Abstract
Studies of diverse phylogenetic lineages reveal that protein disorder increases in concert with organismic complexity but that differences nevertheless exist among lineages. To gain insight into this phenomenology, we analyzed all of the transcription factor (TF) families for which sequences are known for 17 species spanning bacteria, yeast, algae, land plants, and animals and for which the number of different cell types has been reported in the primary literature. Although the fraction of disordered residues in TF sequences is often moderately or poorly correlated with organismic complexity as gauged by cell-type number (r2 < 0.5), an unbiased and phylogenetically broad analysis shows that organismic complexity is positively and strongly correlated with the total number of TFs, the number of their spliced variants and their total disordered residues content (r2 > 0.8). Furthermore, the correlation between the fraction of disordered residues and cell-type number becomes stronger when confined to the TF families participating in cell cycle, cell size, cell division, cell differentiation, or cell proliferation, and other important developmental processes. The data also indicate that evolutionarily simpler organisms allow for the detection of subtle differences in the conserved IDRs of TFs as well as changes in variable IDRs, which can influence the DNA recognition and multifunctionality of TFs through direct or indirect mechanisms. Although strong correlations cannot be taken as evidence for cause-and-effect relationships, we interpret our data to indicate that increasing TF disorder likely was an important factor contributing to the evolution of organismic complexity and not merely a concurrent unrelated effect of increasing organismic complexity.
Keywords: intrinsically disordered protein (IDP), complexity, transcription factors, evolution, cell-type number
Introduction
Significant fractions of eukaryotic genomes encode for proteins with intrinsically disordered regions (IDRs) (Dunker et al. 2000, 2013; Ward et al. 2004; Schlessinger et al. 2011; Yruela and Contreras-Moreira 2012, 2013) that function in signaling and regulatory pathways and dramatically increase protein multifunctionality and nucleic acid interaction through a variety of mechanisms contributing to development (Wright and Dyson 1999; Dyson and Wright 2002, 2005; Levine and Tjian 2003; Liu et al. 2006; Xie et al. 2007; Habchi et al. 2014; van der Lee et al. 2014; Wright and Dyson 2015; Yruela 2015) including the transcriptional regulation of cell differentiation (de Mendoza et al. 2013; Han et al. 2014; Ohtani et al. 2017). Indeed, the massive expansions of transcription factor (TF) families containing significant amounts of IDRs in complex organisms has been used as evidence that IDRs and their alternative splicing and posttranslational modification have been a driving force in the evolution of complex multicellularity (Niklas et al. 2014, 2015; Niklas and Dunker 2016; Dunker et al. 2015; Babu 2016). This proposition is supported by disorder predictions showing that 83–94% of all known TFs possess extended regions of disordered residues and that the degree of intrinsic disorder is significantly higher in eukaryotic than in prokaryotic TFs (Liu et al. 2006; Minezaki et al. 2006).
The goal of this paper is to evaluate the proposition that TFs rich in IDRs could directly contribute to the evolution of complex multicellularity. Clearly, organismic complexity is difficult to quantify objectively since it is likely a multidimensional variable. However, for the purposes of this paper, we have adopted an objective metric—the number of different cell types characterizing an organism—with which to measure complexity. This metric was adopted because: 1) it is applicable to all eukaryotic organisms (Niklas et al. 2014; Chen et al. 2014) since it estimates the extent to which cells are phenotypically differentiated into distinct types independently of an organism’s lineage or level of organization; 2) it can be used to quantify the complexity of unicellular organisms that change their cell phenotype as a result of reproductive or physiological status (Niklas 2014; Duran-Nebreda et al. 2016); 3) it is statistically significantly correlated with proteome size, the total disorder in proteomes, and other metrics (Schad et al. 2011; Nido et al. 2012; Chen et al. 2014); 4) it has been quantified objectively based on an extensive review of the primary literature dealing with all eukaryotic lineages (Bell and Mooers 1997); and 5) it has been used in previous studies analyzing the relationship between genomic features and organism complexity (Xia et al. 2008; Chen et al. 2011; Schad et al. 2011; Nido et al. 2012; Xue et al. 2012).
So as not to bias the results of our study, we analyzed all of the TFs (for which sequences have been documented) for each of 17 phylogenetically diverse species (for which the number of different cell types have been published). This protocol was used because some workers have reported only moderate or poor correlations between IDRs and the number of different cell types in different lineages when using the fraction of disordered residues in entire proteomes, particularly when the data from animals and plants are pooled for regression analysis (i.e., Schad et al. 2011; Xue et al. 2012; Chen et al. 2014). In addition, TFs are known to possess moderate to large IDRs and are also known to play important roles in the regulation of mechanisms implicated in the evolution of multicellularity (i.e., cell division, cell cycle, cell differentiation, or cell proliferation). Accordingly, after reporting the extent to which the IDRs of all TFs are correlated with organismic complexity, we then focus on those TFs that play critical roles in developmental processes.
Materials and Methods
TF Sequences
Protein sequences of all TF families of 17 species were analysed. In particular 66 TF families from nine plant species including four vascular plants (Zea mays [n = 5246], Oryza sativa [n = 3119], Arabidopsis thaliana [n = 2757], Physcomitrella patens [n = 1423]), one bryophyte (Selaginella moellendorffii [n = 949]) and four chlorophytes (Chlamydomonas reinhardtii [n = 367], Chlorella sp. NC64A [n = 321], Micromonas pusilla [n = 309], and Ostreococcus tauri [n = 226]); 51 families from six animal species including model organisms of different categories such as one amphibian (Xenopus tropicalis [n = 1147]), one fish (Danio rerio (n = 2345)), two mammals (Homo sapiens [n = 1691], Mus musculus [n = 1483]), and two other eukaryotes (Drosophila melanogaster [n = 604], Caenorhabditis elegans [n = 706]); 248 regulator proteins from the model yeast Saccharomyces cerevisiae and 304 regulator proteins from the bacteria Escherichia coli.
Protein sequences of plant species were retrieved from the Plant TF Database, http://plntfdb.bio.uni-potsdam.de/v3.0/ (Pérez-Rodríguez et al. 2010). Protein sequences of animal species were retrieved from the Animal TF Database http://bioinfo.life.hust.edu.cn/AnimalTFDB/ (Zhang et al. 2015). Protein sequences from S. cerevisiae and E. coli were retrieved from the Yeast TF Specificity Compendium, http://yetfasco.ccbr.utoronto.ca/ (de Boer and Hughes 2011) and Regulon DB, http://regulondb.ccg.unam.mx/ (Gama-Castro et al. 2016). GO annotations for A. thaliana (Berardini et al. 2004) and H. sapiens were obtained from Gene Ontology Consortium, http://geneontology.org/.
Prediction of Disordered Residues
More than 60 predictors of disorder have been developed (He et al. 2009). A comparison of four well-used predictors and their variants across 1,765 proteomes showed considerable variation in their identification of IDRs (Oates et al. 2013), suggesting that predictor choice is important for this work. In a detailed comparison of 16 commonly used predictors, PONDR VSL2b (Peng et al. 2006) showed the best overall accuracy for long IDRs (Peng and Kurgan 2012). Because such long IDRs are a major feature of TFs (Liu et al. 2006), PONDR VSL2b was selected for our studies. Nevertheless, we also ran DISOPRED3 (Jones and Cozzetto 2015) being overall the results consistent with PONDR VSL2b predictions (supplementary figs. S2 and S3, Supplementary Material online).
Cell-Type Numbers
The number of different cell types for each organism was taken from Bell and Mooers (1997) and Chen et al. (2014). H. sapiens (n = 240), M. musculus (n = 150), X. tropicalis (n = 150), D. rerio (n = 119), Z. mays (n = 100), D. melanogaster (n = 60), O. sativa (n = 44), C. elegans (n = 27), A. thaliana (n = 27), P. patens (n = 20), S. moellendorffii (n = 20), S. cerevisiae (n = 4), C. reinhardtii (n = 2), Chlorella sp. NC64A (n = 2), V. carteri (n = 2), E. coli (n = 2), M. pusilla (n = 1), and O. tauri (n = 1).
Orthologue Searching
Automatic search for orthologues was carried out using the comparative genomic tool EnsemblCompara83 that combines BLAST search and maximum likelihood phylogenetic gene trees, http://useast.ensembl.org/info/genome/compara/homology_method.html (Vilella et al. 2009) and PLAZA, http://bioinformatics.psb.ugent.be/plaza/ (Vandepoele et al. 2013; Proost et al. 2015). For this study we selected Arabidopsis orthologues of TF families, which displayed poor or negative correlation between the fraction of disordered residues and the number of cell types, and the orthologues were represented in at least five organisms including chlorophyte and/or bryophyte, and vascular plants. These TFs were identified in the categories of Gene Ontology of cell differentiation (GO:0030154), cell proliferation (GO:0042127), regulation of cell size (GO:0008361), regulation of cell growth (GO:0030308), cell cycle (GO:0007049), regulation of cell proliferation (GO:0042127), regulation of cell size (GO:0008361), or embryo development (GO:0009793). Complete sequences and the position of typical functional domains were taken from Uniprot (http://www.uniprot.org/). In some cases, if necessary, complete sequences were taken from Phytozome11 (https://phytozome.jgi.doe.gov/).
Data Analysis
Comparative analyses were done using: 1) a set of 66 TF families in 17 species including model organisms of bacteria, yeast, algae, land plants, and animals (see the above TF sequences section); 2) subsets of TFs families represented in selected animal and/or plant species; 3) orthologues associated with the regulation of the cell cycle, cell differentiation, cell proliferation, and cell size (see the above TF sequences section with the constraint that they must be represented in all selected species to provide as broad an evolutionary overview as possible). Statistical analysis was done with Origin Pro8.6. The coefficient of determination, r2, was based on linear regression and determined using standard methods (Anderson-Sprecher 1994). r2 values show the goodness of a fit, and was calculated as:
where RSS is the residual sum of square and TSS is the total sum of square.
Sequence Alignment and Phylogeny Tree
Sequence alignment was done with ClustalOmega (http://www.clustal.org/omega/; Sievers et al. 2011). All of the proteins aligned contain the same conserved domains, which anchors the alignment. Taken as reference the disorder prediction in the consensus sequence of the alignment, three types of disordered residues were calculated, which correspond to: 1) identical disordered residues, where the amino acid sequence and disorder predictions were 100% identity (identical IDRs); 2) similar disordered residues where the disorder predictions were 100% identity but the amino acid sequence varied (similar IDRs); and 3) variable disordered residues where either amino acid sequence or disorder predictions were not conserved (not identical) (variable IDRs) (Bellay et al. 2011). Disordered residues were found using custom Perl scripts. The fraction of each type of disordered region with respect to the total of disordered residues was calculated.
Maximum likelihood trees of selected sequences were calculated using phylogeny.fr tool (http://www.phylogeny.fr/, Dereeper et al. 2008).
Results
General Patterns and Correlations
As noted, it has been suggested in the literature that the evolution of organismic complexity was driven by changes in proteomic disorder. However, there is ample evidence that some functional protein categories play a greater role in driving evolution than others. For example, although the number of TFs in proteomes represents only 0.6–10% of proteomes, members of TF families play key roles in controlling the regulation of the cell cycle, cell division, cell differentiation, cell proliferation or cell size, which are key processes in multicellular organisms. These observations combined with the finding that TFs are predicted to be highly enriched in IDRs makes them good candidates to test the hypothesis whether a positive correlation exists between the number of cell types (see Materials and Methods) and the fraction of disordered residues in TF proteins. This hypothesis was examined at three levels (see Materials and Methods): 1) by analyzing a set of all known TFs across different model organisms including bacteria, yeast, green algae, plants and animals; 2) by examining subsets of TFs families including those that are highly represented in animals and/or plants; and 3) by subsequently analyzing subsets of TF families (i.e., orthologues) associated with the regulation of the cell cycle, cell division, cell differentiation, cell proliferation and cell size with the constraint that they must be found in all selected species to obtain as broad an evolutionary overview as possible (supplementary table S1, Supplementary Material online). Although the subsets of TFs represent only a fraction of ca. 10% of all the TFs examined in this study (fig. 1), they are known to play key roles in the evolution of organismic complexity (Vogel and Chothia 2006). Nevertheless, it is important to note that the TF families examined in our study (as well as the specific members within each of these families) 1) vary in the extent to which they are found in different proteomes (i.e., 277/2757 TFs in A. thaliana and 153/1691 TFs in H. sapiens, fig. 1A and 1B, respectively, and supplementary table S1, Supplementary Material online), 2) participate to different degrees in the aforementioned cellular functions (i.e., 1/111 bZIP and 119/166 MYB in A. thaliana), and 3) are not represented in all lineages (fig. 1), which makes any comparative analysis difficult. For this reason, we selected all well-annotated orthologues of A. thaliana TFs in those families found in all of the 17 phylogenetically representative species.
Fig. 1.
—Distribution of TF protein functions (A and B, left panel) and representative TF families associated with regulation of cell differentiation, cell proliferation, cell cycle, and cell size (A and B, right panel) in A. thaliana (A) and H. sapiens (B). The category “others” refers to poorly represented TF families, with only one protein or only in some taxa.
Analyses using the set of all known TFs indicated that the number of TF proteins correlates significantly and positively with the number of cell types in both plants (r2 = 0.97, P = 1.3E-5) and non-plants (r2 = 0.81, P = 1.4E-3) (supplementary fig. S1, Supplementary Material online). Similarly, a positive linear correlation was observed for the total of disordered residues and the number of different cell types in plants (r2 = 0.82, P = 4.9E-4) and non-plants (r2 = 0.88, P = 3.3E-4) (fig. 2). A strong positive correlation was also observed for the relationship between the fraction of spliced protein variants in TFs and the number of different cell types (r2 = 0.97, P = 5.2E-6) (supplementary fig. S4, Supplementary Material online). However, the fraction of disordered residues in TF sequences did not manifest a significant correlation with the number of different cell types (fig. 3).
Fig. 2.
—Log10-scatter plots of total disordered residues in TF proteins (x axis) versus the number of different cell types in plant (y axis) (A) and non-plant species (B). Disordered residue predictions were made by PONDR VLS2b.
Fig. 3.
—Scatter plot of fraction of disordered residues/total residues in TF proteins (x axis) versus the Log10 of the number of different cell types (y axis) in plant and non-plant species. Disordered residue predictions were made by PONDR VLS2b.
The aforementioned trends varied within TF families. For example, clear differences were found in the bHLH, MYB, and bZIP families (fig. 4). The MYB family had a positive correlation between cell-type number and the fraction of disordered residues for both plants (r2 = 0.60, P = 0.012) and non-plants (r2 = 0.91, P = 0.002) (fig. 4A and 4B and table 1). Conversely, the bHLH and bZIP families had a poor or negative correlation between cell-type number and the fraction of disordered residues (fig. 4C–F and table 1). Moreover, different trends were observed when analyses were done on specific members of these families with potential major implications regarding the evolution of organismic complexity. In the following sections, we illustrate the extent to which this variation is manifest in all of the different TF families for which sequence data and cell number data were available.
Fig. 4.
—Scatter plot of fraction of disordered residues/total residues in TF families versus the Log10 of the number of different cell types in MYB (n = 655 and 98) (A, B), bHLH (n = 862 and 518) (C, D), and bZIP (n = 570 and 249) (E, F) families from plant (A, C, E) and non-plant (B, D, F) species, respectively. Species codes are: Zm, Z. mays; Os, O. sativa; At, A. thaliana; Pp, P. patens; Sm, S. moellendorffii; Ce, C. reinhardtii; Chl, Chlorella sp. NC64A; Mp, M. pusilla; Ot, O. tauri; Hm, H. sapiens; Mm M. musculus; Xt, X. tropicalis; Dr, D. rerio; Dm, D. melanogaster; Ce, C. elegans; Sc, S. cerevisiae; Ec, E. coli. The number of proteins in each species is given between parentheses. Disordered residue predictions were made by PONDR VLS2b.
Table 1.
Regression Parameters of Scatter Plots of Disordered Residues Fraction Versus the Log10 of Number of Cell Types for Several Families and Orthologues of TFs
| Transcription Factor | Species | Proteins | Slope | r2 | Pearson’s r | P Value | F |
|---|---|---|---|---|---|---|---|
| Plants | |||||||
| Families | |||||||
| MYB | 8 | 655 | 10.39 | 0.60 | 0.80 | 0.01 | 11.38 |
| bHLH | 9 | 862 | −2.16 | 0.07 | −0.26 | 0.5 | 0.50 |
| bZIP | 9 | 570 | −9.17 | 0.88 | −0.94 | 1.2E-4 | 58.11 |
| C2H2 zinc finger | 9 | 571 | 0.05 | 0.14 | 0.005 | 0.98 | 1.85E-4 |
| C2C2-GATA | 9 | 179 | 0.78 | 0.13 | 0.07 | 0.85 | 0.03 |
| E2F/DP | 9 | 75 | −4.18 | 0.19 | −0.54 | 0.13 | 2.89 |
| ABI3/VP1 | 9 | 263 | −4.27 | 0.50 | −0.76 | 0.02 | 8.17 |
| SBP | 8 | 200 | −6.95 | 0.55 | −0.78 | 0.02 | 9.48 |
| ARR-B | 9 | 60 | 4.97 | 0.52 | 0.77 | 0.02 | 8.68 |
| NAC | 5 | 541 | 1.56 | 0.35 | 0.72 | 0.17 | 3.17 |
| Orthologues | |||||||
| MYB13 | 6 | 6 | 1.51 | 0.70 | 0.88 | 0.05 | 9.17 |
| CDC5 | 6 | 6 | 6.83 | 0.70 | 0.87 | 0.02 | 12.86 |
| HY5 | 7 | 7 | 4.16 | 0.50 | 0.76 | 0.05 | 6.67 |
| bZIP9 | 5 | 5 | 1.47 | 0.30 | 0.72 | 0.27 | 2.19 |
| bZIP63 | 5 | 5 | 2.39 | 0.84 | 0.93 | 0.01 | 22.21 |
| ZFZ | 7 | 7 | 10.00 | 0.54 | 0.78 | 0.03 | 8.08 |
| GATA9 | 5 | 5 | 5.42 | 0.88 | 0.95 | 0.003 | 38.75 |
| E2F | 8 | 8 | 4.92 | 0.88 | 0.95 | 2.9E-4 | 55.98 |
| ABI3 | 6 | 6 | 3.26 | 0.86 | 0.94 | 0.004 | 32.18 |
| SBP15 | 6 | 6 | 3.20 | 0.86 | 0.94 | 0.004 | 32.23 |
| Animals | |||||||
| Families | |||||||
| MYB | 6 | 98 | 7.36 | 0.91 | 0.96 | 0.002 | 51.18 |
| bHLH | 5 | 518 | 13.63 | 0.39 | 0.73 | 0.1552 | 3.56 |
| bZIP | 6 | 249 | −10.71 | 0.23 | −0.62 | 0.23 | 6.78 |
| E2F | 6 | 46 | −12.21 | 0.50 | −0.77 | 0.06 | 6.08 |
| Orthologues | |||||||
| NHLH1 | 5 | 5 | 3.85 | 0.89 | 0.95 | 0.01 | 33.34 |
| CDC5 | 6 | 6 | 13.20 | 0.81 | 0.92 | 0.008 | 23.23 |
| Dnj11 | 7 | 7 | 4.63 | 0.70 | 0.87 | 0.02 | 12.59 |
| E2F | 5 | 5 | −2.88 | −0.10 | −0.41 | 0.48 | 0.62 |
The bHLH Family
The basic helix-loop-helix bHLH family of TFs is represented in all eukaryotic organisms (n = 862 in plants and n = 518 in animals). The fraction of disordered residues within this group was poorly correlated with cell-type numbers for both plants and non-plants (fig. 4 and table 1). However, subsets of orthologues displayed clear positive correlations with cell-type number (table 1). For instance, the analysis of the TF orthologues encoded by NHLH1 genes (bHLHA35), typical for animals (Lipkowitz et al. 1992; Brown et al. 1992), which belongs to the Gene Ontology category of cell differentiation (GO:0030154), revealed a significant positive relationship between the fraction of disordered residues and the number of cell types (r2 = 0.89, P = 0.01). The sequence alignment shows a gain of disordered residues in vertebrates compared with the invertebrate C. elegans (fig. 5A). Although this gain may reflect the fact that longer sequences are likely to have a higher content of disordered residues, the alignment also shows noticeable variations in the N-terminus and in the typical helix-loop-helix domain. A disordered region connecting two ordered segments within the bHLH domain with 70% identity appears in animals such as D. melanogaster, D. rerio, M. musculus, H. sapiens, but is absent in C. elegans. Additionally, the length of conserved structured segments within this domain and in the N-terminus decreases with increasing organismic complexity as gauged by the number of cell types. The maximum likelihood tree of these orthologues in shown is figure 5C (left panel).
Fig. 5.
—Comparison of orthologues from the bHLH and MYB TF families. Alignments of orthologue proteins encode by (A) NHLH1 gene from H. sapiens (Q02575), M. musculus (Q02576), D. rerio (Q6P0A8), D. melanogaster (O77278) and C. elegans (Q18590) and (B) MYB13 orthologues from Z. mays (K7TYD9), O. sativa (Q6K1S6), Glycine max (Q0PJK2), A. thaliana (Q9LNC9) and S. moellendorffii (D8RNQ0). The number of cell types is given between parentheses on the left side of the sequence. The protein length is written on the right side of the sequence. Sequences are represented by color coded bars representing predicted disorder: disordered residues (red), ordered residues (grey), and alignment gaps (white). (C) Maximum likelihood trees of NHLH1 (left panel) and MYB13 (right panel) sequences used in the alignments. The specie name and the corresponding number of cell types are given in the tree.
The MYB Family
The myeloblastosis MYB family of TFs is also represented in all eukaryotic organisms (n = 655 in plants and n = 98 in animals). Orthologues of the Arabidopsis MYB13 TF, also called AtMYB3R, and members of the plant R2R3-type of the MYB family, which belong to the Gene Ontology category of cell differentiation (GO:0030154), showed a better correlation with cell-type number (r2 = 0.70, P = 0.05) in comparison with the whole family (table 1). The sequence alignment shows that the fraction of disordered residues increases with complexity along the non-conserved C-terminus in land plants. Also the fraction of ordered residues is reduced in the second DNA binding domain (fig. 5B). The maximum likelihood tree of orthologues is shown in figure 5C (right panel).
Similar trends were observed for the CELL DIVISION CYCLE 5 (CDC5) TF (Ohi et al. 1998: Lin et al. 2007; Gräub et al. 2008). Orthologues showed better positive correlations in animals (r2 = 0.81, P = 0.008) and plants (r2 = 0.70, P = 0.02) in comparison with the complete MYB family (table 1). The CDC5 protein is identified in the Gene Ontology category of cell differentiation (GO:0030154). In addition, orthologues of Dnj11 in C. elegans, also called DNAJC2 in H. sapiens, showed a similar positive correlation with cell-type number (r2 = 0.70, P = 0.02). Orthologues of Dnj11 are identified in the Gene Ontology category of regulation of cell growth (GO:0030308).
The bZIP Family
This basic leucine zipper (bZIP) superfamily is represented in both plants (n = 570) and animals (n = 249). It is one of the most ancient plant TF families. It consists of 13 groups, one of which, group H, is the most conserved, being present in all green plant lineages (Guedes-Corrêa et al. 2008). Orthologues of Arabidopsis HY5 (AtbZIP56) (Palme et al. 2016), which belong to group H, showed a significant positive linear correlation between the fraction of disordered residues and the number of cell types (r2 = 0.50, P = 0.05) (table 1). However, across the entire family, a negative correlation was observed (fig. 4E). The HY5 protein has been associated with light-mediated morphogenesis (GO:0010099). Similarly, orthologues of AtbZIP9 and AtbZIP63 TFs (Matiolli et al. 2011), which are related to the Opaque2 family in maize, showed significant positive correlations, especially orthologues of AtbZIP63 (r2 = 0.84, P = 0.01), which plays a role in seed germination. The sequence alignment of orthologues show that overall the fraction of disordered residues increases from green algae and bryophytes to the vascular plants (fig. 6). In the bZIP63 alignment, the structured region close to the conserved bZIP domain (30% identity) is highly reduced in monocots (Z. mays, S. bicolor and O. sativa) compared with the eudicot Arabidopsis and the moss Physcomitrella (fig. 6B). Furthermore, the HY5 alignment shows that the flexible segment involved in COP1 protein interaction (Holm et al. 2001) lacks in green algae compared with vascular plants (fig. 6A). Maximum likelihood trees of orthologues are shown in figure 6C.
Fig. 6.
—Comparison of orthologues from the bZIP TF family. (A) Alignment of HY5 orthologues in Z. mays (B6UEP1), O. sativa (Q0E2Y8), G. max (I1KXV2), A. thaliana (O24646), S. moellendorffii (D8RQ04), C. reinhardtii (A8IM85), and O. lucimarinus (A4RRH6). (B) Alignment of ZIP63 orthologues in Z. mays (B4FJ00), S. bicolor (C5WX70), O. sativa (Q7X9A8), A. thaliana (B9DGI8) and P. patens (A9TD07). The number of cell types is given between parentheses on the left side of the sequence. The protein length is given on the right side of the sequences. Sequences are represented by color coded bars representing predicted disorder: disordered residues (red), ordered residues (grey), and alignment gaps (white). Typical bZIP domain (black box), C3HC4 zinc-finger type domain (black star), and COP1 interacting domain (black triangle) are shown. (C) Maximum likelihood trees of HY5 (left panel) and ZIP63 (right panel) sequences used in the alignment. The specie name and the corresponding number of cell types are given in the tree.
The Zinc-Finger Families
Zinc-finger TFs have been classified in different families in eukaryotes. Here, we report the results obtained with two plant zinc-finger families, the C2H2 (n = 571) and C2C2-GATA (n = 179) families. The data showed no significant linear correlations between the fraction of disordered residues and the number of cell types (r2 = 0.14, P = 0.98 and r2 = 0.13, P = 0.85, respectively) when the complete families were analyzed (table 1). However, AtZFZ (also named REIL2) orthologues, which belong to the C2H2 family (Schmidt et al. 2013), and AtGATA9 orthologues, members of the C2C2-GATA family, showed significant positive correlations with cell-type number (r2 = 0.54, P = 0.03 and r2 = 0.88, P = 0.003, respectively) (table 1). These TFs have been identified in the Gene Ontology categories of ribosomal large subunit biogenesis (GO:0042273) and cell differentiation (GO:0030154), respectively.
The alignment of ZFZ orthologues shows that disordered residue composition varies along the sequences, but that it increases going from the algae (C. reinhardtii and V. carteri) to the monocots (Z. mays and O. sativa) within the C2H2 domains typical of this zinc-finger group (fig. 7A). Note also that the number of C2H2 domains decreases in algae (n = 1) compared with vascular plants (n = 3–4). Similarly, the sequence alignment of GATA9 orthologues shows a disordered residue higher fraction around the conserved typical zinc finger domain in monocots (fig. 7B). Maximum likelihood trees of these orthologues are shown in figure 7C.
Fig. 7.
—Comparison of orthologues from zinc finger TF families. (A) Alignment of FZF orthologues in Z. mays (B6TYD5), O. sativa (Q5Z8K9), A. thaliana (Q9ZQ18), P. patens (A9RG33), S. moellendorffii (D8RTH9), V. carteri (D8UB30) and C. reinhardtii (A8JD88). (B) Alignment of GATA9 orthologues in Z. mays (K7VQ40), M. truncatula (G7LFY2), O. sativa (Q6F2Z7) A. thaliana (O82632) and O. lucimarinus (A4RXG3). The number of cell types is given between parentheses on the left side of the sequence. The protein length is given on the right side of the sequences. Sequences are represented by color coded bars representing predicted disorder: disordered residues (red), ordered residues (grey), and alignment gaps (white). Typical C2H2 and zinc-finger GATA-type domains (black box) are shown. (C) Maximum likelihood trees of the ZFZ (left panel) and GATA9 (right panel) sequences used in the alignments. The specie name and the corresponding number of cell types are given in the tree.
The E2F/DP Family
The E2F/DP family is present in both plants (n = 75) and animals (n = 46). Negative correlations were observed between the fraction of disordered residues and the number of cell types in both kinds of organisms (r2 = 0.19, P = 0.13 and r2 = 0.50, P = 0.06, respectively) (table 1). In contrast, AtE2F-B (also named AtE2F-1) orthologues (Sozzani et al. 2006; Tsai et al. 2008) manifested a strong positive relationship between both variables (r2 = 0.88, P = 2.94E-4) (fig. 8A). It is worth noting that plant and animal orthologues exist and that this protein has been identified in the Gene Ontology category of cell cycle (GO:0007049).
Fig. 8.
—Comparison of orthologues from the E2F/DP TF family. (A) Scatter plot of fraction of disordered residues/total residues (x axis) versus the Log10 of the number of different cell types (y axis) in plants: Z. mays (B4FB61), O. sativa (Q5QL93), A. thaliana (Q9FV71), P. patens (A9RQX0), S. moellendorffii (D8TCC4), V. carteri (Vocar.0001s0396.1), M. pusilla (C1MLR6) and O. tauri (A4RSR6) and animals: M. musculus (P56931), X. tropicalis (F6VW96), D. rerio (A5WUE8), D. melanogaster (O77051) and C. elegans (G5EF11). (B) Alignment of E2F1 orthologues. The number of cell types is given between parentheses on the left side of the sequence. The protein length is given on the right side of the sequences. Sequences are represented by color coded bars representing predicted disorder: disordered residues (red), ordered residues (grey), and alignment gaps (white). Typical DNA-binding and coiled coil domains (black box), cyclin A/CDK2 binding (black star) and retinoblastoma protein binding (black triangle) are shown. (C) The maximum likelihood tree of the E2F1 sequences used in the alignments. The specie name and the corresponding number of cell types are given in the tree.
The sequence alignment displays an increase of disordered residues with increasing organismic complexity that is more pronounced in the case of plants as opposed to animals (fig. 8B). The coil-coiled domain appears more disordered in land plants in comparison with green algae, and it gains increasing flexibility with increasing organismic complexity. Note also that additional interacting motifs involved in the retinoblastoma (Rb) protein binding or in the cyclin A/CDK2 binding appear with increasing complexity. The maximum likelihood tree of orthologues is shown in figure 8C.
Plant Specific TFs
The correlation between the fraction of disordered residues and the number of cell types in specific TFs varied in plant protein families that have no correspondence in animals, as for example the ABI3/VP1, AP2-EREBP, ARR-B, C2C2-CO-like, C2C2-Dof, PBF-2-like/Whirly, SBP-box, and WRKY families, which are present in all plant lineages including the green algae. In general, all of these families showed poor or negative correlations between both variables, with the exception of the ARR-B family (n = 60), which displayed a significant positive correlation (r2 = 0.52, P = 0.02) (table 1). The NAC family (n = 541), one of the largest families of plant-specific TFs and only represented in higher plants, showed a positive but not exceptionally strong correlation (r2 = 0.35, P = 0.17).
Orthologue differences were also found in these families. Two examples are reported here, that are, the ABI3/VP1 family and the SBP-box family.
The complete ABI3/VP1 TF family (n = 263) showed a negative correlation between the fraction of disordered residues and the number of cell types (r2 = 0.50, P = 0.02) (table 1). However, orthologues of AtABI3 (ABSCISIC ACID INSENSITIVE 3) (Delmas et al. 2013) showed a strong positive relationship between both variables (r2 = 0.86, P = 4.0E-3). ABI3 TF has been associated with embryo development (GO:0009793) and plastid organization (GO:0009657).
A negative trend was observed within the complete SBP-box family (n = 200) (r2 = 0.55, P = 0.02). However, orthologues of the Arabidopsis squamosa promoter-binding-like protein SPL15 (Schwarz et al. 2008) showed a strong positive correlation (r2 = 0.86, P = 4.0E-3). AtSPL15 is reported to be involved in the regulation of cell proliferation (GO:0042127) and regulation of cell size (GO:0008361).
Evolution of Disorder in Orthologue Sequences
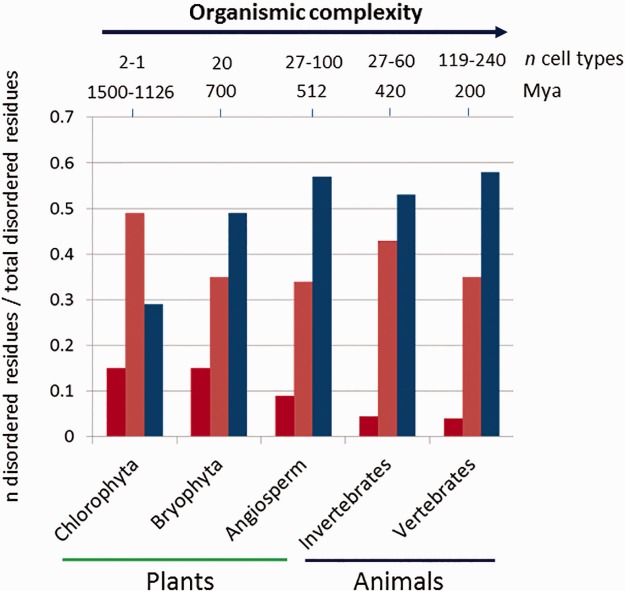
The distribution of IDRs was analyzed in TF orthologues (fig. 9). We defined three different regimes within orthologue alignments: identical IDRs; similar IDRs, and variable IDRs (see Materials and Methods). Analyses indicated that the fraction of identical IDRs is higher in simpler organisms such as the green algae (chlorophyte) and non-vascular land plants (bryophyta) in comparison to the vascular plants and animals. Overall the data for animals showed lower amount of identical IDRs compared to plants. No difference was observed between invertebrates and vertebrates. The amino acid content of similar IDRs and variable IDRs represented a higher proportion or total disordered residues than that of identical IDRs. However, the fraction of less conserved IDRs (variable IDRs) was lower in the green algae (chlorophyte) and the non-vascular plants in comparison to vascular plants and animals.
Fig. 9.
—Bar-plot of the fraction of identical IDRs (dark red), similar IDRs (pink) and variable IDRs (blue) in chlorophyte, bryophyte and angiosperm plants, and invertebrate and vertebrate animals. The data represent the average of nine groups of orthologues in plants and three groups of orthologues in animals.
Discussion
Organism Complexity
Different criteria have been used to measure organismic complexity, for example, genome size, proteome size, protein length, alternative splicing events, protein disorder, and number of cell types (Linch and Conery 2003; Taft and Mattick 2003: Xia et al. 2008; Schad et al. 2011; Chen et al. 2012, 2014; Xue et al. 2012; Niklas et al. 2014). However, not all of these variables show the same degree of correlation with organismic complexity across all lineages and kingdoms. In particular, although the fraction of disordered residues increases significantly between prokaryotes and eukaryotes it has not been established as a significant predictor of organism complexity across eukaryotic clades (Schad et al. 2011; Chen et al. 2014). We attribute this to the fact that many previous studies have only compared complete proteomes, which can obscure trends due to evolutionary changes within specific protein families and functions. When the focus is shifted away from the complete proteome and focused on TFs, particularly those involved in key developmental processes, the relationship between the fraction of disordered residues and organismic complexity becomes far clearer. For example, one previous analysis, focusing on the set of proteins that constitute the centrosome in animal cells, which are particularly involved in asymmetric cell division and which are enriched in coiled-coiled regions or IDRs, reported positive correlations between the fraction of disordered residues and organism complexity (Nido et al. 2012). Indeed, the analyses presented here verify a strong positive correlation between the number of different cell types and the total number of disordered residues in all known TFs.
We interpret the observed positive correlation between these two variables to indicate that increases in the functional versatility of TFs could contributed significantly to the evolution of multicellularity, a conclusion that is consistent with the positive correlation observed previously between the number of cell types and the number of total disordered residues (Niklas et al. 2014). In contrast, our analyses reveal a weaker correlation between the proteome fraction of disordered residues and the number of different cell-types (as reported by Schad et al. 2011; Chen et al. 2014) because the correlation between these two variables of interest differ among different TF families. Some families manifest positive correlations (e.g., MYB and ARR-B), whereas others are poorly correlated (e.g., bHLH, C2H2, and C2C2-GATA) or are even negatively correlated (e.g., bZIP, E2F/DP, ABI3/VP1, and SBP). This phenomenology could indicate that different TF families have experienced different evolutionary processes (Pires and Dolan 2010; Carretero-Paulet et al. 2010; Feller et al. 2011) which probably have obscured the results of some comparative studies.
An additional confounding factor is evolutionary divergence in some but not all TF families. For example, ABI3/VP1, AP2-EREBP, ARR-B, C2C2-CO-like, C2C2-Dof, PBF-2-like/Whirly, ARR-B, SBP-box, and WRKY families took on different functionalities after the Chlorophyta (green algae) and Streptophyta (charophycean algae and the land plants) diverged in the early Paleozoic (Riaño-Pachón et al. 2008) and, in general, they showed poor or negative correlations between both variables. Such phylogenetic divergences can mask or obscure correlations between organismic complexity and the extent to which TFs are disordered. Rapid evolutionary dynamics can drive related proteins to functionally diverge as a consequence of restructuring (Siltberg-Liberles 2011; Dos Santos et al. 2016). Recent proteins often evolve through point mutations, insertion and deletion events, which affect IDRs (Light et al. 2013a, 2013b; Khan et al. 2015). IDRs can rapidly change and undergo rearrangements (and thereby evolve more rapidly) than ordered regions, even under lower selective pressure, particularly when dealing with ancient divergences. In contrast, the retention of functionality can be favored in more recently divergent lineages allowing stronger correlations. For instance, the MYB family, which is enriched in sequence segments involved in cell cycle regulation and cell proliferation (Cominelli and Tonelli 2009), manifest strong positive correlations in both animals and plants (fig. 1).
Another factor that affects the correlation between organismic complexity and the disordered residue fraction in TFs is the distinction between haploid and diploid organisms. For example, when haploid unicellular algae and diploid vascular plant sporophytes are treated together, a poor or even negative correlation is observed, whereas strong correlations are observed among the vascular plants. In this context, it is also worth noting that morphological criteria are used to identify the number of different cell-types (Bell and Mooers 1997) and that this protocol can seriously underestimate the number of different cell types that an unicellular organism produces a result of different physiological cellular states.
Evolution of TFs Disorder
Strong positive correlations between the proportion of disordered TF residues and cell-type number were particularly evident for TF orthologues involved in cell division, cell differentiation, and cell proliferation, which is consistent with the positive correlation between these two variables in centrosome proteins that regulate cell division and differentiation in animal cells (Nido et al. 2012). Furthermore, our data reveal variations in the predicted IDRs in TF sequences (fig. 9), particularly variations in more conserved IDRs (identical IDRs and similar IDRs), which are more prevalent in simpler organisms (e.g., green algae and mosses) compared to late-divergent plant and animal lineages (e.g., angiosperms and mammals). These types of IDRs are strongly associated with diverse multifunctional signaling and regulation pathways (Bellay et al. 2011). Collectively, we interpret our results to indicate that increases in the flexibility/ductility in functional disordered TF domains likely facilitated the innovation of more complex gene regulatory networks involved in cell growth, cell division and cell proliferation, and thus contributed to the evolution of complex multicellularity, for example, the number of functional disordered C2H2 motifs increased in FZF orthologues of vascular plants compared with green algae (fig. 7) and additional functional TF motifs appear in bZIP and E2F/DP TF families of more complex organisms (figs. 6 and 8). On the other hand, variable IDRs, which are not associated to a known specific function (Bellay et al. 2011) but can contribute to increase functionality (Fuxreiter et al. 2011) also increase in complex organisms.
Roles of IDRs in TFs
The experimental exploration of how IDRs and IDPs affect regulation (Dunker et al. 2000) poses a considerable challenge, especially in plants. Nevertheless, some studies have shed light on how IDRs affect TF functionalities. For example, the X-ray crystallographic structure of E2F1 in complex with the retinoblastoma (Rb) C-terminus domain and DP1 (pdb 2AZE) indicates that coiled-coil domains (also called leucine zippers) in E2F1 and DP1 form a heterodimer (Rubin et al. 2005) and interact with high-affinity with the Rb domain. The coiled-coil domains of both E2F1 and EDP1 are predicted IDRs (figs. 8 and 10) and thus likely undergo a disorder-to-order transition on binding (Bracken 2001), a feature that is likely shared by all E2F/DP family members.
Fig. 10.
—Structure of human E2F1 and DP1 proteins in the monomer and heterodimer states. X-ray crystallographic of partial human E2F1-DP1 heterodimer (pdb 2AZE; Rubin et al. 2005) is shown as cartoon. The E2F1 (198–301 residues in pink) and DP1 (196–350 residues in blue) proteins are shown. Predicted disordered residues in monomers by PONDR VLS2b are shown in red line. The three-dimensional cartoons were drawn using PyMol 1.4.1 (Schrodinger LLC).
Note that the Rb domain is absent in unicellular organisms and that the hyperphosphorylation of RbC destabilizes the RbC-E2F-DP complex, indicating that this interaction is regulated by PTMs, a mechanism commonly observed in IDRs (Gavis and Hogness 1991; López and Hogness 1991; Galant et al. 2002; Ronshaugen et al. 2002; Iakoucheva et al. 2004). This mechanism for the regulation of TFs associated with IDRs is probably altered in green algae and invertebrates, which lack conserved phosphorylation sites typical in vascular plants and mammals. For instance, Ser177, Y378 and S379 in A. thaliana (Magyar et al. 2005), and Ser125 and Ser 119 in M. musculus (Wu et al. 2012; Mertins et al. 2014) are absent in sequences of V. carteri, M. pusila, O. tauri and C. elegans, respectively.
The increase of IDR flanking the DNA-binding domains can also influence DNA recognition (Fuxreiter et al. 2011). Our results show that changes in the length and/or amino acid composition of IDRs outside the DNA-binding domains in the bHLH, MYB, bZIP, zinc finger, or E2F/DP TF orthologues correlate positively with increasing complexity. These changes can influence selectivity or binding affinity through dynamic interactions within the DNA-binding site and affect the flexibility/ductility required for specific functions. These IDRs can also promote and modulate the conversion from a non-specific to a specific complex by changes in flexibility and mobility achieved by direct or indirect mechanisms, for example, hydrogen bonding and electrostatic interactions with phosphates (Luscombe et al. 2001; Gromiha et al. 2004). In the Hox family of TFs, IDR outside the DNA-binding site facilitates DNA binding, and recognize specific minor grove sequences (Tóth-Petróczy et al. 2009) and enable rapid sequence search via brachiation (Vuzman and Levy 2010). They also have the dual functionality of inhibiting DNA binding unless bound to a partner TF, thus effectively preventing spurious DNA recognition (Passner et al. 1999). The IDRs in homeobox proteins also affect DNA affinity (Liu et al. 2008) and directly recruit many other proteins (Hsiao et al. 2014). Another example is the DNA recognition domain in ANAC046, a plant-specific NAC transcription factor. This domain forms the RCD1-ANAC046 complex that does not involve folding-upon-binding (O'Shea et al. 2005). Yet more examples are the NPR1-interacting proteins (NIPs) whose conserved disordered leucine zipper domain stimulates DNA-binding (Després et al. 2003).
Distant IDRs also affect DNA binding modulating conformational selection, flexibility and competitive binding (Fuxreiter et al. 2011). The spanning IDRs in the C-terminus and N-terminus of the orthologues in bHLH, MYB, bZIP, zinc-finger, and E2F/DP TF families (variable IDRs) can fine-tune protein and DNA interactions. The specificity/affinity of DNA binding can be also linked to whole protein recognition mechanisms by means of allosteric regulation (Lefstin and Yamamoto 1998; Ma and Nussinov 2009). An example is the disordered segment in the C-terminus PB1 domain of the Arabidopsis Auxin Response Factor 7 (ARF7), which facilitates the interaction with other TFs and phytohormones such as Indole Acetic Acid (IAA) (Guilfoyle and Hagen 2012) to modulate IAA signaling in cell division and plant development (Korasick et al. 2014).
Concluding Remarks
Experimental studies indicate that the conserved functional domains in TFs typically consist of DNA-binding structured motifs linked to IDRs, which modulate TF function in ways including the formation of protein-complexes via disorder-to-order transitions. The recruited protein partners include other TFs and coactivators and corepressors. Although additional research is required to further substantiate this conclusion, we argue that evolutionary variations of IDRs in general contribute to signaling and regulation in fundamental ways those likely increased TF multifunctional domains facilitating the evolution of multicellular organisms within all eukaryotic lineages.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
I.Y. planned the work, performed data analysis, designed figures, discussed the results and wrote the manuscript; C.O. performed IDP and IDR analysis, discussed the results and revised the manuscript; A.K.D. and K.J.N. planned the work, discussed the results and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank to S. Bondos and D. Moreno for comments that helped to improve this manuscript. This work was supported by the Mobility Program for training and research of Spanish Ministry of Education, Culture and Sports (PR2015-00353) and Gobierno de Aragón (DGA-GC B18). Some of these grants were partially financed by the EU FEDER Program.
Literature Cited
- Anderson-Sprecher R. 1994. Model comparisons and R. Am Stat. 48 (2):p113–117. [Google Scholar]
- Babu MM. 2016. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 44:1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Mooers AO.. 1997. Size and complexity among multicellular organisms. Biol J Linnean Soc. 60:345–363. [Google Scholar]
- Bellay J, et al. 2011. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 12:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, et al. 2004. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer CG, Hughes TR.. 2011. YeTFaSCo: a database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res. 40:D169–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C. 2001. NMR spin relaxation methods for characterization of disorder and folding in proteins. J Mol Graph Model. 19:3–12. [DOI] [PubMed] [Google Scholar]
- Brown L, Espinosa R 3rd, Le Beau MM, Siciliano MJ, Baer R.. 1992. HEN1 and HEN2: a subgroup of basic helix-loop-helix genes that are coexpressed in a human neuroblastoma. Proc Natl Acad Sci U S A. 89:8492–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, et al. 2010. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153:1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Lin HY, Pan CL, Chen FC.. 2011. The plausible reason why the length of 5' untranslated region is unrelated to organismal complexity. BMC Res Notes 4:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tovar-Corona JM, Urrutia AO.. 2012. Alternative splicing: a potential source of functional innovation in the eukaryotic genome. Int J Evol Biol. 2012:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Bush SJ, Tovar-Corona JM, Castillo-Morales A, Urrutia AO.. 2014. Correcting for differential transcript coverage reveals a strong relationship between alternative splicing and organism complexity. Mol Biol Evol. 31:1402–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Tonelli C.. 2009. A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 19:1231–1232. [DOI] [PubMed] [Google Scholar]
- Delmas F, et al. 2013. ABI3 controls embryo degreening through Mendel's I locus. Proc Natl Acad Sci U S A. 110:E3888–E3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, et al. 2003. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15:2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos HG, Nunez-Castilla J, Siltberg-Liberles J.. 2016. Functional diversification after gene duplication: paralog specific regions of structural disorder and phosphorylation in p53, p63, and p73. PLoS One 11:e0151961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, et al. 2013. What’s in a name? Why these proteins are intrinsically disordered. Intrinsically Disord Proteins 1(1):e24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Bondos SE, Huang F, Oldfield CJ.. 2015. Intrinsically disordered proteins and multicellular organisms. Semin Cell Dev Biol. 37:44–55. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ.. 2000. Intrinsic protein disorder in complete genomes. Genome Informatics 11:161–171. [PubMed] [Google Scholar]
- Duran-Nebreda S, Bonforti A, Montañez R, Valverde S, Solé R.. 2016. Emergence of proto-organisms from bistable stochastic differentiation and adhesion. J R Soc Interface 13:20160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE.. 2002. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 12:54–60. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE.. 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 6:197–208. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E.. 2011. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66:94–116. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Simon I, Bondos S.. 2011. Dynamic protein-DNA recognition: beyond what can be seen. Trends Biochem Sci. 36:415–423. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, et al. 2016. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 44(D1):D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R, Walsh CM, Carroll SB.. 2002. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129:3115–3126. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Hogness DS.. 1991. Phosphorylation, expression and function of the Ultrabithorax protein family in Drosophila melanogaster. Development 112:1077–1093. [DOI] [PubMed] [Google Scholar]
- Gräub R, et al. 2008. Cell cycle-dependent phosphorylation of human CDC5 regulates RNA processing. Cell Cycle 7:1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha M, Siebers JG, Selvaraj S, Kono H, Sarai A.. 2004. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. J Mol Biol. 337:285–294. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G.. 2012. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 190:82–88. [DOI] [PubMed] [Google Scholar]
- Guedes-Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, et al. 2008. The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3:e2944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S, Uversky VN.. 2014. Introducing protein intrinsic disorder. Chem Rev. 114:6561–6588. [DOI] [PubMed] [Google Scholar]
- Han X, Kumar D, Chen H, Wu S, Kim JY.. 2014. Transcription factor-mediated cell-to-cell signalling in plants. J Exp Bot. 65: 1737–1749. [DOI] [PubMed] [Google Scholar]
- He B, et al. 2009. Predicting intrinsic disorder in proteins: an overview. Cell Res. 19:929–949. [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW.. 2001. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao HC, et al. 2014. The intrinsically disordered regions of the Drosophila melanogaster Hox protein ultrabithorax select interacting proteins based on partner topology. PLoS One 9:e108217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, et al. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32:1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Cozzetto D.. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31:857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T, Douglas GM, Patel P, Nguyen Ba AN, Moses AM.. 2015. Polymorphism analysis reveals reduced negative selection and elevated rate of insertions and deletions in intrinsically disordered protein regions. Genome Biol Evol. 7:1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, et al. 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A. 111:5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR.. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885–888. [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R.. 2003. Transcription regulation and animal diversity. Nature 424:147–151. [DOI] [PubMed] [Google Scholar]
- Light S, Sagit R, Ekman D, Elofsson A.. 2013a. Long indels are disordered: a study of disorder and indels in homologous eukaryotic proteins. Biochim Biophys Acta 1834:890–897. [DOI] [PubMed] [Google Scholar]
- Light S, Sagit R, Sachenkova O, Ekman D, Elofsson A.. 2013b. Protein expansion is primarily due to indels in intrinsically disordered regions. Mol Biol Evol. 30:2645–2653. [DOI] [PubMed] [Google Scholar]
- Lin Z, et al. 2007. AtCDC5 regulates the G2 to M transition of the cell cycle and is critical for the function of Arabidopsis shoot apical meristem. Cell Res. 17:815–828. [DOI] [PubMed] [Google Scholar]
- Linch M, Conery JS.. 2003. The origins of genome complexity. Science 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- Lipkowitz S, et al. 1992. A comparative structural characterization of the human NSCL-1 and NSCL-2 genes. Two basic helix-loop-helix genes expressed in the developing nervous system. J Biol Chem. 267:21065–21071. [PubMed] [Google Scholar]
- Liu Y, Matthews KS, Bondos SE.. 2008. Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J Biol Chem. 283:20874–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. 2006. Intrinsic disorder in transcription factors. Biochemistry 45:6873–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López AJ, Hogness DS.. 1991. Immunochemical dissection of the Ultrabithorax homeoprotein family in Drosophila melanogaster. Proc Natl Acad Sci U S A. 88:9924–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM.. 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29:2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Nussinov R.. 2009. Amplification of signaling via cellular allosteric relay and protein disorder. Proc Natl Acad Sci U S A. 106:6887–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, et al. 2005. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17:2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiolli CC, et al. 2011. The Arabidopsis bZIP gene AtbZIP63 is a sensitive integrator of transient abscisic acid and glucose signals. Plant Physiol. 157:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, et al. 2013. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci U S A. 110:E4858–E4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, et al. 2014. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics 13:1690–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minezaki Y, Homma K, Kinjo AR, Nishikawa K.. 2006. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcription regulation. J Mol Biol. 359:1137–1149. [DOI] [PubMed] [Google Scholar]
- Nido GS, Méndez R, Pascual-García A, Abia D, Bastolla U.. 2012. Protein disorder in the centrosome correlates with complexity in cell types number. Mol Biosyst. 8:353–367. [DOI] [PubMed] [Google Scholar]
- Niklas KJ. 2014. The evolutionary-developmental origins of multicellularity. Am J Bot. 101:6–25. [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Bondos SE, Dunker AK, Newman SA.. 2015. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front Cell Dev Biol. 3:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ, Cobb ED, Dunker AK.. 2014. The number of cell types, information content, and the evolution of complex multicellularity. Acta Soc Bot Pol. 83:337–347. [Google Scholar]
- Niklas KJ, Dunker AK.. 2016. 2 Alternative splicing, intrinsically disordered. Multicellularity: origins and evolution, In Karl J. Niklas, Stuart A. Newman, editors. Cambridge, Massachusetts: MIT Press. pp. 17-40.
- Oates ME, et al. 2013. D2P2: database of disordered protein predictions. Nucleic Acids Res. 41:D508–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, et al. 1998. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol Cell Biol. 18:4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M, Akiyoshi N, Takenaka Y, Sano R, Demura T.. 2017. Evolution of plant conducting cells: perspectives from key regulators of vascular cell differentiation. J Exp Bot. 68:17–26. [DOI] [PubMed] [Google Scholar]
- O'Shea C, et al. 2005. Protein intrinsic disorder in Arabidopsis NAC transcription factors: transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem J. 465:281–294. [DOI] [PubMed] [Google Scholar]
- Palme K, Teale W, Dovzhenko A.. 2016. Plant signaling: HY5 synchronizes resource supply. Curr Biol. 26:R328–R329. [DOI] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK.. 1999. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397:714–719. [DOI] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z.. 2006. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 7:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z-L, Kurgan L.. 2012. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr Protein Pept Sci. 13:6–18. [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez, et al. 2010. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38(1):D822–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N, Dolan L.. 2010. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 27:862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, et al. 2015. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Corrêa LG, Trejos-Espinosa R, Mueller-Roeber B.. 2008. Green transcription factors: a Chlamydomonas overview. Genetics 179:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, McGinnis N, McGinnis W.. 2002. Hox protein mutation and macroevolution of the insect body plan. Nature 415:914–917. [DOI] [PubMed] [Google Scholar]
- Rubin SM, Gall AL, Zheng N, Pavletich NP.. 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F1 release. Cell 123:1093–1106. [DOI] [PubMed] [Google Scholar]
- Schad E, Tompa P, Hegyi H.. 2011. The relationship between proteome size, structural disorder and organism complexity. Genome Biol 12:R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, et al. 2011. Protein disorder: a breakthrough invention of evolution? Curr Opin Struct Biol. 21:412–418. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Dethloff F, Beine-Golovchuk O, Kopka J.. 2013. The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold. Plant Physiol. 163:1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P.. 2008. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 67:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG.. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Sys Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltberg-Liberles J. 2011. Evolution of structurally disordered proteins promotes neostructuralization. Mol Biol Evol. 28:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, et al. 2006. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 140:1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R, Mattick J.. 2003. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biol. 5:P1. [Google Scholar]
- Tóth-Petróczy A, Simon I, Fuxreiter M, Levy Y.. 2009. Disordered tails of homeodomains facilitate DNA recognition by providing a trade-off between folding and specific binding. J Am Chem Soc. 131:15084–15085. [DOI] [PubMed] [Google Scholar]
- Tsai S, et al. 2008. Mouse development with a single E2F activator. Nature 454:1137–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, et al. 2014. Classification of intrinsically disordered regions and proteins. Chem Rev. 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, et al. 2013. pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ Microbiol. 15:2147–2153. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, et al. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Chothia C.. 2006. Protein family expansions and biological complexity. PLoS Comput Biol. 2:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuzman D, Levy Y.. 2010. DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proc Natl Acad Sci U S A. 107:21004–21009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT.. 2004. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 337:635–645. [DOI] [PubMed] [Google Scholar]
- Wu X, et al. 2012. Investigation of receptor interacting protein (RIP3)-dependent protein phosphorylation by quantitative phosphoproteomics. 2012. Mol Cell Proteomics 11:1640–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ.. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 293:321–331. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ.. 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Fu Z, Hou L, Han J-DJ.. 2008. Impacts of protein–protein interaction domains on organism and network complexity. Genome Res. 18:1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, et al. 2007. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 6:1882–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunker AK, Uversky VN.. 2012. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 30:137–149. [DOI] [PubMed] [Google Scholar]
- Yruela I. 2015. Plant development regulation: Overview and perspectives. J Plant Physiol. 182:62–78. [DOI] [PubMed] [Google Scholar]
- Yruela I, Contreras-Moreira B.. 2012. Protein disorder in plants: a view from the chloroplast. BMC Plant Biol. 12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yruela I, Contreras-Moreira B.. 2013. Genetic recombination is associated with intrinsic disorder in plant proteomes. BMC Genomics 14:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-M, et al. 2015. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 43 (D1):D76–D81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.