Abstract
Several prognostic indicators have shown inconsistencies in patients of different genders with lung adenocarcinoma, indicating that these variations may be due to the different genetic background of males and females with lung adenocarcinoma. In this study, we first used the Gene-Cloud of Biotechnology Information (GCBI) bioinformatics platform to identify differentially expressed genes (DEGs) that eliminated gender differences between lung adenocarcinoma and normal lung tissues. Then, we screened out that transcription factor 21 (TCF21) is a hub gene among these DEGs by creating a gene co-expression network on the GCBI platform. Furthermore, we used the comprehensive survival analysis platforms Kaplan-Meier plotter and PrognoScan to assess the prognostic value of TCF21 expression in lung adenocarcinoma patients. Finally, we concluded that decreased mRNA expression of TCF21 is a predictor for poor prognosis in patients with lung adenocarcinoma.
Introduction
Lung cancer is a malignant tumor characterized by uncontrolled cell growth in tissues of the lung and bronchus. Except for prostate cancer in males and breast cancer in females, lung cancer is the second most commonly diagnosed cancer in men and women1. It is the most common cause of cancer death in humans and accounts for more than one-quarter of all cancer deaths1. Moreover, the overall age-adjusted 5-year relative survival for lung cancer is 15.5% in males and 20.3% in females2. Therefore, further researches are needed to develop treatments for lung cancer, including the identification of prognostic factors for lung cancer patients.
Non-small cell lung cancer (NSCLC) accounts for 85% of total lung cancer cases3. According to the latest statistics from the SEER Cancer Statistics Review, from 2006 to 2012, the average 5-year relative survival rate of NSCLC in males is 19.2% and in females is 26.8%4. The major histological phenotypes of NSCLC are classified as adenocarcinoma, squamous cell carcinoma and large cell carcinoma. However, lung adenocarcinoma (approximately 50% of cases) is the most important pathological type of NSCLC5. In recent years, many prognostic factors for the survival of lung adenocarcinoma patients have been reported by researchers6–12. However, some of these indicators have inconsistent prognostic value in patients with lung adenocarcinoma according to different gender13–17. We hypothesized that one of the causes of this inconsistency is due to the different genetic background of lung adenocarcinoma in males and females.
In this study, we used the Gene-Cloud of Biotechnology Information (GCBI) bioinformatics platform to identify differentially expressed genes (DEGs) that eliminated gender differences between lung adenocarcinoma and normal lung tissues. Among these DEGs, we found that transcription factor 21 (TCF21) is a hub gene. Furthermore, using the comprehensive survival analysis platforms Kaplan-Meier plotter and PrognoScan, we showed that the decreased mRNA expression of TCF21 is an unfavorable prognostic factor for lung adenocarcinoma patients.
Results
Major sample characteristics
We used two Gene Expression Omnibus (GEO) datasets (GSE4079118 and GSE1007219) in this study. Both of these datasets were included in the GCBI bioinformatics analysis platform. GSE40791 included 94 lung adenocarcinoma frozen tissues (53 derived from males, 41 derived from females) and 100 non-neoplastic lung samples (58 derived from males, 42 derived from females) (Supplementary Table S1). GSE10072 included 58 tumor samples (35 derived from males, 23 derived from females) and 49 non-tumor tissues (34 derived from males, 15 derived from females) (Supplementary Table S2).
Research design
In our study, we wanted to identify the DEGs that eliminated gender differences between lung adenocarcinoma and normal lung tissues. Therefore, we first divided both lung adenocarcinoma and non-neoplastic lung samples into two groups according to gender. Using the GCBI bioinformatics analysis platform, we then obtained two DEG datasets from males and females. Then, by selecting the intersection of these two datasets, we obtained the co-expressed DEGs that eliminated gender differences. Next, we created a gene co-expression network to screen out hub genes in the co-expressed DEGs. Finally, we verified the prognostic value of core gene expression for patients with lung adenocarcinoma using the comprehensive survival analysis platforms Kaplan-Meier plotter and PrognoScan. The flow diagram of our study design is shown in Fig. 1.
Figure 1.
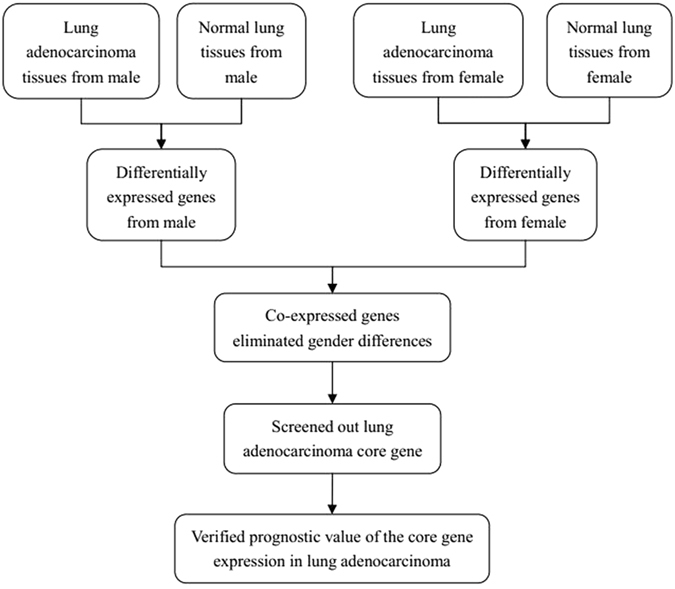
Flow diagram of the study design.
TCF21 is the core gene in the DEGs obtained from GSE40791
We identified 321 potential DEGs from males (Fig. 2a and Supplementary Table S3) and 463 potential DEGs from females (Fig. 2b and Supplementary Table S4). To remove duplicate expressions from the same gene and expressions from unannotated genes, 223 DEGs (Supplementary Table S5) and 330 DEGs (Supplementary Table S6) were selected from the male and female groups. By taking the intersection of these two DEGs datasets, 211 co-expressed DEGs that eliminated gender differences were found (Fig. 2c). Among them, 131 DEGs were incorporated into the gene co-expression network (Fig. 3a). As shown in Table 1 and Fig. 3b, we found that TCF21 had the most gene connections in the gene co-expression network. Therefore, we hypothesized that TCF21 is the core gene in the DEGs between lung adenocarcinoma and normal lung tissues.
Figure 2.
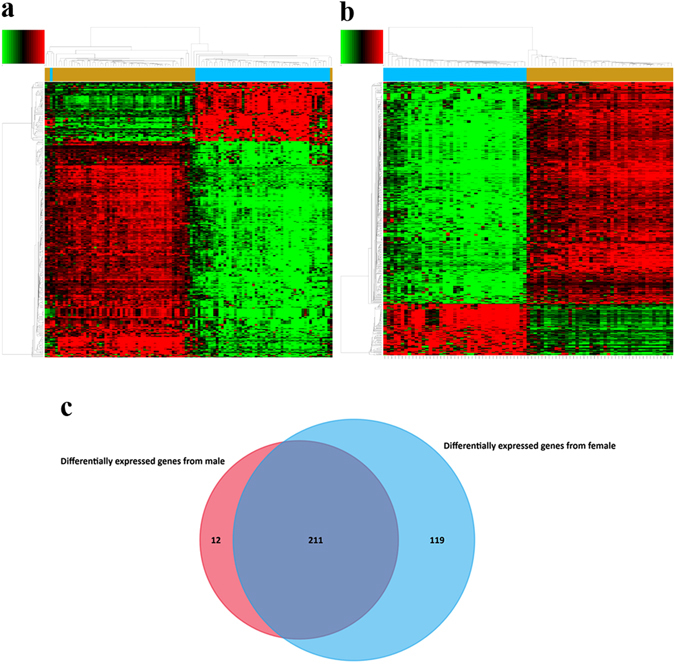
DEGs between lung adenocarcinoma and normal lung tissues for GSE40791. (a) Heat map for potential DEGs (n = 321) from males (lung adenocarcinoma n = 53, in blue; normal lung tissues n = 58, in yellow). (b) Heat map for potential DEGs (n = 463) from females (lung adenocarcinoma n = 41, in blue; normal lung tissues n = 42, in yellow). (c) Duplicate expressions with the same gene symbol and those without a specific gene symbol were removed; the Venn diagram shows DEGs (n = 223) from males, DEGs (n = 330) from females and the co-expressed genes (n = 211) that eliminated gender differences.
Figure 3.
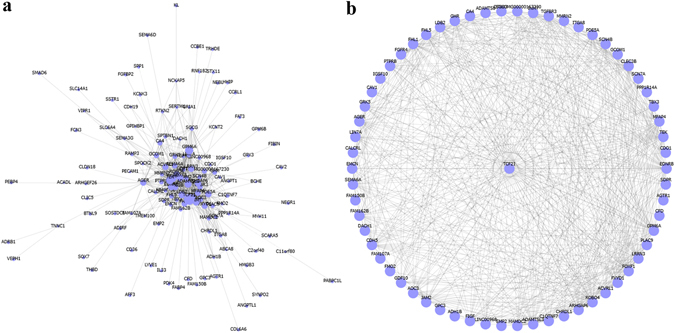
Gene co-expression network of the DEGs that eliminated gender differences for GSE40791. (a) The gene co-expression network. (b) Sub-graph shows that TCF21 has the most gene connections in the gene co-expression network.
Table 1.
Top 10 hub genes in the gene co-expression network of the DEGs that eliminated gender differences for GSE40791.
| Gene Symbol | Degree | Description |
|---|---|---|
| TCF21 | 62 | transcription factor 21 |
| GPM6A | 49 | glycoprotein M6A |
| TEK | 46 | TEK tyrosine kinase, endothelial |
| ADAMTSL3 | 40 | ADAMTS-like 3 |
| FOXF1 | 39 | forkhead box F1 |
| CDH5 | 38 | cadherin 5, type 2 (vascular endothelium) |
| LDB2 | 38 | LIM domain binding 2 |
| EDNRB | 37 | endothelin receptor type B |
| ARHGAP6 | 36 | Rho GTPase activating protein 6 |
| ROBO4 | 35 | roundabout, axon guidance receptor, homolog 4 (Drosophila) |
TCF21 is also the core gene in the DEGs obtained from GSE10072
To verify the results obtained from GSE40791, we then used the same process in GCBI to study the DEGs between lung adenocarcinoma and normal lung tissues from GSE10072. We identified 76 potential DEGs from males (Fig. 4a and Supplementary Table S7) and 87 potential DEGs from females (Fig. 4b and Supplementary Table S8). To remove duplicate expressions from the same gene and expressions from unannotated genes, 52 DEGs (Supplementary Table S9) and 65 DEGs (Supplementary Table S10) were selected from the male and female groups, respectively. By taking the intersection of these two DEGs datasets, 46 co-expressed DEGs that eliminated gender differences were found (Fig. 4c). Among them, 16 DEGs were incorporated into the gene co-expression network (Fig. 5a). As shown in Table 2 and Fig. 5b, we again found that TCF21 has the most gene connections in the gene co-expression network. Consequently, we confirmed that TCF21 is the core gene in the DEGs between lung adenocarcinoma and normal lung tissues.
Figure 4.
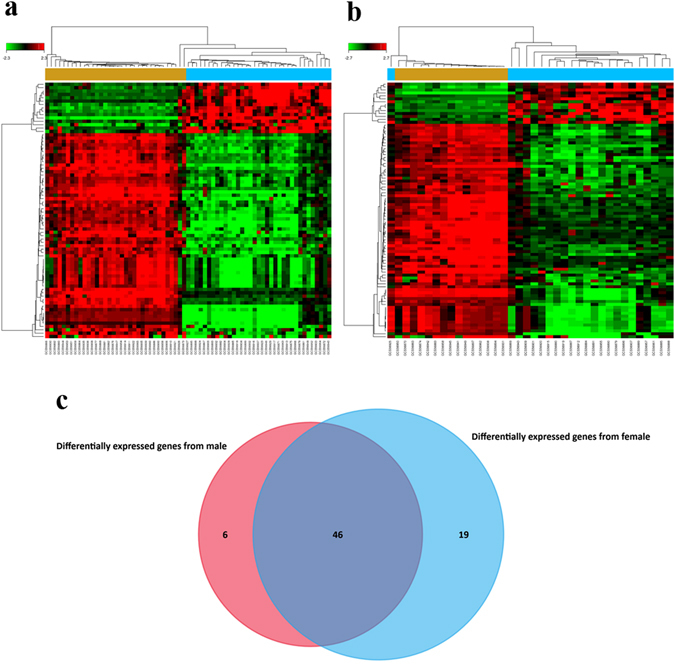
DEGs between lung adenocarcinoma and normal lung tissues for GSE10072. (a) Heat map for potential DEGs (n = 76) from males (lung adenocarcinoma n = 35, in blue; normal lung tissues n = 34, in yellow). (b) Heat map for potential DEGs (n = 87) from females (lung adenocarcinoma n = 23, in blue; normal lung tissues n = 15, in yellow). (c) Duplicate expressions with the same gene symbol and those without a specific gene symbol were removed; the Venn diagram shows DEGs (n = 52) from males, DEGs (n = 65) from females and the co-expressed genes (n = 46) that eliminated gender differences.
Figure 5.
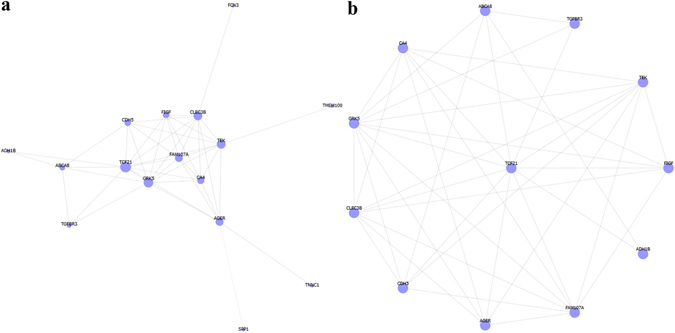
Gene co-expression network of the DEGs that eliminated gender differences for GSE10072. (a) The gene co-expression network. (b) The sub-graph shows that TCF21 has the most gene connections in the gene co-expression network.
Table 2.
Top 10 hub genes in the gene co-expression network of the DEGs that eliminated gender differences for GSE10072.
| Gene Symbol | Degree | Description |
|---|---|---|
| TCF21 | 11 | transcription factor 21 |
| GRK5 | 10 | G protein-coupled receptor kinase 5 |
| CLEC3B | 9 | C-type lectin domain family 3, member B |
| TEK | 9 | TEK tyrosine kinase, endothelial |
| FAM107A | 8 | family with sequence similarity 107, member A |
| AGER | 8 | advanced glycosylation end product-specific receptor |
| CA4 | 7 | carbonic anhydrase IV |
| FIGF | 6 | c-fos induced growth factor (vascular endothelial growth factor D) |
| CDH5 | 6 | cadherin 5, type 2 (vascular endothelium) |
| ABCA8 | 5 | ATP-binding cassette, sub-family A (ABC1), member 8 |
Decreased mRNA expression of TCF21 is an unfavorable prognostic factor for lung adenocarcinoma
Compared with non-neoplastic lung tissues, TCF21 was decreased in the lung adenocarcinoma samples (Supplementary Tables S3–S10). Furthermore, TCF21 is the core gene in the DEGs between lung adenocarcinoma and normal lung tissues (Figs 3 and 5). Accordingly, we hypothesized that TCF21 expression may be a prognostic factor for patients with lung adenocarcinoma. Using the comprehensive Kaplan-Meier survival analysis platform, we discovered that decreased mRNA expression of TCF21 is an unfavorable prognostic factor of overall survival for patients with lung adenocarcinoma (hazard ratio, HR = 0.50; 95% confidence interval, 95% CI: 0.39–0.65; P = 4.7e−08; n = 673) (Fig. 6a). When further considering the gender difference, decreased mRNA expression of TCF21 has similar prognostic value in both males (HR = 0.57; 95% CI: 0.41–0.81; P = 0.0014; n = 328) and females (HR = 0.47; 95% CI: 0.31–0.72; P = 0.00045; n = 287) (Fig. 6b and c). Simultaneously, results generated from PrognoScan based on the GEO dataset of GSE31210 indicated that decreased mRNA expression of TCF21 is not only an unfavorable prognostic factor for lung adenocarcinoma patients’ relapse-free survival (HR = 0.59; 95% CI: 0.46–0.76; P = 0.000032; n = 204) (Fig. 7a) but also for overall survival (HR = 0.71; 95% CI: 0.55–0.91; P = 0.006584; n = 204) (Fig. 7b). However, also using the comprehensive survival analysis platforms Kaplan-Meier plotter and PrognoScan, we found that TCF21 expression has no prognostic value in patients with lung squamous cell carcinoma (data not shown).
Figure 6.
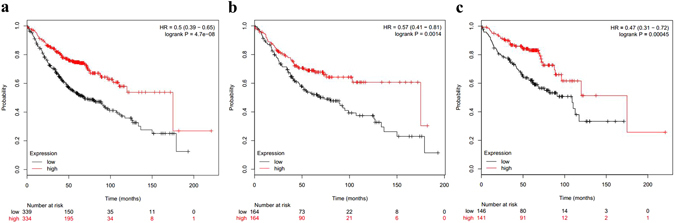
Kaplan–Meier survival curves generated from Kaplan-Meier plotter for TCF21 mRNA expression in patients with lung adenocarcinoma. Overall survival curves for lung adenocarcinoma patients that ignored gender differences (a), in male patients (b) and in female patients (c). HR = hazard ratio.
Figure 7.
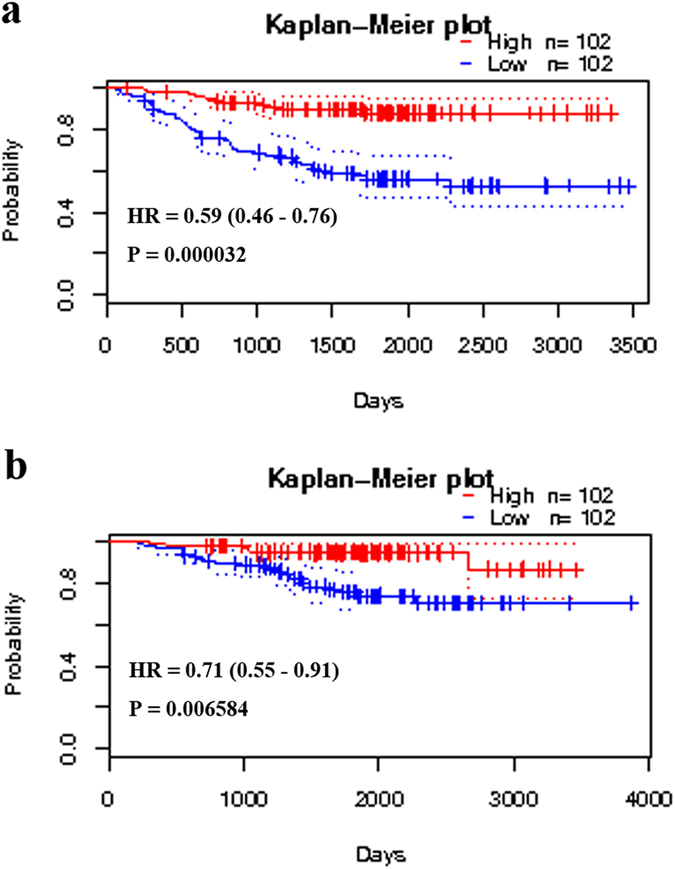
Kaplan–Meier survival curves generated from PrognoScan for TCF21 mRNA expression in patients with lung adenocarcinoma. Relapse-free survival curves (a) or overall survival curves (b) for lung adenocarcinoma patients in the GEO dataset GSE31210. HR = hazard ratio.
Discussion
To assess the prognostic value of gene expression for lung cancer patient survival, the expression of the genes was normally detected by immunohistochemistry in protein level20–22. However, the expression level of mRNA detected by polymerase chain reaction also performed reliable prognostic value for lung cancer patients23, 24. In this study, we extracted gene expression array data from GEO datasets using the GCBI bioinformatics analysis platform and further assessed these data using the comprehensive survival analysis platform Kaplan-Meier plotter and PrognoScan. We finally found that decreased TCF21 mRNA expression is an unfavorable prognostic factor for patients with lung adenocarcinoma. Our study indicated that comprehensive utilization of bioinformatics analyses is a good strategy to study prognosis of lung adenocarcinoma and may be useful to assess prognosis for other cancer types.
Previous studies did not consider the effect of gender differences in DEGs between lung and normal tissues at the design stage. Because of the different genetic background between males and females, we considered that lung adenocarcinomas with different gender origins may also have significant genetic difference. In our study, to eliminate the impact of gender differences, we divided both lung adenocarcinoma and non-neoplastic lung samples into two groups according to gender. Using comprehensive bioinformatics analyses, we reported that the decreased mRNA expression of TCF21 is an unfavorable prognostic factor for patients with lung adenocarcinoma. To our knowledge, this is the first study to investigate the prognostic value of TCF21 in patients with this specific lung cancer type.
TCF21 (also known as Capsulin, Pod-1 or Epicardin) is a member of the basic-helix-loop-helix transcription factor family25–27. During embryogenesis, TCF21 is crucial for the development of a number of cell types in the spleen28, heart29, kidney30 and lung30. However, TCF21 is also important in cancer development. In vitro, it can affect tumor cell cycle balance31 and suppress cancer cell proliferation, migration32 and invasion32, 33. Additionally, mouse model experiments showed that TCF21 can significantly reduce tumor growth in vivo 34, 35. Consequently, TCF21 is considered to be a potential tumor suppressor34, 36.
Unfortunately, TCF21 expression is commonly deregulated by promoter hypermethylation in different types of cancer32, 33, 37–40. This phenomenon is detrimental for cancer patients. Study reported that TCF21 promoter methylation is correlated with decreased survival in patients with metastatic skin melanoma33. Low expression of TCF21 is an independent prognostic factor for poor survival in patients with clear cell renal cell carcinoma41 or gastric cancer42.
For lung cancer, previous studies demonstrated that aberrant TCF21 promoter methylation existed in 9 of 10 lung cancer cell lines39 and the majority of lung cancer tissues (70–81%)34, 38, 39. Richards KL and colleagues38 reported that 84% of non-small cell lung cancer samples showed decreased TCF21 protein expression. TCF21 expression was not correlated with gender and was lower in adenocarcinoma than in squamous cell carcinoma38, indicating that TCF21 is a crucial tumor suppressor for lung adenocarcinoma in both males and females. We corroborated these findings in the present study. Recently, Wu H and colleagues35 found that low TCF21 expression is associated with poor survival for lung cancer using the survival analysis platform Kaplan-Meier plotter. In our study, we comprehensively used both Kaplan-Meier plotter and PrognoScan to first conclude that decreased mRNA expression of TCF21 is an unfavorable prognostic factor in lung adenocarcinoma patients without gender difference and that TCF21 expression showed no prognostic value in patients with lung squamous cell carcinoma. We consider that maybe TCF21 is a specific prognostic factor in lung adenocarcinoma rather than in lung squamous cell carcinoma, for which this point has not taken into account in previous studies.
We know that TCF21 is a crucial tumor suppressor, and TCF21 expression is commonly deregulated by aberrant promoter methylation in different types of cancer. Additionally, decreased expression of TCF21 is a poor prognostic factor for cancer patients. However, we know little about the tumor suppressor mechanism of TCF21. Therefore, in the future, more studies should be conducted to elucidate the mechanisms of TCF21 tumor suppressor function. In addition, the bioinformatics analysis platforms of GCBI, Kaplan-Meier plotter and PrognoScan used in this study were based on the method of unsupervised analysis of gene expression profiles (named the “one-step-clustering” approach by Li J et al.43). Because of the extremely high variability in gene expression profiles between individual tumors and because “passenger signals” may mask the “real” cancer gene signals, our results derived from this “one-step-clustering” approach may be less robust and accurate43, 44.
In summary, we first used the GCBI bioinformatics analysis platform to identify DEGs that eliminated gender differences between lung adenocarcinoma and normal lung tissues, which showed that TCF21 is the hub gene. Then, using the comprehensive survival analysis platforms of Kaplan-Meier plotter and PrognoScan, we concluded that decreased mRNA expression of TCF21 is a poor prognostic factor in patients with lung adenocarcinoma.
Methods
Gene-Cloud of Biotechnology Information (GCBI)
GCBI (Shanghai, China, https://www.gcbi.com.cn) is a platform that combines a variety of research findings, genetic information, sample information, data algorithms and bioinformatics to create a “gene knowledge base,” which encompasses biology, medicine, informatics, computer science, mathematics, graphics and other disciplines. GCBI includes more than 120 million copies of genomic samples, approximately 90,000 copies of tumor samples and more than 17 million copies of genetic information. Therefore, GCBI is a good bioinformatics analysis platform and has provided data analysis support for many studies on cancer research45–49. In this study, we used GCBI to identify DEGs between lung adenocarcinoma and normal lung tissues and finally screened out that TCF21 is the hub gene in the DEG co-expression network. In the Differential Gene Expression Analysis module on the GCBI platform (Supplementary File), we chose a fold expression change >5 at cutoff values Q < 0.05 and P < 0.05 to screen out DEGs. Then, we selected the Gene Co-expression Network module on the GCBI platform to create a gene co-expression network for the DEGs that eliminated gender differences.
Gene Expression Omnibus (GEO) DataSets
GEO DataSets (https://www.ncbi.nlm.nih.gov/gds) is the public repository for storing high throughput gene expression datasets at National Center of Biotechnology Information. In this study, we selected datasets according to the following inclusion criteria: (1) Human lung cancer specimens containing the pathological type of adenocarcinoma; (2) Normal lung tissues used as the controls; (3) Specimens had gender information; (4) Number of samples no less than 100; (5) Expression profiling by array and raw data had the CEL format. First, we used the search terms “Lung cancer [MeSH Terms] AND Homo sapiens [Organism] AND Adenocarcinoma [Description] CEL AND [Supplementary Files] AND Expression profiling by array [DataSet Type]” in the GEO DataSets to identify potential datasets. Then, we further screened these datasets according to the above inclusion criteria. Finally, two GEO datasets, GSE40791 and GSE10072, were included into our study.
Kaplan-Meier plotter
The Kaplan-Meier plotter (http://kmplot.com/analysis/) is a comprehensive online platform that can assess the effect of 54,675 genes on survival based on 10,293 cancer samples. These include 2,437 lung cancer patients with a mean follow-up of 49 months. With a meta-analysis of published microarray datasets, this online tool is suitable for in silico validation of new biomarkers related to survival for patients with non-small cell lung cancer50. We used the Kaplan-Meier plotter to assess the prognostic value of TCF21 expression in patients with lung adenocarcinoma or squamous cell carcinoma.
PrognoScan
PrognoScan (http://www.prognoscan.org/) is a comprehensive platform for evaluating potential tumor biomarkers and therapeutic targets. Based on a large collection of cancer microarray datasets with clinical annotation, PrognoScan is a useful online tool for assessing the association between specific gene expression and prognosis in patients with cancer51. We used the PrognoScan platform to validate the prognostic value of TCF21 expression in patients with lung adenocarcinoma or squamous cell carcinoma.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81572284) and the Important Research and Development Plan of Hunan Provincial Science and Technology Department (Grant No. 2015SK20662). We thank Donglin Cheng, Lang Ma and Qingqing LYU from Gene-Cloud of Biotechnology Information Center for assisting with the data analysis.
Author Contributions
Conceived and designed the study: Q.C. and J.X.; Searched databases: A.B.L. and X.X.L.; Data analyses: W.L., X.C. and J.X.; Preparation of tables and figures: S.Y.H., B.X.H. and J.X.; Wrote and revised the manuscript: J.X. and Q.C.; All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02290-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Eberle A, et al. Lung cancer survival in Germany: A population-based analysis of 132,612 lung cancer patients. Lung Cancer. 2015;90:528–33. doi: 10.1016/j.lungcan.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–94. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howlader, N. et al. SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2013/, based on November 2015 SEER data submission, posted to the SEER web site, April 2016.
- 5.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–46. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoda MA, et al. High circulating activin A level is associated with tumor progression and predicts poor prognosis in lung adenocarcinoma. Oncotarget. 2016;7:13388–99. doi: 10.18632/oncotarget.7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, et al. SIRT1 expression is associated with poor prognosis of lung adenocarcinoma. Onco Targets Ther. 2015;8:977–84. doi: 10.2147/OTT.S82378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P, et al. Long Noncoding RNA RGMB-AS1 Indicates a Poor Prognosis and Modulates Cell Proliferation, Migration and Invasion in Lung Adenocarcinoma. PLoS One. 2016;11:e0150790. doi: 10.1371/journal.pone.0150790. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Thomas A, et al. High mesothelin expression in advanced lung adenocarcinoma is associated with KRAS mutations and a poor prognosis. Oncotarget. 2015;6:11694–703. doi: 10.18632/oncotarget.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, et al. Elevated nuclear CCND1 expression confers an unfavorable prognosis for early stage lung adenocarcinoma patients. Int J Clin Exp Pathol. 2015;8:15887–94. [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng YZ, et al. ROR1 is a novel prognostic biomarker in patients with lung adenocarcinoma. Sci Rep. 2016;6:36447. doi: 10.1038/srep36447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi YX, et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7:85235–85243. doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto A, et al. Loss of TSLC1 expression in lung adenocarcinoma: relationships with histological subtypes, sex and prognostic significance. Cancer Sci. 2005;96:480–6. doi: 10.1111/j.1349-7006.2005.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okano Y, et al. SNP (−617C > A) in ARE-like loci of the NRF2 gene: a new biomarker for prognosis of lung adenocarcinoma in Japanese non-smoking women. PLoS One. 2013;8:e73794. doi: 10.1371/journal.pone.0073794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193–8. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka K, et al. Prognostic significance of aromatase and estrogen receptor beta expression in EGFR wild-type lung adenocarcinoma. Am J Transl Res. 2016;8:81–97. [PMC free article] [PubMed] [Google Scholar]
- 17.Yang CJ, et al. Poorer prognosis in Taiwanese female ever smokers with stage IV lung adenocarcinoma who were readministered a tyrosine kinase inhibitor. Onco Targets Ther. 2016;9:1511–8. doi: 10.2147/OTT.S100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest. 2012;122:4362–74. doi: 10.1172/JCI63084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landi MT, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husni RE, et al. DNMT3a expression pattern and its prognostic value in lung adenocarcinoma. Lung Cancer. 2016;97:59–65. doi: 10.1016/j.lungcan.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Tang Z, et al. Elevated expression of FABP3 and FABP4 cooperatively correlates with poor prognosis in non-small cell lung cancer (NSCLC) Oncotarget. 2016;7:46253–46262. doi: 10.18632/oncotarget.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang G, et al. High ABCG4 Expression Is Associated with Poor Prognosis in Non-Small-Cell Lung Cancer Patients Treated with Cisplatin-Based Chemotherapy. PLoS One. 2015;10:e0135576. doi: 10.1371/journal.pone.0135576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz-Martinez M, et al. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget. 2016;7:51515–51524. doi: 10.18632/oncotarget.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mecklenburg I, Sienel W, Schmid S, Passlick B, Kufer P. A Threshold of Systemic MAGE-A Gene Expression Predicting Survival in Resected Non-Small Cell Lung Cancer. Clin Cancer Res. 2017;23:1213–1219. doi: 10.1158/1078-0432.CCR-16-0557. [DOI] [PubMed] [Google Scholar]
- 25.Hidai H, Bardales R, Goodwin R, Quertermous T, Quertermous EE. Cloning of capsulin, a basic helix-loop-helix factor expressed in progenitor cells of the pericardium and the coronary arteries. Mech Dev. 1998;73:33–43. doi: 10.1016/S0925-4773(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 26.Miller CL, et al. Disease-related growth factor and embryonic signaling pathways modulate an enhancer of TCF21 expression at the 6q23.2 coronary heart disease locus. PLoS Genet. 2013;9:e1003652. doi: 10.1371/journal.pgen.1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quaggin SE, Vanden Heuvel GB, Igarashi P. Pod-1, a mesoderm-specific basic-helix-loop-helix protein expressed in mesenchymal and glomerular epithelial cells in the developing kidney. Mech Dev. 1998;71:37–48. doi: 10.1016/S0925-4773(97)00201-3. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, et al. The basic helix-loop-helix transcription factor capsulin controls spleen organogenesis. Proc Natl Acad Sci USA. 2000;97:9525–30. doi: 10.1073/pnas.97.17.9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acharya A, et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–49. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quaggin SE, et al. The basic-helix-loop-helix protein pod1 is critically important for kidney and lung organogenesis. Development. 1999;126:5771–83. doi: 10.1242/dev.126.24.5771. [DOI] [PubMed] [Google Scholar]
- 31.Franca MM, Ferraz-de-Souza B, Lerario AM, Fragoso MC, Lotfi CF. POD-1/TCF21 Reduces SHP Expression, Affecting LRH-1 Regulation and Cell Cycle Balance in Adrenocortical and Hepatocarcinoma Tumor Cells. Biomed Res Int. 2015;2015:841784–9. doi: 10.1155/2015/841784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Y, et al. Down-regulation of TCF21 by hypermethylation induces cell proliferation, migration and invasion in colorectal cancer. Biochem Biophys Res Commun. 2016;469:430–6. doi: 10.1016/j.bbrc.2015.09.109. [DOI] [PubMed] [Google Scholar]
- 33.Arab K, et al. Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis. 2011;32:1467–73. doi: 10.1093/carcin/bgr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LT, et al. Epigenetic regulation of the tumor suppressor gene TCF21 on 6q23-q24 in lung and head and neck cancer. Proc Natl Acad Sci USA. 2006;103:982–7. doi: 10.1073/pnas.0510171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H, et al. Curcumin increases exosomal TCF21 thus suppressing exosome-induced lung cancer. Oncotarget. 2016;7:87081–87090. doi: 10.18632/oncotarget.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arab K, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55:604–14. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 37.Gooskens SL, et al. TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget. 2015;6:15828–41. doi: 10.18632/oncotarget.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richards KL, et al. Methylation of the candidate biomarker TCF21 is very frequent across a spectrum of early-stage nonsmall cell lung cancers. Cancer. 2011;117:606–17. doi: 10.1002/cncr.25472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shivapurkar N, et al. Differential methylation of a short CpG-rich sequence within exon 1 of TCF21 gene: a promising cancer biomarker assay. Cancer Epidemiol Biomarkers Prev. 2008;17:995–1000. doi: 10.1158/1055-9965.EPI-07-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss D, Stockmann C, Schrodter K, Rudack C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 2013;36:213–24. doi: 10.1007/s13402-013-0129-5. [DOI] [PubMed] [Google Scholar]
- 41.Ye YW, et al. Down-regulation of TCF21 is associated with poor survival in clear cell renal cell carcinoma. Neoplasma. 2012;59:599–605. doi: 10.4149/neo_2012_076. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Li DM, Xie Q, Dai DQ. Protein expression and promoter methylation of the candidate biomarker TCF21 in gastric cancer. J Cancer Res Clin Oncol. 2015;141:211–20. doi: 10.1007/s00432-014-1809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun. 2010;1:34. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013;340:261–9. doi: 10.1016/j.canlet.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 45.Feng A, Tu Z, Yin B. The effect of HMGB1 on the clinicopathological and prognostic features of non-small cell lung cancer. Oncotarget. 2016;7:20507–19. doi: 10.18632/oncotarget.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang YZ, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016;76:2105–14. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 47.Kong F, et al. Hepatitis B virus X protein promotes interleukin-7 receptor expression via NF-kappaB and Notch1 pathway to facilitate proliferation and migration of hepatitis B virus-related hepatoma cells. J Exp Clin Cancer Res. 2016;35:172. doi: 10.1186/s13046-016-0448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G, et al. Simvastatin induces cell cycle arrest and inhibits proliferation of bladder cancer cells via PPARgamma signalling pathway. Sci Rep. 2016;6:35783. doi: 10.1038/srep35783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, et al. MARCKS contributes to stromal cancer-associated fibroblast activation and facilitates ovarian cancer metastasis. Oncotarget. 2016;7:37649–37663. doi: 10.18632/oncotarget.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


