A kelch-containing repressor complex limits expression of the floral antagonist and meristem size promoter SELF PRUNING, thus restricting doming of the shoot apical meristem to the floral transition.
Abstract
Enlargement and doming of the shoot apical meristem (SAM) is a hallmark of the transition from vegetative growth to flowering. While this change is widespread, its role in the flowering process is unknown. The late termination (ltm) tomato (Solanum lycopersicum) mutant shows severely delayed flowering and precocious doming of the vegetative SAM. LTM encodes a kelch domain-containing protein, with no link to known meristem maintenance or flowering time pathways. LTM interacts with the TOPLESS corepressor and with several transcription factors that can provide specificity for its functions. A subgroup of flowering-associated genes is precociously upregulated in vegetative stages of ltm SAMs, among them, the antiflorigen gene SELF PRUNING (SP). A mutation in SP restored the structure of vegetative SAMs in ltm sp double mutants, and late flowering was partially suppressed, suggesting that LTM functions to suppress SP in the vegetative SAM. In agreement, SP-overexpressing wild-type plants exhibited precocious doming of vegetative SAMs combined with late flowering, as found in ltm plants. Strong flowering signals can result in termination of the SAM, usually by its differentiation into a flower. We propose that activation of a floral antagonist that promotes SAM growth in concert with floral transition protects it from such terminating effects.
INTRODUCTION
The shoot apical meristem (SAM) maintains a fairly constant size despite initiation of lateral organs continuously from its peripheral cells. The balance between the proliferation of SAM cells and their differentiation into organs ensures SAM maintenance and is regulated primarily by the WUSCHEL-CLAVATA (WUS-CLV) signaling pathway (Clark et al., 1997; Fletcher et al., 1999; Schoof et al., 2000). In contrast with the stable vegetative SAM size, the transition to flowering is usually associated with dramatic SAM changes; overall enlargement and altered dimensions, shifting from a flat top to a domed shape, are common to many plants (Bernier, 1988). SAM doming during the floral transition has been reported for both monocot and dicot species (Metcalf et al., 1975; Lyndon and Battey, 1985) and for plants with diverse growth habits. For most plants, the first visible change toward the transition to flowering is an increase in height relative to width (i.e., doming), followed by broadening of the meristem (Bernier et al., 1981). Such physical changes are reminiscent of the enlarged SAM found in mutants of the meristem maintenance pathway (Clark et al., 1993; Taguchi-Shiobara et al., 2001; Xu et al., 2015); however, there are no reports that changes in these mutants affect floral transition or flowering time.
Why the floral transition is coupled with SAM doming is not understood, and the possibility that the domed meristem is an intermediate stage of the transition from a meristem to a flower has not been determined. This may be the case with particular growth patterns, as in sunflower (Helianthus annuus) plants, where a capitulum type of inflorescence is formed (Steeves et al., 1969). However, SAM doming during floral transition was also reported in monopodial plants such as rice (Oryza sativa; Asai et al., 2002; Tamaki et al., 2015) and Arabidopsis thaliana (Hempel and Feldman, 1994; Hempel et al., 1998), where the apical meristem is maintained after floral transition and does not terminate with a flower. Thus, SAM doming may be an integral part of meristem development, irrespective of the fate of the meristem itself. Some evidence suggest that doming is not essential for flowering and does not trigger floral transition; in some plants, such as Impatiens balsamina, the meristem does not dome during floral transition (Battey and Lyndon, 1984), and in Humulus lupus, the meristem even becomes smaller (Bernier et al., 1981). Furthermore, mutants of the CLV signaling pathway that show precocious SAM doming are not fast-flowering plants (Clark et al., 1993; Xu et al., 2015). Moreover, doming is not the only SAM change that is associated with floral transition. In both monopodial and sympodial plants, floral transition is coupled with loss of apical dominance (Bernier et al., 1981). As clv mutants that have precocious doming were not reported to have altered apical dominance, and meristem size mutants were never reported following screens for altered branching, we assume that doming per se is not the cause for this physiological change. Hence, although they often occur simultaneously, SAM doming per se and floral transition might not be mechanistically linked.
As doming in tomato (Solanum lycopersicum) is coupled with floral transition, we chose here to focus on a novel late-flowering tomato mutant we named late termination (ltm) that also displays precocious doming. We show that LTM represses SELF PRUNING (SP), an inhibitor of floral transition and promoter of SAM doming. Previous studies showed that SP is a CETS (CENTRORADIALIS, TFL1, SP) protein that functions as an “antiterminator” or a floral inhibitor; tomato sp plants show accelerated floral termination of sympodial meristems (Pnueli et al., 1998). A related SP family member is SINGLE FLOWER TRUSS (SFT), an ortholog of Arabidopsis FLOWERING LOCUS T, which encodes the flowering signal florigen (Kardailsky et al., 1999; Kobayashi et al., 1999; Lifschitz et al., 2006). Genetic and grafting experiments in tomato showed that endogenous SFT/SP ratios regulate local growth-termination equilibria in all meristems of the shoot system (Lifschitz and Eshed, 2006). In addition, the ratio of these two proteins (or of their respective homologs) is involved in various developmental process such as leaf complexity and shape, stem thickness, pedicel abscission zone formation, and overall shoot architecture in a variety of plants, such as tomato, maize (Zea mays), cotton (Gossypium hirsutum), and Arabidopsis (Shalit et al., 2009; Danilevskaya et al., 2011; Pin and Nilsson, 2012; Park et al., 2014; McGarry et al., 2016). Florigenic CETS proteins have also been shown to control the differentiation of potato (Solanum tuberosum) stolon meristems into tubers (Navarro et al., 2011). Here, we show that expression of SP in the tomato SAM plays a role in meristem doming, acting in parallel to the WUS-CLV pathway. The coordination of doming with floral transition by LTM and the possible roles of CETS proteins in meristem functions are discussed.
RESULTS
ltm Displays Precocious SAM Doming but Late Flowering
Pronounced doming of the apical meristem has been documented in various plant species shortly before or in conjunction with the floral transition (Nougarède, 1967; Kanchanapoom and Thomas, 1987; Thomas and Kanchanapoom, 1991). To determine whether doming is an abrupt process, or represents an endpoint of a gradual change in the SAM as the plant approaches flowering, we used tomato with its easily accessible apical shoot meristems. Flowering in tomato is largely insensitive to classical environmental cues (daylength, vernalization, heat) and under our greenhouse conditions, anthesis occurred 35 to 40 d after germination, whereas maximal doming and first signs of flower formation were evident 8 to 12 d after germination. Upon germination, the seedling had two visible cotyledons and the small flat primary shoot meristem (PSM) contained two to three leaf primordia (Supplemental Figure 1). Analysis of the size of the PSM every 2 d postgermination showed a slight and continuous expansion of the vegetative SAM. This expansion dramatically accelerated after the development of seven to eight leaves. Shortly after enhancement of PSM growth, it reached a point that we will refer to as a transition meristem (TM; Figure 1A). In essence, a TM is the stage 2 d before the first visible signs of initiation of a new flowering branch at the basal PSM part. This branching is the earliest indication that the PSM has turned into a flower, as it precedes initiation of sepals and it is coupled to the release of the sympodial meristem from apical dominance (Figure 1A). These observations suggest that while classical SAM maintenance programs act to restrict its growth (Xu et al., 2015), the tomato SAM continues to gradually grow with age.
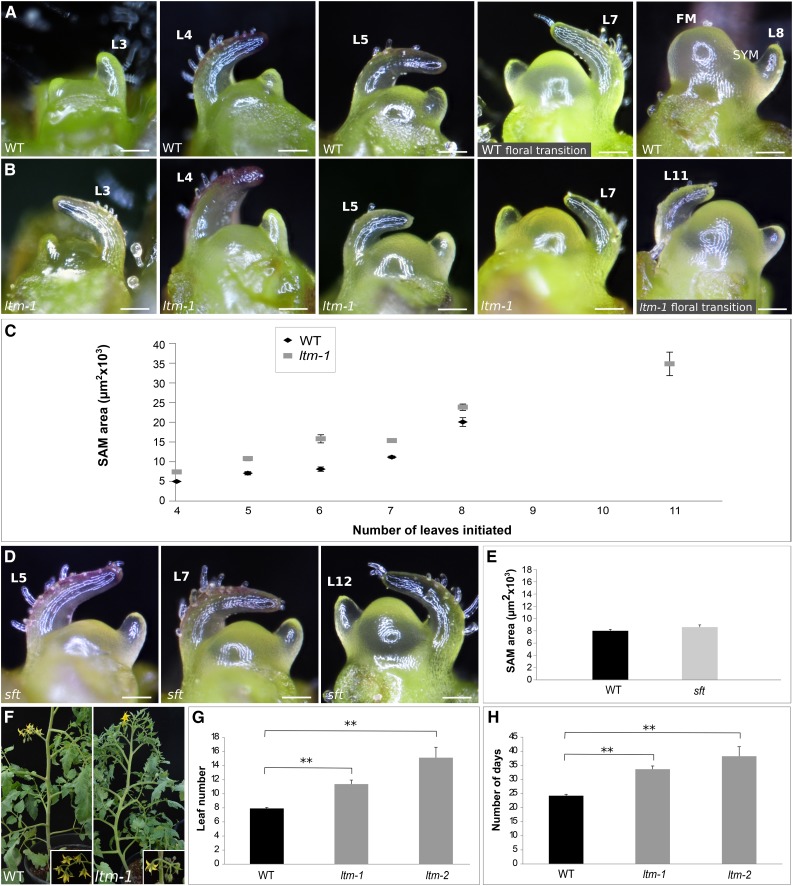
Figure 1.
ltm Plants Show a Domed Vegetative PSM and Late Floral Termination.
(A) A developmental series of wild-type (WT) PSM, from the vegetative shoot with four leaves (L) to floral transition and flower meristem (FM) and sympodial meristem (SYM) formation.
(B) A developmental series of ltm PSM, from the vegetative shoot to floral transition.
(C) Average 2D PSM size (±se) of developing vegetative wild-type and ltm shoot apices (n = 4).
(D) Developmental series of vegetative sft PSM after producing 6, 8, and 12 (TM) leaves.
(E) Average 2D PSM size (±se) of wild-type and sft shoot apices after production of six leaves (n = 4). Using Tukey HSD, the sizes of the two PSMs were not significantly different.
(F) Flowering wild-type and ltm-1plants grown together. Inset: wild-type zigzag and ltm branched inflorescence containing a leaf.
(G) Mean number of leaves (±se) produced by the PSM before flowering; wild type (n = 8), ltm-1 (n = 8), and ltm-2 (n = 39). **P < 0.01 (Tukey HSD).
(H) Mean days (±se) after germination until first visible macroscopic flower of the plants shown in (G). **P < 0.01 (Tukey HSD).
Bars = 100 μm in (A), (B), and (D).
To further understand the link between floral transition and SAM enlargement, we surveyed the apices of late-flowering mutants identified in a large mutant screen (Menda et al., 2004). These mutants were backcrossed three to four times to our standard wild-type line, the isogenic indeterminate M82 (SP/SP), before detailed characterization. Surprisingly, apices of one late-flowering mutant, which we termed ltm, showed precocious doming. Detailed examination of two mutant alleles of ltm, ltm-1 and ltm-2, showed a similar effect on SAM size; compared with the wild type, ltm plants had larger domed apical meristems throughout the PSM vegetative stage. As ltm plants are late-flowering, after the development of eight leaves, their PSM is as domed as wild-type plants are at the TM stage, even though ltm meristems will still produce at least three more leaves before reaching TM (Figure 1B). To capture the ontogeny of the PSM, it was exposed and photographed at successive time points and its projected area was calculated. Quantification over time revealed that a gradual change in size is maintained in both the wild type and ltm (Figure 1C). In comparison, normal doming, which occurred just before the delayed floral transition, was observed in the late-flowering mutant sft (Lifschitz et al., 2006) (Figures 1D and 1E). Thus, precocious PSM doming in the ltm mutant is not caused merely by a delay in floral transition but, rather, uncouples the timing of the physical change in meristem dimensions from that of the floral transition process.
In both ltm mutant alleles, the PSM and the first sympodial shoot meristem flowered significantly later than the wild type. In addition, their inflorescence was slightly branched and often contained one or a few small leaves (Figure 1F). Wild-type tomato plants developed, on average, seven to eight leaves before flowering. In comparison, ltm plants (of both mutant alleles) flowered a few days later and produced more leaves before undergoing floral transition (Figures 1G and 1H). While most ltm plants flowered late, some plants did not flower for an exceptionally long period of time (up to 3 to 5 months). This variation was not heritable, as progenies of extremely late flowering plants showed the same range of flowering times as progenies of regular ltm plants. When ltm-2 and wild-type plants were grown under the same conditions for more than 2 months, only 89% of ltm-2 plants flowered. The ltm-2 plants that flowered produced on average (±se) 16.37 ± 2.9 leaves before the first flower, while the remaining plants produced 29.4 ± 1.1 leaves 65 d after germination and still did not flower. This partially penetrant “extreme late-flowering” phenotype was common to both ltm alleles; however, we could not pinpoint the specific environmental conditions that enhanced or suppressed its penetrance.
LTM Is Not Part of the Classical SAM Maintenance Program
Precocious, floral-independent PSM doming was previously described for several tomato mutants that are part of the WUS-CLV signaling pathway; the fab (CLV1) and fin (arabinosyltransferase) mutants both show PSM enlargement and doming compared with the wild type at all developmental stages (Xu et al., 2015). Likewise, both locule number (disrupted in a WUS cis-element) and fasciated (fas; disrupted in CLV3), found in most large-fruited varieties, have large PSMs (Muños et al., 2011; Xu et al., 2015). When examined in the same genetic background as the other mutants (M82), both fab and fas showed precocious doming of the vegetative PSM compared with the wild type (Figure 2A). To understand the possible contribution of the WUS-CLV signaling pathway to floral transition doming, the expression dynamics of WUS was examined before and during floral transition. WUS expression in the vegetative PSM of wild-type plants was detected two to three cells below the apex summit. During meristem doming and floral transition, the WUS expression domain shifted up and expanded (Figure 2B). RNA sequencing data of three developmental time points of wild-type tomato apices (transcriptome analyses described below) showed a clear trend of significantly increased WUS expression in domed apices, nearly 2 times higher than its expression in wild-type vegetative PSM (Figure 2C). The same trend was documented in an independent analysis of gene expression dynamics in meristems of gradually older tomato plants (Park et al., 2012; Solyc02g083950; http://tomatolab.cshl.edu/efp/cgi-bin/efpWeb.cgi). In summary, WUS expression was upregulated and its domain shifted upward during PSM floral transition and SAM doming.
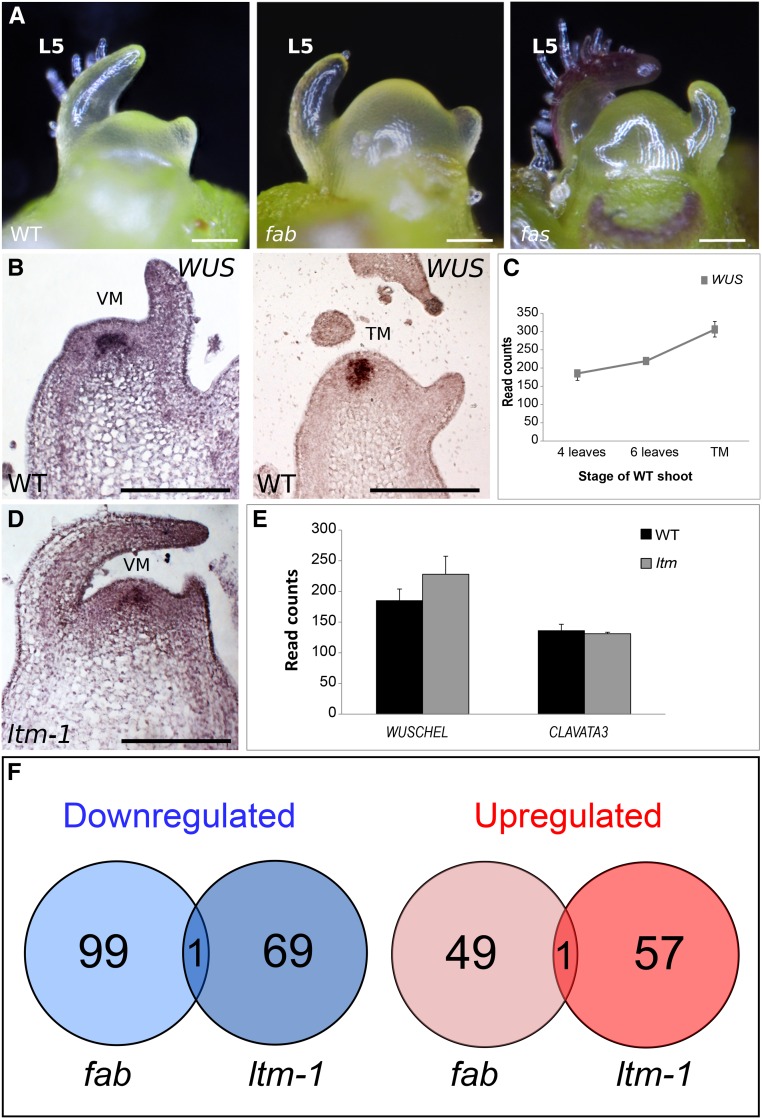
Figure 2.
Precocious Doming in ltm Is Not Accompanied by Altered WUS-CLV Regulation.
(A) Domed vegetative fab and fas PSM shoots with six leaves.
(B) Distribution of WUS RNA in vegetative and transitional apices of wild-type detected by RNA in situ hybridization.
(C) Expression levels of WUS in two vegetative developmental stages and during floral transition, detected by RNA in situ hybridization. VM, vegetative meristem.
(D) Distribution of WUS RNA in vegetative apex of ltm.
(E) Expression levels of WUS and CLV3, as determined by RNA sequencing of wild-type and ltm vegetative apices (n = 2).
(F) Overlap between genes differentially expressed in fab and ltm mutant apices.
Bars = 100 μm in (A) and 200 μm in (B).
To determine whether precocious SAM doming in ltm plants is a result of misregulation of WUS-CLV signaling, WUS in situ hybridization was performed on apices of ltm-1 seedlings, 2 d after germination. Although ltm PSMs were larger and more domed, no change in WUS expression was detected (Figure 2D). For a broader view of the WUS signaling pathway, gene expression profiles of vegetative ltm apices were compared with those of same-age wild-type plants. In agreement with the spatial expression pattern, expression levels of WUS and of CLV3 were similar in ltm and wild-type vegetative apices (Figure 2E), in contrast to the differential expression observed between wild-type and fab and fin apices (Xu et al., 2015).
To determine whether LTM can impact downstream WUS-CLV targets, a list of genes up- and downregulated in ltm mutants, as determined by a transcriptome comparison between ltm and the wild type, was generated and compared with a list of genes differentially expressed in wild-type versus fab plants (Xu et al., 2015). Seventy genes were found to be downregulated and 58 were upregulated (fold change ≥2 and P < 0.05) in ltm-1 apices compared with the wild type. Of the 50 genes that were upregulated in fab apices, only one (Solyc05g005130) was also upregulated in ltm-1. Similarly, of the 100 genes downregulated in fab apices compared with the wild type, only one (Solyc09g092750) was also downregulated in ltm-1 apices (P < 0.242). Thus, distinct transcriptional signatures characterize the two mutants (Figure 2F; Supplemental Data Set 1), suggesting that their similar doming behavior is of different molecular origins.
LTM Is a Unique Nuclear Kelch Repeat-Containing Protein
Genetic and expression profiling analyses suggest that LTM is a novel regulator that affects SAM maintenance independently of the classical factors involved in this process. To identify the causal mutation, the ltm-1 mutant was crossed with Solanum pimpinellifolium and 125 F2 plants were analyzed. LTM was mapped to an area spanning 1.7 Mb of the tomato chromosome 1. Illumina sequencing of ltm-1 genomic DNA identified three changes in protein-coding regions, one of which caused an amino acid change in a gene encoding a protein containing five kelch domains (Solyc01g100600). ltm-2 sequencing revealed an 8-bp insertion, resulting in a frame-shift mutation and a premature stop codon in the same gene (Figure 3A). LTM belongs to a family of kelch repeat proteins, a motif forming β-propeller domains that mediate protein-protein interactions. Such proteins are found in multiprotein complexes and participate in a wide range of cellular activities (Adams et al., 2000). The first LTM kelch domain was unique, in that it contained the five-amino acid motif LVLNL, which can function as an EAR domain via the consensus sequence pattern LxLxL (Ohta et al., 2001; Hiratsu et al., 2004). By aligning multiple LTM homologs, we identified three additional motifs conserved in all LTM proteins, termed here LTM boxes 1 to 3 (Figure 3B). These motifs were only found in highly related LTM homologs, which are only present once in most tested genomes and were not detected in any Brassicaceae genome (analyzed by the Plaza dicots 3.0 database) (Figure 3C). Notably, box 1 usually contained another LxLxL EAR domain; however, this was not the case for the tomato LTM. Transient expression of a chimeric RFP-LTM gene under the 35S promoter in Nicotiana benthamiana leaves revealed accumulation of RFP signals in the nucleus (Figure 3D).
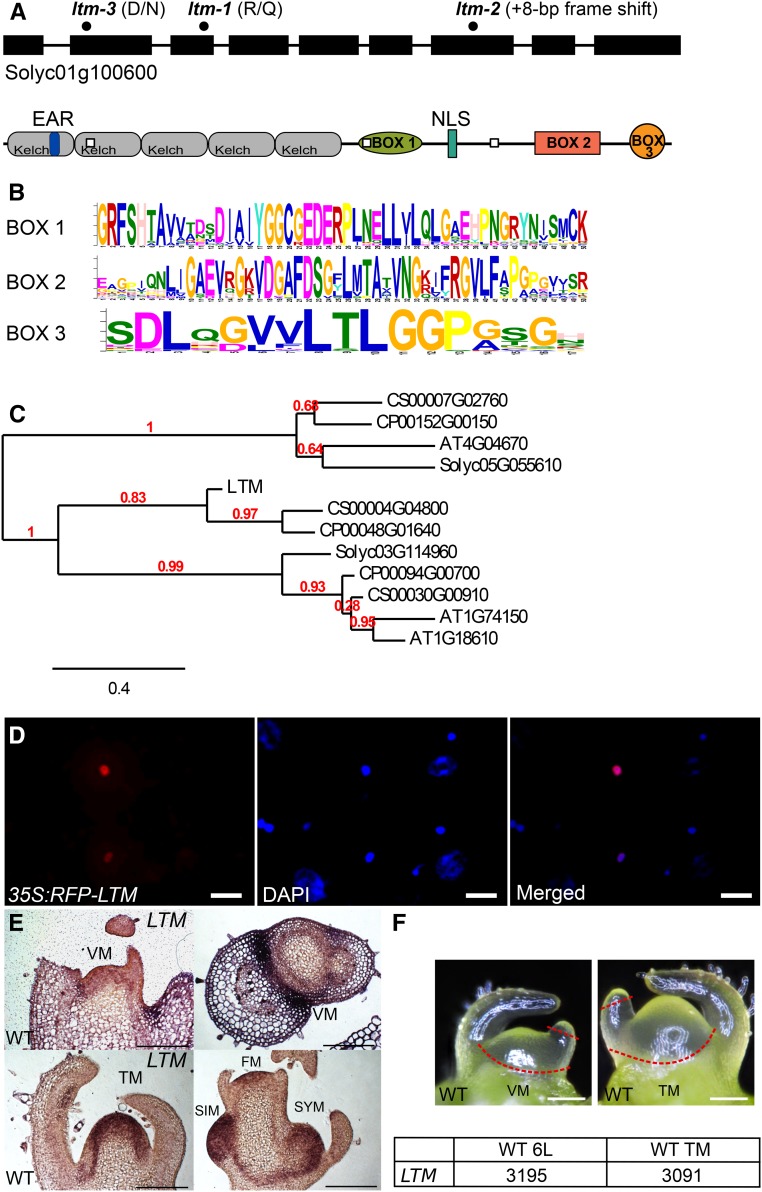
Figure 3.
Protein Motifs, Cellular Localization, and RNA Distribution of LTM Gene Products.
(A) Genomic organization, motif distribution, and lesions in three different ltm alleles (black dots on DNA and white dots on protein). Kelch domains are indicated by gray rectangles. EAR domains are marked in dark blue. Nuclear localization signal (NLS) is marked by light blue.
(B) Amino acid sequence of the three conserved LTM boxes identified by MEME (see Methods).
(C) Phylogenetic tree of the most related LTM-like genes from four plant species: tomato (Solyc), orange (Citrus x sinensis, CS), papaya (Carica papaya, CP), and Arabidopsis (AT). BLAST analysis using Plaza 3.0 database.
(D) Colocalization (pink) of RFP-LTM (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) in tobacco leaf epidermal cells. Bar =20 μm.
(E) RNA in situ localization of LTM on wild-type seedlings: young (2 d after germination), before (14 d after germination), at and shortly after floral transition. Bar = 200 μm.
(F) Illustration of the meristems used for PSM transcriptome profiling. Dashed lines indicate the tissues removed. Expression of LTM (Solyc01g100600) at these two stages is the normalized mean of two biological replicas.
VM, vegetative meristem; FM, flower meristem; SIM, sympodial inflorescence meristem; SYM, sympodial meristem; L, leaves produced by the PSM prior to collection.
In young vegetative meristems, LTM was weakly and uniformly expressed at the SAM but more prominently in a boundary domain between the apex and leaf primordia (Figure 3E). LTM expression was also detected in axillary meristems, be it the sympodial meristem or regular axillary buds. In a domed SAM undergoing floral transition, LTM expression at the SAM proper was stronger and, shortly after, in the flower meristem too. RNA sequencing data of wild-type tomato apices at two developmental time points (transcriptome analyses described below) showed that LTM is expressed, at similar levels, in both vegetative and transitional apices (Figure 3F). Furthermore, analysis of the tomato meristem maturation data set (Park et al., 2012) showed that LTM expression is equally expressed in tomato meristems of different plant ages. Overall, these results show that LTM is found primarily in meristems and meristem periphery and encodes for a nuclear protein.
The LTM EAR Domain Facilitates Its Interaction with TOPLESS
While kelch motifs are usually associated with diverse protein-protein interactions, the postulated EAR domain with the consensus sequence motif LxLxL may facilitate physical interaction with the plant corepressor TOPLESS (TPL) (Long et al., 2006; Szemenyei et al., 2008). To test this possibility, a yeast two-hybrid assay using full-length LTM and the N terminus of TPL, which mediates the interaction with EAR-containing proteins (Szemenyei et al., 2008; Ke et al., 2015), was conducted. LTM and TPL interacted when fused with either an activation domain or with a DNA binding domain (Figure 4A). No such interactions with other plant corepressors such as LEUNIG (LUG) and SAP18 (Conner and Liu, 2000; Song and Galbraith, 2006) were detected (Figure 4A). Similarly, LTM did not interact with the adaptor repressor protein SEUSS, which mediates many of the LUG interactions (Franks et al., 2002). Recently, the crystal structure of the TPL domain of rice TOPLESS RELATED2 in complexes with EAR motif proteins from Arabidopsis was described. The structure indicated that all LxLxL-type EAR motifs share a common mode of interaction with TPL, which is mediated by a key hydrophobic interaction between the three conserved leucine residues of the EAR and highly conserved hydrophobic and positively charged cleft residues of TPL (Ke et al., 2015). When the postulated EAR domain of LTM LVLNL was mutated to VVVNL, interaction with TPL was abolished (Figure 4A). Thus, the LVLNL motif of LTM functions as a bona fide EAR domain that facilitates the interaction with TPL. Cotransfection of the LTM and TPL protein pair (adopted for bimolecular fluorescence complementation [BiFC] assay) into leaf epidermal cells of N. benthamiana resulted in nuclear fluorescence. In contrast, cotransfection of LTM with WUS or TPL with COMPOUND INFLORESCENCE (S) provided no fluorescent signal (Figure 4B). Lastly, in an in vitro coimmunoprecipitation assay involving bacterially expressed His-tagged TPL and FLAG-tagged LTM, although significant LTM degradation occurred, the two proteins successfully pulled each other down (Figure 4C). Taken together, the interaction between LTM and TPL through an EAR domain may facilitate repression of target genes.
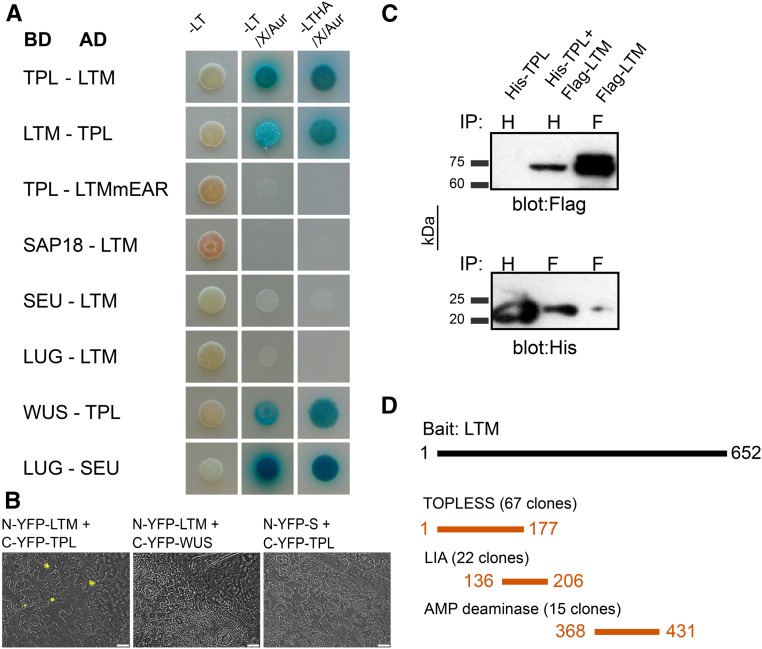
Figure 4.
LTM Interacts with the Plant Corepressor TPL.
(A) A yeast two-hybrid assay detecting positive interaction of LTM and TPL, but not of LTMmEAR and TPL. L, leucine; T, tryptophan; H, histidine; A, adenine; X, X-α-gal; Aur, aurebasidine. Dropout medium is indicated by a minus symbol.
(B) A split YFP complementation assay in tobacco leaf epidermal cells. BiFC complementation was achieved when LTM and TPL were cotransfected. No complementation was obtained when LTM was cotransfected with WUS, and TPL with S (bar = 20 μm).
(C) Coimmunoprecipitation of TPL-His and LTM-flag in Escherichia coli cells.
(D) A yeast two-hybrid interaction screen using full-length LTM as bait. Amino acids of selected interaction domain, shared by all prey fragments matching the same protein, are shown for strongest LTM-interacting proteins.
A yeast two-hybrid screen of a library of tomato meristem RNA was performed using LTM as bait, in search of a DNA component that may bind such repressor complex. The screen resulted in identification of 51 independent proteins, which were categorized into four classes (A–D) according to the number of independent clones found for each interactor (Supplemental Data Set 2). An AMP deaminase (Solyc09g014770), an AP2 domain transcription factor (Solyc03g123430), and the TPL gene product (Solyc03g117360) described above (Figure 4D) were categorized as having the strongest interaction with LTM. To determine whether expression of these factors overlaps with LTM, the cDNA clones of TPL and the LTM-INTERACTING-AP2-CONTAINING were cloned from cDNA made exclusively from tomato apical meristems. A similar interaction, in which an adaptor protein can link TPL with a DNA binding protein, was shown in jasmonic acid signaling in Arabidopsis. In this case, the JAZ repressor function relies on interaction with NINJA, which mediates the recruitment of TPL via its EAR domain (Pauwels et al., 2010).
A Unique Set of Floral Transition Genes Are Precociously Activated in ltm Apices
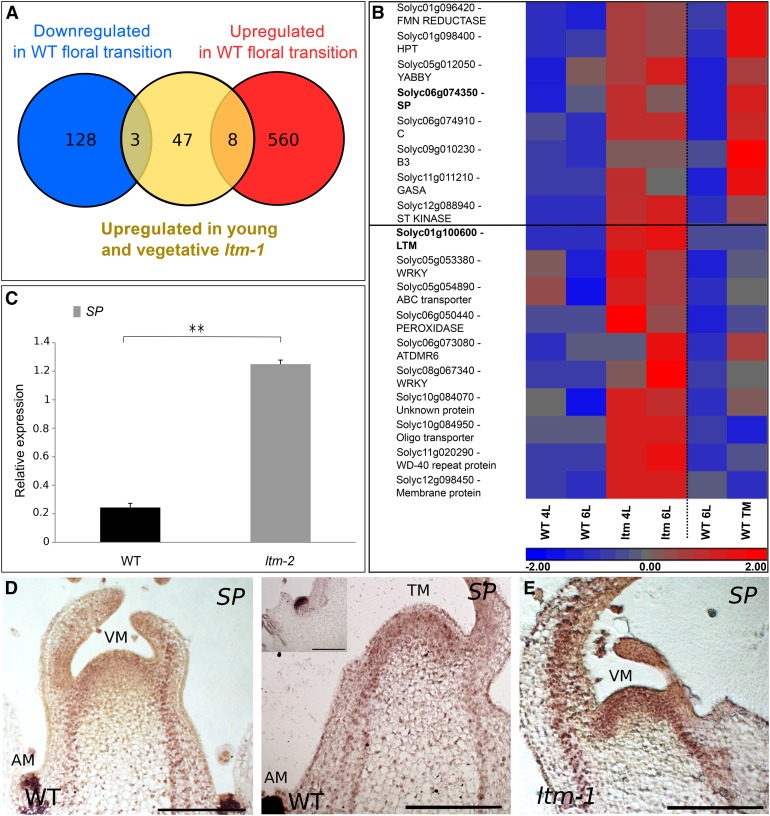
To characterize processes and genes regulated by LTM-containing complexes and which orchestrate doming with the floral transition, global expression profiles of vegetative ltm apices were compared with those of same-age wild-type apices. As ltm apices display delayed flowering, we speculated that floral suppressors or genes associated with precocious doming would be identified among the genes upregulated in its domed PSM. Likewise, promoters of the floral transition were expected to be among the genes downregulated in the domed ltm apices. To test this hypothesis, transcriptomes of PSM from vegetative wild-type plants that produced six leaves and from plants at floral transition were assayed. A comparison of the two time points identified 568 upregulated (floral-activated genes) and 131 downregulated genes at the TM stage of the wild type (Figure 5A).
Figure 5.
Precocious Expression of Flowering-Associated Genes in Vegetative ltm Apices.
(A) Schematic demonstration of the overlap (calculated using the hypergeometric distribution) between genes upregulated in young and vegetative ltm-1 apices (marked in yellow) and genes differentially expressed in wild-type PSM undergoing floral transition. Downregulated genes are marked in blue, and upregulated genes are marked in red.
(B) A heat map of genes upregulated in vegetative ltm-1 apices. The log2-normalized counts were standardized to have a zero mean and unit sd for each gene (replicates were averaged). The expression profile is accompanied by a color bar indicating the standardized log2 counts. Upper panel shows the eight genes that are also floral activated. Lower panel shows genes upregulated ≥5-fold in ltm over wild type but are not floral induced. Separate experiments are divided by a dashed line (n = 2 in both). L, leaves produced by the PSM prior to collection.
(C) Relative expression of SP as determined by RT-qPCR on cDNA from young and vegetative wild-type and ltm-2 apices. Values (SP-to-TUBULIN ratios) are means of three biological replicates. Error bars show se, **P < 0.01, compared with the wild type (Tukey HSD).
(D) RNA in situ hybridization of SP in 2-d-old seedlings of the wild type and ltm-1. Longitudinal sections are shown. Expression in the PSM was only detected in ltm-1 apices. Inset: Axillary meristem from the same shoot apex captured at a different section. AM, axillary meristem. Bar = 200 μm.
Even though ltm plants are late flowering, several genes with increased expression in the wild type at the floral transition were highly expressed in young ltm vegetative apices; out of the 58 genes upregulated in young ltm, eight were floral-activated genes (P < 4.37 × 10−5; Figures 5A and 5B; Supplemental Data Set 1). Additional genes that were not floral induced were strongly upregulated in ltm compared with same-age wild type, most significantly, the ltm transcript itself, which was upregulated by 9-fold in the mutant apices (Figure 5B), indicating at an auto-negative feedback regulation.
To reexamine the unexpected precocious elevation of floral-activated genes, the tomato meristem maturation atlas (Park et al., 2012), with documentation of meristems approaching TM at four successive ages, was used to query the LTM-modified genes. Cluster analysis revealed that out of 2924 genes differentially expressed in wild-type apices, 1213 genes showed a trend of upregulation toward floral transition. Out of the 58 genes upregulated in ltm, 10 were among these 1213 genes (P < 3.937 × 10−4; Supplemental Data Set 1). Thus, in two independent sets of gradually more mature apices, precocious upregulation of floral-activated genes was detected in young vegetative ltm apices. These floral-activated genes might contribute to the precocious doming of ltm PSM and may account for the tight association between doming and wild-type floral transition.
Precocious Activation of SP Contributes to Early Doming of ltm Plants
One of the floral-activated genes upregulated in vegetative ltm-1 apices was SP. The precocious SP expression was independently examined by RT-qPCR of RNA samples collected from young ltm-2 and wild-type apices. A 5-fold increase in SP expression was found in young ltm-2 apices compared with the wild type (Figure 5C). SP encodes a CETS protein that is upregulated in the primary apical meristem toward floral transition and in axillary meristems, from their inception, where it functions antagonistically to the flowering process (Pnueli et al., 1998; Lifschitz and Eshed, 2006; Lifschitz et al., 2014). RNA in situ hybridization demonstrated diffuse SP expression at the apex of vegetative young ltm-1 meristems but not in those of the wild type. In axillary meristems of both ltm-1 and the wild type, SP expression was strong to similar degrees (Figure 5D; Supplemental Figure 2).
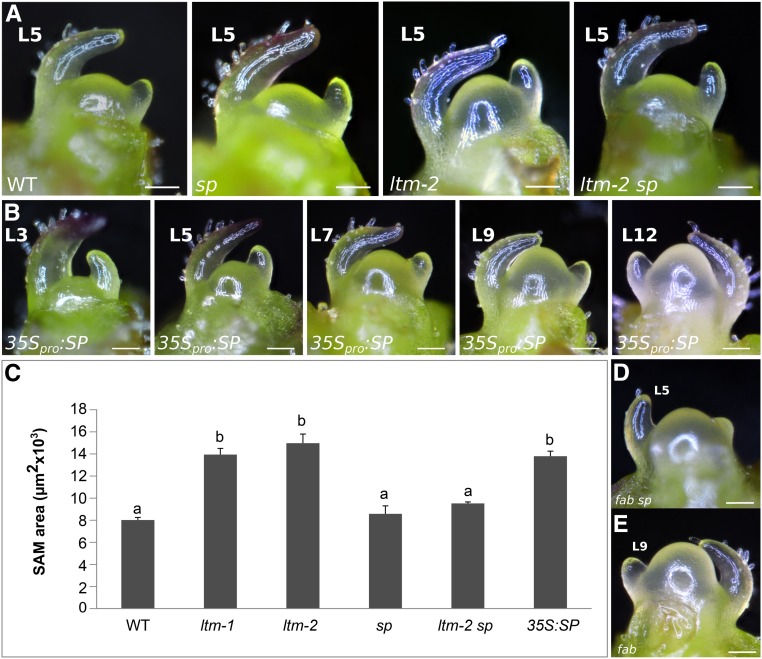
To test the contribution of SP to ltm mutant phenotypes, we characterized the PSM size of ltm-2, sp, and ltm-2 sp plants. Strikingly, ltm sp double mutant plants lacked doming of the vegetative PSM and their apical meristem was the same size and shape as that of the wild type (Figure 6A). In agreement, apices of 35S:SP-MYC plants were domed long before floral transition compared with the wild type (Figure 6B). As with ltm apices, the vegetative PSM of 35S:SP-MYC plants was also larger with age and its PSM size after the production of six leaves was similar to those of ltm-1 and ltm-2 and significantly larger than those of the wild type and sp at the same developmental stage (Figure 6C). Ectopic expression of SP in the PSM, under the pTCS promoter (Steiner et al., 2016), which is strongly expressed in the SAM, also resulted in doming during the vegetative phase (Supplemental Figure 3A).
Figure 6.
SP Is a Floral Repressor That Also Regulates SAM Size.
(A) Shape of wild-type, sp, ltm-2, and ltm-2 sp vegetative PSM. L5, primordia of the 5th leaf. Bar = 100 μm.
(B) Ontogeny of 35S:SP-MYC vegetative PSM showing precocious doming.
(C) PSM size (average ± se) of wild-type, ltm-1, ltm-2, sp, ltm-2 sp, and 35S:SP vegetative PSM after production of six leaves (n = 4). Different letters mark statistically significant differences (Tukey HSD).
(D) Vegetative fab sp PSM.
(E) Transitional meristem of fab.
In contrast with its effects in the ltm background, SP did not contribute to the precocious doming of fab plants (Figure 2A); fab apices are as domed as fab sp (Figure 6D). We thus argue that ltm and fab are not part of the same regulatory circuit and that SP is not the sole driver of SAM doming. Indeed, when undergoing floral transition, the fab PSM is larger and more domed than during the vegetative stage (Figure 6E).
Precocious Activation of SP Contributes to Late Flowering of ltm Plants
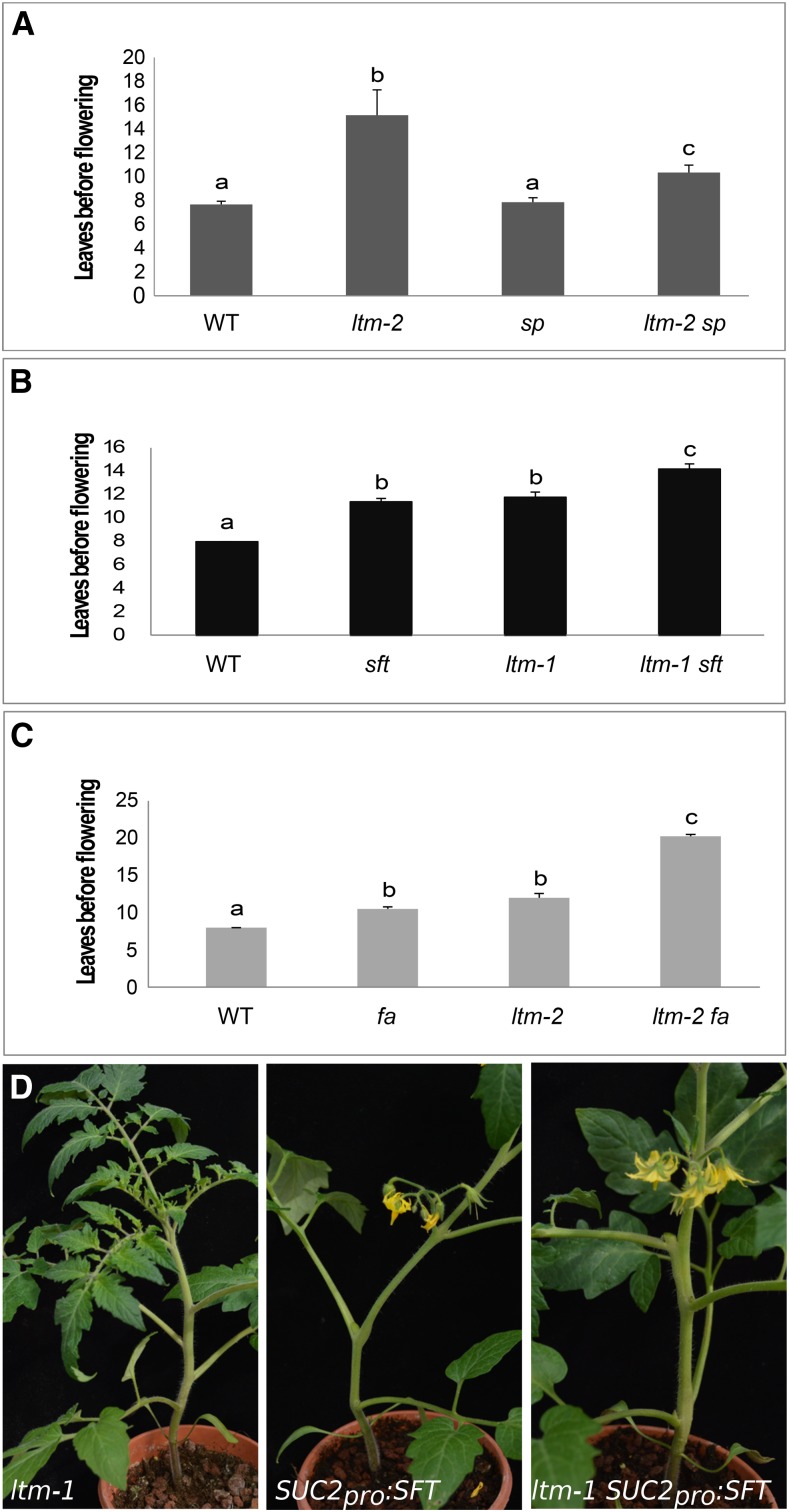
SP is a known antagonist of the flowering hormone florigen (Lifschitz and Eshed, 2006; Shalit et al., 2009) and 35S:SP-MYC plants, which are, as expected, late flowering and produce 11.6 ± 0.2 leaves (n = 18) before the floral transition. To test whether precocious SP expression in ltm apices contributes to its late flowering, double mutant plants were examined (Figure 7A). Wild-type plants flowered after of 7.6 ± 0.2 leaves and sp plants flowered after producing 7.8 ± 0.3 leaves, demonstrating that SP has no detectable role in the PSM floral transition in the wild-type background. In contrast, ltm-2 plants flowered after the production of 15 ± 2.1 leaves, whereas ltm-2 sp plants flowered after producing only 10.3 ± 0.6 leaves (P < 0.01). In addition to its effect on flowering time in ltm plants, the sp mutation also had a marked effect on the “extreme late-flowering” syndrome; all 50 ltm-2 sp double mutant plants flowered in <30 d (27.06 ± 0.61 d after germination). In fact, in all the four years that plants were grown, no extreme late-flowering ltm sp plant was ever found. Taken together, both late-flowering and precocious PSM doming of ltm are caused primarily, but not solely, by precocious SP expression in the vegetative apices.
Figure 7.
LTM Promotes Flowering Independently of Florigen and FA.
(A) Number of leaves produced by the PSM before flowering (average ± se). Wild type (n = 18), ltm-2 (n = 39), sp (n = 21), and ltm-2 sp (n = 50). Different letters mark statistically significant differences based on Tukey HSD.
(B) Number of leaves (average ± se) produced by the PSM before flowering; n = 5. Samples with different letter are significantly different by Tukey HSD.
(C) Number of leaves (average ± se) produced by the PSM before flowering; n = 4. Samples with different letter are significantly different by Tukey HSD.
(D) ltm-1, pSUC2:SFT, and ltm-1 pSUC2:SFT grown together. ltm-1 plants had eight visible leaves and no flowers, while plants ectopically expressing SFT flowered after producing three to four leaves.
LTM Regulates Flowering Time Independently of the Florigen and FALSIFLORA Pathways
The ltm plants are significantly late-flowering, much like sft, a tomato plant bearing a mutant in the florigen gene (Kerr, 1982; Lifschitz et al., 2006) and falsiflora (fa), a mutant in the tomato ortholog of LEAFY (Molinero-Rosales et al., 1999). The vegetative sft PSM shows normal doming, which occurs just before the delayed floral transition (Figure 1D), suggesting that ltm is not part of the SFT signaling pathway. Indeed, ltm sft plants flowered after the production of 14.2 ± 0.4 leaves, significantly later than either of the single mutants (11.8 ± 0.4 and 11.4 ± 0.2, respectively; Figure 7B). In tomato, FA also promotes flowering in an SFT-independent manner as fa sft double mutants never flower (Molinero-Rosales et al., 2004). However, the double mutant ltm fa showed an additive effect on flowering time (Figure 7C), suggesting that LTM is distinct from the known floral regulation pathways. In agreement, expression of pSUC2:SFT in wild-type and ltm plants resulted in early floral termination after production of a similar number of leaves (Figure 7D).
DISCUSSION
Traditionally, floral transition studies are performed with plant models that strongly respond to environmental cues. Here, we characterized the floral transition process in the day-neutral tomato, which enabled specific focus on the growth and development of the SAM upon exposure to gradually increased levels of floral stimulants produced in mature leaves (Lifschitz et al., 2006). Changes in SAM size were shown to be gradual and age-dependent. Then, the role of the adaptor protein LTM in preventing precocious doming of the vegetative PSM, via restriction of a subset of floral-induced genes, was demonstrated. The precocious activation of SP in ltm PSM proved essential to precocious doming and late flowering of the mutant apices. Thus, by regulating SP, and other proteins, LTM acts to coordinate doming with floral transition.
The Tomato SAM Grows Slowly with Shoot Age and Swells Rapidly toward Floral Transition
The easily accessible PSM of tomato enabled monitoring of SAM size changes starting from 2 d after germination (after the production of four leaves) and onward (Figure 1; Supplemental Figure 1). Although the meristem maintenance pathways act to restrict SAM size and constantly balance cell proliferation with organ differentiation (Schoof et al., 2000), we showed here that there is a gradual increase in SAM size during vegetative shoot development. Similarly, the SAM dimensions of Arabidopsis shoots grown in short days show a slight increase with age (Hempel and Feldman, 1994; Hempel et al., 1998), as do the SAM of rice plants grown in 28°C under long days (Asai et al., 2002). Do SAMs of shoots of long-lived trees keep on growing? Also, what happens to plants that lack seasonal and/or internal changes in florigen levels? A careful characterization is required before generalizations can be made.
LTM Coordinates Floral Transition with the Meristem Doming Process
Previous studies of floral transition regulation in late-flowering tomato mutants defined two main genetic pathways that act in parallel. The florigen pathway involves the systemic and graft transmittable SFT (Lifschitz and Eshed, 2006) and its meristematic partner SPGB1/SUPRESSOR OF SP (SSP; Park et al., 2014), while the tomato LFY ortholog FA controls flowering time, inflorescence architecture, and the fate of flower meristems (Molinero-Rosales et al., 1999). The sft fa double mutant plants do not flower, suggesting that the two are part of distinct regulatory pathways (Molinero-Rosales et al., 2004). However, both sft ltm and fa ltm double mutants showed an additive late-flowering effect (Figure 7), indicating that LTM represents an independent floral transition pathway. While misregulation of the floral antagonist SP is a main cause for late flowering and precocious doming in ltm plants, ltm sp plants are still weak late-flowering, suggesting that one or more additional floral inhibitors are precociously activated in ltm apices. Indeed, several other floral-activated genes showed the same expression trend as SP and were precociously upregulated in vegetative ltm apices (Figure 5; Supplemental Data Set 1).
Why Do Apical Meristems Dome toward the Floral Transition?
Doming of apical meristems during floral transition is a highly conserved developmental process, which raises questions with regards to its developmental role. After studying the apical dome of the short-day plant Chrysanthemum morifolium, Horridge and Cockshull (1979) suggested that flower initiation is an irreversible consequence of exceeding a critical size of the apical meristem. However, this suggestion was refuted by observations in other species, where the transition to flowering occurs over a wide range of apical dome sizes and, rarely, a decrease in size can even be found (Battey and Lyndon, 1984). In addition, meristems of nutrient-starved plants commonly flower at a reduced size (Dale and Wilson, 1979; Bernier et al., 1981; Battey and Lyndon, 1984). Thus, the reason why most plant meristems dome during the transition to flowering remains unknown.
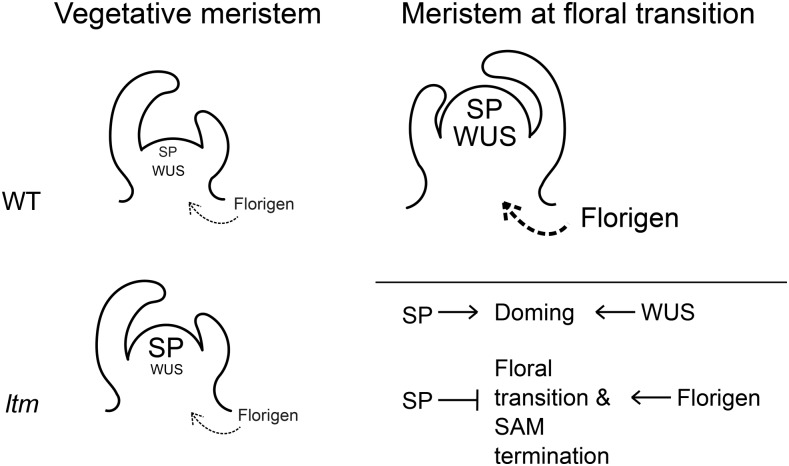
A prime and universal booster of floral transition is florigen. Florigen is a general growth hormone, and promotion of meristem termination and flowering is only one of its functions (Shalit et al., 2009; Pin and Nilsson, 2012). Negative feedback, whereby antagonists are rapidly induced, is a common response to plant hormones (Santner and Estelle, 2009). Such regulatory circuits provide both transient and quantitatively balanced signal outputs. The florigen antagonist and floral repressor SP (Pnueli et al., 1998; Lifschitz et al., 2014) is upregulated at the PSM toward the floral transition (Figure 5; Park et al., 2012). Examination of global expression data sets showed that this is true for the Arabidopsis SP homolog TERMINAL FLOWER1 (TFL1) as well (Schmid et al., 2005; Klepikova et al., 2015). Here, we showed that in addition to its floral inhibiting functions, SP plays a promoting role in regulation of PSM size. When meristems of sp plants were exposed to strong flowering signals, as in a strong 35S:SFT sp line, the PSM terminated early after producing 0 to 2 flowers; sympodial branching was suppressed as well. After their release, lateral meristems grew and terminated with a single flower or a blind apex (Shalit et al., 2009). Such strong termination of sp 35S:SFT meristems was not observed in SP 35S:SFT sibs. Taken together with the results of this study, a plausible reason for the required PSM size increase toward floral transition would be to maintain a functioning meristem under the strong terminating signals of florigen. Several factors can participate in such a SAM size-increasing process: SP and its signaling targets, other LTM-repressed genes that are activated at floral transition, as well as the meristem maintenance regulator WUS and its targets (Figure 8). The division of labor among these factors awaits a more detailed dissection.
Figure 8.
Synchronization of PSM Doming with Floral Transition.
A model for meristem doming during floral transition. Once florigen signals reach the PSM, they drive both the transition to flowering and SAM termination. Expression of SP and WUS are elevated in the transitional PSM, leading to a physical change in its dimensions. The change is restricted to the TM stage by maintaining vegetative SP suppression via an LTM-containing complex. Gene expression level is illustrated by font size.
What Is the Significance of Regulatory Programs That Are Dispensable in Particular Lineages?
LTM is a kelch protein that contains an EAR repression motif in addition to three conserved boxes with an unidentified function. A BLAST survey of LTM in the Plaza database revealed LTM homologs in many plant species, but not in plants of the Brassicaceae family (Figure 3C). In a further search for Arabidopsis kelch proteins, we failed to identify a kelch protein that contains an EAR motif. These findings suggest that LTM is absent in the Arabidopsis genome. Using BLAST analysis, we found several other elements of the LTM regulation system, either genes upregulated in ltm or LTM-interacting partners, which seem to lack an Arabidopsis ortholog (Supplemental Data Set 3). We thus suggest that Arabidopsis lost this particular regulatory pathway. In contrast, the function of SP and of its orthologous floral inhibitors was studied in many plant species and is highly conserved (Pnueli et al., 1998; Jensen et al., 2001; Elitzur et al., 2009; Mohamed et al., 2010; McGarry et al., 2016). However, AT-CENTRORADIALIS, the Arabidopsis ortholog of SP, has the potential to inhibit flowering in the SAM, but its expression is detected in roots, the hypocotyl, and mature leaves (Mimida et al., 2001; Huang et al., 2012). In this plant, TFL1 fulfills the floral inhibition functions needed in the SAM, and in its absence, SAM cells terminate by a flower (Alvarez et al., 1992).
We suggest that LTM-driven transcriptional regulation of SP and other targets is dedicated to the protection of meristems under strong floral-inducing signals. In tomato, we have shown that ectopic expression of SP leads to SAM doming. However, SP upregulation in the transitional meristem is not the sole cause of the observed doming phenotype; apices of sp plants also dome while undergoing floral transition (Supplemental Figure 3B). Shoots of plants with precocious doming, such as ltm and 35S:SP, show larger, more domed meristems at floral transition. Furthermore, genetic evidence clearly shows that the WUS-CLV SAM maintenance pathway also regulates doming of vegetative meristems (Xu et al., 2015). Indeed, the fab sp TM is domed and, much like 35S:SP plants, the fab TM becomes larger and more domed. The upregulation of both WUS and SP transcripts at the TM indicates that both genes have the potential to take part in this developmental change of the meristem during floral transition. In Arabidopsis, TFL1 is the floral antagonist upregulated in the SAM during floral transition, and future work will be necessary to determine whether TFL1 is involved in SAM doming or if other factors provide for meristem protection during floral transition in this “derived” plant system.
METHODS
Plant Material
All tomato (Solanum lycopersicum) lines were in the CV. M82 background. Mutants (generated by either ethyl methane sulfonate or fast neutron) were isolated as previously described (Menda et al., 2004). Plants were grown in the Weizmann Institute greenhouses. For transgenic lines, constructs were subcloned into the pART27 binary vector and were introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation. Transgenic lines were generated by cotyledon transformation according to McCormick (1997).
Microscopy
Live images were taken using either a Nikon D3200 SLR camera or a Nikon SMZ18 stereomicroscope. In some cases, plants were dissected in order to expose the SAM and put on a damp black cloth. Images were analyzed using ImageJ or NIS-elements software. For some images, the NIS-elements EDF (extended depth of focus) module was used. Fluorescence was imaged using an Olympus IX71S8F-3 inverted fluorescence microscope (light source of Lumen 200 Pro and ProScan III Controller) equipped with an UPLSAPO40X2/0.95 objective. Chroma filters used were as follows: 4′,6-diamidino-2-phenylindole (excitation, D350/50×; emission, ET460/50m), RFP (excitation, ET560/40×; emission, ET630/75m), and YFP (excitation, ET500/20×; emission, ET535/30m). Images were captured using an ExiBlue (Q Imaging) camera and processed using Olympus cellSens Dimension (v1.11) software. RNA in situ hybridization images were captured using a Nikon eclipse E800 microscope, equipped with a Nikon color digital sight DS-5Mc camera. Images were processed using NIS-elements BR 3.2 software.
RNA in Situ Hybridization
The antisense cRNA probes were produced by in vitro transcription with digoxigenin-11-UTP (Roche) using an RNA in vitro reverse transcription kit (CellScript; cat. no. C-AS2607), according to the manufacturer’s protocol, from PCR fragment templates containing a T7 promoter sequence (TTTGCGGTAATACGACTCACTATAGGGCGAATTGGGTACC) flanking the full-length sense/antisense LTM, WUS, and SP cDNAs. Shoot apices from 2- and 10-d-old tomato (M82) plants were fixed in PFA (3.8% PFA in 1× PBS, pH 7.0, by H2SO4), gradually transferred to ethanol and then to K-clear plus (Kaltek), and embedded in Paraplast Plus (Leica Biosystems). Eight-micrometer-thick tissue sections were produced and mounted on Superfroset Plus slides (Thermo Scientific). Slides were successively treated with K-clear plus, an ethanol series, diethylpyrocarbonate-treated double distilled water, 2× SSC, proteinase K (1 μg/mL) in 100 mM Tris-HCl, pH 8.0, and 50 mM EDTA at 37°C, glycine (2 mg/mL) in PBS, twice with PBS, 4% paraformaldehyde in PBS, twice with PBS, triethanolamine (0.1 M, with stirring), twice with PBS, and increasing ethanol concentrations, up to 100% ethanol. For hybridization, slides were incubated with sense or antisense cRNA probes in hybridization buffer (0.3 M NaCl, 10 mM Tris-HCl, pH 8, 10 mM sodium phosphate buffer, pH 6.8, 5 mM EDTA, 50% [v/v] deionized formamide, 10% [w/v] dextran sulfate, 1× Denhardt’s solution, and 200 μg tRNA) overnight at 55°C. Following hybridization, slides were successively washed twice with 0.2× SSC at 55°C. Then, slides were blocked with 1% fresh Boehringer block (Roche) in 100 mM Tris-HCl, pH 7.5, and 150 mM NaCl and then with 1% BSA solution (1% BSA, 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.3% Triton X-100). Blocked slides were incubated with antidigoxigenin antibodies (Roche) for 2 h at room temperature and then washed three times with 1% BSA solution and three times with detection buffer (100 mM Tris-HCl, pH 9.5, and 100 mM NaCl). The slides were then incubated with NBT/BCIP color development substrate (Promega) for 24 h, washed with double distilled water followed by increasing ethanol concentrations, and then mounted and analyzed.
Yeast Two-Hybrid Analysis
Protein interaction assays in yeast were performed according to the protocol for the Matchmaker Gold Yeast Two-Hybrid System (GAL4 based; Clontech). The coding sequences for bait proteins were cloned into the pGBKT7 vector, and the resulting vectors were transformed into the Y2HGold yeast strain. The coding sequences for prey proteins (SSP, ssp-2129, ssp-610, and 14-3-3 proteins) were cloned into the pGADT7 activation domain vector, which was then transformed into the Y187 yeast strain. After mating the two yeast strains expressing bait and prey proteins, diploid yeast cells were selected and grown on dropout medium free of leucine and tryptophan. Clones were then selected on both double-dropout medium containing X-α-gal and Aureobasidin toxin and on a quadruple-dropout medium free of leucine, tryptophan, adenine, and histidine and X-α-gal and Aureobasidin toxin for 3 d at 30°C to assay protein-protein interactions. The screen was performed by Hybrigenics Services.
The coding sequence for tomato LTM (XM_004231246.2) was PCR amplified and cloned into pB27 as a C-terminal fusion to LexA (LexA-LTM). The construct was checked by sequencing the entire insert and used as a bait to screen a random-primed tomato meristem cDNA library constructed into pP6. pB27 and pP6 derive from the original pBTM116 (Vojtek and Hollenberg, 1995) and pGADGH (Bartel et al., 1993) plasmids, respectively.
Sixty-six million clones (7-fold the complexity of the library) were screened using a mating approach with YHGX13 (Y187 ade2-101::loxP-kanMX-loxP, matα) and L40ΔGal4 (mata) yeast strains, as previously described (Fromont-Racine et al., 1997). A total of 251 His+ colonies were selected on medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (NCBI) using a fully automated procedure. A confidence score (predicted biological score) was assigned to each interaction as previously described (Formstecher et al., 2005). The predicted biological score relies on two different levels of analysis. First, a local score considers the redundancy and independency of prey fragments, as well as the distribution of reading frames and stop codons in overlapping fragments. Second, a global score takes into account the interactions found in all the screens performed at Hybrigenics using the same library. This global score represents the probability of an interaction being nonspecific. For practical use, the scores were divided into four categories, from A (highest confidence) to D (lowest confidence). A fifth category (E) specifically flags interactions involving highly connected prey domains previously found several times in screens performed on libraries derived from the same organism. The predicted biological scores have been shown to positively correlate with the biological significance of interactions (Rain et al., 2001; Wojcik et al., 2002).
BiFC Assays and Subcellular Localization
Protein interaction assays in planta were performed by transient expression in N. benthamiana leaf epidermal cells (Sparkes et al., 2006) using the BiFC system previously described (Citovsky et al., 2006). Cloning was performed using the pSAT-nEYFP and pSAT-cEYFP vectors and the pRSC2 binary vector (Chung et al., 2005).
Pull-Down Assay
BL21(DE3) cells were cotransformed with His-TPLpET28 and Flag-LTMpETDuet plasmids. In parallel, BL21(DE3) cells were transformed with the individual plasmids His-TPLpET28 or with Flag-LTMpETDuet plasmids, as controls. Bacterial culture cells of the individual plasmids or the coexpression plasmids were grown at 37°C in Luria-Bertani medium with the appropriate antibiotics, and protein expression was induced at OD600 = 0.7 to 0.8, with 200 μM isopropyl-1-thio-β-d-galactopyranoside at 15°C for 20 h. Two identical cultures were prepared for the His-TPL and Flag-LTM coexpression plasmids. Bacteria were lysed by sonication and the soluble fraction was isolated by centrifugation. The soluble proteins were incubated with Ni-beads or on anti-Flag M2 beads to capture coexpressed His-TPL and Flag-LTM or expression of each alone, respectively. The beads were washed extensively and bound proteins were eluted with 500 mM imidazole from the Ni-beads or with 150 μg/mL of Flag peptide from the anti-Flag M2 beads. Eluted proteins were subjected to immunoblot analysis with anti-His or anti-Flag M2 monoclonal antibodies.
Phylogenetic Tree Analysis
BLAST analysis and protein sequences of LTM orthologs were performed using the Plaza 3.0 project (Proost et al., 2015). Analysis of phylogenetic relationships between protein sequences and construction of a phylogenetic tree were performed using the Phylogeny.fr web service tool, which uses MUSCLE for sequence alignment, Gblocks for curation of the alignment, and PhyML for analysis of phylogenetic relationships (Dereeper et al., 2008, 2010). Text files corresponding to the alignments are provided as supplemental files.
Protein Domains
Analysis of conserved protein domains between LTM orthologs was performed using the MEME suite 4.11.1 bioinformatics online tool (Bailey et al., 2009).
RT-qPCR Analysis
RT-qPCR analysis was performed using the Absolute Blue qPCR SYBR Green ROX Mix (AB-4162/B) kit (Thermo Fisher Scientific). Reactions were performed using a Rotor-Gene 6000 cycler (Corbett Research). A standard curve was obtained for each gene, using dilutions of a cDNA sample. Each gene was quantified using Corbett Research Rotor-Gene software. At least three independent technical repeats were performed for each cDNA sample. Relative expression of each sample was calculated by dividing the expression level of the analyzed gene by that of TUBULIN. Gene-to-TUBULIN ratios were then averaged. Primers were as follows: for SP (forward, ATGGCTTCCAAAATGTGTGA; reverse, CAGACATCTTAACACTTGGACAGAA) and for TUBLIN (forward, CACATTGGTCAGGCCGGTAT; reverse, CGCGAGATGAGATAAACCA).
DNA Library Construction and Sequencing
PCR-free DNA-seq was performed as previously described (Blecher-Gonen et al., 2013) with the following modifications: 1 µg of tomato DNA was sheared using the Covaris S220 sonicator. End repair was performed in 80-μL reaction at 20°C for 30 min followed by Agencourt AmPURE XP beads cleanup (Beckman Coulter) in a ratio of 0.75× beads/DNA volume. A base in the 3′ end of both strands was added and two adapters (NexTflex PCR free barcodes; BioScientific) were ligated, followed by SPRI beads cleanup in a ratio of 0.75× beads/DNA volume. The sample preparation was done without the PCR step. Libraries were evaluated by qPCR. Sequencing libraries were constructed with barcodes to allow multiplexing of two samples in one lane of Illumina HiSeq 2500 instrument rapid mode. Per sample, 21 to 25 million 100-bp paired end reads were sequenced.
cDNA Library Construction and Sequencing
Total RNA (0.5 μg) was processed using the TruSeq RNA Sample Preparation Kit v2 protocol (Illumina). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of eight samples on one lane. Twenty to twenty-five million single-end 60-bp reads were sequenced per sample on an Illumina HiSeq 2500 V4 instrument.
Sequence Data Analysis
TopHat (v2.0.10) (Kim et al., 2013) was used to align the reads to the tomato genome sequence SL2.50 (downloaded from the Sol genomics network; http://solgenomics.net/organism/Solanum_lycopersicum/genome). Counting reads on ITAG2.4 genes (downloaded from Sol genomics network) was done with HTSeq-count (version 0.6.1p1) (Anders et al., 2013, 2015). Differential expression analysis was performed using DESeq2 (1.6.3) (Anders et al., 2013; Love et al., 2014). To find differentially expressed genes in the ltm mutant, a two-factor model including the strain (wild type or ltm) and the time (early or mid) was built using DESeq2, and the difference between ltm versus the wild type was tested. Genes that had an absolute fold change >2, a P value < 0.05, and a count of at least 25 reads were considered differentially expressed. The RNA-seq data were deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE95117 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95117).
Raw fastq files of the time-course [EVM1-2, MVM1-2, LVM1-2 and TM1-2, (Park et al., 2012)] and for fab [fabvm1, fabvm2, M82vm1 and M82vm2 (Xu et al., 2015)] were downloaded and analyzed. All samples were subjected to the same workflow described above. For the time-course data, genes that were differentially expressed at least in one of the six possible pair-wise comparisons, by a fold-change of at least 1.5 and p-value below 0.05, were clustered. 3451 genes that passed these criteria were clustered to eight clusters using Pearson dissimilarity distance measure. The significance of the overlap between 2 sets of genes was calculated using the hypergeometric distribution.
Accession Numbers
Sequence data from this article can be found in GenBank under the following accession numbers: LTM, KX346231; TPL, KY348751; and LTM-INTERACTING-AP2-CONTAINING, KY348752.
Supplemental Data
Supplemental Figure 1. Prefloral development of wild-type and ltm seedlings.
Supplemental Figure 2. Expression of SP in axillary meristems.
Supplemental Figure 3. The role of SP in meristem doming.
Supplemental Data Set 1. Genes with differential expression between regular and domed meristems.
Supplemental Data Set 2. LTM interacting proteins.
Supplemental Data Set 3. Cladistic analyses of LTM associated genes.
Supplemental File 1. LTM Solyc01g100600 amino acid sequences used for phylogenetic analysis in Figure 3C.
Supplemental File 2. Solyc03g123430 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplemental File 3. Solyc06g074910 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplemental File 4. Solyc07g062810 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplemental File 5. Solyc11g011210 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplemental File 6. Solyc11g071290 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplemental File 7. Solyc12g088940 sequences used for phylogenetic analysis in Supplemental Data Set 3.
Supplementary Material
Acknowledgments
We thank David Weiss, Idan Efroni, and Zach Lippman for their helpful discussions, as well as members of the Eshed laboratory for their valuable input. We also thank Zach Lippman, Bruno Muller, Akiva Shalit, and Eliezer Lifschitz for materials provided and Ziva Amsellem, Sagie Brodsky, Shai Torgeman, and Evyatar Steiner for technical assistance. This work was supported by Israel Science Foundation Research Grant 1788/14 and BARD Grant IS-4818-15 to Y.E. G.F. is the Incumbent of the David and Stacey Cynamon Research Fellow Chair in Genetics and Personalized Medicine. Y.E. is an incumbent of the Mimran Family Professorial Chair.
AUTHOR CONTRIBUTIONS
L.T. and Y.E. designed the research and analyzed data. L.T. performed the research. T.U. and N.S.G. designed and performed the protein coimmunoprecipitation assay. S.G. constructed libraries and performed next-generation sequencing. G.F. did all bioinformatics analyses. L.T. and Y.E. wrote the manuscript with input from all other authors.
Glossary
- SAM
shoot apical meristem
- PSM
primary shoot meristem
- TM
transition meristem
- BiFC
bimolecular fluorescence complementation
References
- Adams, J., Kelso, R., and Cooley, L. (2000). The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10: 17–24. [DOI] [PubMed] [Google Scholar]
- Alvarez J., Guli C.L., Yu X.-H., Smyth D.R. (1992). terminal flower: a gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2: 103–116. [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., Robinson M.D. (2013). Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8: 1765–1786. [DOI] [PubMed] [Google Scholar]
- Asai K., Satoh N., Sasaki H., Satoh H., Nagato Y. (2002). A rice heterochronic mutant, mori1, is defective in the juvenile-adult phase change. Development 129: 265–273. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202– W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P.L., Chien C.-T., Sternglanz R., Fields S. (1993). Using the two- hybrid system to detect protein-protein interactions. In Cellular Interactions in Development: A Practical Approach, Hartley D.A., ed (Oxford, UK: Oxford University Press; ), pp. 153–179. [Google Scholar]
- Battey N.H., Lyndon R.F. (1984). Changes in apical growth and phyllotaxis on flowering and reversion in Impatiens balsamina L. Ann. Bot. (Lond.) 54: 553–567. [Google Scholar]
- Bernier G., Sachs R.M., Kinet J.-M. (1981). The Physiology of Flowering: Transition to Reproductive Growth. (Boca Raton, FL: CRC Press). [Google Scholar]
- Bernier G. (1988). The control of floral evocation and morphogenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 175–219. [Google Scholar]
- Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I. (2013). High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8: 539–554. [DOI] [PubMed] [Google Scholar]
- Chung S.M., Frankman E.L., Tzfira T. (2005). A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 10: 357–361. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Running M.P., Meyerowitz E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397–418. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J.E., Wilson R.G. (1979). The effects of photoperiod and mineral nutrient supply on growth and primordia production at the stem apex of barley seedlings. Ann. Bot. (Lond.) 44: 537–546. [Google Scholar]
- Danilevskaya O.N., Meng X., McGonigle B., Muszynski M.G. (2011). Beyond flowering time: pleiotropic function of the maize flowering hormone florigen. Plant Signal. Behav. 6: 1267–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Audic S., Claverie J.M., Blanc G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitzur T., Nahum H., Borovsky Y., Pekker I., Eshed Y., Paran I. (2009). Co-ordinated regulation of flowering time, plant architecture and growth by FASCICULATE: the pepper orthologue of SELF PRUNING. J. Exp. Bot. 60: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Formstecher E., et al. (2005). Protein interaction mapping: a Drosophila case study. Genome Res. 15: 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks R.G., Wang C., Levin J.Z., Liu Z. (2002). SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129: 253–263. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain J.C., Legrain P. (1997). Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282. [DOI] [PubMed] [Google Scholar]
- Hempel F.D., Feldman L.J. (1994). Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192: 276–286. [Google Scholar]
- Hempel F.D., Zambryski P.C., Feldman L.J. (1998). Photoinduction of flower identity in vegetatively biased primordia. Plant Cell 10: 1663–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Mitsuda N., Matsui K., Ohme-Takagi M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321: 172–178. [DOI] [PubMed] [Google Scholar]
- Horridge J.S., Cockshull K.E. (1979). Size of the Chrysanthemum shoot apex in relation to inflorescence initiation and development. Ann. Bot. 44: 547–556. [Google Scholar]
- Huang N.C., Jane W.N., Chen J., Yu T.S. (2012). Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J. 72: 175–184. [DOI] [PubMed] [Google Scholar]
- Jensen C.S., Salchert K., Nielsen K.K. (2001). A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 125: 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanapoom M.L., Thomas J.F. (1987). Stereological study of ultrastructural changes in the shoot apical meristem of Nicotiana tabacum during floral induction. Am. J. Bot. 74: 152–163. [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Ke J., Ma H., Gu X., Thelen A., Brunzelle J.S., Li J., Xu H.E., Melcher K. (2015). Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors. Sci. Adv. 1: e1500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr E. (1982). Single flower truss ‘sft’appears to be on chromosome 3. Tomato Genetics Cooperative Reports 32: 31. [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova A.V., Logacheva M.D., Dmitriev S.E., Penin A.A. (2015). RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genomics 16: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57: 3405–3414. [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Ayre B.G., Eshed Y. (2014). Florigen and anti-florigen - a systemic mechanism for coordinating growth and termination in flowering plants. Front. Plant Sci. 5: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyndon R.F., Battey N.H. (1985). The growth of the shoot apical meristem during flower initiation. Biol. Plant. 27: 339–349. [Google Scholar]
- McCormick, S. (1997). Transformation of tomato with Agrobacterium tumefaciens. In Plant Tissue Culture Manual, K. Lindsey, ed (Dordrecht, The Netherlands: Springer), pp. 311–319. [Google Scholar]
- McGarry R.C., Prewitt S.F., Culpepper S., Eshed Y., Lifschitz E., Ayre B.G. (2016). Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol. 212: 244–258. [DOI] [PubMed] [Google Scholar]
- Menda N., Semel Y., Peled D., Eshed Y., Zamir D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38: 861–872. [DOI] [PubMed] [Google Scholar]
- Metcalf R., Fernandez A., Williams R. (1975). The genesis of form in bulrush millet (Pennisetum americanum (L.) K. Schum.). Aust. J. Bot. 23: 761–773. [Google Scholar]
- Mimida, N., Goto, K., Kobayashi, Y., Araki, T., Ahn, J.H., Weigel, D., Murata, M., Motoyoshi, F., and Sakamoto, W. (2001). Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336. [DOI] [PubMed]
- Mohamed R., Wang C.T., Ma C., Shevchenko O., Dye S.J., Puzey J.R., Etherington E., Sheng X., Meilan R., Strauss S.H., Brunner A.M. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62: 674–688. [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N., Latorre A., Jamilena M., Lozano R. (2004). SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218: 427–434. [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N., Jamilena M., Zurita S., Gómez P., Capel J., Lozano R. (1999). FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 20: 685–693. [DOI] [PubMed] [Google Scholar]
- Muños S., Ranc N., Botton E., Bérard A., Rolland S., Duffé P., Carretero Y., Le Paslier M.C., Delalande C., Bouzayen M., Brunel D., Causse M. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156: 2244–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122. [DOI] [PubMed] [Google Scholar]
- Nougarède, A. (1967). Experimental Cytology of the Shoot Apical Cells during Vegetative Growth and Flowering. In International Review of Cytology, G.H. Bourne and J.F. Danielli, eds (New York: Academic Press), pp. 203–351. [DOI] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Schatz M.C., Lippman Z.B. (2012). Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 109: 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin P.A., Nilsson O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35: 1742–1755. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Carmel-Goren L., Hareven D., Gutfinger T., Alvarez J., Ganal M., Zamir D., Lifschitz E. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989. [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Vaneechoutte D., Van de Peer Y., Inzé D., Mueller-Roeber B., Vandepoele K. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43: D974–D981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J.C., et al. (2001). The protein-protein interaction map of Helicobacter pylori. Nature 409: 211–215. [DOI] [PubMed] [Google Scholar]
- Santner A., Estelle M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506. [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.P., Galbraith D.W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60: 241–257. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Steeves T.A., Hicks M.A., Naylor J.M., Rennie P. (1969). Analytical studies on the shoot apex of Helianthus annuus. Can. J. Bot. 47: 1367–1375. [Google Scholar]
- Steiner E., Livne S., Kobinson-Katz T., Tal L., Pri-Tal O., Mosquna A., Tarkowská D., Mueller B., Tarkowski P., Weiss D. (2016). The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171: 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F., Yuan Z., Hake S., Jackson D. (2001). The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Tsuji H., Matsumoto A., Fujita A., Shimatani Z., Terada R., Sakamoto T., Kurata T., Shimamoto K. (2015). FT-like proteins induce transposon silencing in the shoot apex during floral induction in rice. Proc. Natl. Acad. Sci. USA 112: E901–E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.F., Kanchanapoom M.L. (1991). Shoot meristem activity during floral transition in Glycine max (L.) Merr. Bot. Gaz. 152: 139–147. [Google Scholar]
- Vojtek A.B., Hollenberg S.M. (1995). Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255: 331–342. [DOI] [PubMed] [Google Scholar]
- Wojcik J., Boneca I.G., Legrain P. (2002). Prediction, assessment and validation of protein interaction maps in bacteria. J. Mol. Biol. 323: 763–770. [DOI] [PubMed] [Google Scholar]
- Xu C., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47: 784–792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.