Abstract
Bovine serum albumin (BSA) is widely used as an additive in polymerase chain reaction (PCR)-based microfluidic devices to passivate reactors and alleviate nucleic-acid amplification. BSA is available commercially in two types: either acetylated or non-acetylated. A survey of literature indicates that both types of BSA are used in PCR-based microfluidic devices. Our study results reveal that the use of acetylated BSA in PCR micro-devices leads to differential inhibition of PCR, compared to non-acetylated BSA. This result is noticed for the first time, and the differential inhibition generally goes un-noticed, as compared to complete PCR inhibition.
I. INTRODUCTION
Miniaturization of polymerase chain reaction (PCR)-based biochip have the advantages of reduced consumption of nucleic-acid template and reagents, reduced PCR cycling time by efficient heat transfer to the PCR solution, precise control of thermocycling parameters, and potential towards a high-throughput platform.1 However, the effect of surface on PCR dominates in such platforms leading to PCR inhibition, and hence, it is difficult to perform efficient PCR with uniformity across a high-throughput biochip. Miniaturization leads to an increase in the surface area-to-volume ratio (SVR) of the chamber. In high SVR chambers, the effect related to the non-specific adsorption of PCR components, especially Taq DNA polymerase and/or template DNA to chip surface becomes significant.2 PCR inhibition in microfluidic devices is attributed to the choice of material,3–7 reagents (interaction of specific material and polymerase enzyme source),8 and SVR.2 Material-based PCR inhibition is possible by two mechanisms: either ions/inhibitory components from the material might leach into PCR mixture or adsorption of critical PCR components onto the material surface. For adsorption of PCR components, there are numerous reports of Taq polymerase adsorption in chip fabricated using silicon,4,6 glass-silicon,5 poly-dimethyl siloxane (PDMS),9 and low temperature co-fired ceramic (LTCC).7
To perform PCR efficiently on the microfluidic chips, the surface of the chips requires PCR compatible coating. There are two major approaches to achieve PCR compatible coatings: either by coating the chip surface with materials that restrain non-specific adsorption of proteins9 or pre-treat the channels/chambers with proteins (static coatings). As an alternate strategy, additives such as Bovine Serum Albumin (BSA), polyethylene glycol (PEG), polyvinylpyrrolidone (PVP), and gelatin are added to the PCR solution (dynamic) to prevent adsorption of Taq polymerase onto the biochip substrates and ultimately achieve a PCR compatible coating.10 BSA can be used in various ways such as a carrier protein, stabilizing agent in enzymatic reactions, and surface blocking agent. BSA is included in the PCR mix to reduce the problem of the abstraction of Taq polymerase and possible abstraction of other PCR components on material surface. The addition of BSA may not be required for tube-based (macro-volume) reactions, but it is absolutely required for PCR-based microdevices.5 BSA is widely used as a PCR inhibition alleviating agent in microfluidic devices in both static11–15 and dynamic passivation modes.16–18 Also, there are many reports, where combinations of static and dynamic surface passivation using BSA have been applied to PCR micro-devices.4,19–25 It is known that surface adsorption of BSA onto cured PDMS renders the PDMS surface hydrophilic (contact angle of water changes from 109° to 53°),26 and such passivation is suitable for capillary-driven microfluidics.27 However, static passivation leads to extra step during chip fabrication, and hence dynamic passivation is preferred over static passivation. In addition, efficiency of static coating depends on the pH of BSA solution and possible functional groups on the material surface, and this might require optimization based on the selected chip material. For example, Qin et al. showed efficient coating of BSA in the pH 4.0 solution, when compared to pH 7.0 in a chip fabricated using Norland Optical Adhesive 81 (NOA81).28
BSA is available commercially in two types: either acetylated or non-acetylated. The two types of BSA with different quality (molecular biology, histochemical, and cell culture grade) are available from different vendors (Sigma Aldrich, NEB, Ambion, Affymetrix, Fisher Scientific, and Wako pure chemicals). The acetylation process used to remove nucleases during BSA preparation changes the BSA's binding characteristics. The selection of BSA type and PCR optimization based on the material surface and SVR is not clearly defined. However, there are four well documented reports on the inhibitory effect of common microfluidic materials on PCR.8,10,29,30 Potrich et al. reported an interesting observation that polymerase differentially adsorbs on plasma-enhanced chemical vapor deposition-silicon oxide (PECVD-SO) and reactive-ion-etched silicon oxide (RIE-SO) surfaces in a polymerase source dependent manner.8 Christensen et al. showed that PCR inhibition due to Taq polymerase adsorption on silicon, glass, and SU-8 surface can be alleviated by pretreatment with BSA.10 Kolari et al. spiked real time PCR solution (in tubes) with various micro-electro mechanical system (MEMS) materials such as oxidized silicon, fused silica, pyrex, PDMS, and poly-methyl-methacrylate (PMMA) to show that PCR inhibition was due to abstraction of polymerase enzyme, and the addition of BSA was insignificant in alleviating PCR inhibition.29 Kodzius et al. performed a systematic and thorough study of PCR inhibition by at least 20 materials.30 The authors used a pre-PCR incubation strategy to study the inhibitory effects of material on PCR due to polymerase adsorption, and reported a list of PCR-friendly materials. For these materials, the addition of BSA did not significantly improve the PCR yield. With the exception of SU-8, all other materials that showed slight or strong inhibition were alleviated by the addition of BSA. In the case of SU-8, the addition of BSA inhibited the PCR. Although the four publications provided a wealth of knowledge about PCR inhibition mechanism with respect to various materials, it is hard to reproduce these works as the BSA type and the vendor were not reported. Also, from these works, it is hard to conclude, if the differential inhibition of PCR is due to the effect of material surface/SVR or possible use of acetylated BSA? Hence, we evaluated the BSA types (acetylated and non-acetylated) with an aim to clearly define the BSA type requirement for use in PCR microfluidic devices.
II. MATERIALS AND METHODS
A. SYBR Green-based PCR
A BNI-1 gene fragment (189 bp) of SARS cDNA was PCR amplified in the PDMS-glass hybrid chip. Based on optimized conditions, the PCR mixture contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, and 0.1% Triton X-100, 0.2 mM each of dATP, dCTP, dTTP and dGTP (PCR Nucleotide Mix, Promega, Madison, P/N C1141), 3 mM MgCl2 (Invitrogen by Life Technologies, Carlsbad, P/N 10966), 0.3 μM each of forward and reverse gene specific primer (1st BASE, Singapore), 0.1 U/μl of hot-start Platinum Taq DNA polymerase (Invitrogen by Life Technologies, Carlsbad, P/N 10966), various concentration and types of bovine serum albumin (BSA) (Sigma Aldrich, P/N B 8667; Promega P/N R3961), 1× SYBR Green I (Cambrex Biosciences, Maine, USA), and 0.1 ng/μl (3 × 107 copies) of plasmid pGEM-3Z DNA template.
B. TaqMan probe-based PCR
A TaqMan-based PCR is performed in a polypropylene (PP) tube on the Rotorgene 3000 real-time PCR system. The PCR solution (20 μl) contained 1× LuminoCt qPCR ReadyMix (Sigma Aldrich, Singapore), 1× ROX internal reference dye (Sigma Aldrich, Singapore), TaqMan probe for the nuc gene at 250 nM, forward and reverse primers at 900 nM each, 1 μg/μl of either acetylated BSA (from two vendors: Ambion AM2614 and Promega P/N R3961) or non-acetylated BSA (Sigma-Aldrich P/N B8667), and 1 ng/μl of genomic DNA isolated from Staphylococcus aureus subsp. aureus ATCC 33591 (MRSA, strain 328; ATCC). The LuminoCt qPCR ReadyMix contains optimized concentrations of Tris-HCl (pH 8.3), KCl, dNTPs, stabilizers, MgCl2, and Jumpstart Taq DNA polymerase.
C. SYBR Green-based PCR in acid washed glass capillary and PDMS-glass microfluidic devices
Glass capillaries were cleaned by soaking them in the piranha solution (acid) overnight. The piranha solution is a volatile mixture of 70% H2SO4 and 30% H2O2. This washing step removed all organic impurities, which may interfere with DNA amplification. Following the piranha solution treatment, the capillaries were extensively washed (3×) with ultra-pure water. For details on fabrication and performance of PCR in capillary-driven PDMS-glass microfluidic devices, the readers are directed to our earlier publication.16
III. RESULTS AND DISCUSSION
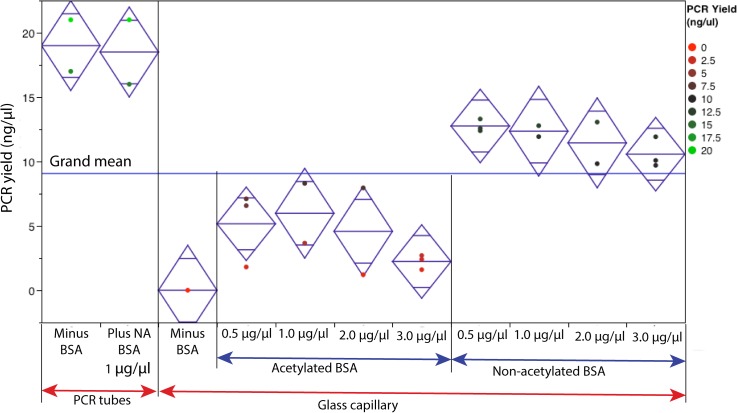
Experiments were performed to understand the effect of SVR (with and without BSA) on PCR yield. Both acetylated and non-acetylated BSA were tested at various concentrations (0.5 to 3.0 μg/μl). In order to have a proper control on the SVR, PCR was performed in a borosilicate glass capillary (acid-washed). A 32 mm long glass capillary (Drummond Scientific Company, USA), which can hold 1 μl of PCR solution was used for this experiment. The SVR for this glass capillary is estimated to be 20 mm−1. The SYBR-green based PCR without BSA failed in glass capillary with 20 mm−1 SVR (Fig. 1). As a control reaction, macro-volume PCR was performed in the widely used polypropylene (PP) tube. The SVR for PP tube is ∼1.5 mm−1.31 The yield of PCR in the PP tube is ∼19 ng/μl. With the addition of acetylated BSA at 0.5 μg/μl, PCR was successful in glass capillary (SVR: 20 mm−1), but the yield was ∼4 lower compared to the PCR yield in the PP tube (SVR: 1.5 mm−1). Increasing the concentration of acetylated BSA (up to 3.0 μg/μl) did not improve the yield significantly. The addition of non-acetylated BSA at 0.5 μg/μl drastically improved the PCR yield from glass capillary. The PCR yield from reactions with 0.5 μg/μl non-acetylated BSA is only ∼1.5 lower compared to the PCR yield (without BSA) in the PP tube. The addition of 1 μg/μl of non-acetylated BSA to reaction in the PP tube did not improve the PCR yield. Hence, BSA is required for successful PCR in microstructures with 20 mm−1 SVR and may not be required when performing PCR in PP tubes with low SVR. Based on the result from Fig. 1, it is concluded that acetylated BSA differentially inhibits SYBR-green based PCR. In literature, there are reports of PCR inhibition by BSA in a concentration dependent manner.4,5,28,32 The optimal BSA concentration depend on many parameters such as SVR, material, enzyme source, and our results from this work indicate that it also depends on the type of BSA. Yu et al. reported PCR inhibition by BSA additive in a Tween 20 treated PDMS-glass device.32 However, they attributed this inhibition to deleterious interaction of Tween 20 and BSA, and failed to report the type of BSA used in this study.
FIG. 1.
Effect of BSA type and concentration on the PCR yield. The PCR was performed in a glass capillary and compared to the macro-volume PCR in a polypropylene tubes. Diamond illustrates a sample mean (center line) and 95% confidence interval (height).
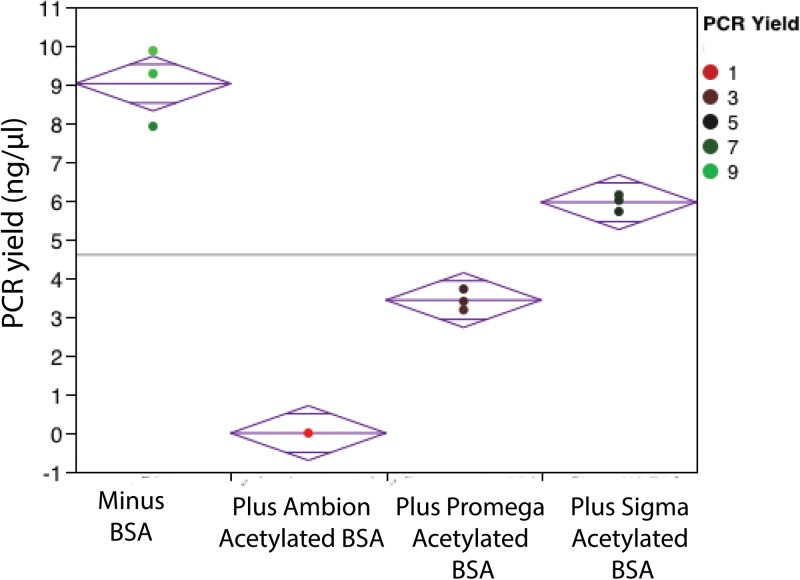
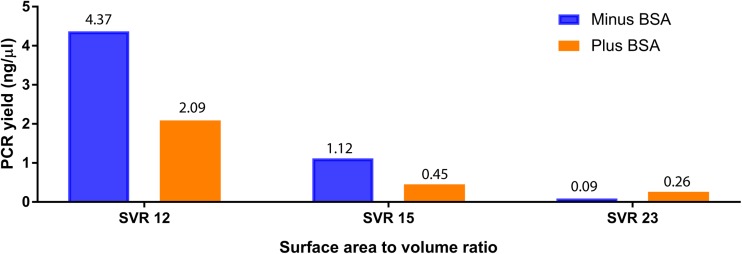
Further experiment was performed to understand if acetylated BSA showed a similar effect on TaqMan-based PCR in polypropylene tubes? For macro-volume PCR (1.5 mm−1), the acetylated BSA from different vendors showed a differential effect on the PCR yield (Fig. 2). The acetylated BSA from Ambion (AM2614) completely inhibited TaqMan-based PCR, whereas acetylated BSA from Promega (P/N R3961) partially inhibited TaqMan-based PCR (∼2.6 lower compared to the PCR yield without BSA). Again, non-acetylated BSA showed PCR yield higher than the yield of PCR with acetylated BSA. It is also noted that for macro-volume PCR with 1 μg/μl non-acetylated BSA, the yield is ∼1.5 lower compared to the PCR yield (without BSA) from the PP tube. We also evaluated the benefit of non-acetylated BSA (1 μg/μl) addition in PCR performed in capillary-driven microfluidic chips fabricated with chambers (PDMS-glass) at ∼12 mm−1, ∼15 mm−1, and ∼23 mm−1 SVR. For chambers with low SVR (∼12 mm−1, ∼15 mm−1), the addition of non-acetylated BSA did not improve the PCR yield, but at ∼23 mm−1, the addition of BSA alleviated the amplification in the PDMS-glass-based PCR chamber (Fig. 3).
FIG. 2.
Effect of BSA type on the TaqMan-based PCR yield. Acetylated BSA from Ambion and Promega, non-acetylated BSA from Sigma were tested at 1 μg/μl concentration in polypropylene tubes. Diamond illustrates a sample mean (center line) and 95% confidence interval (height).
FIG. 3.
Effect of the surface area to volume ratio (SVR) on PCR (with non-acetylated BSA) yield.
In literature, many researchers have used acetylated BSA for both micro- and pico-liter volume PCR.33–36 Results from this study indicate that the use of non-acetylated BSA might have improved the PCR yield in such studies. McKeown et al. reported complete inhibition of PCR (end-point mode) in tubes due to acetylated BSA.37 Our results differ from the results reported by McKeown et al. The surface-to-volume (SVR) of PCR tubes (generally polypropylene PP) is very low (1.5 mm−1), when compared to PCR chambers in microdevices. In microdevices, the effect of surface on PCR is dominant, when compared to reactions in a PP tube. Even at very low concentration of 0.113 μg/μl acetylated BSA, the authors reported complete inhibition of PCR. Complete inhibition is easy to detect experimentally (presence or absence of desired bands in agarose gels). In contrast, we report differential inhibition of acetylated BSA at different concentrations (0.5, 1, 2, and 3 μg/μl). This differential inhibition generally goes un-noticed. McKeown et al. tested different manufacturing batches of BSA from GIBCO (one vendor), whereas we showed that acetylated BSA from different vendors shows differential inhibition (Ambion vs Promega). Finally, in contrast to McKeown et al., we report differential inhibition in the real-time PCR mode (both intercalating and TaqMan probe based).
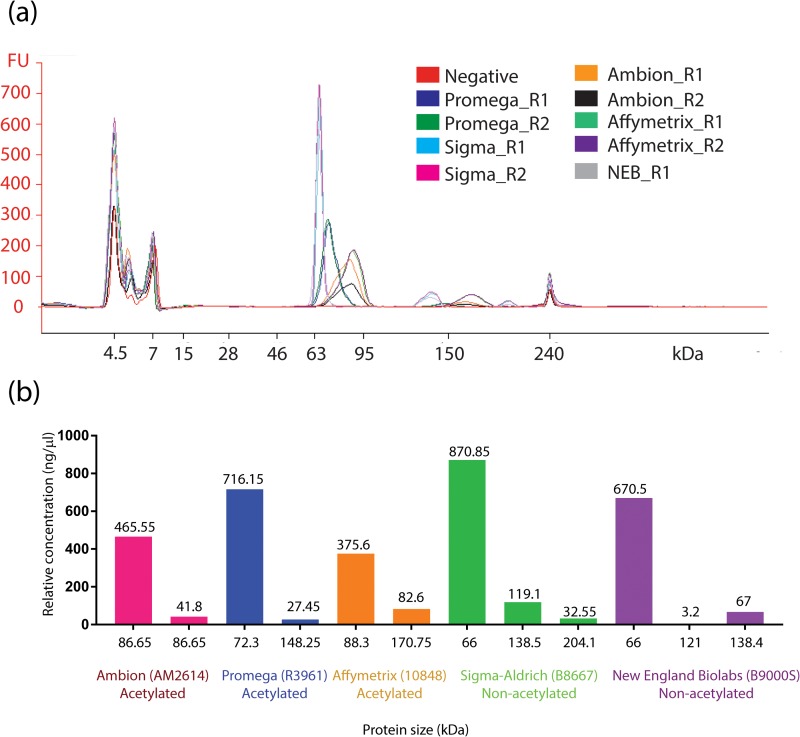
Lastly, we established a quantitative and qualitative method to understand the purity (qualitative) and acetylated BSA (quantitative) components of BSA from different vendors. We evaluated acetylated BSA from Ambion (AM2614), Promega (R3961), Affymetrix (10848), and non-acetylated BSA from Sigma-Aldrich (B8667), NEB (B9000S). The BSA from different vendors was diluted to 1 μg/μl using 1× DPBS (GIBCO 14190–144), and purity was determined by using Protein 230 kit series II on 2100 Bioanalyzer instrument (Agilent Technologies). Fig. 4(a) shows electropherogram of different BSA protein types. Non-acetylated BSA showed a sharp band ∼66 KDa, which is close to the molecular weight value (66 KDa) reported in literature,38 whereas Ac-BSA oligos runs at approximately 120–260 KDa.39 Acetylated BSA (ac-BSA) from both Ambion and Promega showed diffused band with varying degree of modifications. Note that acetylated BSA from Ambion shows a more diffused band compared to acetylated BSA from Promega. Based on our estimation (Fig. 4(b)), the ac-BSA component is higher in Ambion (∼8% of total) when compared to Promega (∼3.7% of total). The higher percentage of ac-BSA component in Ambion (AM2614) possibly contributes to complete inhibition of TaqMan-based PCR (Fig. 2), when compared to partial inhibition of Promega (P/N R3961).
FIG. 4.
(a) Electropherogram of BSA proteins tested using Protein 230 chip on 2100 Bioanalyzer instrument. R1 and R2 are replicates. (b) Relative concentration of acetylated and non-acetylated components estimated from the lab-on-a-chip capillary electrophoresis of BSA proteins.
IV. CONCLUSIONS
Based on this study, it is highly recommended to use non-acetylated BSA in microfluidic devices. Partial inhibition due to acetylated BSA generally goes un-noticed in PCR microfluidic devices. In order to compare performance across PCR-based microfluidic devices, the authors should reveal the vendor name and part-number of the BSA used in their study. A quick review of literature indicates that most authors indicate the vendor's name. But, there are different types of BSA from the same vendor. This work calls for guidelines for performing PCR in microdevices, similar to Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines.40 Although MIQE guidelines include reporting additives used in PCR, there are other parameters such surface area-to-volume ratio (SVR), additive types, reaction volume, ramp rates, amplification speed, template and amplicon length, and limit of detection (LOD), which will be more useful in evaluating PCR-based microfluidic devices, when analysed as a set of parameters.
References
- 1. Zhang C. and Xing D., Nucl. Acids Res. 35(13), 4223–4237 (2007). 10.1093/nar/gkm389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krishnan M., Burke D. T., and Burns M. A., Anal. Chem. 76(22), 6588–6593 (2004). 10.1021/ac0488356 [DOI] [PubMed] [Google Scholar]
- 3. Shoffner M. A., Cheng J., Hvichia G. E., Kricka L. J., and Wilding P., Nucl. Acids Res. 24(2), 375–379 (1996). 10.1093/nar/24.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor T. B., Winn-Deen E. S., Picozza E., Woudenberg T. M., and Albin M., Nucl. Acids Res. 25(15), 3164–3168 (1997). 10.1093/nar/25.15.3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ivan Erill S. C., Erill N., Barbé J., and Aguiló J., Sens. Actuators B: Chem. 96(3), 685–692 (2003). 10.1016/S0925-4005(03)00522-7 [DOI] [Google Scholar]
- 6. Wang W., Wang H. B., Li Z. X., and Guo Z. Y., J. Biomed. Mater. Res. A 77(1), 28–34 (2006). 10.1002/jbm.a.30627 [DOI] [PubMed] [Google Scholar]
- 7. Besecker J., Cornell K. A., and Hampikian G., Sens. Actuators B: Chem. 176, 118–123 (2013). 10.1016/j.snb.2012.09.110 [DOI] [Google Scholar]
- 8. Potrich C., Lunelli L., Forti S., Vozzi D., Pasquardini L., Vanzetti L., Panciatichi C., Anderle M., and Pederzolli C., Eur. Biophys. J. 39(6), 979–986 (2010). 10.1007/s00249-009-0466-5 [DOI] [PubMed] [Google Scholar]
- 9. Liu Y., Zhang L., Wu W., Zhao M., and Wang W., Biomicrofluidics 10(2), 024126 (2016). 10.1063/1.4946870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Christensen T. B., Pedersen C. M., Gröndahl K. G., Jensen T. G., Sekulovic A., Bang D. D., and Wolff A., J. Micromech. Microeng. 17(8), 1527 (2007). 10.1088/0960-1317/17/8/015 [DOI] [Google Scholar]
- 11. Burns M. A., Mastrangelo C. H., Sammarco T. S., Man F. P., Webster J. R., Johnsons B. N., Foerster B., Jones D., Fields Y., Kaiser A. R., and Burke D. T., Proc. Natl. Acad. Sci. U. S. A. 93(11), 5556–5561 (1996). 10.1073/pnas.93.11.5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cady N. C., Stelick S., Kunnavakkam M. V., and Batt C. A., Sens. Actuators B: Chem. 107(1), 332–341 (2005). 10.1016/j.snb.2004.10.022 [DOI] [Google Scholar]
- 13. Rodriguez I., Lesaicherre M., Tie Y., Zou Q., Yu C., Singh J., Meng L. T., Uppili S., Li S. F., Gopalakrishnakone P., and Selvanayagam Z. E., Electrophoresis 24(1-2), 172–178 (2003). 10.1002/elps.200390010 [DOI] [PubMed] [Google Scholar]
- 14. Zhi Qiang N., Wen Yuan C., Shi Yi S., Xiao Yu J., and Wei Ping Z., J. Micromech. Microeng. 16(2), 425 (2006). 10.1088/0960-1317/16/2/029 [DOI] [Google Scholar]
- 15. Crabtree H. J., Lauzon J., Morrissey Y. C., Taylor B. J., Liang T., Johnstone R. W., Stickel A. J., Manage D. P., Atrazhev A., Backhouse C. J., and Pilarski L. M., Microfluid. Nanofluid. 13(3), 383–398 (2012). 10.1007/s10404-012-0968-9 [DOI] [Google Scholar]
- 16. Ramalingam N., Liu H.-B., Dai C.-C., Jiang Y., Wang H., Wang Q., M Hui K., and Gong H.-Q., Biomed. Microdevices 11(5), 1007–1020 (2009). 10.1007/s10544-009-9318-4 [DOI] [PubMed] [Google Scholar]
- 17. Friedman N. A. and Meldrum D. R., Anal. Chem. 70(14), 2997–3002 (1998). 10.1021/ac971303n [DOI] [PubMed] [Google Scholar]
- 18. Lagally E., Simpson P. C., and Mathies R. A., Sens. Actuators B 63(3), 138–146 (2000). 10.1016/S0925-4005(00)00350-6 [DOI] [Google Scholar]
- 19. Hong J. W., Fujii T., Seki M., Yamamoto T., and Endo I., Electrophoresis 22(2), 328–333 (2001). [DOI] [PubMed] [Google Scholar]
- 20. Khandurina J., McKnight T. E., Jacobson S. C., Waters L. C., Foote R. S., and Ramsey J. M., Anal. Chem. 72(13), 2995–3000 (2000). 10.1021/ac991471a [DOI] [PubMed] [Google Scholar]
- 21. Matsubara Y. K. K., Kobayashi M., Yamanura S., Morita Y., and Tamiya E., Biosens. Bioelectron. 20, 1482–1490 (2005). 10.1016/j.bios.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 22. Obeid P. J., Christopoulos T. K., Crabtree H. J., and Backhouse C. J., Anal. Chem. 75(2), 288–295 (2003). 10.1021/ac0260239 [DOI] [PubMed] [Google Scholar]
- 23. Schneegass I., Brautigam R., and Kohler J. M., Lab Chip 1(1), 42–49 (2001). 10.1039/B103846J [DOI] [PubMed] [Google Scholar]
- 24. Trau D., Lee T. M., Lao A. I., Lenigk R., Hsing I. M., Ip N. Y., Carles M. C., and Sucher N. J., Anal. Chem. 74(13), 3168–3173 (2002). 10.1021/ac020053u [DOI] [PubMed] [Google Scholar]
- 25. Ranjit Prakash A., Adamia S., Sieben V., Pilarski P., Pilarski L. M., and Backhouse C. J., Sens. Actuators B: Chem. 113(1), 398–409 (2006). 10.1016/j.snb.2005.03.049 [DOI] [Google Scholar]
- 26. Nikcevic I., Bange A., Peterson E. T. K., Papautsky I., Heineman W. R., Halsall H. B., and Seliskar C. J., Proc. SPIE 5718, 159–167 (2005). 10.1117/12.591781 [DOI] [Google Scholar]
- 27. Ramalingam N., Warkiani M. E., Ramalingam N., Keshavarzi G., Hao-Bing L., and Hai-Qing T. G., Biomed Microdevices 18(4), 68 (2016). 10.1007/s10544-016-0099-2 [DOI] [PubMed] [Google Scholar]
- 28. Qin K., Lv X., Xing Q., Li R., and Deng Y., Anal. Methods 8(12), 2584–2591 (2016). 10.1039/C5AY03233D [DOI] [Google Scholar]
- 29. Kolari K., Satokari R., Kataja K., Stenman J., and Hokkanen A., Sens. Actuators B: Chem. 128(2), 442–449 (2008). 10.1016/j.snb.2007.06.034 [DOI] [Google Scholar]
- 30. Kodzius R., Xiao K., Wu J., Yi X., Gong X., Foulds I. G., and Wen W., Sens. Actuators B: Chem. 161(1), 349–358 (2012). 10.1016/j.snb.2011.10.044 [DOI] [Google Scholar]
- 31. Wilding P., Shoffner M. A., Cheng J., Hvichia G., and Kricka L. J., Clin. Chem. 41(9), 1367–1368 (1995). [Google Scholar]
- 32. Yu C., Liang W., Kuan I., Wei C., and Gu W., J. Chin. Inst. Chem. Eng. 38(3–4), 333–339 (2007). 10.1016/j.jcice.2007.05.001 [DOI] [Google Scholar]
- 33. Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L., Jarvie T. P., Jirage K. B., Kim J. B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., and Rothberg J. M., Nature 437(7057), 376–380 (2005) 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang Z., Drijber R. A., Lee D. J., Dwiekat I. M., Harris S. D., and Wedin D. A., Soil Biol. Biochem. 40(4), 956–966 (2008). 10.1016/j.soilbio.2007.11.016 [DOI] [Google Scholar]
- 35. Chen J., Shen C., and Ko Y., Biomed. Microdevices 15(2), 261–278 (2013). 10.1007/s10544-012-9728-6 [DOI] [PubMed] [Google Scholar]
- 36. Phaneuf C., Oh K., Pak N., Saunders D. C., Conrardy C., Landers J., Tong S., and Forest C., Biomed. Microdevices 15(2), 221–231 (2013). 10.1007/s10544-012-9720-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McKeown B. J., Biotechniques 17(2), 246–248 (1994). [PubMed] [Google Scholar]
- 38. Hirayama K., Akashi S., Furuya M., and Fukuhara K., Biochem. Biophys. Res. Commun. 173(2), 639–646 (1990). 10.1016/S0006-291X(05)80083-X [DOI] [PubMed] [Google Scholar]
- 39.See http://www.cytoskeleton.com/acetylated-bsa-aax01 for Chemically Acetylated Bovine Serum Albumin (BSA) Cat. # AACX1 product sheet, 2017.
- 40. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., and Wittwer C. T., Clin. Chem. 55(4), 611–622 (2009) 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]