Recent genomic and quantitative work is transforming our understanding of quantitative resistance in plants.
Abstract
Molecular plant pathology has focused on studying large-effect qualitative resistance loci that predominantly function in detecting pathogens and/or transmitting signals resulting from pathogen detection. By contrast, less is known about quantitative resistance loci, particularly the molecular mechanisms controlling variation in quantitative resistance. Recent studies have provided insight into these mechanisms, showing that genetic variation at hundreds of causal genes may underpin quantitative resistance. Loci controlling quantitative resistance contain some of the same causal genes that mediate qualitative resistance, but the predominant mechanisms of quantitative resistance extend beyond pathogen recognition. Indeed, most causal genes for quantitative resistance encode specific defense-related outputs such as strengthening of the cell wall or defense compound biosynthesis. Extending previous work on qualitative resistance to focus on the mechanisms of quantitative resistance, such as the link between perception of microbe-associated molecular patterns and growth, has shown that the mechanisms underlying these defense outputs are also highly polygenic. Studies that include genetic variation in the pathogen have begun to highlight a potential need to rethink how the field considers broad-spectrum resistance and how it is affected by genetic variation within pathogen species and between pathogen species. These studies are broadening our understanding of quantitative resistance and highlighting the potentially vast scale of the genetic basis of quantitative resistance.
INTRODUCTION
Plants must defend themselves from a variety of potential opportunistic microbial pathogens that employ a variety of virulence strategies. For example, biotrophic pathogens feed off resources derived from living host tissue, and successful biotrophs have found methods to avoid or interfere with the host immune response (Dangl and Jones, 2001; Jones and Dangl, 2006; Bent and Mackey, 2007). Biotrophic pathogens appear to derive evolutionarily from commensal microbial species commonly found on plants in microbiome studies (Lundberg et al., 2012; Ortega et al., 2016). By contrast, necrotrophic pathogens actively attempt to destroy living host tissue and use the dead tissue for nutrients (Mengiste, 2012). Necrotrophic pathogens appear to be evolutionarily derived from saprophytic microbes that feed off naturally senesced plant tissue in the environment (Kohler et al., 2015). In addition to feeding lifestyle (necrotroph versus biotroph), pathogens can be described according to their host range, either as specialists or generalists, whereby specialist pathogens are adapted to a very narrow host range and typically have robust means of overcoming the defenses for that range of hosts (Fan et al., 2011). By contrast, generalist pathogens are adapted to a broad range of host plants, but the mechanism(s) enabling this broad host range are unknown. To survive, a plant must accurately detect and appropriately respond to each of these differential pathogenic strategies.
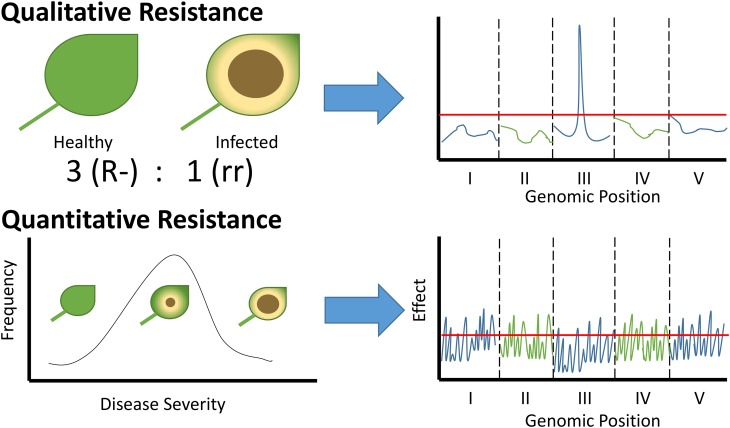
Qualitative plant resistance traits produce discrete classes of resistant and susceptible individuals and segregate as simple Mendelian loci; quantitative plant-pathogen interactions produce a continuous distribution of phenotypes from susceptible to resistant, so individuals do not group into discrete classes. A significant body of current plant pathology research has focused on examples of qualitative resistance, where the underlying genetic architecture relies on a few genes of large effect that are amenable to detailed molecular analysis (Figure 1). By contrast, the genetic architecture underlying quantitative resistance is thought to involve many genes with small to moderate effects (Figure 1) (Poland et al., 2009; St Clair, 2010; Roux et al., 2014; Niks et al., 2015). However, quantitative resistance governs the outcome of the vast majority of host-pathogen interactions. Many quantitative disease resistance loci have been mapped over the past decades, but the total number of genes controlling variation in quantitative resistance remains unknown in any system.
Figure 1.
Illustration Showing the Expected Difference in Phenotypes between Qualitative and Quantitative Resistance.
Qualitative resistance results in phenotypes that fit into distinct categories according to Mendelian ratios. Genetic mapping of qualitative resistance results in a single genetic locus that often maps to genes encoding RLK or NLR proteins. Alternatively, quantitative resistance cannot be easily categorized into distinct groups where resistance adheres more to a continuous distribution of susceptible and resistant individuals. Genetic mapping of qualitative resistance will result in either a large number of genomic loci or no genomic loci associated with resistance depending on the power and stringency of statistical testing within the experimental design.
The biological complexity of quantitative resistance and the technical complexity of identifying the underlying genes have hindered detailed molecular analysis of the causal genes. To address this knowledge gap, systematic studies are investigating the genetic underpinnings of quantitative resistance using modern mapping populations with improved power and resolution, such as in Southern maize leaf blight (Bipolaris maydis) and gray leaf spot (Cercospora zeae-maydis) in maize (Zea mays) (Kump et al., 2011; Benson et al., 2015), Sclerotina sclerotiorum in soybean (Glycine max) (Kim and Diers, 2000), Alternaria solani in tomato (Solanum lycopersicum) (Foolad et al., 2002; Zhang et al., 2003), and Botrytis cinerea in Arabidopsis thaliana (Denby et al., 2004; Rowe and Kliebenstein, 2008; Corwin et al., 2016b). These studies are developing a better understanding of the genetic architecture driving quantitative resistance and identifying the causal loci controlling this variation.
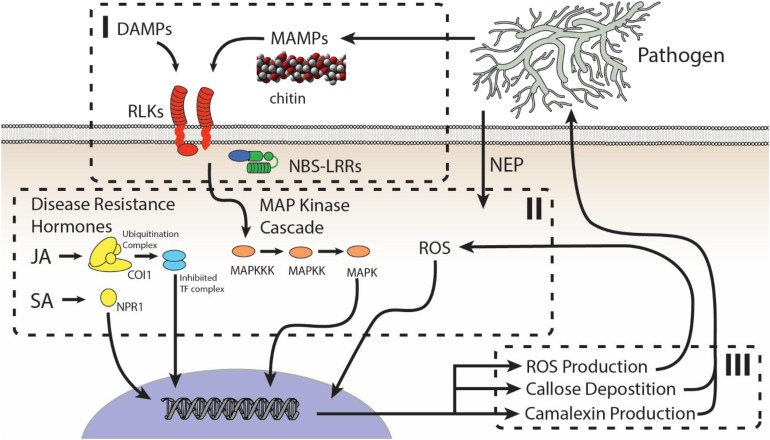
Tests that directly assess if the genes driving quantitative resistance include or resemble known components of the innate immune system are illuminating key aspects of quantitative resistance (Jones and Dangl, 2006; French et al., 2016). Typically, these innate immunity genes consist of plasma membrane-localized receptor-like kinases (RLKs) and/or cytoplasmic Nod-like receptors (NLRs) that detect the presence of a pathogenic microbe and initiate the appropriate immune response (Figure 2, I). Alternatively, quantitative resistance genes may have functions not previously associated with disease resistance (Figure 2, II and III) (Poland et al., 2009). Characterizing the interplay of innate immunity and quantitative resistance is critical to understand the evolutionary selective pressures that shape quantitative resistance. For example, this knowledge would allow us to evaluate if variation in quantitative resistance loci is primarily driven by endemic pathogens constantly present within the host’s environment or if these genes are predominantly responding to the transient presence of epidemic pathogens. Currently, evolutionary studies on resistance genes focus on qualitative resistance genes linked to epidemic pathogens that have predictable boom-and-bust disease cycles. Understanding the evolutionary pressures on quantitative resistance loci is necessary for the development of crops with durable resistance to some pathogens and to broaden our understanding of plant-microbe interactions. Here, we review studies that are beginning to illuminate how the different levels of defense, perception, signaling, and output contribute to quantitative resistance (Figure 2). Additionally, we discuss existing limitations and potential solutions that may give finer precision and more inclusive answers to the issues described above.
Figure 2.
General Model for the Plant Innate Immune System.
The model can be split into (I) perception, (II) signal transduction, and (III) defense response. (I) Plant perception of damage-associated molecular patterns (DAMPs) and MAMPs or PAMPs are detected either apoplastically via RLKs or symplastically via cytoplasmic NBS-LRRs. (II) Signal transduction of DAMPs and MAMPs is performed by the MAP kinase cascade and a series of transcription factors, including members of the WRKY family (not depicted). (III) Signal transduction drives the production of specific defense responses, including reactive oxygen species (ROS) production, callose deposition, and other specialized metabolism (represented here as camalexin). Importantly, the type of defense response can be shaped by either salicylic acid (SA) to drive responses to biotrophic pathogens or JA to drive responses to necrotrophic pathogens and herbivores. In addition, ROS may play a role, both as a defense response (III) and a signal for shaping defense responses (II). NEP, necrosis and ethylene-inducing proteins.
POLYGENIC ARCHITECTURE OF QUANTITATIVE RESISTANCE
Quantitative resistance is by definition polygenic since the continuous distribution of heritable phenotypes must result from combinations of genetic loci. In this review, we use the purely quantitative definition of quantitative resistance, where resistance is a quantifiable trait that typically displays a continuous distribution. This is in contrast to the alternative quantitative resistance terminologies like partial resistance, where resistance is phenotypically incomplete allowing some pathogen growth, or durable, where resistance is stable over longer evolutionary time frames (Niks et al., 2015). The purely quantitative definition is inherently agnostic to the mechanisms underlying the trait or potential outcomes. Additionally, the term partial resistance has an implication that total resistance is an achievable state, via biotechnology or evolution, which may not be possible for some pathogens. Illustrating the difficulty with the partial versus total resistance terminology is a qualitative resistance to B. cinerea within Solanum that is actually generated by a polygenic quantitative resistance architecture (Finkers et al., 2007a, 2007b, 2008).
Part of this discrepancy in terminology arises from the fact that developing a true picture of the depth and breadth of the genetic architecture driving quantitative resistance has been largely limited by the mapping populations used to study the trait. These mapping populations typically have been small, generally consisting of <500 lines, and focused on discrete germplasm chosen for studying specific resistance mechanisms, such as biparental populations (St Clair, 2010). These small populations (<500 lines) are greatly limited in their power to detect variable loci, especially in cases where the underlying genetic architecture is highly polygenic and likely contains epistatic loci (Falconer and Mackay, 1996; Mackay, 2001, 2014; Manolio et al., 2009). This leads to an underappreciated false-negative discovery rate that leads to an underestimation of the true number of causal loci (Chan et al., 2011; Joseph et al., 2014).
Higher power mapping populations are being developed to alleviate this potential problem in two ways. The first is the use of large multiparent populations, such as multiparent advanced generation intercross (MAGIC) and nested association mapping (NAM) populations, which address the power issue by greatly increasing the number of lines within a population to 5000 or more (Stich et al., 2007; Buckler et al., 2009; McMullen et al., 2009). The second is the use of genome-wide association (GWA) mapping populations that use more lines and also utilize the increased number of meiotic generations to provide increased recombination and potentially increased mapping resolution (Nordborg et al., 2002, 2005; Nordborg and Weigel, 2008; Atwell et al., 2010; Alonso-Blanco et al., 2016).
The 5000-line maize NAM population has been applied to the study of quantitative resistance for a number of pathogens, including southern leaf blight, and these studies are beginning to illuminate a recurring pattern of resistance loci (Kump et al., 2011; Belcher et al., 2012). This analysis identified a minimum of 32 quantitative trait loci (QTLs) using a single pathogen isolate. Extending this study to investigate how the genetic map and the quality of the phenotyping data influence the ability to detect loci found that while the specific loci identified were sensitive to the quality of the genetic map and the care taken in phenotyping, the general pattern of total loci found and mechanisms affected was unaltered. A similar number of loci were found to control northern leaf blight in the same NAM population (Poland et al., 2011). Analysis of these loci showed that they controlled the majority of variance in the trait, but a significant fraction of heritable variance had no detectable loci. The combination of unexplained heritability and locus sensitivity to the map suggests that the loci found in these studies are a fraction of the total number of loci actually controlling the resistance. It remains to be seen if the identified loci represent the majority of causal loci.
Interestingly, Bian et al. (2014) found an association between flowering time and southern leaf blight resistance among lines within the NAM population. Controlling for this association did not dramatically affect the number or identity of loci found associated with resistance, but it raises the question of pleiotropy and potential phenotypic trade-offs among traits in the measurement of quantitative disease resistance. For instance, age-related resistance demonstrates a trade-off where a plant’s resistance to specific pathogens changes with the age of the plant. In an agronomic setting where optimal flowering time is predetermined by agronomic practices, ontogenic changes linked to disease resistance are not an option for breeding and are typically removed from the analysis. For example, the analysis of quantitative resistance to scab in apples was complicated by age-related resistance that hindered some of the mapping (Calenge et al., 2004). However, in wild plants, linking disease resistance to ontogeny may be a key adaptation to pathogen attack in the wild, where the frequency of specific pathogen species or possibly even specific genotypes of given pathogens may fluctuate seasonally. For example, in grapes (Vitis vinifera), the occurrence of specific genotypes of the generalist necrotrophic fungal pathogen, B. cinerea, changes across seasons with distinct populations occurring in different seasons and tissues (Fournier and Giraud, 2008; Johnston et al., 2014). However, even with the predominance of studies on agricultural pathogens, little is known about the potential role of ontogenic variation in quantitative resistance in the wild and how this affects selection on the underlying loci. However, this does suggest that the loci identified when mapping at a single life stage and a single environment are merely a subset of the total possible loci controlling a specific quantitative resistance trait.
Similar to multiparent intercross populations, GWA mapping of genes controlling variation in quantitative resistance of Arabidopsis to B. cinerea showed that the genetic architecture of resistance was highly polygenic (Corwin et al., 2016b). Previous mapping of quantitative resistance loci in small biparental mapping populations using Arabidopsis and B. cinerea had generated the standard image of a few moderate-effect loci that were highly dependent upon the specific mapping population (Denby et al., 2004; Rowe and Kliebenstein, 2008). Extending this with GWA mapping, Corwin et al. (2016b) suggested that there were likely thousands of causal genes underlying the variation in quantitative resistance to B. cinerea within Arabidopsis (Corwin et al., 2016b). Resistance to B. cinerea among the Arabidopsis accessions involved predominantly small-effect loci with no evidence of major-effect resistance loci. This is in contrast to GWA mapping studies in other Arabidopsis pathosystems containing known large-effect loci that participate in gene-for-gene interactions (Atwell et al., 2010). In these qualitative resistance pathosystems, the same large effect loci were identified using biparental and GWA populations (Aranzana et al., 2005; Atwell et al., 2010). The population for the Arabidopsis/B. cinerea GWA study was more powerful than existing biparental populations, but it is still largely underpowered given the number of accessions and the effects of residual population structure (Chan et al., 2010; Platt et al., 2010; Brachi et al., 2015). Therefore, in combination with the NAM studies and the presence of ontogenic and environmentally conditional loci, we still do not have the ability to identify all of the genes shaping quantitative disease resistance and that future studies with even more powerful populations are required.
GENOME-WIDE EXPRESSION QTL AND META-QTL MAPPING EXPAND VIEWS OF QUANTITATIVE RESISTANCE CAUSALITY
The extensive polygenic nature of quantitative resistance complicates cloning and validation of the associated genes to fully describe the functional mechanisms driving quantitative resistance. To accomplish this with standard map-based cloning approaches requires moving a small-effect locus into an otherwise homogeneous background for fine mapping of the individual locus (Zhang et al., 2006). In systems with dozens to thousands of underlying causal loci, this locus-by-locus approach is a major impediment and may not make financial sense for any but the most economically relevant crops. One approach to identify the potential causal genes and/or mechanisms underlying quantitative disease resistance is to combine genetic mapping with systems biology approaches that attempt to identify multiple causal genes and/or mechanisms in parallel (Chen et al., 2010). One example of this systems approach to understand quantitative disease resistance is the comparison of phenotypic resistance QTLs to gray leaf spot to expression QTLs (eQTLs) mapped simultaneously using a whole-transcriptome analysis of the maize recombinant inbred line population (Christie et al., 2017). eQTLs are QTLs that control the accumulation of specific transcripts within the mapping population and can be used to link cause and effect within QTL mapping populations for a variety of traits (Hansen et al., 2008; Keurentjes et al., 2008; Kliebenstein, 2009a). eQTL analysis in the maize/gray leaf spot pathosystem identified eQTL hot spots, regions with an overabundance of QTLs controlling the accumulation of diverse transcripts, that overlapped with the disease resistance QTLs. Analysis of the transcripts affected by these hot spots showed that they were predominantly transcripts for potential downstream defense mechanisms like metabolism and defense proteins. One of the eQTL hot spot networks had a major hub gene encoding a maize ortholog of Arabidopsis CORONATINE INSENSITIVE1 (COI1), suggesting that this network affects the regulation of the jasmonic acid (JA) defense signaling pathway (Benedetti et al., 1995; Xie et al., 1998). The COI1 transcript was affected by this QTL in trans, meaning that the COI1 gene was not located within the physical boundaries of the QTL. Therefore, COI1 is not the causal gene for this locus and an unknown mechanism alters the function of the JA signaling pathway. Thus, this approach allowed the identification of a mechanism associated with a quantitative resistance locus but identification of the causal gene will require further effort.
A mechanistic comparison between qualitative and quantitative resistance was generated using a comparative transcriptomic study of how barley (Hordeum vulgare) resists stem rust (Druka et al., 2008; Moscou et al., 2011). The authors designed this study to include qualitative and quantitative resistance loci, enabling an explicit comparison of the mechanisms underlying resistance loci with different effect magnitudes. A quarter of the transcriptome significantly responded to pathogen attack, and these responses were dependent upon the underlying disease resistance loci. Interestingly, the vast majority of these transcripts were controlled by genetic variation at the quantitative resistance loci rather than the qualitative resistance locus. The authors proposed a model whereby the quantitative resistance loci primarily modulate the transcriptome to shape the response in a way that optimized the function of the qualitative resistance locus. This model is supported by studies that mapped large-effect disease resistance loci and frequently found associated small-effect quantitative resistance loci that epistatically affected the major-effect locus (Debener et al., 1991; Martin et al., 1993).
The above systems approaches yielded mechanistic insight into the quantitative resistance loci, but they had a difficult time finding the causal genes because of a lack of recombination for fine-mapping. One approach to fix this difficulty is to increase the population size by developing new populations or adding to existing populations. This solution requires that the new individuals added to a population are independent from the existing individuals; for GWA studies, this may not feasible if there is strong population structure or selection. An alternative approach is to conduct a meta-analysis of existing data. The agricultural importance of several pathosystems has motivated the development of independent mapping populations to map resistance to a specific pathogen. These populations are independent samples of genetic variation within the species that can be combined to boost the available recombination and thus boost mapping resolution, similar to the NAM concept (McMullen et al., 2009). In wheat (Triticum aestivum), many studies have mapped QTL for resistance to powdery mildew (Blumeria graminis; Alam et al., 2011). Marone et al. (2013) conducted a meta-analysis that combined these studies into a single QTL mapping study. In this analysis, the authors used a variety of approaches to identify genetic markers allowing them to combine the maps and thus compare the underlying loci within the populations. This greatly increased the genetic mapping resolution for the underlying loci and allowed 24 QTLs to be positioned within smaller mapping windows than was available in any individual mapping study. This approach of reusing existing data to increase the available recombination allows researchers to generate QTL lists that are reproducible and mapped to finer scale resolution. These QTLs will aid targeted breeding efforts and studies of the molecular mechanisms of disease resistance.
RELATIONSHIP OF ALLELIC EFFECT SIZE AND MOLECULAR FUNCTION IN THE PATHOGEN RESPONSE OF PLANTS
One key question is which mechanisms play a role in quantitative resistance. This question arises because most loci controlling plant/pathogen interactions are quantitative, but few have been cloned, in contrast to the numerous large-effect loci that have been cloned using QTL approaches. These large-effect loci often have sufficiently large effects that they gray the boundary between qualitative and quantitative resistance and they typically encode upstream signaling components in the plant pathogen response (Figure 2, I). For example, a major-effect locus controlling Arabidopsis resistance to Xanthomonas campestris, RKS1, was found to encode a kinase that appears to function in the signaling pathway linking pathogen perception to defense response (Huard-Chauveau et al., 2013). Cloning of two large-effect loci controlling resistance to Fusarium oxysporum in Arabidopsis identified one as a kinase and the other as a signaling peptide (Diener and Ausubel, 2005; Shen and Diener, 2013). Similarly, mapping of large-effect loci in wheat identified a novel kinase (Fu et al., 2009). Supporting these observations that medium-to-large effect loci appear to function in perception and/or signaling are the large number of studies that investigated the quantitative genetics of rice resistance to rice blast (Magnaporthe oryzae; Ballini et al., 2008; Miah et al., 2013; Kang et al., 2016; Raboin et al., 2016). The vast majority of cloned loci encode RLKs or NLRs, suggesting that the majority of causal loci are in genes controlling the detection of the pathogen. However, it is important to note that these large-effect RLKs and NLRs drive the immune response through a network of downstream genes, many of which likely contain polymorphisms with some impact on their function.
NAM and GWA mapping approaches are providing methods for identifying small-to-moderate effect loci and yielding more detail about the underlying mechanisms. In contrast to the largely upstream mechanisms (i.e., perception and signaling) described above for moderate-to-large effect loci, studies on quantitative resistance of predominantly small-effect loci in maize suggested that the majority of potential causal genes affected downstream resistance mechanisms (Kump et al., 2011; Belcher et al., 2012) (Figure 2). The predominant class of genes linked to these loci as potential causal genes were those involved in downstream defense mechanisms including defensins, pathogenesis-related proteins, and secondary metabolite enzymes. The next largest group of causal loci encoded potential transcription factors that can influence the response to defense hormones like salicylic acid and ethylene. Interestingly, the smallest class of identified loci was in potential microbe- or pathogen-associated molecular pattern (MAMP/PAMP) signaling components, implying that quantitative disease resistance in maize is a function of a variety of cellular processes and not simply pathogen detection. Similarly, a quantitative disease resistance locus in maize against gray leaf spot was linked to a flavin mono-oxygenase (FMO) that was hypothesized to be a potential detoxification enzyme to alleviate the virulence effects of cercosporin, a toxin produced by the pathogen (Benson et al., 2015). However, FMOs typically add an oxygen to a nitrogen or sulfur and there is no obvious molecular reaction for the FMO with cercosporin, suggesting that it may have a role in the plant defense response like FMO1 in Arabidopsis (Bartsch et al., 2006; Mishina and Zeier, 2006; Hansen et al., 2007). While this together suggests that there is a divergence in the mechanisms altered by large-effect and small-effect quantitative resistance loci in maize, there is currently no extensive molecular validation on these results.
Similar observations about small-to-moderate effect loci were found using GWA mapping in the Arabidopsis/B. cinerea pathosystem (Corwin et al., 2016b). This analysis showed that there was a slight but significant enrichment in likely quantitative resistance causal genes being in MAMP/PAMP signaling mechanisms. However, this accounted for only a tiny fraction of the associated causal genes. Instead, the vast majority of causal genes encoded proteins involved in intermediate signal transduction and downstream defense response mechanisms. Interestingly, the majority of these genes had not been previously linked to B. cinerea resistance using traditional biparental populations or Mendelian mutation genetics and represent potential new defense mechanisms. This included vesicle-associated genes required to move defense compounds, primary metabolism genes likely involved in reprogramming plant metabolism, genes in cell wall synthesis and function, and sugar metabolism. Importantly, the study was able to show that these causal gene predictions validate with a nearly 60% frequency using insertional mutants. The candidate genes chosen for validation were randomly selected, suggesting that there should be a similar rate of validation for the full candidate gene list. This same approach for calling candidate genes was previously shown to have a greater than 60% success rate, while a corresponding analysis of genes with no GWA evidence showed almost no effect on the traits analyzed (Chan et al., 2011; Francisco et al., 2016). Thus, while the full gene list is not yet validated, the experimental evidence supports the general observations from this list. Together with the maize NAM results, this indicates that quantitative resistance genes are predominantly altering specific defense mechanisms like defense metabolite or cell wall synthesis with less of a role for variation in the detection of specific pathogens and corresponding signaling mechanisms to coordinate the response.
QUANTITATIVE BASIS OF PATHOGEN PERCEPTION
Even with the above NAM and GWA studies, there is still a significant power issue hindering the identification all of the causal genes underlying quantitative disease resistance variation. This is illustrated by comparisons of power analysis within the 5000 line NAM population to the actual results for flowering time mapping whereby the statistical power analysis shows that there is likely a significant false negative error rate leading to missed loci within this population (Buckler et al., 2009; McMullen et al., 2009). Similarly, a meta-analysis showed that it is not possible to estimate the number of causal loci that vary between any two accessions even when using Arabidopsis populations up to 500 lines (Joseph et al., 2014). A further complication arises from the observation that when crossing two individuals there is often significant transgressive segregation for quantitative resistance. Thus, even a resistant genotype does not solely contain alleles that positively contribute to resistance. Instead, an individual genome contains a blend of genes with some genes containing the sensitive allele while other genes contain a resistance allele. Thus, the phenotype is the sum of the positive and negative alleles across myriad genes and mechanisms. One avenue to simplify the study of quantitative resistance is to focus on one specific mechanism that contributes to the phenotype and to analyze variation in that mechanism. For example, general studies of genetic diversity have suggested that while there is significant genetic variation in resistance genes in planta, the associated signaling genes are potentially under purifying selection and not playing a role in quantitative resistance (Bakker et al., 2006, 2008). However, this is based solely on sequence variation in the absence of empirical testing within one or more defined pathosystems.
A recent study directly tested if quantitative disease resistance may be caused by genetic variation in components downstream of MAMP/PAMP perception by applying MAMPs to a plant and measuring the quantitative response to these MAMPs (Vetter et al., 2016). The authors applied the well-studied MAMPs, EF-Tu and flagellin, to Arabidopsis accessions and mapped the downstream response (Boller and Felix, 2009; Zipfel, 2014; Vetter et al., 2016). In addition to mapping natural variation, this allowed the authors to test if the downstream signaling pathways are identical between the two MAMPs as is the general assumption. There was extensive natural variation in the response to both MAMPs suggesting that signaling components linking growth to the response to pathogen signals may partly control quantitative disease resistance. Furthermore, the quantitative growth response to both of these MAMPs was highly polygenic in the accession and the loci underlying this differential response were largely unique to the one or the other MAMP, suggesting that the MAMPs target different pathways to alter growth in Arabidopsis (Vetter et al., 2016). Validation of a subset of these loci confirmed the independence of the signaling pathways controlling the response to these different MAMPs. This is in contrast to the common assumption that MAMP signaling pathways should be canalized to create a common response to diverse input stimuli.
Most studies on the detection of pathogens have focused on MAMP/PAMP signals found as a part of a large effect gene-for-gene resistance pathosystem. Extending these studies to other detection events has been facilitated by the advent of modern genomics approaches where genomics surveys have identified suites of bacterial virulence effectors independent of a specific virulence function (Lee et al., 2012). These effector surveys have shown that pathogens send a large suite of signals into the plant that if each was sensed and responded to by an independent suite of polymorphic genes could lead to a highly polygenic response system. Thus, it is possible to find new effectors and directly test if there is quantitative variation in the response to these pathogen signals that could control quantitative disease resistance by altering how the plant perceives the pathogen. Using a delivery disarmed Pseudomonas syringae, a study was conducted whereby a single effector, HopAM1, was delivered to Arabidopsis and used to identify quantitative natural variation in the response to this effector (Iakovidis et al., 2016). This showed that the plant’s response to this quantitative effector was controlled by a large number of variable loci. This was equally true when testing GWA mapping populations as well as when conducting QTL mapping in several recombinant inbred line populations. Interestingly, work in maize is identifying some of the genes underlying this polygenic response by finding that some downstream components of the effector detection are enzymes in defense metabolite pathways (Wang et al., 2015; Wang and Balint-Kurti, 2016). Thus, dissecting the quantitative basis of individual downstream mechanisms can aid in identifying the causal genes underlying this response, but even the response to single smaller effect pathogen signals is quantitative and polygenic in nature.
PATHOGEN GENETICS AND ITS IMPACT ON BROAD-SPECTRUM RESISTANCE
A common assumption and justification for studying quantitative resistance loci is that it may be a source of resistance that is more stable than the large-effect loci, i.e., durable resistance (Poland et al., 2009; St Clair, 2010; Roux et al., 2014; Niks et al., 2015). This assumption is associated with a common inference that the smaller to moderate effect loci found in quantitative resistance confer broad-spectrum resistance either to multiple genotypes of the same pathogen or to multiple pathogens. For example, studies of Arabidopsis quantitative resistance to F. oxysporum showed that the variable loci controlled resistance to an array of races from this pathogen (Diener and Ausubel, 2005; Shen and Diener, 2013). Similarly, Arabidopsis quantitative resistance loci against X. campestris were also effective against diverse pathogen isolates (Debieu et al., 2016). However, these studies illustrate a conundrum in the literature about the term “broad spectrum.” Most quantitative resistance studies with natural variation limit the term to a broad spectrum of isolates of a single pathogen and not necessarily to multiple pathogens (Hutin et al., 2015; Zhang et al., 2015a).
However, a contrasting image is obtained when using generalist pathogens such as B. cinerea that show elevated genetic and phenotypic diversity in an array of virulence-associated phenotypes (Rowe and Kliebenstein, 2007; Staats et al., 2007; Williamson et al., 2007; Amselem et al., 2011; Atwell et al., 2015; Corwin et al., 2016a). Mapping of B. cinerea quantitative resistance loci within multiple plant species shows that the identified loci are highly dependent upon the specific pathogen genotype utilized and typically not broad spectrum (Denby et al., 2004; Rowe and Kliebenstein, 2008; Rowe et al., 2010; Corwin et al., 2016a; Finkers et al., 2007b, 2008; Zhang et al., 2016). An isolate dependency on the identification of quantitative disease resistance has also been found for the generalist S. sclerotiorum when studying sunflower (Helianthus annuus; Davar et al., 2011). This dependency on pathogen genotype is not limited to generalist necrotrophs as it has also been found in specialist biotrophs. For example, Arabidopsis and tomato quantitative resistance to diverse P. syringae lines was largely dependent on the pathogen genotype (Thapa et al., 2015; Kover and Schaal, 2002; Kover and Cheverud, 2007). Thus, quantitative or partial resistance is not inherently broad spectrum and durable, and it remains to be determined how genes underlying quantitative resistance relate to broad-spectrum resistance.
Considering the different axes of diversity in the host, pathogen, and microbial community, we may have to reconsider how we define broad-spectrum resistance. The observation that genes for specific resistance mechanisms like secondary metabolism may predominantly control quantitative disease resistance loci raises the possibility that quantitative resistance and quantitative virulence are linked. An example of this arises within P. syringae where Brassica-specialized isolates contain specific genes, SURVIVAL IN ARABIDOPSIS EXTRACTS (SAX), that allow them to detoxify Brassica-specific defense compounds called glucosinolates (Sønderby et al., 2010; Fan et al., 2011). These defense compounds are highly polymorphic in both structure and content within the Brassica including Arabidopsis (Wentzell et al., 2007; Kliebenstein, 2009b). This generates the possibility that variation in the P. syringae SAX loci can interact with the plants genetic variation in the associated defense compound to generate quantitative resistance within the interaction. These same defense compounds provide resistance to a number of other pathogens including nonhost microbes that each may also vary in their sensitivity to the compounds (Mithen et al., 1986, 1987; Stotz et al., 2011; Zhang et al., 2015b; Bednarek and Osbourn, 2009; Bednarek et al., 2009; Clay et al., 2009). This generates a potential system whereby a plant has variation in defense compounds while a multitude of pathogens also have genetic variation in the ability to detoxify specific plant defense metabolites when investigated (Pedras and Khan, 1997; Pedras and Ahiahonu, 2002, 2005; Pedras et al., 2011). This suggests that we need a new thesis that moves beyond the categorical classification of plant/pathogen interactions such that nonhost, host, and broad-spectrum resistance are intricately linked and simply revolve around the ability of a host to create a defense and a pathogen to counter that defense. Within this model, because there is genetic variation in the defense and the counter-defense, broad spectrum quantitative resistance becomes the ability to resist specific isolates from a broad array of pathogens but not the ability to resist all genotypes of a set of pathogen species. It will be interesting to develop a system that can directly test the role of host and pathogen variation in shifting quantitative resistance across pathogen species.
CONCLUSIONS AND PERSPECTIVES
The above studies are rapidly illuminating the genetic basis of quantitative resistance and how the genetic variation is distributed across the entirety of the plant’s disease response from detection of the pathogen to signaling to specific output resistance mechanisms. Together with analysis of the pathogen, this is developing a more integrated understanding of what is and is not possible in generating disease resistance against one or more pathogens in a single plant genotype. For example, the manipulation of a specific defense metabolite may provide resistance to specific genotypes of a diverse array of host and nonhost pathogens, but given the presence of diverse detoxification mechanisms across all possible pathogens, it will not provide the long sought broad-spectrum, stable resistance. In a natural setting, this suggests that quantitative resistance genes are responding to the blend of pathogens in a specific environment rather than a single predominant pathogen and that it may be better to consider the evolution and variation at these loci in that light.
While the mechanisms for quantitative resistance are gradually coming into focus, the true scale of how many genes may causally influence the trait is still out of focus. This is especially true when one considers the polygenic nature of variation in specific defense mechanisms that combine to create the final resistance output. To address this question, the community needs to develop tools and concepts that can fully encapsulate the possibility that quantitative resistance to a specific pathogen may involve 10% or more of the genes in a given organism. While this may not be immediately necessary or useful in the applied context of the agronomic field and crop improvement, this is likely how selection has shaped quantitative resistance in wild plants including crop progenitors. Thus, it is essential to develop a complete understanding of the full polygenic nature of quantitative resistance both using models with tens to thousands of genes and with new mapping populations that have the possibility to test what is the finite limit to the number of causal genes within a species for resistance to a pathogen.
Supplementary Material
Acknowledgments
This work was funded by National Science Foundation Award IOS 1339125 to D.J.K., by the USDA National Institute of Food and Agriculture, Hatch project number CA-D-PLS-7033-H to D.J.K., and by a Danish National Research Foundation (DNRF99) grant to D.J.K.
AUTHOR CONTRIBUTIONS
Both authors contributed to writing the article.
References
- Alam M.A., Xue F., Wang C., Ji W. (2011). Powdery mildew resistance genes in wheat: identification and genetic analysis. J. Mol. Biol. Res. 1: 20–39. [Google Scholar]
- Alonso-Blanco C., et al.; 1001 Genomes Consortium (2016). 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana M.J., et al. (2005). Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., Corwin J.A., Soltis N.E., Subedy A., Denby K.J., Kliebenstein D.J. (2015). Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front. Microbiol. 6: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E.G., Toomajian C., Kreitman M., Bergelson J. (2006). A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18: 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E.G., Traw M.B., Toomajian C., Kreitman M., Bergelson J. (2008). Low levels of polymorphism in genes that control the activation of defense response in Arabidopsis thaliana. Genetics 178: 2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E., Morel J.B., Droc G., Price A., Courtois B., Notteghem J.L., Tharreau D. (2008). A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21: 859–868. [DOI] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P., Osbourn A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324: 746–748. [DOI] [PubMed] [Google Scholar]
- Bednarek P., Pislewska-Bednarek M., Svatos A., Schneider B., Doubsky J., Mansurova M., Humphry M., Consonni C., Panstruga R., Sanchez-Vallet A., Molina A., Schulze-Lefert P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106. [DOI] [PubMed] [Google Scholar]
- Belcher A.R., Zwonitzer J.C., Santa Cruz J., Krakowsky M.D., Chung C.L., Nelson R., Arellano C., Balint-Kurti P.J. (2012). Analysis of quantitative disease resistance to southern leaf blight and of multiple disease resistance in maize, using near-isogenic lines. Theor. Appl. Genet. 124: 433–445. [DOI] [PubMed] [Google Scholar]
- Benedetti C.E., Xie D., Turner J.G. (1995). Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J.M., Poland J.A., Benson B.M., Stromberg E.L., Nelson R.J. (2015). Resistance to gray leaf spot of maize: genetic architecture and mechanisms elucidated through nested association mapping and near-isogenic line analysis. PLoS Genet. 11: e1005045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Bian Y., Yang Q., Balint-Kurti P.J., Wisser R.J., Holland J.B. (2014). Limits on the reproducibility of marker associations with southern leaf blight resistance in the maize nested association mapping population. BMC Genomics 15: 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Brachi B., Meyer C.G., Villoutreix R., Platt A., Morton T.C., Roux F., Bergelson J. (2015). Coselected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 4032–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E.S., et al. (2009). The genetic architecture of maize flowering time. Science 325: 714–718. [DOI] [PubMed] [Google Scholar]
- Calenge F., Faure A., Goerre M., Gebhardt C., Van de Weg W.E., Parisi L., Durel C.E. (2004). Quantitative trait loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology 94: 370–379. [DOI] [PubMed] [Google Scholar]
- Chan E.K., Rowe H.C., Corwin J.A., Joseph B., Kliebenstein D.J. (2011). Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.K.F., Rowe H.C., Kliebenstein D.J. (2010). Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185: 991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., et al. (2010). An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS One 5: e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie N., et al. (2017). Systems genetics reveals a transcriptional network associated with susceptibility in the maize-gray leaf spot pathosystem. Plant J. 89: 746–763. [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin J.A., Subedy A., Eshbaugh R., Kliebenstein D.J. (2016a). Expansive phenotypic landscape of Botrytis cinerea shows differential contribution of genetic diversity and plasticity. Mol. Plant Microbe Interact. 29: 287–298. [DOI] [PubMed] [Google Scholar]
- Corwin J.A., Copeland D., Feusier J., Subedy A., Eshbaugh R., Palmer C., Maloof J., Kliebenstein D.J. (2016b). The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 12: e1005789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Davar R., Darvishzadeh R., Majd A. (2011). Genotype-isolate interaction for resistance to Sclerotinia sclerotiorum in sunflower. Phytopathol. Mediterr. 50: 442–449. [Google Scholar]
- Debener T., Lehnackers H., Arnold M., Dangl J.L. (1991). Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1: 289–302. [DOI] [PubMed] [Google Scholar]
- Debieu M., Huard-Chauveau C., Genissel A., Roux F., Roby D. (2016). Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol. Plant Pathol. 17: 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby K.J., Kumar P., Kliebenstein D.J. (2004). Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana Plant J. 38: 473–486. [DOI] [PubMed] [Google Scholar]
- Diener A.C., Ausubel F.M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171: 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A., et al. (2008). Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp. tritici in barley. Theor. Appl. Genet. 117: 261–272. [DOI] [PubMed] [Google Scholar]
- Falconer D.S., Mackay T.F.C. (1996). Introduction to Quantitative Genetics. (Essex, UK: Longman, Harlow; ). [Google Scholar]
- Fan J., Crooks C., Creissen G., Hill L., Fairhurst S., Doerner P., Lamb C. (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331: 1185–1188. [DOI] [PubMed] [Google Scholar]
- Finkers R., Bai Y.L., van den Berg P., van Berloo R., Meijer-Dekens F., ten Have A., van Kan J., Lindhout P., van Heusden A.W. (2008). Quantitative resistance to Botrytis cinerea from Solanum neorickii. Euphytica 159: 83–92. [Google Scholar]
- Finkers R., van Heusden A.W., Meijer-Dekens F., van Kan J.A., Maris P., Lindhout P. (2007a). The construction of a Solanum habrochaites LYC4 introgression line population and the identification of QTLs for resistance to Botrytis cinerea. Theor. Appl. Genet. 114: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkers R., van den Berg P., van Berloo R., ten Have A., van Heusden A.W., van Kan J.A.L., Lindhout P. (2007b). Three QTLs for Botrytis cinerea resistance in tomato. Theor. Appl. Genet. 114: 585–593. [DOI] [PubMed] [Google Scholar]
- Foolad M.R., Zhang L.P., Khan A.A., Nino-Liu D., Lin G.Y. (2002). Identification of QTLs for early blight (Alternaria solani) resistance in tomato using backcross populations of a Lycopersicon esculentum x L. hirsutum cross. Theor. Appl. Genet. 104: 945–958. [DOI] [PubMed] [Google Scholar]
- Fournier E., Giraud T. (2008). Sympatric genetic differentiation of a generalist pathogenic fungus, Botrytis cinerea, on two different host plants, grapevine and bramble. J. Evol. Biol. 21: 122–132. [DOI] [PubMed] [Google Scholar]
- Francisco M., Joseph B., Caligagan H., Li B., Corwin J.A., Lin C., Kerwin R.E., Burow M., Kliebenstein D.J. (2016). Genome wide association mapping in Arabidopsis thaliana identifies novel genes involved in linking allyl glucosinolate to altered biomass and defense. Front. Plant Sci. 7: 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French E., Kim B.S., Iyer-Pascuzzi A.S. (2016). Mechanisms of quantitative disease resistance in plants. Semin. Cell Dev. Biol. 56: 201–208. [DOI] [PubMed] [Google Scholar]
- Fu D., Uauy C., Distelfeld A., Blechl A., Epstein L., Chen X., Sela H., Fahima T., Dubcovsky J. (2009). A novel kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323: 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B.G., Kliebenstein D.J., Halkier B.A. (2007). Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50: 902–910. [DOI] [PubMed] [Google Scholar]
- Hansen B.G., Halkier B.A., Kliebenstein D.J. (2008). Identifying the molecular basis of QTLs: eQTLs add a new dimension. Trends Plant Sci. 13: 72–77. [DOI] [PubMed] [Google Scholar]
- Huard-Chauveau C., Perchepied L., Debieu M., Rivas S., Kroj T., Kars I., Bergelson J., Roux F., Roby D. (2013). An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 9: e1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M., Sabot F., Ghesquière A., Koebnik R., Szurek B. (2015). A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84: 694–703. [DOI] [PubMed] [Google Scholar]
- Iakovidis M., Teixeira P.J., Exposito-Alonso M., Cowper M.G., Law T.F., Liu Q., Vu M.C., Dang T.M., Corwin J.A., Weigel D., Dangl J.L., Grant S.R. (2016). Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204: 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P.R., Hoksbergen K., Park D., Beever R.E. (2014). Genetic diversity of Botrytis in New Zealand vineyards and the significance of its seasonal and regional variation. Plant Pathol. 63: 888–898. [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Joseph B., Atwell S., Corwin J.A., Li B., Kliebenstein D.J. (2014). Meta-analysis of metabolome QTLs in Arabidopsis: trying to estimate the network size controlling genetic variation of the metabolome. Front. Plant Sci. 5: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., et al. (2016). Dissection of the genetic architecture of rice resistance to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 17: 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes J.J.B., Koornneef M., Vreugdenhil D. (2008). Quantitative genetics in the age of omics. Curr. Opin. Plant Biol. 11: 123–128. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Diers B.W. (2000). Inheritance of partial resistance to Sclerotinia stem rot in soybean. Crop Sci. 40: 55–61. [Google Scholar]
- Kliebenstein D. (2009a). Quantitative genomics: analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu. Rev. Plant Biol. 60: 93–114. [DOI] [PubMed] [Google Scholar]
- Kliebenstein D.J. (2009b). A quantitative genetics and ecological model system: understanding the aliphatic glucosinolate biosynthetic network via QTLs. Phytochem. Rev. 8: 243–254. [Google Scholar]
- Kohler A., et al.; Mycorrhizal Genomics Initiative Consortium (2015). Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47: 410–415. [DOI] [PubMed] [Google Scholar]
- Kover P.X., Schaal B.A. (2002). Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc. Natl. Acad. Sci. USA 99: 11270–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover P.X., Cheverud J. (2007). The genetic basis of quantitative variation in susceptibility of Arabidopsis thaliana to Pseudomonas syringae (Pst DC3000): evidence for a new genetic factor of large effect. New Phytol. 174: 172–181. [DOI] [PubMed] [Google Scholar]
- Kump K.L., Bradbury P.J., Wisser R.J., Buckler E.S., Belcher A.R., Oropeza-Rosas M.A., Zwonitzer J.C., Kresovich S., McMullen M.D., Ware D., Balint-Kurti P.J., Holland J.B. (2011). Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 43: 163–168. [DOI] [PubMed] [Google Scholar]
- Lee J., Teitzel G.M., Munkvold K., del Pozo O., Martin G.B., Michelmore R.W., Greenberg J.T. (2012). Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 158: 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg D.S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F.C. (2001). The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay T.F.C. (2014). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T.A., et al. (2009). Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone D., Russo M.A., Laido G., De Vita P., Papa R., Blanco A., Gadaleta A., Rubiales D., Mastrangelo A.M. (2013). Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: from consensus regions to candidate genes. BMC Genomics 14: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.B., Brommonschenkel S.H., Chunwongse J., Frary A., Ganal M.W., Spivey R., Wu T., Earle E.D., Tanksley S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262: 1432–1436. [DOI] [PubMed] [Google Scholar]
- McMullen M.D., et al. (2009). Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50: 267–294. [DOI] [PubMed] [Google Scholar]
- Miah G., Rafii M.Y., Ismail M.R., Puteh A.B., Rahim H.A., Asfaliza R., Latif M.A. (2013). Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol. Biol. Rep. 40: 2369–2388. [DOI] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141: 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen R.F., Lewis B.G., Fenwick G.R. (1986). In vitro activity of glucosinolates and their products against Leptosphaeria maculans. Mycol. Res. 87: 433–440. [Google Scholar]
- Mithen R.F., Lewis B.G., Heaney R.K., Fenwick G.R. (1987). Resistance of leaves of Brassica species to Leptosphaeria maculans. Trans. Br. Mycol. Soc. 88: 525–531. [Google Scholar]
- Moscou M.J., Lauter N., Steffenson B., Wise R.P. (2011). Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PLoS Genet. 7: e1002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niks R.E, Qi X.Q., Marcel T.C. (2015). Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53: 445–470. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Weigel D. (2008). Next-generation genetics in plants. Nature 456: 720–723. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Borevitz J.O., Bergelson J., Berry C.C., Chory J., Hagenblad J., Kreitman M., Maloof J.N., Noyes T., Oefner P.J., Stahl E.A., Weigel D. (2002). The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Nordborg M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega R.A., Mahnert A., Berg C., Müller H., Berg G. (2016). The plant is crucial: specific composition and function of the phyllosphere microbiome of indoor ornamentals. FEMS Microbiol. Ecol. 92: fiw173. [DOI] [PubMed] [Google Scholar]
- Pedras M.S.C., Khan A.Q. (1997). Unprecedented detoxification of the phytoalexin camalexin by a root rot pathogen. Bioorg. Med. Chem. Lett. 7: 2255–2260. [Google Scholar]
- Pedras M.S.C., Ahiahonu P.W.K. (2002). Probing the phytopathogenic stem rot fungus with phytoalexins and analogues: unprecedented glucosylation of camalexin and 6-methoxycamalexin. Bioorg. Med. Chem. 10: 3307–3312. [DOI] [PubMed] [Google Scholar]
- Pedras M.S.C., Ahiahonu P.W.K. (2005). Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry 66: 391–411. [DOI] [PubMed] [Google Scholar]
- Pedras M.S.C., Hossain S., Snitynsky R.B. (2011). Detoxification of cruciferous phytoalexins in Botrytis cinerea: spontaneous dimerization of a camalexin metabolite. Phytochemistry 72: 199–206. [DOI] [PubMed] [Google Scholar]
- Platt A., Vilhjálmsson B.J., Nordborg M. (2010). Conditions under which genome-wide association studies will be positively misleading. Genetics 186: 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.A., Bradbury P.J., Buckler E.S., Nelson R.J. (2011). Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. Proc. Natl. Acad. Sci. USA 108: 6893–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.A., Balint-Kurti P.J., Wisser R.J., Pratt R.C., Nelson R.J. (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14: 21–29. [DOI] [PubMed] [Google Scholar]
- Raboin L.M., Ballini E., Tharreau D., Ramanantsoanirina A., Frouin J., Courtois B., Ahmadi N. (2016). Association mapping of resistance to rice blast in upland field conditions. Rice (N.Y.) 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F., Voisin D., Badet T., Balagué C., Barlet X., Huard-Chauveau C., Roby D., Raffaele S. (2014). Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15: 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.C., Kliebenstein D.J. (2007). Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol. Plant Microbe Interact. 20: 1126–1137. [DOI] [PubMed] [Google Scholar]
- Rowe H.C., Kliebenstein D.J. (2008). Complex genetics control natural variation in Arabidopsis thaliana resistance to Botrytis cinerea. Genetics 180: 2237–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H.C., Walley J.W., Corwin J., Chan E.K.-F., Dehesh K., Kliebenstein D.J. (2010). Deficiencies in jasmonate-mediated plant defense reveal quantitative variation in Botrytis cinerea pathogenesis. PLoS Pathog. 6: e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Diener A.C. (2013). Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 9: e1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby I.E., Geu-Flores F., Halkier B.A. (2010). Biosynthesis of glucosinolates--gene discovery and beyond. Trends Plant Sci. 15: 283–290. [DOI] [PubMed] [Google Scholar]
- St Clair D.A. (2010). Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48: 247–268. [DOI] [PubMed] [Google Scholar]
- Staats M., van Baarlen P., Schouten A., van Kan J.A.L., Bakker F.T. (2007). Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Genet. Biol. 44: 52–63. [DOI] [PubMed] [Google Scholar]
- Stich B., Yu J., Melchinger A.E., Piepho H.P., Utz H.F., Maurer H.P., Buckler E.S. (2007). Power to detect higher-order epistatic interactions in a metabolic pathway using a new mapping strategy. Genetics 176: 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H.U., Sawada Y., Shimada Y., Hirai M.Y., Sasaki E., Krischke M., Brown P.D., Saito K., Kamiya Y. (2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 67: 81–93. [DOI] [PubMed] [Google Scholar]
- Thapa S.P., Miyao E.M., Michael Davis R., Coaker G. (2015). Identification of QTLs controlling resistance to Pseudomonas syringae pv. tomato race 1 strains from the wild tomato, Solanum habrochaites LA1777. Theor. Appl. Genet. 128: 681–692. [DOI] [PubMed] [Google Scholar]
- Vetter M., Karasov T.L., Bergelson J. (2016). Differentiation between MAMP triggered defenses in Arabidopsis thaliana. PLoS Genet. 12: e1006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.F., Balint-Kurti P.J. (2016). Maize homologs of CCoAOMT and HCT, two key enzymes in lignin biosynthesis, form complexes with the NLR Rp1 protein to modulate the defense response. Plant Physiol. 171: 2166–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.F., He Y., Strauch R., Olukolu B.A., Nielsen D., Li X., Balint-Kurti P.J. (2015). Maize homologs of hydroxycinnamoyltransferase, a key enzyme in lignin biosynthesis, bind the nucleotide binding leucine-rich repeat Rp1 proteins to modulate the defense response. Plant Physiol. 169: 2230–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell A.M., Rowe H.C., Hansen B.G., Ticconi C., Halkier B.A., Kliebenstein D.J. (2007). Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet. 3: 1687–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B., Tudzynski B., Tudzynski P., van Kan J.A.L. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8: 561–580. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Zhang F., Huang L.Y., Zhang F., Ali J., Cruz C.V., Zhuo D.L., Du Z.L., Li Z.K., Zhou Y.L. (2015a). Comparative transcriptome profiling of a rice line carrying Xa39 and its parents triggered by Xanthomonas oryzae pv. oryzae provides novel insights into the broad-spectrum hypersensitive response. BMC Genomics 16: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.P., Lin G.Y., Nino-Liu D., Foolad M.R. (2003). Mapping QTLs conferring early blight (Alternaria solani) resistance in a Lycopersicon esculentum x L. hirsutum cross by selective genotyping. Mol. Breed. 12: 3–19. [Google Scholar]
- Zhang W., Kwon S.T., Chen F., Kliebenstein D.J. (2016). Isolate dependency of Brassica rapa resistance QTLs to Botrytis cinerea. Front. Plant Sci. 7: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huai D., Yang Q., Cheng Y., Ma M., Kliebenstein D.J., Zhou Y. (2015b). Overexpression of three glucosinolate biosynthesis genes in Brassica napus identifies enhanced resistance to Sclerotinia sclerotiorum and Botrytis cinerea. PLoS One 10: e0140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ober J.A., Kliebenstein D.J. (2006). The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18: 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35: 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.