respectively, whereas nonradioactive U1 snRNA does not compete with U4 snRNA. The lack of competition between U1 and U4 snRNA is consistent with the previous observation that the SMN complex has at least two distinct high-affinity RNA binding sites (56). Furthermore, nonradioactive HSUR1, HSUR3, and HSUR4 minimally compete with labeled U1 even at the highest concentration but more effectively compete with labeled U4 snRNA better than U4 competes with itself (data not shown), suggesting that the HSURs might bind to a similar domain as U4 snRNA on the SMN complex. It is important to note that these experiments were not performed under equilibrium conditions due to the extensive washing of the beads after binding competition. Therefore, we can only assess the possibility that the RNAs share a common binding site, but not the relative affinities of the various RNAs for the SMN complex. The possibility remains that a more complicated or overlapping arrangement of binding sites on the SMN complex might allow the presence of more than one U4 snRNA or HSUR at one time; however, it is clear that all HSURs tested effectively compete U4 snRNA for binding to the SMN complex.
HSURs compete with U snRNAs for Sm core assembly.
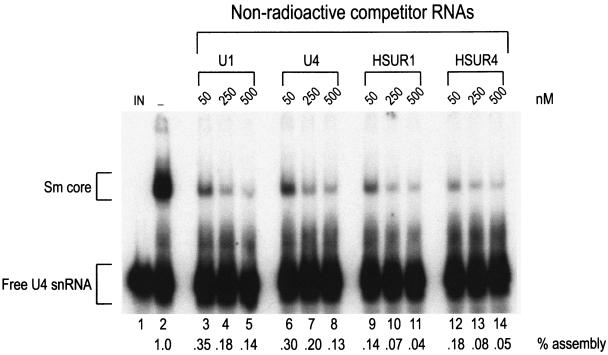
We further investigated the capacity of HSURs to compete with U snRNAs in Sm core assembly. 32P-labeled U4 snRNA and various nonsaturating concentrations of nonradioactive U1 and U4 snRNAs, HSUR1 and HSUR4 (50, 250, and 500 nM) were incubated in HeLa extracts to allow snRNP assembly, and the assembly reaction products were analyzed on native polyacrylamide gels (Fig. 6). At all concentrations tested, nonradioactive HSUR1 and HSUR4 more effectively competed for the assembly of U4 snRNP than cold U1 snRNA or U4 snRNA itself (compare lanes 9 to 14 to lanes 3 to 8). In addition, nonradioactive HSUR1 and HSUR4 were more effective than U4 snRNA in competition for assembly of U1 snRNP as well (data not shown). These data suggest that HSUR1 and HSUR4 are able to outcompete U4 snRNA for access to SMN complex and Sm core assembly on U snRNAs.
FIG. 6.
HSURs compete with U snRNAs for Sm core assembly. A total of 10,000 cpm of [32P]UTP-labeled U4 snRNA was incubated with buffer (IN), HeLa cytoplasmic extract (−), or HeLa cytoplasmic extract plus increasing amounts (50, 250, or 500 nM) of nonradioactive U1, U4, HSUR1, or HSUR4 for 1 h at 30°C. Assembly reaction products were analyzed by electrophoresis on 6% native polyacrylamide gels and quantitated by using a phosphorimager. Sm cores and free RNAs are each indicated by brackets, and relative percentages of assembly are indicated below each lane.
HSURs show a similar cellular localization to U snRNAs.
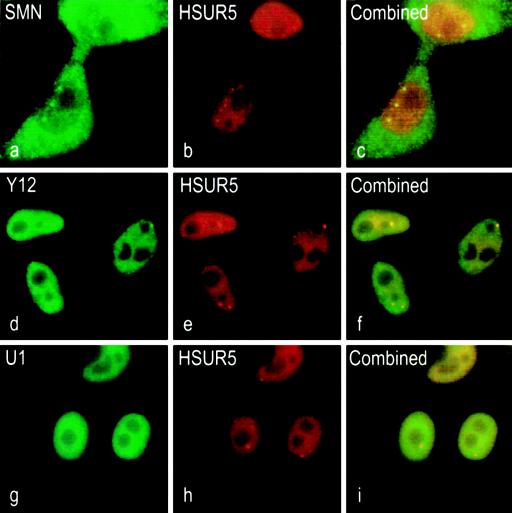
Properly assembled snRNPs show a characteristic localization in the nucleoplasm and a distinct concentration in Cajal bodies, often merged with gems. To investigate the subcellular localization of HSURs, we performed in situ hybridization for HSUR5. HSUR5 was used for these experiments because it contains a single-stranded region that lacks the HuR protein-binding motif present at the 5′ end of HSURs 1, 2, and 5 (43). In situ hybridization of HSUR5 showed that the majority of HSUR5 is found throughout the nucleoplasm and is highly concentrated in discrete nuclear bodies (Fig. 7b). Subsequently, the same cells were also indirectly immunostained for endogenous SMN, which showed the expected pattern of diffuse cytoplasmic staining with distinct nuclear gems (Fig. 7a). The combined images (Fig. 7c) indicate an overlap of a subset of the nuclear dots that contain HSUR5 with the gems. This suggests that HSURs and SMN complex components can be found within the same subnuclear compartment. We also examined whether HSUR5 (Fig. 7e) colocalizes with endogenous Sm proteins by costaining the cells with Y12 antibody. Figure 7d shows the diffuse nuclear pattern with distinct speckles of the Y12 immunofluorescence. Again, the combined images (Fig. 7f) show that HSUR5 colocalizes with Sm proteins. Also, to confirm that HSUR5 expressed in cells localizes in a similar pattern to U snRNAs, we performed a double in situ hybridization for endogenous U1 snRNA, which is localized throughout the nucleus in a speckled pattern (Fig. 7g). As seen in Fig. 7i, the merge of U1 snRNA with HSUR5 illustrates that the two RNAs exist within the same nuclear compartments. These observations suggest that HSUR5 utilizes the endogenous snRNP biogenesis pathway to acquire an Sm core.
FIG. 7.
HSUR5 colocalizes with SMN and U snRNAs. (a) Indirect immunofluorescence of SMN protein with anti-SMN monoclonal antibody (2B1) and a FITC-conjugated secondary antibody showing localization in nuclear gems. (b) In situ hybridization of HSUR5 with a biotinylated antisense probe and an antibiotin secondary antibody conjugated to Texas red. (c) Combined image of panels a and b showing colocalization of HSUR5 in nuclear gems. (d) Indirect immunofluorescence of Sm proteins with anti-Sm monoclonal antibody (Y12) and an FITC-conjugated secondary antibody showing nuclear speckles. (e) Same as panel b. (f) Combined image of panels d and e showing colocalization of HSUR5 with Sm proteins. (g) In situ hybridization of endogenous U1 snRNA with an FITC-conjugated antisense probe. (h) Same as panel b. (i) Combined image of panels g and h showing colocalization of U1 snRNA with HSUR5.
DISCUSSION
Here we show that the HVS-encoded HSURs use the SMN complex to assemble Sm cores. Using in vitro snRNP assembly assays, we demonstrate that the SMN complex is both necessary and sufficient for Sm core assembly on the HSURs. Furthermore, the HSURs bind to the SMN complex directly with an affinity at least similar to, or higher than, that of the U snRNAs. Two distinct types of U snRNA binding sites have been identified in the SMN complex: one for U1 snRNA and one, the U4 snRNA-type site, for the other major host Sm site-containing snRNAs (56). In both secondary structure and Sm site sequence, the HSURs appear to more closely resemble U4 snRNA. The competition of HSURs with U4 snRNA for binding to the SMN complex is consistent with this. Previously, we have determined the equilibrium dissociation constant for SMN complex binding to U4 snRNA to be ca. 17 nM (56). In the present study, we report the apparent Kd values for SMN complex binding to HSUR4 and HSUR1 to be 7 and 108 nM, respectively. The competition experiments suggest that the binding affinities of the other HSURs for the SMN complex are also in the same range. These remarkably high affinities explain how the HSURs can effectively outcompete host U snRNAs for Sm core assembly.
Although HSURs are the most abundant viral gene products expressed in latently infected T cells (41), they are not required for viral replication or transformation of T cells in vitro and their function remains unknown (14, 41, 42). The fact that the HSURs have been conserved among various HVS strains (5, 26, 42, 52, 54) and in the closely related Herpesvirus ateles (1) supports the conclusion that these RNAs perform some critical function for the virus. Most likely, the assembly of an Sm core is an essential part of the life cycle of the HSURs that may provide protection from degradation and determine their subcellular localization.
HVS-transformed cells produce, at most, about 20,000 copies of HSURs (11), compared to 105 to 106 copies of endogenous U snRNAs (4). It is possible that acute infection in some cell types results in much higher levels of HSUR expression than those observed in the HVS-transformed lymphocytes. Furthermore, this relatively low HSUR copy number represents cellular steady-state levels and likely underestimates the actual load that HSURs place on the SMN complex. HSURs 1, 2, and 5 have been shown to contain destabilizing AU-rich elements (AREs) at their 5′ ends that interact with the ARE-binding protein HuR (43) and result in reduced steady-state levels of HSUR1 in HVS-transformed T cells (11). It has not been determined whether the degradation of the ARE-containing HSURs occurs prior to or after Sm core assembly. The possibility remains that newly transcribed HSURs place a much greater burden on the SMN complex than their steady-state levels suggest.
Reduced levels of functional SMN cause motor neuron degeneration that often results in death. It is likely that cellular invasion by a foreign or infectious agent that engages the SMN complex would be deleterious to cells. The observations reported here highlight the general utility of the SMN complex for RNP assembly for both host cells and viruses. These findings suggest that viruses can engage the SMN complex, possibly leading to a reduction in available SMN complex for host functions. Because reduced levels of functional SMN cause spinal muscular atrophy, we suggest that infectious agents that engage the SMN complex may cause cellular damage especially to motor neurons, the target cells in SMA. SMA patients may be particularly susceptible in such a scenario, but it is also conceivable that SMN complex-usurping agents play a role in the etiology of other motor neuron degenerative diseases for which a genetic cause has not been found.
Acknowledgments
We are grateful to Ronald C. Desrosiers and Joan A. Steitz for generously providing constructs for HVS and the HSURs. We thank the members of our laboratory, especially Amelie Gubitz, for helpful discussions and comments on the manuscript and Jin Wang for providing S5 cells. We are also grateful to Gina Daly for secretarial assistance.
This study was supported by the Association Française Contre les Myopathies and by a grant from the National Institute of Health. T.J.G. is a Predoctoral Fellow of the Howard Hughes Medical Institute. G.D. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Albrecht, J. C. 2000. Primary structure of the Herpesvirus ateles genome. J. Virol. 74:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., and B. Fleckenstein. 1992. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 20:1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baccon, J., L. Pellizzoni, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 277:31957-31962. [DOI] [PubMed] [Google Scholar]
- 4.Baserga, S. J., and J. A. Steitz. 1993. The diverse world of small ribonucleoproteins, p. 359-381. In R. F. Gesteland and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 5.Biesinger, B., J. J. Trimble, R. C. Desrosiers, and B. Fleckenstein. 1990. The divergence between two oncogenic Herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology 176:505-514. [DOI] [PubMed] [Google Scholar]
- 6.Branlant, C., A. Krol, J. P. Ebel, E. Lazar, B. Haendler, and M. Jacob. 1982. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1:1259-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhler, D., V. Raker, R. Luhrmann, and U. Fischer. 1999. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 8:2351-2357. [DOI] [PubMed] [Google Scholar]
- 8.Charroux, B., L. Pellizzoni, R. A. Perkinson, A. Shevchenko, M. Mann, and G. Dreyfuss. 1999. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charroux, B., L. Pellizzoni, R. A. Perkinson, J. Yong, A. Shevchenko, M. Mann, and G. Dreyfuss. 2000. Gemin4: a novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 148:1177-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cifuentes-Diaz, C., T. Frugier, and J. Melki. 2002. Spinal muscular atrophy. Semin. Pediatr. Neurol. 9:145-150. [DOI] [PubMed] [Google Scholar]
- 11.Cook, H. L., H. E. Mischo, and J. A. Steitz. 2004. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 24:4522-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coovert, D. D., T. T. Le, P. E. McAndrew, J. Strasswimmer, T. O. Crawford, J. R. Mendell, S. E. Coulson, E. J. Androphy, T. W. Prior, and A. H. Burghes. 1997. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 6:1205-1214. [DOI] [PubMed] [Google Scholar]
- 13.Crawford, T. O., and C. A. Pardo. 1996. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 3:97-110. [DOI] [PubMed] [Google Scholar]
- 14.Ensser, A., A. Pfinder, I. Muller-Fleckenstein, and B. Fleckenstein. 1999. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J. Virol. 73:10551-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fickenscher, H., and B. Fleckenstein. 2001. Herpesvirus saimiri. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:545-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023-1029. [DOI] [PubMed] [Google Scholar]
- 17.Friesen, W. J., and G. Dreyfuss. 2000. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN). J. Biol. Chem. 275:26370-26375. [DOI] [PubMed] [Google Scholar]
- 18.Friesen, W. J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111-1117. [DOI] [PubMed] [Google Scholar]
- 19.Friesen, W. J., S. Paushkin, A. Wyce, S. Massenet, G. S. Pesiridis, G. Van Duyne, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277:8243-8247. [DOI] [PubMed] [Google Scholar]
- 21.Gubitz, A. K., Z. Mourelatos, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277:5631-5636. [DOI] [PubMed] [Google Scholar]
- 22.Iannaccone, S. T., S. A. Smith, and L. R. Simard. 2004. Spinal muscular atrophy. Curr. Neurol. Neurosci. Rep. 4:74-80. [DOI] [PubMed] [Google Scholar]
- 23.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, K. W., K. Gorzynski, C. M. Hales, U. Fischer, F. Badbanchi, R. M. Terns, and M. P. Terns. 2001. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 276:38645-38651. [DOI] [PubMed] [Google Scholar]
- 25.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375-387. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. I., S. C. Murthy, J. J. Trimble, R. C. Desrosiers, and J. A. Steitz. 1988. Four novel U RNAs are encoded by a herpesvirus. Cell 54:599-607. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S. I., and J. A. Steitz. 1990. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J. Virol. 64:3905-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155-165. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, S., P. Burlet, Q. Liu, S. Bertrandy, O. Clermont, A. Munnich, G. Dreyfuss, and J. Melki. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16:265-269. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Q., and G. Dreyfuss. 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15:3555-3565. [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Q., U. Fischer, F. Wang, and G. Dreyfuss. 1997. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013-1021. [DOI] [PubMed] [Google Scholar]
- 32.Luhrmann, R. 1990. Functions of U-snRNPs. Mol. Biol. Rep. 14:183-192. [DOI] [PubMed] [Google Scholar]
- 33.Luhrmann, R., B. Kastner, and M. Bach. 1990. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087:265-292. [DOI] [PubMed] [Google Scholar]
- 34.Matera, A. G., and D. C. Ward. 1993. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 121:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meister, G., D. Buhler, B. Laggerbauer, M. Zobawa, F. Lottspeich, and U. Fischer. 2000. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 9:1977-1986. [DOI] [PubMed] [Google Scholar]
- 36.Meister, G., D. Buhler, R. Pillai, F. Lottspeich, and U. Fischer. 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3:945-949. [DOI] [PubMed] [Google Scholar]
- 37.Meister, G., C. Eggert, D. Buhler, H. Brahms, C. Kambach, and U. Fischer. 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990-1994. [DOI] [PubMed] [Google Scholar]
- 38.Meister, G., and U. Fischer. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal U snRNPs. EMBO J. 21:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mourelatos, Z., L. Abel, J. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murthy, S., J. Kamine, and R. C. Desrosiers. 1986. Viral-encoded small RNAs in herpesvirus saimiri induced tumors. EMBO J. 5:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murthy, S. C., J. J. Trimble, and R. C. Desrosiers. 1989. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J. Virol. 63:3307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myer, V. E., S. I. Lee, and J. A. Steitz. 1992. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. USA 89:1296-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai, K., Y. Muto, D. A. Pomeranz Krummel, C. Kambach, T. Ignjatovic, S. Walke, and A. Kuglstatter. 2001. Structure and assembly of the spliceosomal snRNPs. Biochem. Soc. Trans. 29:15-26. [DOI] [PubMed] [Google Scholar]
- 45.Pellizzoni, L., J. Baccon, B. Charroux, and G. Dreyfuss. 2001. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11:1079-1088. [DOI] [PubMed] [Google Scholar]
- 46.Pellizzoni, L., J. Baccon, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 277:7540-7545. [DOI] [PubMed] [Google Scholar]
- 47.Pellizzoni, L., B. Charroux, and G. Dreyfuss. 1999. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. USA 96:11167-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellizzoni, L., B. Charroux, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 152:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pellizzoni, L., N. Kataoka, B. Charroux, and G. Dreyfuss. 1998. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95:615-624. [DOI] [PubMed] [Google Scholar]
- 50.Pellizzoni, L., J. Yong, and G. Dreyfuss. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298:1775-1779. [DOI] [PubMed] [Google Scholar]
- 51.Stark, H., P. Dube, R. Luhrmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409:539-542. [DOI] [PubMed] [Google Scholar]
- 52.Trimble, J. J., D. A. Regier, and R. C. Desrosiers. 1990. Herpesvirus saimiri U RNA sequence variation. Nucleic Acids Res. 18:6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, J., and G. Dreyfuss. 2001. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 276:9599-9605. [DOI] [PubMed] [Google Scholar]
- 54.Wassarman, D. A., S. I. Lee, and J. A. Steitz. 1989. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 17:1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 56.Yong, J., T. J. Golembe, D. J. Battle, L. Pellizzoni, and G. Dreyfuss. 2004. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 24:2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yong, J., L. Pellizzoni, and G. Dreyfuss. 2002. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 21:1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]