Abstract
The cyclin-dependent kinase inhibitor p27Kip1 (p27) also behaves as a transcriptional repressor. Data showing that the p300/CBP-associated factor (PCAF) acetylates p27 inducing its degradation suggested that PCAF and p27 could collaborate in the regulation of transcription. However, this possibility remained to be explored. We analyzed here the transcriptional programs regulated by PCAF and p27 in the colon cancer cell line HCT116 by chromatin immunoprecipitation sequencing (ChIP-seq). We identified 269 protein-encoding genes that contain both p27 and PCAF binding sites being the majority of these sites different for PCAF and p27. PCAF or p27 knock down revealed that both regulate the expression of these genes, PCAF as an activator and p27 as a repressor. The double knock down of PCAF and p27 strongly reduced their expression indicating that the activating role of PCAF overrides the repressive effect of p27. We also observed that the transcription factor Pax5 interacts with both p27 and PCAF and that the knock down of Pax5 induces the expression of p27/PCAF target genes indicating that it also participates in the transcriptional regulation mediated by p27/PCAF. In summary, we report here a previously unknown mechanism of transcriptional regulation mediated by p27, Pax5 and PCAF.
INTRODUCTION
The protein p27Kip1 (p27) is a member of the Cip/Kip family of cyclin dependent kinase (Cdk) inhibitors that also includes p21Cip1 and p57Kip2 (1). Its classical role is to inhibit the activity of most cyclin–Cdk complexes involved in cell cycle progression. It interacts with both cyclin and Cdk subunits by the KID domain sited on its amino moiety (2). The KID consists of three subdomains: the cyclin-binding subdomain (D1), the Cdk-binding subdomain (D2) and a linker subdomain that joins D1 and D2 (3). Phosphorylation of specific tyrosine residues in the D2 subdomain (Y74 and Y88), by members of the Src family of kinases, allows the partial activation of the Cdk in the trimeric complexes formed by cyclin, Cdk and p27 (4, 5). Thus, p27 behaves as a dual regulator of cyclin–Cdk complexes by inhibiting their activities in the non-tyrosine phosphorylated form and allowing Cdk to be active when phosphorylated at these specific tyrosine residues.
p27 directly interacts with the acetyltransferase and transcriptional co-activator p300/CBP associated factor (PCAF) by a region (aa 91–120) containing a proline rich domain (PRD, aa 91–96) included in the D2 subdomain of p27 (6). PCAF interacts with p27 by its catalytic domain and acetylates p27 at Lysine 100, very close to the PRD. This acetylation induces its ubiquitylation and the subsequent degradation via proteasome (6). Thus, Src kinases and PCAF regulate cell cycle progression by generating post-translational modifications of p27 that lead to the activation of cyclin–Cdk complexes.
p27 also plays a role as a transcriptional regulator (7–9). It associates with a number of gene promoters through E2F4/p130 complexes behaving as a transcriptional repressor of these genes (9). The specific role of p27 on these repressor complexes is to recruit cyclin D2/D3–Cdk4 complexes needed for p130 phosphorylation at early-mid G1 phase (10). This possibility is supported by the fact that the interaction of p27 with E2F4 and p130 is through its carboxyl-moiety (9) indicating that the KID subdomain in the NH2 region will be free to associate with cyclin–Cdk complexes when they appear at early G1. The role of p27 as a transcriptional repressor mediated by E2F4/p130 complexes has been confirmed by a recent work showing that p27 represses transcription of SOX2 through this mechanism (11).
p27 deficiency is associated with tumorigenesis and reduced p27 levels are frequently observed in human cancers in association with tumor aggressiveness and poor clinical outcome (12–14). In most of the cases, low p27 levels are due to post-transcriptional mechanisms that induce its proteasome-dependent degradation (15, 16). Interestingly, the over-expression of p27-target genes (p27-TGs) in human tumors correlates with poor clinical outcome suggesting that the transcriptional regulatory function of p27 plays an important role in tumor development (9). Thus, to understand how p27 regulates transcription is an important goal to be achieved not only to better define the mechanisms involved in cell cycle regulation but also the role of p27 in tumorigenesis.
The evidence that the transcriptional co-activator PCAF acetylates the repressor p27 inducing its degradation (6) led us to investigate whether these two proteins collaborate in the regulation of gene expression. We performed chromatin immunoprecipitation sequencing (ChIP-seq) analysis to identify the putative transcriptional programs regulated by these two proteins. We selected a number of targets common to both proteins and analyzed the mechanism by which p27 and PCAF regulate the transcription of these genes. Interestingly, we found that the transcription factor PAX5 interacts with both p27 and PCAF and also participates in the transcriptional regulation of p27/PCAF target genes.
MATERIALS AND METHODS
Antibodies
Antibodies against p27 (ChIP and IP) (sc-528), PAX-5 (WB) (sc-13146), PAX-5 (WB) (sc-1974), UNC5D (WB) (anti-UNC5H4, sc-135262), PCAF (WB) (sc-13124), SPAG 9 (WB) (anti- JIP-4, sc-134972) and ROBO1 (WB) (sc-25672) were from Santa Cruz. p27 antibody (WB) (610242) was from BD Transduction Laboratories. Antibodies against PCAF (WB) (P7493) and tubulin (WB) (T9026) were from Sigma. Antibodies against PCAF (ChIP/IP) (ab12188) and PAX-5 (ChIP/IP, WB) (ab15164) were from Abcam. GRIN3A antibodies (WB) (MA1-10881) were from Thermo Fisher Scientific and actin antibody (WB) (0869100) was obtained from MP Biomedicals.
Cell cultures
HCT116 cells were cultured in Dulbecco's modified Eagle's medium (DMEM)–HAM F12 (1:1). MCF-7 cells and mice embryonic fibroblasts (MEFs) WT and p27−/− were cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, 1% non-essential amino acids, 1 mM pyruvic acid and 5% penicillin/streptomycin. Cultures were maintained at 37°C and 5% CO2.
Determination of generation time
Generation time was determined in HCT116 Control, p27 and PCAF cells transfected with the CRISPR plasmid as described in CRISPR/Cas9 transfection section. Cells were seeded at a density of 300 000 cells per 60 mm dish to determine their proliferation. We calculated the rate of growth of each cell line by counting the total number of viable cells in duplicate dishes for 24, 48, 72 and 96 h with the Scepter™ 2.0 Cell Counter (Millipore).
Immunoprecipitation
Asynchronously growing HCT116 cells were scraped and washed twice with PBS. Pellets were lysed in 1 ml of immunoprecipitation (IP) buffer (PBS containing 0.5% Triton X-100, 1 mM EDTA, 100 μM sodium orthovanadate, 0.25 mM PMSF, 20 mM β glycerol phosphate, complete protease inhibitor mixture by Roche Applied Science, and 1/25 vol of DNAse I) (Sigma). Cell lysates were incubated for 15 min rotating at RT and 15 min on ice. Then, they were centrifuged at 3000 rpm at 4°C for 5 min. After centrifugation, samples were subsequently quantified using the Lowry method (17). 1 mg of total protein was incubated overnight at 4°C with 2–4 μg of antibodies (PAX5, p27 or PCAF). Proteins and antibody were incubated with protein A-Dynabeads (Invitrogen) for 1 h and the immunocomplexes were extensively washed with IP buffer. Subsequently they were eluted by boiling at 100°C in Laemmli buffer for western blot (WB) analysis. As a control, lysates were incubated with irrelevant rabbit IgG.
Chromatin immunoprecipitation (ChIP)
HCT116 cells were asynchronously grown till achieve an 80% confluence. ChIP assays were then performed as previously described (18). Briefly, cells were lysed and chromatin from cross-linked cells was sonicated. Chromatin was incubated with 5 μg of anti-p27, anti-PCAF or anti-PAX5 antibodies in RIPA buffer (50 mM Tris−HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.1 mM Na3VO4, 0.5 μg/μl aprotinin, 10 μg/μl leupeptin) adding 20 μl of Magna ChIP Protein A magnetic beads (Millipore). Samples were incubated in rotation overnight at 4°C. Beads were washed with low salt buffer, high salt buffer, LiCl buffer and TE buffer. Subsequent elution and purification of the immunoprecipitated DNA-proteins complexes was performed using the IPure kit (Diagenode) according to manufacturer's protocol. Samples were analyzed by qPCR. The association of p27 and PCAF to their respective binding sites of the gene Grin3A, SPAG9, ROBO1 and UNC5D was performed by RT-PCR using the primers listed in Supplementary Table S1.
ChIP–seq
HCT116 cells were grown to confluence and made quiescent by serum starvation during 48 h for doing the ChIP-seq for p27. In contrast, asynchronously growing HCT116 cells were used for the ChIP-seq studies using anti-PCAF. After DNA isolation, ChIP samples, together with the input, were sequenced. Libraries were prepared using the NEBNext® ChIP-Seq Library Prep Reagent Set for Illumina® kit (ref. E6200S) according to the manufacturer's protocol. Briefly, 10 ng of input and ChIP enriched DNA were subjected to end repair, addition of ‘A’ bases to 3΄ ends and ligation of PE adapters. All purification steps were performed using Qiagen PCR purification columns (refs. 50928106 and 50928006). Library size selection was done with 2% low-range agarose gels. Library amplification was performed by PCR on the size selected fragments. Final libraries were analyzed using Agilent DNA 1000 chip to estimate the quantity and check size distribution, and were then quantified by qPCR using the KAPA Library Quantification Kit (ref. KK4835, KapaBiosystems) prior to amplification with Illumina's cBot. Sequencing was done on the HiSeq2000, Single Read, 50 nts.
ChIP-seq data analysis
Illumina pipeline analyzed short reads were uniquely aligned allowing at best two mismatches to the UCSC (The Genome Sequencing Consortium) human genome version hg18, using the program BOWTIE (19). Peak caller algorithm MACS (version 1.4) was used to determine enriched peak region with parameters: –nomodel, –tsize = 46, –bw = 300. Shift size were determined using Pyicos (20) strand correlation method. Enriched peaks were annotated to nearest EnsEMBL (21) gene (EnsEMBL Biomart version 54) using Bioconductor package ChIPpeakAnno (22). Distribution of enriched reads along the genomes and transcription start site (TSS) of RefSeq genes were determined using CEAS (23). Data from ChIP-seq experiments have been deposited in GEO (accession number GSE93975).
De novo motif discovery
To determine the over-represented short sequence motifs in enriched peaks, we used total 100nt sequences (from peak summit, 50 bp down and 50 bp up). We used top 1000 peaks (based on fold change) for finding motifs using both WEEDER (24) and MEME (25) programs.
Functional enrichment analysis
Functional annotation of target genes is based on Gene Ontology (GO) (26) Consortium, 2000; (http://www.geneontology.org) as extracted from EnsEMBL (21) and KEGG pathway database (27). Accordingly, all genes are classified into the ontology categories biological process (GOBP) and pathways when possible. We have taken only the GO/pathway categories that have at least 10 genes annotated. We used Gitools for enrichment analysis and heat map generation (28) (http://www.gitools.org). Resulting P-values were adjusted for multiple testing using the Benjamin and Hochberg's method of False Discovery Rate (29). Enrichment analysis was also performed using the public DAVID tools (Database for Annotation, visualization and integrated discovery v 6.7) (http://david.abcc.ncifcrf.gov) (30).
Putative transcription factor binding
Possible occurrence of transcription factor (TF) motifs in peak regions (150 bp around peak summit), or in target gene promoters were predicted with STORM algorithm (31) with a P-value cutoff determined based on the size of the input sequence as (1/100 × sequence-size), and using position frequency matrices (PFM) from TRANSFAC database (professional version release 2009.4).
Co-occurrence and enrichment of TFs around enriched peaks
Possible occurrence of TF motifs in peak regions (150 bp around peak summit) and overrepresentation of these TFs is based on the tool Pscan (32) with parameter: mixed background hg18, TRANSFAC matrix. Pscan evaluates local enrichment, comparing with a t-test mean and standard deviation of the score of the best matching oligonucleotides in the input regions to mean and standard deviation of the best match in the genomic regions flanking the input ones. Local enrichment can be used to identify motifs with significant preference for binding within the regions, that is, the motif corresponding to the ChIP’ed TF, as well as other TFs likely to interact with it and binding in its neighborhood. Local enrichment P-value (L.PV): describing whether the motif is over- or underrepresented in the 150 bp input regions with respect to the genomic regions flanking them, indicating whether the motif is overrepresented.
Overlap analysis
Overlap of genomic position range data was done using BedTools (33) and significance of overlap was analyzed by Hypergeometric test. However, Venn-diagram generation and overlap of target genes analysis was performed using in-house R programming language script. Significance of overlap calculated based on Chi-square test. Unless stated, peak overlap is defined as overlap by at least 1 bp.
RNA extraction, reverse transcription-PCR and quantitative PCR (qPCR) for gene expression analysis
Total RNA from cells was extracted using High Pure RNA Isolation kit (Roche). cDNA was obtained from 1 μg of RNA using SuperScript VILO cDNA synthesis (Invitrogen) according to manufacturer's instructions. Gene expression was analyzed by real-time PCR, using Express SYBR GreenER qPCR supermix (Invitrogen), corrected by GADPH expression and expressed as relative units. Primer sequences used for qPCR assessment of mRNA levels of target genes are listed in Supplementary Table S2. Primer sequences used for qPCR assessment of the primary non-spliced transcripts of target genes are listed in Supplementary Table S3.
ShRNA lentiviral infection
HCT116 or MCF-7 cells were infected with MISSION shRNA control vector and specific p27 or PCAF shRNA (Sigma Aldrich) with the following sequences:
p27: 5΄-CCGGGTAGGATAAGTGAAATGGATACTCGAGTATCCATTTCACTTATCCTACTTTTTG-3΄
PCAF:5΄-CCGGGCAGACTTACAGCGAGTCTTTCTCGAGAAAGACTCGCTGTAAGTCTGCTTTT-3΄
The protocol for viral particles production and cell infections has been described elsewhere (34). Twenty four hours after infection, cells were selected with 2 mg/ml of Puromycin (Sigma Aldrich) for five days. shRNA-mediated down-regulation was tested by WB with specific antibodies.
CRISPR/Cas9 transfection
HCT116 cells were co-transfected with Control CRISPR/Cas9 (sc-418922), p27 CRISPR/Cas9 KO (sc-400074) or PCAF CRISPR/Cas9 KO (sc-400753) plasmids from Santa Cruz and with pLPCX plasmid (K1061-1, Clontech) using Lipofectamine 2000 transfection reagent (Invitrogen P/N 52887) according to manufacturer's protocol. Transfected cells were subsequently selected with media containing 2 mg/ml of Puromycin (Sigma Aldrich) for 3–5 days. Successful transfection was confirmed by WB with specific antibodies. For the stable generation of a p27KO cell line we used the CRISPR/Cas9 system transfecting HCT116 cells with p27 Double Nickase Plasmid (sc-400074-NIC) and with Control Double Nickase (sc-437281) for the control cell line. Individual GFP positive colonies were isolated by cell sorting (FACSAriaII) and seeded in 96-well plates and grown for 2–3 weeks until the positive colonies reached confluence when they were transferred to 24-well plates. When the latter reached confluence, cells were trypsinized and an aliquot was kept for protein screening by WB and mRNA levels validation. The remaining cells were frozen or expanded in 60 mm dishes for additional analyses.
Expression microarrays
Asynchronously growing HCT116 cells were washed with PBS and RNA was isolated from 3 different sets of cells transfected with shRNA control or shRNAPCAF. Total RNA was extracted using the RNeasy Kit (Qiagen, Iberia, Madrid, Spain). RNA quality control was done with Agilent 2100 bioanalyzer. Samples were then hybridized with Affymetrix Human Gene 1.0 ST arrays. Dataset obtained by Expression Console (Affymetrix) were analyzed by DNA-ChIP Analyzer (dChIP) software and tools. The normalization of the arrays was performed using the robust multi-array average (RMA) method as implemented in the Affymetrix package of R bioconductor. Quality control measurements were obtained with ArrayQualityMetrics, while the statistical analysis of significant hits with Limma (35). Data from expression microarray experiments have been deposited in GEO (accession number GSE93694).
RESULTS
Differential distribution of PCAF and p27 chromatin binding sites in HCT116 cells
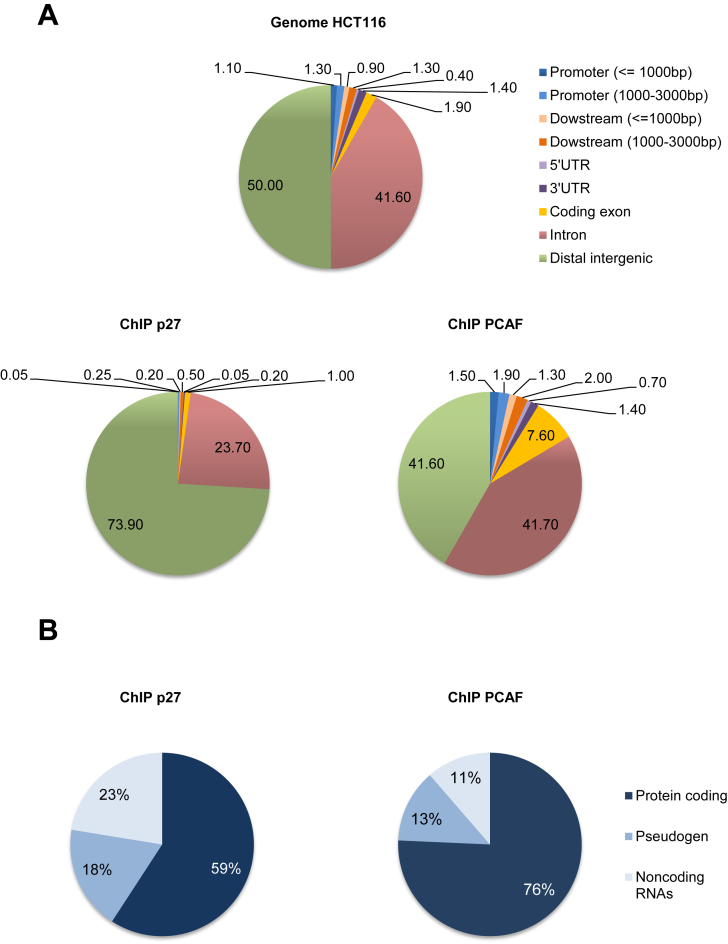
To analyze the transcriptional programs regulated by p27, ChIP-seq experiments were performed in HCT116 cells. Results showed that p27 significantly binds to the chromatin at 1981 sites that corresponded to 1012 genes. Analysis of p27-binding sites (p27-BSs) distribution overall genome indicates that it mostly associates to distal intergenic regions (∼74%) and to intronic sequences (∼24%). In contrast, association with proximal and exonic regions was minimal (Figure 1A). ChIP-seq analyses were also performed to identify the putative PCAF-BSs. Results showed that PCAF associates to the chromatin at 10.878 specific sites that corresponded to 6.131 genes. PCAF mostly associates with distal intergenic (∼42%), intronic (∼42%) and exonic (∼8%) sequences. Interestingly, its association to proximal regions, although low (∼4%), was much higher than p27 (Figure 1A). Because neither p27 nor PCAF interact directly with DNA, their association with chromatin has to be mediated by chromatin associated proteins.
Figure 1.
Distribution of p27- and PCAF-BSs on the chromatin. (A) Distribution of different chromatin regions overall genome of HCT116 cells (upper panel), p27-BSs (Bottom panel, left) and PCAF-BSs (Bottom panel, right) obtained by ChIP-seq experiments. Results are represented as percentage. (B) The different biotypes of genes putatively regulated by p27 (left) or PCAF (right) are represented as percentage.
We also analyzed the distribution of p27- and PCAF-BSs over chromosomes. As shown in Supplementary Figure S1, PCAF-BSs significantly accumulated in chromosomes 8, 10, 16, 17, 19, 20, 21 and 22. Interestingly, the distribution of p27-BSs was quite similar. They accumulated in chromosomes 8, 10, 16, 17, 21 and Y. It worth's noting that the presence of both p27 and PCAF- BSs in chromosome X was extremely reduced. Finally, we also analyzed the biotype profile of the genes potentially regulated by p27 or PCAF. In the case of PCAF the 76% of the genes were encoding for proteins, the 13% for pseudogenes and the remaining 11% corresponded to non-coding RNAs (ncRNAs) (Figure 1B). Intriguingly, the 41% of the genes putatively regulated by p27 corresponded to nc-RNAs (23%) and to pseudogenes (18%) and only the 59% were genes encoding for proteins (Figure 1B). These results indicate that both, p27 and PCAF, but p27 in a higher extension, can play a significant role in the expression of nc-RNAs and pseudogenes.
Gene ontology analysis of p27- BSs and PCAF-BSs
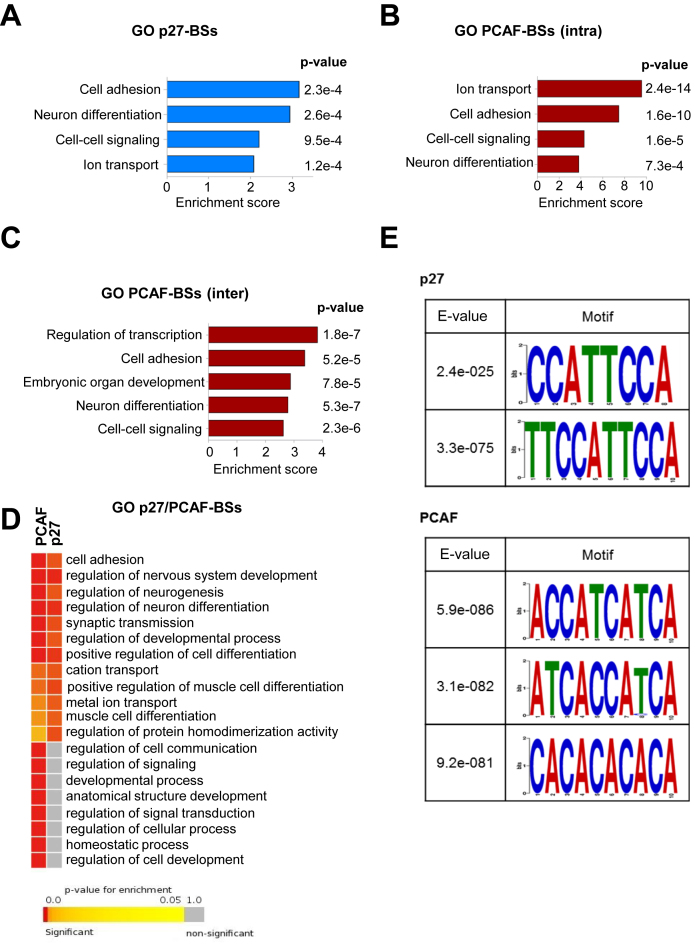
Gene ontology (GO) analysis of the protein encoding genes potentially regulated by p27 was performed using the DAVID program. Results revealed that the four more significant biological processes were: cell adhesion, neuron differentiation, cell-cell signaling and ion transport (Figure 2A). In the case of PCAF, due the high number of peaks, the analyses had to be done in two separated groups. One group contained the genes with PCAF-BSs in intragenic regions and the other included genes with PCAF-BSs in intergenic regions. Interestingly, the four biological processes enriched in p27-target genes (p27-TGs) were also significantly enriched (Figure 2B and C). We observed that around 50% of p27-TGs in these four groups also contained PCAF-BSs (Supplementary Figure S2). These results suggest that p27 and PCAF can collaborate in the transcriptional regulation of genes involved in these cellular functions. To further confirm this data, GO analysis using the Gitools program was additionally performed. Results showed that three of the biological processes (cell adhesion, neuron differentiation and ion transport) shared by p27 and PCAF-TGs were also found to be significantly enriched using this more restrictive program (Figure 2D).
Figure 2.
Gene ontology analysis of protein encoding genes with p27- or PCAF-BSs in their proximity. The Database for Annotation, Visualization and Integrated Discovery (DAVID) program was used to define biological processes enriched in the protein encoding genes putatively regulated by p27 (A), PCAF (intragenic BSs) (B) and PCAF (intergenic BSs) (C). (D) The Gitools program was used to define biological processes enriched in the protein encoding genes putatively regulated by p27 or PCAF. (E) MEME programs were used to identify short sequence motifs enriched in the p27-BSs (upper panel) or PCAF-BSs (bottom panel).
We next aimed to identify DNA consensus sequences that could interact with p27 or with PCAF using the MEME tool. These analyses allowed the identification of two consensus sequences for p27 and three other sequences for PCAF (Figure 2E).
Identification of protein encoding genes containing p27- and PCAF- binding sites
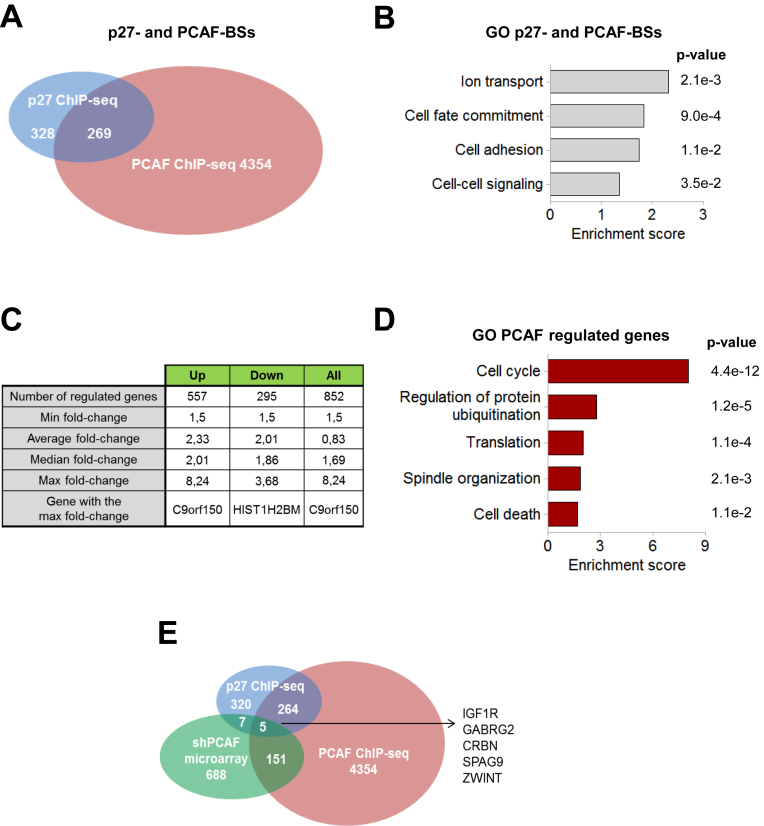
By comparing the p27 and PCAF ChIP-seq data we identified 269 protein encoding genes that contain BSs for both proteins (Figure 3A). These genes represent the 45% of all the protein encoding p27-TGs, a percentage similar to that observed in the four biological processes most significantly regulated by these two proteins according to the GO analysis (Supplementary Figure S2). Interestingly, in most cases, the chromatin region able to associate with PCAF is different to that interacting with p27. GO analysis of these 269 genes revealed that the biological processes ionic transport, cell adhesion, and cell-cell signaling were significantly enriched (Figure 3B).
Figure 3.
Identification of genes putatively regulated by p27 and PCAF. (A) Venn diagram representing the number of genes encoding for proteins identified by ChIP-seq with anti-PCAF antibodies (pink) or with anti-p27 (blue). (B) The biological processes enriched in the group of genes sharing p27 and PCAF-BSs (269 genes) were identified by using the DAVID program. (C) Number of genes differentially expressed in PCAF-KD versus control HCT116 cells identified by expression microarray analysis and main characteristics of the data. (D) The biological processes enriched in the group of genes differentially expressed in PCAF-KD cells versus control were identified using the DAVID program. (E) Venn diagram representing the number of genes encoding for proteins identified by ChIP-seq with anti-PCAF antibodies (pink), with anti-p27 (blue) or differentially expressed in PCAF-KD cells (green).
Expression microarrays in HCT116 cells knocked down for PCAF
HCT116 cells were knocked down (KD) for PCAF using a specific shRNA (Supplementary Figure S3A). Then, expression microarray analyses, in PCAF-KD cells versus control, were performed. Results showed that 852 genes were differentially expressed in these cells being 557 genes up-regulated and 295 down-regulated (Figure 3C). GO analysis revealed that the most significantly enriched group was cell cycle (Figure 3D). Interestingly, GO analysis of down-regulated genes revealed that most of these cell cycle regulatory genes were down-regulated in PCAF-KD cells (Supplementary Figure S3B). These results suggest that PCAF regulates cell cycle progression by controlling the expression of a number of relevant cell cycle regulatory genes. Genes up-regulated are mainly involved in translation and apoptosis (Supplementary Figure S3C).
On comparing ChIP-seq and expression microarrays data we observed that 156 genes from those differentially expressed in the PCAF-KD cells also showed PCAF-BSs suggesting that PCAF participates in the regulation of the expression of these genes by associating with regulatory protein complexes at these specific sites (Figure 3E). The rest of the genes could be genes indirectly regulated by PCAF. GO analysis of these 156 genes showed that a significant number of them are involved in cellular functions related to cell adhesion and neurogenesis (Supplementary Figure S3D). We also observed that 5 of these genes (IGF1R, GABRG2, CRBN, SPAG9 and ZWINT) also contain p27-BSs (Figure 3E).
Role of p27 and PCAF in the regulation of transcription
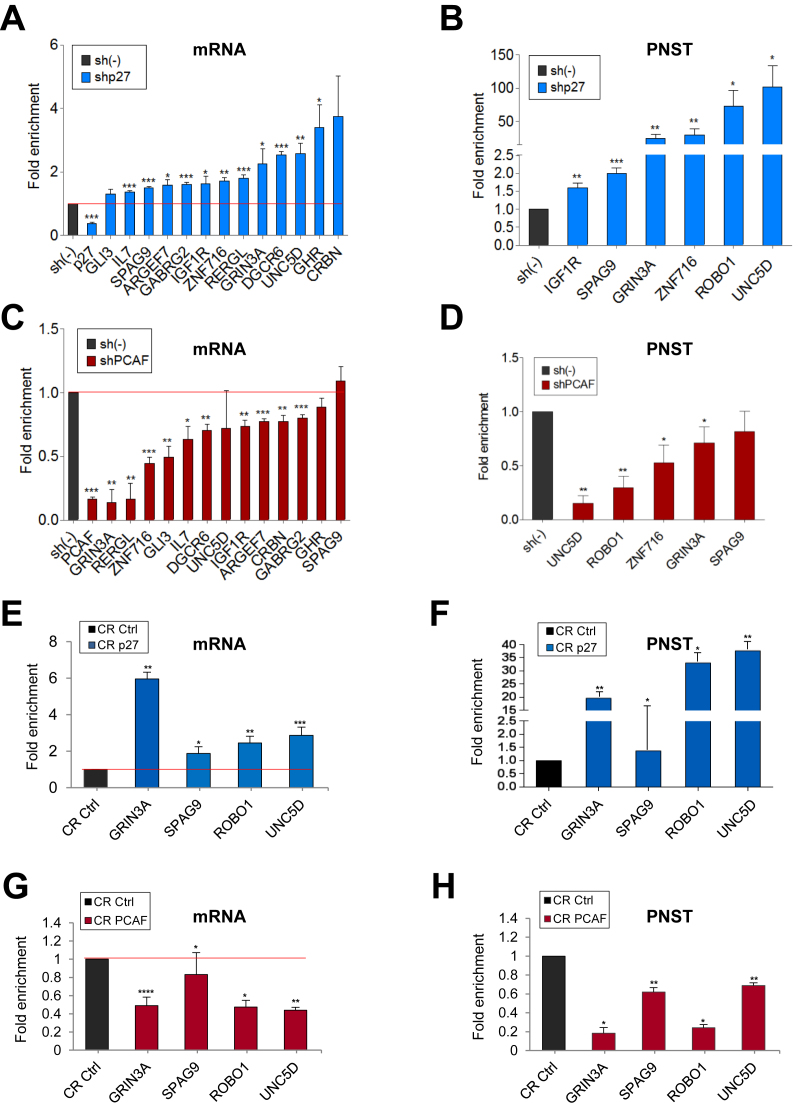
To define the role of p27 and PCAF on the expression of TGs we analyzed the effect of knocking down p27 or/and PCAF on transcription of a number of selected common targets (Table 1) using specific shRNAs (Supplementary Figure S3A). Determinations were performed in asynchronously proliferating cells (mostly in G1) thus minimizing the cell cycle progression differences between p27-KD cells and PCAF-KD cells (both proliferating slightly faster than controls) as observed by determining the generation time (Supplementary Figure S4A and B). We found that in p27-KD cells the mRNA levels of most TGs were increased (Figure 4A). To investigate whether these higher levels of mRNA were due to increased rates of transcription we determined the amount of the primary non-spliced transcripts (PNST) by qPCR. Results revealed that these transcripts were elevated in p27-KD cells indicating that p27 represses transcription of these genes (Figure 4B).
Table 1. List of selected genes that have both p27 and PCAF- binding sites (BSs) in their vicinity. The table includes localization and extension of the p27- and PCAF-BSs and the intensity of the binding (Fold-enrichment).
| ChIP-seq | Cromosome | Start | End | Fold-enrichment | Inside feature | Distance to feature | |
|---|---|---|---|---|---|---|---|
| GABRG2 | p27 | chr5 | 161583970 | 161584252 | 9.91 | Downstream | 156675 |
| PCAF | chr5 | 161580341 | 161580611 | 11.44 | Downstream | 153046 | |
| GHR | p27 | chr5 | 42434475 | 42434749 | 8.49 | Upstream | -25308 |
| PCAF | chr5 | 42267322 | 42267561 | 10.01 | Upstream | -192461 | |
| GRIN3A | p27 | chr9 | 103520403 | 103520670 | 8.49 | Inside | 20280 |
| PCAF | chr9 | 103539425 | 103539691 | 8.58 | Inside | 1258 | |
| IGF1R | p27 | chr15 | 97121406 | 97121648 | 9.91 | Inside | 111104 |
| PCAF | chr15 | 97135969 | 97136244 | 11.86 | Inside | 125667 | |
| PCAF | chr15 | 96961148 | 96961421 | 8.46 | Upstream | -49154 | |
| ROBO1 | p27 | chr3 | 79257543 | 79257776 | 9.83 | Upstream | -163 |
| PCAF | chr3 | 80182644 | 80183002 | 11.44 | Upstream | -460893 | |
| PCAF | chr3 | 80060831 | 80061238 | 10.15 | Upstream | -339080 | |
| UNC5D | p27 | chr8 | 35345752 | 35346062 | 8.49 | Inside | 132907 |
| p27 | chr8 | 35652578 | 35652839 | 8.49 | Inside | 131126 | |
| PCAF | chr8 | 35828412 | 35828659 | 8.58 | Downstream | 306960 | |
| IL7 | p27 | chr8 | 80047436 | 80047703 | 11.32 | Upstream | -167123 |
| p27 | chr8 | 80007304 | 80007574 | 9.91 | Upstream | -126991 | |
| PCAF | chr8 | 79892345 | 79892640 | 7.98 | Upstream | -12032 | |
| ARHGEF7 | p27 | chr13 | 110645835 | 110646168 | 6.97 | Upstream | -9802 |
| PCAF | chr13 | 110492766 | 110493092 | 10.15 | Upstream | -73017 | |
| RERGL | p27 | chr12 | 18151836 | 18152131 | 11.32 | Upstream | -17455 |
| PCAF | chr12 | 17993973 | 17994265 | 12.87 | Downstream | 140408 | |
| SPAG9 | p27 | chr17 | 46415723 | 46416022 | 8.9 | Inside | 63547 |
| PCAF | chr17 | 46406076 | 46406443 | 8.87 | Inside | 73194 | |
| CRBN | p27 | chr3 | 3355488 | 3355749 | 9.64 | Upstream | -159098 |
| p27 | chr3 | 3328299 | 3328565 | 11.32 | Upstream | -131909 | |
| PCAF | chr3 | 3417081 | 3417428 | 11.44 | Upstream | -220691 | |
| GLI3 | p27 | chr7 | 42180561 | 42180853 | 11.32 | Inside | 48859 |
| PCAF | chr7 | 42190975 | 42191217 | 8.12 | Inside | 38445 | |
| ZNF716 | p27 | chr7 | 57555151 | 57555705 | 7.79 | Downstream | 41320 |
| PCAF | chr7 | 57555035 | 57555523 | 7.7 | Downstream | 41204 | |
| DGCR6 | p27 | chr22 | 17263635 | 17264251 | 9.48 | Upstream | -10144 |
| PCAF | chr22 | 17263972 | 17264283 | 8.79 | Upstream | –9807 |
Figure 4.
Effect of knocking down p27 or PCAF on the expression of target genes. (A) HCT116 cells were infected with a specific shRNA for p27 (shp27) or with a control shRNA (sh(–)). Then, mRNA levels of different common target genes (see Table 1) were determined by qPCR. In these cells, the levels of primary non spliced transcripts (PNST) were also determined by qPCR (B). (C) HCT116 cells were infected with a specific shRNA for PCAF (shPCAF) or with a control shRNA (sh(–)) and the levels of mRNA of different common target genes were subsequently determined by qPCR. In these cells, the levels of primary non spliced transcripts (PNST) were also determined by qPCR (D). Levels of mRNA (E) or of primary non spliced transcripts (PNST) (F) from different common target genes were determined in p27 knocked down cells using the CRISPR/Cas9 (CR p27) methodology and in control cells (CR Ctrl). Levels of mRNA (G) or of primary non spliced transcripts (PNST) (H) from different common target genes were determined in PACF knocked down cells using the CRISPR/Cas9 (CR PCAF) methodology and in control cells (CR Ctrl). In all experiments results are the mean value ± SEM of four independent experiments and are represented as fold enrichment versus control. Statistical analyses were performed using the t-student's test. *P < 0.05, **P < 0.01 and ***P < 0.001.
We also found that the mRNA levels of common TGs were significantly lower in PCAF-KD cells than in control cells (Figure 4C) and that this reduction was due to a decreased rate of transcription (Figure 4D). This indicates that PCAF activates transcription of these TGs. To confirm the transcriptional regulatory role of p27 and PCAF we performed similar experiments using the CRISPR/Cas9 methodology. Thus, after knocking out p27 (Supplementary Figure S4A) we observed that the levels of mRNA and primary non-spliced transcripts were increased (Figures 4E and 4F). In contrast in PCAF-KD cells the levels of mRNA and primary non-spliced transcript were found to be decreased (Figure 4G and H). We analyzed the expression of specific target genes for only p27 or PCAF as controls (Supplementary Figure S5A). The protein levels in p27KO or PCAF-KD cells were also determined (Supplementary Figure S5B).
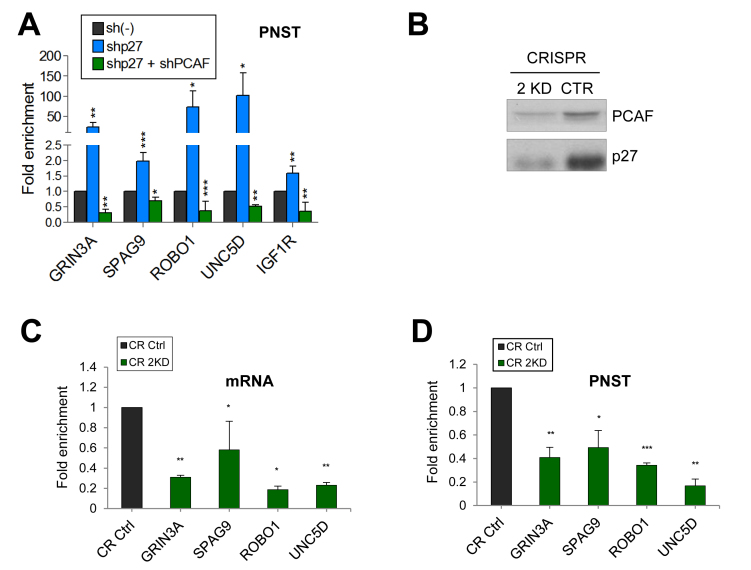
We also investigated the effect of the double KD (2KD) of p27 and PCAF on transcription. In 2KD-cells obtained using shRNAs we observed that the amount of the primary non-spliced transcripts was reduced in all the cases indicating that the co-activator effect of PCAF overrides the repressor role of p27 in all the cases (Figure 5A). These results were corroborated by generating 2KD cells using CRISP/Cas9 (Figure 5B). In these cells the levels of mRNA (Figure 5C) primary non-spliced transcripts (Figure 5D) and protein (Supplementary Figure S5C) were significantly reduced. Additionally, KD experiments using the MCF-7 breast cancer cell line gave similar results: whereas the KD p27 increased transcription of TGs, the KD of PCAF reduced their expression (Supplementary Figure S6).
Figure 5.
Effect of knocking down both, p27 and PCAF on the expression of target genes. (A) HCT116 cells were infected with a specific shRNA for p27 alone, or with shRNAs for p27 and PCAF or with a control shRNA (sh(–)). Then, the levels of primary non spliced transcripts (PNST) were determined by qPCR. (B). Cells were Knocked down for p27 and PCAF (double knockout) (2KD) using the CRISPR/Cas9 methodology. Then, the levels of PCAF and p27 were determined by WB. Levels of mRNA (C) or of primary non spliced transcripts (PNST) (D) from different common target genes were determined in double knock down cells (CR 2KD) and in control cells (CR Ctrl) by qPCR. In all experiments results are the mean value ± SEM of four independent experiments and are represented as fold enrichment versus control. Statistical analyses were performed using the t-student's test. *P < 0.05, **P < 0.01 and ***P < 0.001.
PAX5 collaborates with p27 and PCAF in the regulation of transcription
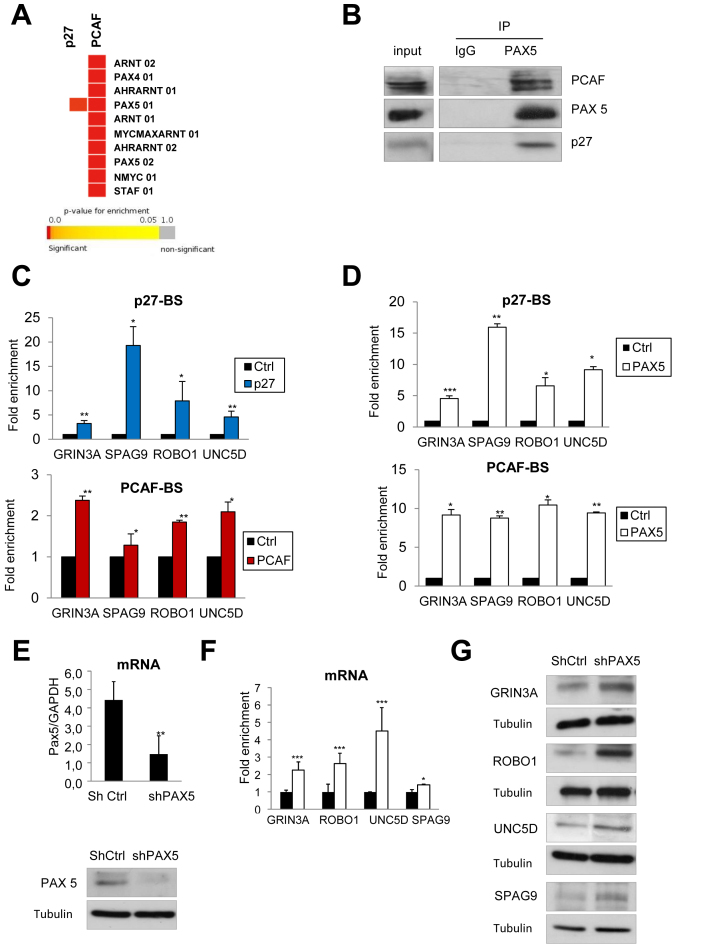
The identification of TFs that could bind to p27- and PCAF-BSs was performed by studying 150 bp regions of these peaks (75 bp up the maximum and 75 bp down) by using the STORM tool for the scanning of the peaks and the TF-BSs motif position weight matrix from TRANSFAC. The ten TFs more significantly identified have been represented in a heat map that compares results from p27 and PCAF (Figure 6A). Interestingly, only PAX5 appears to be able to significantly associate to the p27-BSs. In contrast, a number of different TFs can putatively associate to the PCAF-BSs and among these different candidates PAX5 was one of the most significant (Figure 6A). Interestingly, The consensus sequence for Pax5 (Supplementary Figure S7) is quite similar to the sequences found for p27 and PCAF (Figure 2E and Supplementary Figure S7). Interestingly, the sequence analysis of the chromatin peaks for p27 and PCAF of a number of genes (Table 1) also revealed that these sequences also include regions of high similarity with the PAX5 consensus sequence (data not shown). Hence, we aimed to analyze the putative interaction of PAX5 with both p27 and PCAF by IP. We observed that PAX5 levels are constant along cell cycle (Supplementary Figure S8A). IP experiments using anti-PAX5 were performed in asynchronously growing HCT116 cells. Results revealed that this protein interacts with both p27 and PCAF (Figure 6B). Also reciprocal IPs using anti-p27 and anti-PCAF showed this interaction (Supplementary Figure S8B and C). Interestingly, the absence of PCAF did not affect the interaction between PAX5 and p27 (Supplementary Figure S8D). Similarly, the absence of p27 did not alter the association between PAX5 and PCAF (Supplementary Figure S8E). These results suggest that PAX5 is able to bind to both (p27 and PCAF), most probably through different binding sites because in the absence of p27 the levels of PCAF associated to PAX5 are quite similar to those associated in its presence (Supplementary Figure S8E). The interaction of p27 with its specific binding site in the GRIN3A, SPAG9, ROBO1 and UNC5D genes was validated by ChIP in HCT116 cells (Figure 6C, upper panel). Additionally, the specificity of the interaction was validated by ChIP in HCT116 p27KO cells (Supplementary Figure S9A). Moreover, the interaction of PCAF with its specific binding site in the same genes was also validated by ChIP (Figure 6C bottom panel). Next, ChIP analysis with anti-PAX5 showed the binding of PAX5 to the p27- and PCAF-BSs of the same group of genes (Figure 6D) indicating that PAX5 is able to associate to both p27 and PCAF-BSs. Finally, ChIP experiments performed in HCT116 cells knocked down for PAX5 revealed a significant decreased association of p27 and PCAF to their respective binding sites, suggesting that PAX5 mediates the interaction of both proteins with DNA (Supplementary Figure S9B). Interestingly, we observed that the mRNA and the protein levels of several target genes were increased in PAX5-KD cells (Figure 6E, F and G). All these data indicate that PAX5 collaborates in the transcriptional regulation of specific p27 and PCAF target genes playing a role as a repressor.
Figure 6.
PAX5 collaborates in the regulation of p27- and PCAF-target genes. (A) Enrichment analysis of the putative transcription factors that could significantly associate to p27- or PCAF-BSs was performed using GiTools, the STORM tool for the scanning of the peaks and the TF-BSs motif position weight matrix from TRANSFAC. (B) IP experiments were performed using anti-PAX5 antibodies or IgG as a control. The presence of p27, PAX5 and PCAF in the inputs and in the immunoprecipitates was determined by WB. (C) The interactions of p27 (upper panel) or PCAF (bottom panel) with their specific chromatin binding sites in several target genes were performed by ChIP using anti-p27, anti-PCAF or no-antibodies as a control. Results are represented relative to the control. (D) The interaction of PAX5 with the p27- BSs (upper panel) and PCAF-BSs (bottom panel) of the target genes was analyzed by ChIP using anti-PAX5 antibodies or no-antibodies as a control. Results are represented relative to the control (E) Cells were infected with a specific shRNA for PAX5 or a control shRNA. Then, the levels of PAX5 mRNA (upper panel) and PAX5 protein (bottom panel) were determined by qPCR or WB, respectively. (F) The mRNA levels of different target genes were determined by qPCR in cells infected with a specific shRNA for PAX5 (white bars) or a control shRNA (black bars). Results are represented as a fold change relative to the control. In all cases, results are the mean value ± SD of three independent experiments. Statistical analyses were performed using the t-student's test. *P < 0.05, **P < 0.01 and ***P < 0.001. (G) Protein levels of the different TGs were determined by WB in PAX5-KD cells. Tubulin was used as a loading control.
DISCUSSION
PCAF is a member of the GNAT (GCN5-related N-acetyltransferases) family of acetylases that participates in large multi-subunit complexes involved in transcriptional regulation (36). It also possesses an intrinsic ubiquitin E3 ligase activity that allows specific ubiquitylation of target proteins (37). PCAF behaves as a transcriptional co-activator by acetylating a number of different substrates, the most well-known being histones, although non-histones proteins are also substrates of PCAF (36). These include TFs as c-myc, E2F1, GATA-2 and Myo-D (38–41), other proteins involved in transcription as Cdk9 and some cell cycle regulatory proteins as p27, cdk2 and cyclin A (6,42–44).
The evidence showing that PCAF acetylates p27 inducing its degradation (6) and that p27 behaves as a transcriptional repressor (9,11) suggested the possibility of the cooperation between both proteins in the regulation of transcription. We analyzed this possibility by performing ChIP-seq analysis in HCT116 cells using both anti-PCAF and anti-p27 antibodies. The first interesting observation was that the distribution of the binding sites of both proteins overall genome was very different. Intriguingly, only a small number of p27-BSs were on gene promoters while most of them were in distal intergenic (74%) or in intronic regions (24%). These results expand the previous information of the identification of p27-BSs performed by ChIP on chip in NIH3T3 cells in which only the association of p27 with gene promoters was analyzed (9) and reveals for the first time that the role of p27 in the regulation of transcription is mainly produced by associating to chromatin regions distal from the promoters. In contrast, around the 60% of the PCAF-BSs were on the gene body (intron and exon regions) or in proximal regions.
Another attractive observation was that a significant number of the p27-BSs (40%) were close to pseudogenes or sequences of non-coding RNAs. Similar results were observed for PCAF although the proportion was lower (25%). This observation suggests a putative role of both proteins in the regulation of ncRNA and pseudogene expression. Due to the relevance of these types of RNAs in multiple cellular functions and their involvement in different pathologies, as for instance cancer, these data reveal a putative new role of p27 and PCAF as transcriptional regulators of these RNAs (45–48).
Results reported here also yield up that both p27 and PCAF proteins regulate the expression of similar functional groups of genes as cell adhesion, neuronal differentiation, ion transport and cell signaling. It's worth noting that the 45% of protein-encoding p27-TGs also contain PCAF binding sites indicating that this transcriptional co-activator could be a very important partner of p27 in the regulation of transcription. In fact, we demonstrate here the cooperation between these two proteins in the regulation of transcription of a number of common targets. Specifically, by knocking down p27, PCAF or both we have seen that PCAF operates as a co-activator and p27 as a repressor and that PCAF is needed for the expression of p27-TGs because in the double knock-down cells gene expression is blocked. Curiously, in this cooperative role each one of these proteins associates to different regulatory sites of the chromatin.
The evidence that PCAF acetylates K100 sited in D2 subdomain of p27 and that this acetylation induces p27 ubiquitylation and the subsequent degradation (6) together with results reported here allows us to postulate a model of how p27 and PCAF can cooperate in the regulation of transcription. In this model, p27 associated to specific regulatory domains of the chromatin represses transcription of its target genes. To activate transcription PCAF (associated to a different regulatory domain) interacts with p27, possible by a change in chromatin conformation. Then, PCAF acetylates p27 inducing its degradation. Whether the p27-ubiquitylating enzyme is the own PCAF still remains unknown but this is an attractive possibility. Degradation of p27 would eliminate the repressor role of this protein and would allow PCAF to act as a transcriptional co-activator. That PCAF mediated acetylation occurs on p27 associated to the regulatory chromatin BSs still remains to be demonstrated. However, a similar mechanism has been reported to operate during the activation of the Hedgehog pathway (49). Authors described that cells subjected to a genotoxic stress induce the p53-dependent transcriptional activation and accumulation of PCAF (50). Then, PCAF associates with and acetylates the TF GLI1 that is subsequently ubiquitylated by the intrinsic ubiquitin E3 ligase of PCAF that induced its degradation. As a consequence, GLI1 targets cannot be transcribed.
We also report here that PAX5 participates in the transcriptional regulation mediated by p27 and PCAF. PAX5 belongs to the PAX family of TFs that include 9 members that are involved in the control of development (51). Specifically, PAX5 is essential for commitment of lymphoid progenitors to the B lymphocyte lineage. It activates the expression of B-lineage specific genes but represses a subset of genes not needed for this process of differentiation (52). Interestingly, mice lacking PAX5 in mature B cells develop aggressive lymphomas (53). We report here that PAX5 interacts with both p27 and PCAF, that it associates to p27- and PCAF-BSs and that the knockdown of PAX5 increased transcription of p27/PCAF-TGs indicating that it behaves as a transcriptional repressor of these genes together with p27. An attractive speculation is that PAX5, similarly to p27, could be a PCAF substrate and that PAX5 acetylation could lead to its inactivation as a transcriptional repressor. However, this possibility remains to be explored. In summary, we describe here a novel mechanism of transcriptional regulation mediated by p27, PAX5 and PCAF that might be relevant for the control of several important biological processes and that can participate in the development of different pathological situations as for instance tumorigenesis.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Economía y Competitividad (MINECO) [SAF2012-38078 to O.B. and BFU2015-65311-R to R.A.]; Instituto de Salud Carlos III [RD12/0036/0054 to O.B. and CB16/12/00244 (CIBERONC) to A.B.]; ‘La Caixa’ Foundation 2016-052407 (to R.A.). Funding for open access charge: MInisterio the Economia y Competitividad, Spain La Caixa Foundation. Spain.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sherr C.J., Roberts J.M.. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999; 13:1501–1512. [DOI] [PubMed] [Google Scholar]
- 2. Russo A.A., Jeffrey P.D., Patten A.K., Massagué J., Pavletich N.P.. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996; 382:325–331. [DOI] [PubMed] [Google Scholar]
- 3. Otieno S., Grace C.R., Kriwacki R.W.. The role of the LH subdomain in the function of the Cip/Kip cyclin-dependent kinase regulators. Biophys. J. 2011; 100:2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chu I., Sun J., Arnaout A., Kahn H., Hanna W., Narod S., Sun P., Tan C.-K., Hengst L., Slingerland J.. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007; 128:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grimmler M., Wang Y., Mund T., Cilensek Z., Keidel E.-M., Waddell M.B., Jäkel H., Kullmann M., Kriwacki R.W., Hengst L.. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007; 128:269–280. [DOI] [PubMed] [Google Scholar]
- 6. Pérez-Luna M., Aguasca M., Perearnau A., Serratosa J., Martínez-Balbas M., Jesús Pujol M., Bachs O.. PCAF regulates the stability of the transcriptional regulator and cyclin-dependent kinase inhibitor p27 Kip1. Nucleic Acids Res. 2012; 40:6520–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nguyen L., Besson A., Heng J.I.-T., Schuurmans C., Teboul L., Parras C., Philpott A., Roberts J.M., Guillemot F.. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006; 20:1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Acosta J.C., Ferrándiz N., Bretones G., Torrano V., Blanco R., Richard C., O’Connell B., Sedivy J., Delgado M.D., León J.. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol. Cell. Biol. 2008; 28:7286–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pippa R., Espinosa L., Gundem G., García-Escudero R., Dominguez a, Orlando S., Gallastegui E., Saiz C., Besson a, Pujol M.J. et al. . p27Kip1 represses transcription by direct interaction with p130/E2F4 at the promoters of target genes. Oncogene. 2012; 31:4207–4220. [DOI] [PubMed] [Google Scholar]
- 10. Orlando S., Gallastegui E., Besson A., Abril G., Aligué R., Pujol M.J., Bachs O.. p27Kip1 and p21Cip1 collaborate in the regulation of transcription by recruiting cyclin–Cdk complexes on the promoters of target genes. Nucleic Acids Res. 2015; doi:10.1093/nar/gkv593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H., Collado M., Villasante A., Matheu A., Lynch C.J., Cañamero M., Rizzoti K., Carneiro C., Martínez G., Vidal A. et al. . p27 (Kip1) directly represses Sox2 during embryonic stem cell differentiation. Cell Stem Cell. 2012; 11:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Porter P.L., Malone K.E., Heagerty P.J., Alexander G.M., Gatti L.A., Firpo E.J., Daling J.R., Roberts J.M.. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat. Med. 1997; 3:222–225. [DOI] [PubMed] [Google Scholar]
- 13. Tsihlias J., Kapusta L., Slingerland J.. The prognostic significance of altered cyclin-dependent kinase inhibitors in human cancer. Annu. Rev. Med. 1999; 50:401–423. [DOI] [PubMed] [Google Scholar]
- 14. Chu I.M., Hengst L., Slingerland J.M.. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008; 8:253–267. [DOI] [PubMed] [Google Scholar]
- 15. Pagano M., Tam S.W., Theodoras A.M., Beer-Romero P., Del Sal G., Chau V., Yew P.R., Draetta G.F., Rolfe M.. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995; 269:682–685. [DOI] [PubMed] [Google Scholar]
- 16. Catzavelos C., Bhattacharya N., Ung Y.C., Wilson J.A., Roncari L., Sandhu C., Shaw P., Yeger H., Morava-Protzner I., Kapusta L. et al. . Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer. Nat. Med. 1997; 3:227–230. [DOI] [PubMed] [Google Scholar]
- 17. Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J.. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951; 193:265–275. [PubMed] [Google Scholar]
- 18. Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., Bigas A.. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16537–16542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Althammer S., González-Vallinas J., Ballaré C., Beato M., Eyras E.. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics. 2011; 27:3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hubbard T.J.P., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T. et al. . Ensembl 2007. Nucleic Acids Res. 2007; 35:D610–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shin H., Liu T., Manrai A.K., Liu X.S.. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009; 25:2605–2606. [DOI] [PubMed] [Google Scholar]
- 24. Pavesi G., Mereghetti P., Mauri G., Pesole G.. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004; 32:W199–W203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 26. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. . Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T. et al. . KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008; 36:D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perez-Llamas C., Lopez-Bigas N.. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One. 2011; 6:e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I.. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001; 125:279–284. [DOI] [PubMed] [Google Scholar]
- 30. Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C. et al. . DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007; 35:W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schones D.E., Smith A.D., Zhang M.Q.. Statistical significance of cis-regulatory modules. BMC Bioinformatics. 2007; 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zambelli F., Pesole G., Pavesi G.. PscanChIP: finding over-represented transcription factor-binding site motifs and their correlations in sequences from ChIP-Seq experiments. Nucleic Acids Res. 2013; 41:W535–W543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gallastegui E., Millán-Zambrano G., Terme J.-M., Chávez S., Jordan A.. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 2011; 85:3187–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004; 3:Article3. [DOI] [PubMed] [Google Scholar]
- 36. Nagy Z., Tora L.. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007; 26:5341–5357. [DOI] [PubMed] [Google Scholar]
- 37. Linares L.K., Kiernan R., Triboulet R., Chable-Bessia C., Latreille D., Cuvier O., Lacroix M., Le Cam L., Coux O., Benkirane M.. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat. Cell Biol. 2007; 9:331–338. [DOI] [PubMed] [Google Scholar]
- 38. Sartorelli V., Puri P.L., Hamamori Y., Ogryzko V., Chung G., Nakatani Y., Wang J.Y., Kedes L.. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell. 1999; 4:725–734. [DOI] [PubMed] [Google Scholar]
- 39. Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.-J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S. et al. . The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004; 24:10826–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hayakawa F., Towatari M., Ozawa Y., Tomita A., Privalsky M.L., Saito H.. Functional regulation of GATA-2 by acetylation. J. Leukoc. Biol. 2004; 75:529–540. [DOI] [PubMed] [Google Scholar]
- 41. Martínez-Balbás M.A., Bauer U.M., Nielsen S.J., Brehm A., Kouzarides T.. Regulation of E2F1 activity by acetylation. EMBO J. 2000; 19:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sabò A., Lusic M., Cereseto A., Giacca M.. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell. Biol. 2008; 28:2201–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mateo F., Vidal-Laliena M., Canela N., Busino L., Martinez-Balbas M.A., Pagano M., Agell N., Bachs O.. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009; 28:2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mateo F., Vidal-Laliena M., Canela N., Zecchin A., Martínez-Balbás M., Agell N., Giacca M., Pujol M.J., Bachs O.. The transcriptional co-activator PCAF regulates cdk2 activity. Nucleic Acids Res. 2009; 37:7072–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Groen J.N., Capraro D., Morris K. V. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int. J. Biochem. Cell Biol. 2014; 54:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris K.V, Mattick J.S.. The rise of regulatory RNA. Nat. Rev. Genet. 2014; 15:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cowie P., Hay E.A., MacKenzie A.. The noncoding human genome and the future of personalised medicine. Expert Rev. Mol. Med. 2015; 17:e4. [DOI] [PubMed] [Google Scholar]
- 48. Ricciuti B., Mecca C., Crinò L., Baglivo S., Cenci M., Metro G.. Non-coding RNAs in lung cancer. Oncoscience. 2014; 1:674–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Infante P., Canettieri G., Gulino A., Di Marcotullio L.. Yin-Yang strands of PCAF/Hedgehog axis in cancer control. Trends Mol. Med. 2014; 20:416–418. [DOI] [PubMed] [Google Scholar]
- 50. Mazzà D., Infante P., Colicchia V., Greco A., Alfonsi R., Siler M., Antonucci L., Po A., De Smaele E., Ferretti E. et al. . PCAF ubiquitin ligase activity inhibits Hedgehog/Gli1 signaling in p53-dependent response to genotoxic stress. Cell Death Differ. 2013; 20:1688–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blake J.A., Ziman M.R.. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014; 141:737–751. [DOI] [PubMed] [Google Scholar]
- 52. Cobaleda C., Schebesta A., Delogu A., Busslinger M.. Pax5: the guardian of B cell identity and function. Nat. Immunol. 2007; 8:463–470. [DOI] [PubMed] [Google Scholar]
- 53. Cobaleda C., Jochum W., Busslinger M.. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007; 449:473–477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.