Abstract
A mutation in gene MPN142 (orf6) was identified in the Mycoplasma pneumoniae cytadherence mutant III-4. MPN142 encodes virulence-specific proteins P90 and P40 (proteins B and C, respectively). Analysis of MPN142 in a cytadhering revertant and complementation using a recombinant wild-type allele confirmed the role of this mutation in the cytadherence defect.
The cell wall-less bacterium Mycoplasma pneumoniae is a significant cause of bronchitis and atypical pneumonia in humans. Early in the course of disease the bacteria establish an intimate association with the host respiratory epithelium (cytadherence), mediated largely by a differentiated polar structure, the attachment organelle. A number of proteins have been identified that are associated with the attachment organelle and involved in cytadherence, some directly but most by means other than a classical receptor-binding role. These accessory proteins are essential to the architecture of the attachment organelle, providing a scaffolding for the localization and/or maturation of other proteins such as the major adhesin, P1. This large transmembrane protein is densely clustered, although not exclusively, at the attachment organelle of wild-type M. pneumoniae cells (9).
Passage of M. pneumoniae in broth culture leads to the spontaneous loss of cytadherence at a high frequency (11). One such cytadherence mutant, designated III-4, lacks the three virulence-specific proteins A, B, and C (72, 85, and 37 kDa, respectively), originally described by comparing virulent and avirulent strains of M. pneumoniae by two-dimensional polyacrylamide gel electrophoresis (PAGE) (6). Proteins B and C appear to correspond to the 90- and 40-kDa products of MPN142 (orf6), respectively. The predicted product of MPN142 is 130 kDa, but antisera raised against recombinant protein fragments derived from regions at the 5′ and 3′ ends of MPN142 detected proteins of 40- and 90-kDa, respectively, in wild-type M. pneumoniae (22), indicating that the gene product undergoes a processing event (14). P90 and P40 are integral membrane proteins located at the attachment organelle in wild-type M. pneumoniae (3) in proximity to P1 (15). In mutant III-4 and the apparently similar noncytadherent strain M5 (13), which also lacks the 90- and 40-kDa proteins, P1 is located at a focus in one or more but not all branched structures (19, 20). Mutants III-4 and M5 and an avirulent high broth-passage strain (6, 12, 16) lack the 90- and 40-kDa proteins detected with antibodies to the MPN142 gene products, suggesting that B and C correspond to the 90- and 40-kDa proteins, respectively. Despite the availability of the M. pneumoniae genome sequence, the identity of protein A or its gene remains unknown.
The genetic defect responsible for the cytadherence phenotype of mutant III-4 has not been previously determined. Here we identify a frameshift mutation in MPN142 of mutant III-4 and demonstrate that a spontaneously arising, cytadhering revertant of III-4 (III-4-R1) restores the MPN142 reading frame. Furthermore, a recombinant wild-type MPN142 was engineered and introduced into mutant III-4, restoring a wild-type phenotype.
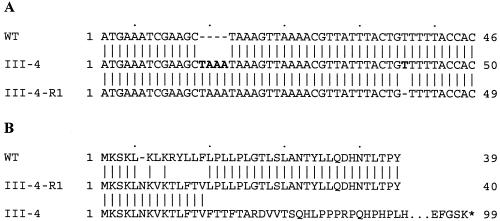
Using primers corresponding to the published genome sequence (7) and genomic DNA from mutant III-4 as templates, regions of MPN142 and flanking sequences were amplified to yield five overlapping PCR fragments. These were cloned into pCRII using a TA cloning kit (Invitrogen, Carlsbad, Calif.) and sequenced for both strands (Integrated Biotech Laboratories, University of Georgia). A direct repeat of the sequence TAAA was identified at nucleotide 14 in the coding sequence of MPN142 in mutant III-4 but not in wild-type M. pneumoniae, causing a frameshift (Fig. 1). This difference from the published sequence was confirmed by repeated amplification and sequencing. The resulting gene encodes a peptide of 98 residues, compared to 1,218 residues for wild-type MPN142.
FIG. 1.
Sequence comparisons of MPN142 of wild-type M. pneumoniae, cytadherence mutant III-4, and revertant III-4-R1. (A) Nucleotide sequence. Nucleotide 1 is the annotated start of the gene. The TAAA acquired in mutant III-4 and the T lost in the reversion of III-4 to III-4-R1 are shown in bold. (B) Deduced amino acid sequence. Dashes show an alignment gap. The ellipsis denotes a break in the representation. The asterisk denotes a translational stop. Note that the order of the strains differs between panels A and B for convenience in alignment.
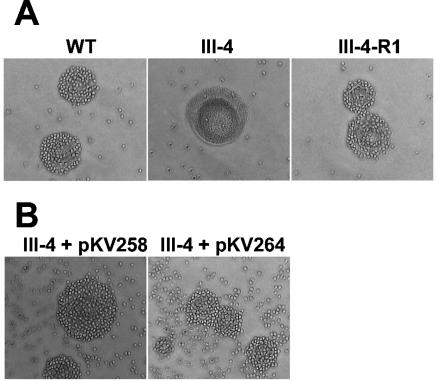
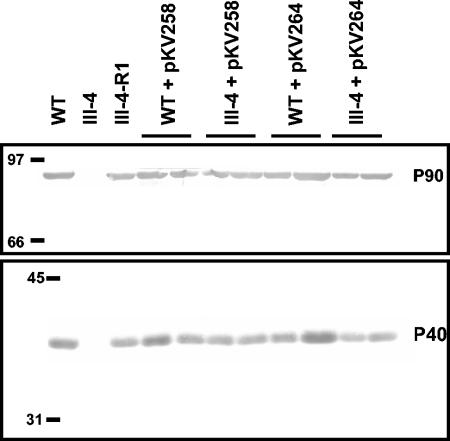
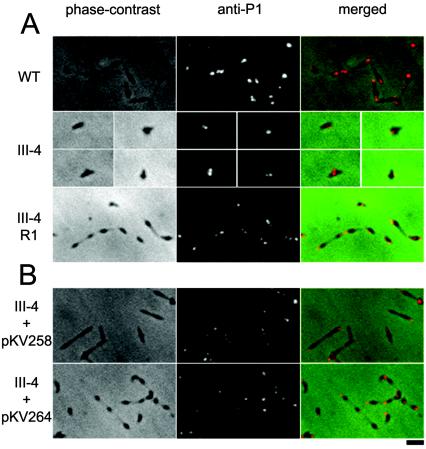
In previous studies the identification of cytadhering revertants of M. pneumoniae adherence mutants required enrichment by attachment to erythrocytes (10) or inert surfaces (18). A cytadhering revertant of mutant III-4 (III-4-R1) was obtained by repeatedly enriching for attachment to plastic. The attachment of erythrocytes to M. pneumoniae colonies (hemadsorption) is a convenient model for cytadherence (21), and III-4-R1 was hemadsorption positive (Fig. 2A) according to the qualitative hemadsorption assay performed as described previously (4) except that sheep blood was used. The MPN142 gene of III-4-R1 was sequenced as described for III-4, revealing that the reading frame was restored by a single deletion in a stretch of five thymine bases starting at nucleotide 37 in the coding sequence of wild-type MPN142. The resulting deduced amino acid sequence differed from the wild type by the nine residues defined by the sites of the original and second-site mutations (Fig. 1). In addition to a hemadsorption-positive phenotype, III-4-R1 produced wild-type levels of P90 and P40 (Fig. 3) and exhibited a wild-type cellular morphology and P1 localization pattern (Fig. 4A) when examined using methods described previously (1).
FIG. 2.
Qualitative hemadsorption screening of M. pneumoniae colonies. (A) Wild-type (WT), mutant (III-4), and revertant (III-4-R1). (B) Transformants (III-4 plus pKV258 or pKV264).
FIG. 3.
Western immunoblot analysis of wild-type (WT), mutant (III-4), revertant (III-4-R1), and transformant M. pneumoniae lysates. Two independent transformants are shown for each transposon-background combination. Forty micrograms of protein was used per lane. Anti-P90 (upper) and anti-P40 (lower) sera were used at a dilution of 1:1,000.
FIG. 4.
Phase-contrast, anti-P1 immunofluorescence, and merged micrographs of M. pneumoniae. (A) Wild-type (WT), mutant (III-4), and revertant (III-4-R1). Due to the poor growth of mutant III-4 and its sparse distribution on the slide, individual cells are shown in separate panels. (B) Mutant III-4 transformed with pKV258 or pKV264. Bar, 2 μm.
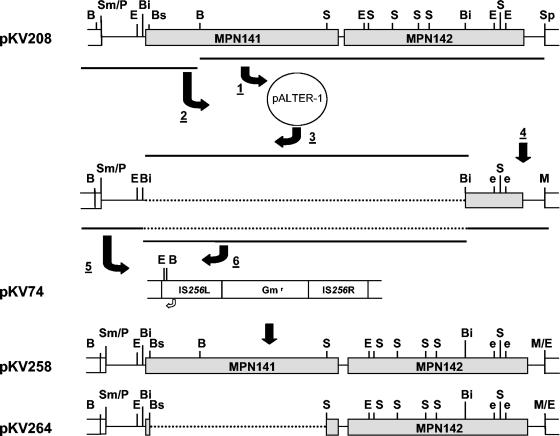
The genetic tools available for M. pneumoniae are limited. Neither allelic exchange nor suitable plasmid vectors have been described, but complementation is possible by recombinant transposon delivery (4). Further, complementation studies with mutant III-4 have been difficult because of the complexity of the operon that includes MPN142. The MPN142 gene lacks an obvious ribosome-binding site, and a frameshift mutation in the upstream MPN141 gene encoding P1 also results in the loss of proteins B and C (23), raising the possibility of translational coupling between P1 and the B/C precursor. Therefore, a construct was engineered with MPN141 and MPN142 in Tn4001mod (5) under the transcriptional control of the Pout promoter (2) of the transposon and with certain restriction sites removed to facilitate future studies. Briefly, a 9.1-kb fragment beginning 345 bp upstream of MPN141 and extending through MPN142 was excised from pCOS_E07 (24) using PmeI and SphI (Fig. 5) and cloned into pGEM-7zf+ (Promega, Madison, Wis.) to yield pKV208. The construction of pKV258 from pKV208 is illustrated in Fig. 5.
FIG. 5.
Creation of pKV258 and pKV264. 1. The 7.5-kb SphI-BamHI fragment from pKV208 was subcloned into the corresponding sites of pALTER-1 (Promega). 2. Into this was cloned the 1.5-kb BamHI fragment spanning the 5′ end of MPN141. 3. A 7.7-kb BsiWI fragment was removed to reduce the size of the plasmid temporarily for efficient silent mutagenesis. 4. The second and third EcoRI sites in MPN142 were silently changed to GGATTC using an AlteredSites II mutagenesis kit (Promega), and the SphI site was replaced with an MfeI site using complementary synthetic linkers (5′-CAATTGTCATG and 5′-ACAATTGCATG). 5. The resulting 1.4-kb BamHI-MfeI fragment was cloned into the BamHI and EcoRI sites of the Tn4001mod-bearing plasmid pKV74 (5). 6. The 7.7-kb BsiWI fragment was then restored to yield pKV258. To create pKV264, the 4.6-kb BsaBI-StuI fragment in MPN141 was removed to create a massive internal in-frame deletion, as described in the text. B, BamHI; Bi, BsiWI; Bs, BsaBI; E, EcoRI; e, destroyed EcoRI site; S, StuI; Sm/P, SmaI/PmeI junction; M/E, MfeI/EcoRI junction at the former SphI site. Open arrow from Tn4001mod in pKV74 shows the location and direction of the Pout promoter. Note that the inserts are inverted when cloned into Tn4001mod. The figure is not created to scale.
In order to engineer a construct that would not introduce a second copy of wild-type MPN141, we created a deletion derivative of pKV258. Briefly, a plasmid containing a 5.7-kb EcoRI fragment bearing MPN141 in pUC19 (a gift from J. Baseman) was digested with BsaBI and StuI and religated. This resulted in an in-frame internal deletion of 95% of MPN141 (88 residues compared to 1,627 in the wild type). The 1.1-kb EcoRI fragment bearing the altered MPN141 was exchanged with the 5.7-kb EcoRI fragment of pKV258 containing MPN141 to yield pKV264, which contains the altered MPN141 and wild-type MPN142 (Fig. 5). We hypothesized that this altered MPN141 would allow the production of MPN142 at wild-type levels if translational coupling were required, while removing potential effects of higher MPN141 gene dosage when transformed into the M. pneumoniae mutant III-4 background.
Plasmids pKV258 and pKV264 were transformed into wild-type M. pneumoniae and mutant III-4. Transformants were isolated and immunoblot and hemadsorption analyses were performed as previously described (4). Several transformants were examined for each transposon-background combination to control for potential variability associated with the site of transposon insertion. Wild-type levels of P90 and P40 were observed in wild-type and mutant III-4 M. pneumoniae transformed with pKV258 or pKV264 when examined by sodium dodecyl sulfate-PAGE and immunoblotting with P90- or P40-specific antisera; as expected, P90 or P40 were not observed in mutant controls (Fig. 3). The transformants attached to plastic (data not shown), were hemadsorption positive (Fig. 2B), and exhibited a wild-type P1 localization pattern (Fig. 4B). Mutant III-4 transformed with pKV258 had a pronounced extended cellular morphology compared to wild-type M. pneumoniae and mutant III-4 transformed with pKV264 (Fig. 4B). This phenotype was consistent in independent transformants, and we speculated it might be due to the increased gene dosage of MPN141 in mutant III-4 with pKV258. However, when an extra copy of MPN141 without MPN142 was added to wild-type M. pneumoniae, transformant cells appeared normal (data not shown), and the explanation and importance of this observation are not known.
Several lines of evidence suggest that the virulence-specific proteins B and C described originally by Hansen et al. (6) correspond to the P90 and P40 protein products of MPN142, but a definitive relationship has not been previously established. Thus, the possibility remained that the cytadherence defect in mutant III-4 for example was not the direct result of the absence of proteins B and C. Furthermore, the inability to identify the gene for protein A has contributed some measure of uncertainty regarding cause and effect. Here we demonstrate conclusively that the genetic defect responsible for the loss of cytadherence in mutant III-4 is a frameshift in the gene MPN142. This defect can be rescued by a second-site mutation that restores the MPN142 reading frame or by complementation with the wild-type MPN142 allele.
Our attempts to determine the identity of the protein A spot, which is seen in wild-type M. pneumoniae only by two-dimensional PAGE using isoelectric focusing in the first dimension (6) and is absent in mutant III-4 (11), were unsuccessful. While the identity of protein A remains unclear, we speculate that its loss may be a secondary consequence of the mutation in MPN142, perhaps in the same manner that loss of cytadherence-associated protein HMW2 is accompanied by reduced steady-state levels of HMW1, HMW3 (17), and P65 (8).
Finally, the construction of pKV258 and pKV264 and the ability to express their recombinant proteins in and complement a mutant background provide opportunities for more-detailed studies into the nature of proteins B, C, and P1. For example, recombinant derivatives of each can be examined in wild-type and mutant M. pneumoniae for structure-function analyses of these proteins, their interrelationships, and the mechanisms of their processing and maturation.
Acknowledgments
We thank Richard Herrmann for the kind gift of P90- and P40-specific antisera in addition to pCOS_E07. The plasmid containing MPN141 was a gift from Joel Baseman. Kyungok Lee provided invaluable technical assistance in the construction of pKV258.
This work was supported by the Public Health Service research grant AI23362 from the National Institute of Allergy and Infectious Diseases (D.C.K.) and a National Science Foundation Research Training Grant in Prokaryotic Diversity (NSF BIR9413235) (R.H.W.).
REFERENCES
- 1.Balish, M. F., S. M. Ross, M. Fisseha, and D. C. Krause. 2003. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol. Microbiol. 50:1507-1516. [DOI] [PubMed] [Google Scholar]
- 2.Fisseha, M., H. W. Gohlmann, R. Herrmann, and D. C. Krause. 1999. Identification and complementation of frameshift mutations associated with loss of cytadherence in Mycoplasma pneumoniae. J. Bacteriol. 181:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franzoso, G., P. C. Hu, G. A. Meloni, and M. F. Barile. 1993. The immunodominant 90-kilodalton protein is localized on the terminal tip structure of Mycoplasma pneumoniae. Infect. Immun. 61:1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hahn, T. W., K. A. Krebes, and D. C. Krause. 1996. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol. Microbiol. 19:1085-1093. [DOI] [PubMed] [Google Scholar]
- 5.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen, E. J., R. M. Wilson, and J. B. Baseman. 1979. Two-dimensional gel electrophoretic comparison of proteins from virulent and avirulent strains of Mycoplasma pneumoniae. Infect. Immun. 24:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan, J. L., K. M. Berry, M. F. Balish, and D. C. Krause. 2001. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J. Bacteriol. 183:7387-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 10.Krause, D. C., D. K. Leith, and J. B. Baseman. 1983. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect. Immun. 39:830-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause, D. C., D. K. Leith, R. M. Wilson, and J. B. Baseman. 1982. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 35:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kufuor, N. K., C. Huang, M. F. Barile, and P. Hu. 1994. Molecular basis for the absence of the 40kDa and 90kDa proteins in the avirulent Mycoplasma pneumoniae strain M129-B169. IOM Lett. 3:673. [Google Scholar]
- 13.Layh-Schmitt, G., and M. Harkenthal. 1999. The 40- and 90-kDa membrane proteins (ORF6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesin) association with the Triton shell. FEMS Microbiol. Lett. 174:143-149. [DOI] [PubMed] [Google Scholar]
- 14.Layh-Schmitt, G., and R. Herrmann. 1992. Localization and biochemical characterization of the ORF6 gene product of the Mycoplasma pneumoniae P1 operon. Infect. Immun. 60:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layh-Schmitt, G., and R. Herrmann. 1994. Spatial arrangement of gene products of the P1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 62:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipman, R. P., W. A. J. Clyde, and F. W. Denny. 1969. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 100:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popham, P. L., T. W. Hahn, K. A. Krebes, and D. C. Krause. 1997. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc. Natl. Acad. Sci. USA 94:13979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobeslavsky, O., B. Prescott, and R. M. Chanock. 1968. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J. Bacteriol. 96:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperker, B., P. Hu, and R. Herrmann. 1991. Identification of gene products of the P1 operon of Mycoplasma pneumoniae. Mol. Microbiol. 5:299-306. [DOI] [PubMed] [Google Scholar]
- 23.Su, C. J., A. Chavoya, and J. B. Baseman. 1989. Spontaneous mutation results in loss of the cytadhesin (P1) of Mycoplasma pneumoniae. Infect. Immun. 57:3237-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenzel, R., and R. Herrmann. 1989. Cloning of the complete Mycoplasma pneumoniae genome. Nucleic Acids Res. 17:7029-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]