Abstract
As Cpf1 cleaves double-stranded DNA in a staggered way, it can be used in DNA assembly. However, the Cpf1 cleavage was found to be inaccurate, which may cause errors in DNA assembly. Here, the Cpf1 cleavage sites were precisely characterized, where the cleavage site on the target strand was around the 22nd base relative to the protospacer adjacent motif site, but the cleavage on the non-target strand was affected by the spacer length. When the spacer length was 20 nt or longer, Cpf1 mainly cleaved around the 14th and the 18th bases on the non-target strand; otherwise, with a shorter spacer (i.e. 17–19 nt), Cpf1 mainly cleaved after the 14th base, generating 8-nt sticky ends. With this finding, Cpf1 with a 17-nt spacer crRNA were employed for in vitro substitution of the actII-orf4 promoter in the actinorhodin biosynthetic cluster with a constitutively expressing promoter. The engineered cluster yielded more actinorhodin and produced actinorhodin from an earlier phase. Moreover, Taq DNA ligase was further employed to increase both the ligation efficiency and the ligation accuracy of the method. We expect this CCTL (Cpf1-assisted Cutting and Taq DNA ligase-mediated Ligation) method can be widely used in in vitro editing of large DNA constructs.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is an acquired immunity system in prokaryotic organism and CRISPR-associated protein Cas9 has been widely applied in genome editing and many other applications (1–3). Another CRISPR associated protein Cpf1, which belongs to the class II type V CRISPR system and has similar genome editing efficiency but with lower off-target effect, has great potential to be utilized in clinic applications (4–7). Cpf1 is guided by a single crRNA and utilizes T-rich protospacer adjacent motif (PAM) sequences to cleave double-stranded DNA (dsDNA) targets (6). Cas9 uses the HNH and RuvC endonuclease domains to cleave the target and non-target DNA strands, respectively (8); however, Cpf1 lacks the HNH nuclease domain and contains the RuvC and Nuc (or UK) domains (9,10), where the Nuc domain may serve as a new nuclease domain and cleave the target strand (9,10). Cpf1 was also reported as an RNase responsible for processing the precursor-crRNA to mature crRNA (11). Although structures have been obtained for the complex of Cpf1/crRNA/target DNA, no electron density was observed for regions near the cleavage sites in both the crRNA and the target DNA strand, and these regions may be flexible and disordered in the crystal structure (10,12).
Unlike Cas9 which introduces double-stranded breaks near the PAM site and produces blunt ends (8), Cpf1 cleaves target DNA distal to the PAM site and produces 5-nt sticky ends (6). This characteristic makes Cpf1 a useful tool for DNA assembly in vitro, and a DNA assembly standard namely C-Brick, which is based on Cpf1 digestion and T4 DNA ligase-mediated ligation, was recently established (13). Notably, C-Brick standard both recognizes long DNA sequences and produces short scars between parts, and may have great potential to be widely used.
With the rapid development of DNA assembly techniques, DNA fragments can now be seamlessly assembled up to several hundreds of kilobases or even a whole genome (14–20). However, there is still no powerful technology to conveniently change a biological part in an existing construct. Although Cpf1 can be designed to excise a target DNA region, generating sticky ends and allowing for the insertion of foreign DNA sequences, its cleavage sites have been found to be inaccurate (13), which may generate errors during assembly and hinders its further application. Here, the Cpf1 cleavage sites were precisely characterized and were found to be influenced by the spacer length in crRNAs. When the spacer was shorter than 20 nt, Cpf1 mainly cleaved after the 14th base on the non-target strand and 22nd base on the target strand, generating 8-nt long sticky ends. This characteristic of Cpf1 was further employed to develop a novel method for highly efficient replacement of a promoter region in a large antibiotic biosynthetic cluster.
MATERIALS AND METHODS
In vitro transcription
The transcription template was prepared as previously described (13). Briefly, 200 ng template DNA was used to transcribe the corresponding crRNA in a 20 μl reaction mix according to the protocol provided by the manufacture (Thermo Fisher Scientific). crRNA was purified using the RNA Clean-up & Concentration kit (Zymo Research), and quantified by both NanoDrop 2000C and TBE-buffered polyacrylamide gel electrophoresis (PAGE).
Purification of Cpf1 proteins
Protein FnCpf1 (WP_003040289) and AsCpf1 (WP_021736722) were purified as described previously (13). LbCpf1 (WP_051666128) from Lachnospiraceae bacterium ND2006 firstly synthesized with codon optimization by Tolo Biotech and then cloned into expression vector (pET28a-TEV (21)). Expression and purification of LbCpf1 was performed following the same procedure as those of FnCpf1 and AsCpf1.
Cleavage of short dsDNA targets
Short dsDNA targets were prepared through annealing of two complementary oligonucleotides (Supplementary Table S1). Paired oligonucleotides (0.8 μM) were annealed in 1 × PCR (polymerase chain reaction) buffer (Takara) in a total volume of 50 μl, followed by running the annealing program: initial denaturation at 95°C for 5 min and then cool down from 95°C to 20°C with 1°C decrease per min using a thermocycler.
Cleavage of double stranded oligonucleotides were performed at 37°C in NEB buffer 3 for 1 h, employing 250 nM Cpf1, 500 nM synthesized crRNA, 40 nM target DNA and 10 U RNase inhibitor (Takara) in a 20 μl volume. Reactions were stopped by heating at 98°C for 10 min, followed by immediately chilling on ice before further analysis through urea PAGE.
Electrophoresis analysis by denaturing urea PAGE
FAM-labeled DNA were firstly digested by Cpf1 and then heated at 98°C for 10 min after the addition of loading buffer to stop the reactions. Heated samples were immediately chilled on ice, followed by being loaded on 10–15% denaturing polyacrylamide gels containing 7 M urea (urea PAGE). Electrophoresis was performed by running at 1800 V (about 40 v/cm) for about 70–90 min using the Sequi-Gen GT Sequencing Cell system (Bio-Rad). Gels were scanned using a FLA-7000 phosphoimager (FujiFilm Corporation).
Plasmids construction and optimization of the ligation conditions
Plasmid pSY1A2-DNMT1-3 was constructed using the Ezmax One-Step Cloning kit (Tolo Biotech.). Briefly, the DNMT1-3 fragment was PCR amplified with primers of DNMT1-795-F and DNMT1-795-R, using the HEK 293T genomic DNA as the template. Meanwhile, the vector was PCR amplified with primers of pSB1A2-DNMT1-795-F and pSB1A2-DNMT1-795-R, employing pSB1A2 (iGEM) as the template. Notably, 20-bp sequences homologous to the ends of the DNMT1-3 fragment were introduced to each end of the vector fragment during the PCR process. Then, two fragments were seamlessly assembled following the manufacture's instruction (Ezmax for One-step Cloning Tolo Biotech.).
To substitute the promoter region of actII-orf4 with the constitutively expressing promoter of the erythromycin resistance gene (ermP), the ermP fragment was prepared with PCR using primers of Ermp-F1 and Ermp-R1, which was subsequently cloned into pEASY-Blunt-zero (Transgen Biotech.) to generate plasmid pEASY-ermP. Plasmids pHIW (15) and pEASY-ermP were digested by FnCpf1 under the guide of act-crRNA1 and act-crRNA2 in NEB buffer 3. Digested pHIW was dephosphorylated by FastAP (Thermo Fisher Scientific) and directly cleaned up with the Wizard SV Gel and PCR Clean-Up system (Promega), while the Cpf1-digested ermP fragment was purified by gel electrophoresis followed by gel purification using the Wizard SV Gel and PCR Clean-Up system. The HIW and ermP fragments were mixed together with a molar ratio of 1:20 and ligated with Taq DNA ligase (Tolo Biotech.). Different reaction temperatures and time courses were tested to determine the most optimal reaction conditions. The ligation product was transformed into the Escherichia coli DH10B competent cells to generate the object plasmid pHIW-ermP. To determine the cloning efficiency, clones were firstly confirmed by clone PCR using primers of actII-orf4CKF and actII-orf4CKR. Positive clones (with 700-bp PCR products) were then confirmed by Sanger sequencing using the primer of actII-orf4CKF and further analyzed by StuI digestion.
Characterization of pHIW-ermP in Streptomyces sp. 4F
Plasmid pHIW-ermP was firstly transformed into E. coli ET12567 (22), and then conjugated into Streptomyces sp. 4F, following the same procedure as previously described (15,23). The wild-type Streptomyces sp. 4F, strain 4F integrated with pHIW-ermP and strain 4F integrated with pHIW were grown on solid R2YE medium (24) at 30°C, and the production of actinorhodin was observed every 24 h. Meanwhile, 4F (pHIW-ermP) and 4F (pHIW) were cultured with liquid R2YE medium and grown at 30°C. The actinorhodin yield was measured every 12 h, following the same protocol as previously reported (25). Total RNA was extracted using the ZR Fungal/Bacterial RNA MiniPrep™ kit (Zymo Research) following the manufacture's instruction. Further RNA reverse transcription and quantitative realtime PCR analysis were the same as previously described (26).
RESULTS
Cpf1 cleaves more specifically on target DNA with a shorter crRNA spacer
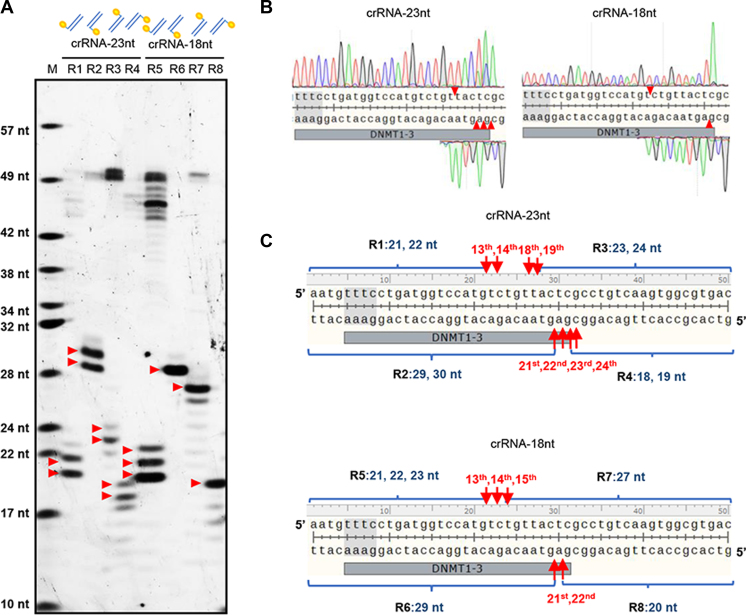
Cpf1 was known to cleave after the 18th base downstream of the PAM site on the non-target strand and the 23rd base on the target strand (6). However, we recently found the Cpf1 cleavage sites vary among different targets and different Cpf1 origins (Ref 13 and Supplementary Figure S1). To precisely determine the Cpf1 cleavage sites, denaturing urea PAGE was employed to analyze the Cpf1 digested products. 5(6)-carboxyfluorescein (FAM)-labeled short DNA fragments of target DNMT1-3 (individually labeled on either 5΄- or 3΄-ends of the target or non-target strands) were digested by Cpf1 using crRNA-DNMT1-3 with the full length spacer (23 nt), and subsequently analyzed by urea PAGE. To our surprise, we found the FnCpf1 (from Francisella tularensis) cleavage sites were after the 13th, 14th, 18th and 19th bases on the non-target strand and from the 21st to 24th bases on the target strand (Figure 1). Similar cleavage results were obtained when using either target DNMT1-3 with another two Cpf1s (AsCpf1 (from Acidaminococcus sp. BV3L6) and LbCpf1 (from Lachnospiraceae bacterium ND2006)) or FnCpf1 with other six targets (Supplementary Figure S2a and b). Notably, the fact that FnCpf1 cleaves after the 14th base can also be deduced from the urea PAGE result in a recent study (11).
Figure 1.
Precise characterization of the FnCpf1 cleavage sites. (A) FAM-labeled short double stranded DNA fragments were digested by FnCpf1 and analyzed with urea PAGE. Two crRNAs of DNMT1-3 with different spacer length were employed, i.e. 23 nt (crRNA-23nt) and 18 nt (crRNA-18nt). The FAM labeling was indicated by yellow circles, e.g. R1 and R3 were labeled on the 5΄-end and 3΄-end of the non-target strand, respectively. (B) Identification of the FnCpf1 cleavage sites by Sanger DNA sequencing. Plasmid pSB1A2-DNMT1-3 was digested with FnCpf1 and the products were purified for sequencing, following the procedure as previously described (6). The cleavage sites were indicated by red triangles according to the sequencing results. (C) Schematic illustration of the digested fragments in panel A. The precise cleavage sites were labeled and indicated by orange arrows. The length of the labeled fragments after FnCpf1 digestion was also indicated. For example, the length of R1 labeling is 21 or 22 nt, which was equivalent to the cleavage after the 13th and 14th bases relative to the PAM site. M, a DNA ladder made through mixing the synthesized FAM-labeled oligonucleotides of different length. The Cpf1 PAM sequences were indicated by gray background in panels B and C.
To test the influence of the spacer length of crRNA on the cleavage sites of Cpf1, we cleaved a target plasmid with the spacer length ranging from 14 to 30 nt, and found that crRNAs with 17-nt or longer spacer could enable Cpf1 with complete double-stranded break activity (Supplementary Figure S3a). Then, the Cpf1-digested products were analyzed by the Sanger sequencing method (6) (Supplementary Figure S3b and Figure 1). Based on the sequencing results, when the length of the spacer was 20 nt or longer, Cpf1 mainly cleaved after the 18th base on the non-target strand and near the 23rd bases on the target strand downstream of the PAM site, which was consistent with the previous findings (6) and our analyses above. However, when the spacer length was shorter than 20 nt, Cpf1 no longer cleaved after the 18th base but mainly cleaved after the 14th base on the non-target strand (Figure 1B and Supplementary Figure S3b). Besides, the cleavage sites were more specific on the target strand after the 22nd base.
Then, the urea PAGE method was used to analyze the cleavage of a short target DNA with a crRNA of 18-nt spacer (crRNA-18 nt). Similarly, Cpf1 mainly cleaved after the 13th, 14th and 15th bases (without the 18th base) on the non-target strand and the 21st and 22nd bases on the target strand. Although the cleavage sites on short DNA targets were slightly different from those on longer DNA targets (plasmid DNA), both showed that the Cpf1 cleavage sites were affected by the spacer length and Cpf1 cleaved the target more specifically with a shorter spacer crRNA.
Cpf1-generated long sticky ends facilitate the replacement of actII-orf4 promoter in act cluster
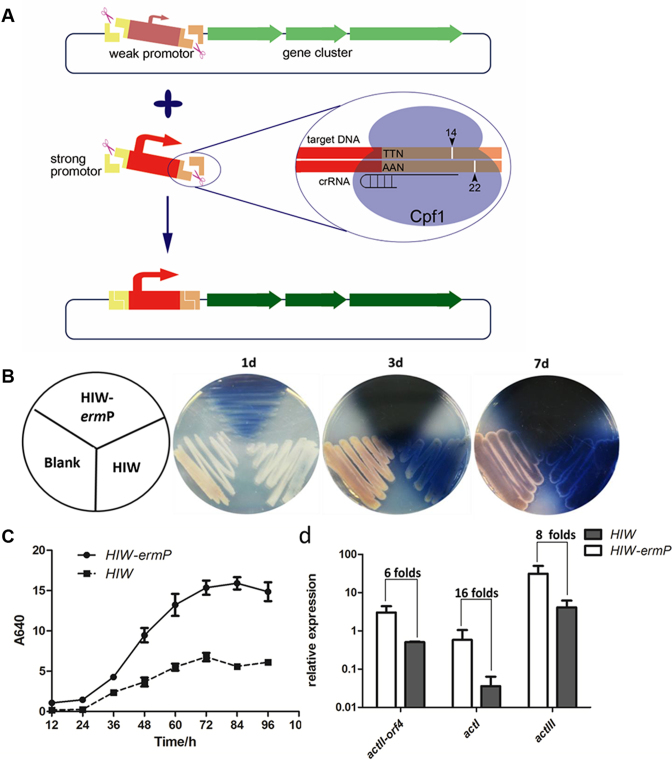
With the availability of suitable PAM sites, Cpf1 could cleave target DNA at objective sites under the guide of specifically designed crRNAs, which characteristic can be used to replace or modify a defined region in a large cluster. As crRNAs with spacer length less than 20 nt could direct Cpf1 a more specific cleavage, crRNAs with 17-nt spacers were chosen for substitution of a DNA part in act cluster. As actII-orf4 encodes a pathway specific regulator for act (27) and increase of its expression may lead to higher actinorhodin production, its promoter (orf4P) was then replaced with a constitutively expressing promoter from the erythromycin resistance gene ermE (ermP). Plasmid pHIW containing the whole act cluster was digested by Cpf1, which was guided by two 17-nt spacer crRNAs specifically targeting the flanked DNA sequences of orf4P, removing the orf4P. Similarly, the ermP fragment was cut from plasmid pEASY-ermP by Cpf1, carrying 8-nt complementary sticky ends (Figure 2A), which allowed for the subsequent ligation of ermP and pHIW to obtain pHIW-ermP.
Figure 2.
Cpf1-assisted substitution of actII-orf4P with ermP in act cluster. (A) Schematic chart for substitution of the actII-orf4 promoter, employing Cpf1-assisted cleavage. A strong promoter was PCR amplified, containing the same flanking sequences as the actII-orf4 promoter. A pair of 17-nt spacer sequences were chosen nearby the promoter region, and both promoters were then cleaved by Cpf1 in vitro, followed by DNA ligation to form an engineered cluster. (B and C) The production of actinorhodin in both solid (B) and liquid (C) R2YE medium. HIW-ermP and HIW represented Streptomyces sp. 4F harboring pHIW-ermP and pHIW expression vector, while blank represented the wild-type Streptomyces sp. 4F. (D) Transcriptional levels of actII-orf4 and the target genes of actI and actIII in HIW-ermP and HIW strains, respectively.
In consideration of the long sticky ends, Taq DNA ligase was used for DNA ligation. Different temperatures (i.e. 45°C, 55°C and 65°C) and time courses (i.e. 10 min, 1 h and 2 h) were tested to determine the optimal ligation conditions. And based on both the ligation efficiency and the positive rates, a ligation condition of 10 min at 45°C was taken as the reaction condition (Table 1 and Supplementary Figure S4a). To confirm the ligation accuracy, 16 positive clones were selected and a region covering the ligation sites was sequenced to find that all clones had correct sequences (Supplementary Figure S4b). Then, a positive clone was further verified by StuI digestion, employing plasmid pHIW as a control, and the cleavage patterns were the same as predicted (Supplementary Figure S4c).
Table 1. Influence of the ligation conditions on the positive rates with the CCTL method.
| 10 min | 60 min | 120 min | |
|---|---|---|---|
| 45°C | 78 ± 5% (87 ± 36)a | 72 ± 12% (70 ± 10) | 66 ± 16% (62 ± 10) |
| 55°C | 57 ± 7% (88 ± 23) | b | |
| 65°C | 66 ± 14% (66 ± 9) |
aBoth the positive rates and the number of clones in bracket were shown in the form of mean ± SD. Positive clones were determined by colony PCR (details in ‘Materials and Methods’ section) and the results were obtained from three independent experiments.
bNot tested in this study.
Both plasmid pHIW-ermP and the control plasmid pHIW were conjugated into Streptomyces sp. 4F and then cultured on solid R2YE medium. Compared to strain 4F (pHIW), the actinorhodin yield in 4F (pHIW-ermP) was remarkably increased and the production of actinorhodin started from a much earlier stage (Figure 2B). Similar results could be obtained when strains were grown in liquid R2YE medium, and the production of actinorhodin was 2-fold higher in 4F (pHIW-ermP) than in 4F (pHIW) (Figure 2C), which was consistent with the higher transcriptional level of actII-orf4 and its target genes of actI and actIII in the engineered cluster (Figure 2D).
DISCUSSION
The CCTL method is an efficient tool for in vitro modification of large DNA pieces
Dozens of DNA assembly technologies have emerged or been further developed in the past few years, among which both the in vitro Gibson assembly and the in vivo yeast TAR (transformation-associated recombination) technology are widely used for one-step assembly of multiple DNA fragments (18,28). With careful design, the assembled constructs could be seamless and can be directly used for further test of their biological functions. Unlike DNA assembly, no powerful technology can now be taken for efficient change of a designated part within a large DNA construct.
In some situations, site-specific recombination technologies (e.g. SSRTA (29)) that facilitate both the assembly of multiple DNA parts and the replacement of a designated part from a construct in a sequence-independent way, but they usually leave integration sites (e.g.attL and attR sites in SSRTA) between parts. Therefore, for some accurately designed constructs that require seamless DNA assembly, de novo assembly may be needed. Alternatively, the aim can be achieved by the combination of CRISPR-Cas9 and Gibson assembly (i.e. similar to CATCH (30)), where Cas9 is firstly used to linearize DNA at designated sites and Gibson assembly allows for the introduction of foreign DNAs. However, because Gibson assembly requires homologous sequences between DNA parts, the method might be inefficient when dealing with complicated DNA sequences. For example, when dealing with DNA sequences with high G+C content, the Gibson assembly efficiency would dramatically decrease (31), while the assembly efficiency of CCTL was not affected (i.e. the up to 70% G+C content in act cluster).
In this study, we developed a novel DNA substitution method (namely CCTL), which was based on the Cpf1-assisted DNA cutting and Taq DNA ligase-assisted DNA ligation. Although typical restriction and ligation method can be used to change a piece of DNA within a construct, the experiment must be well designed to choose suitable restriction sites for type II restriction endonucleases, which is actually mission impossible when the target is a large DNA piece. By contrast, the cleavage sites of Cpf1 are determined by the spacer sequences in crRNAs. Considering the wide spread of the PAM sites (TTN), the Cpf1 cleavage sites can be easily changed for different target sequences. Besides of the substitution of DNA parts, CCTL also can be used for other applications of DNA modification (e.g. correction of mutations, in-frame deletion of a gene sequence and insertion of genes) and the method is especially useful for large constructs. Moreover, because CCTL uses 8-nt (or 9-nt) sticky ends instead of the 20-nt (or longer) homologous sequences in Gibson assembly, CCTL can be used for the treatment of complicated DNA sequences (e.g. sequences with repeats), which therefore makes the CCTL a sequence-independent method.
Approaches to deal with the inaccuracy of Cpf1 cleavage
As reported, the Cpf1 cleavage sites are inaccurate and can be affected by different target sequences (13). In this study, we employed both the Sanger DNA sequencing method and the urea PAGE method to precisely identify the Cpf1 cleavage sites. Interestingly, the spacer length also affects the cleavage of Cpf1, and Cpf1 with short spacer (i.e. <20 nt) shows a different profile of the cleavage sites. It is possible that a shorter spacer crRNA causes an incomplete conformational change of Cpf1, which then leads to a distinct cleavage behavior. However, because the structure of Cpf1/crRNA/target DNA complex was not stable near the cleavage activity sites (10,12), further studies are needed to reveal the mechanisms involved in the change of Cpf1 cleavage by different spacer lengths.
In addition, the Cpf1 cleavages was also found to be slightly influenced by the target length in this study (Figure 1), where Cpf1 cleaved longer targets (plasmid DNA) with more accuracy (Figure 1). It is possible that the complex of Cpf1 with shorter targets is more flexible, which leads to the inaccurate Cpf1 cleavage.
Although longer targets usually lead to more accurate cleavage when the spacer is less than 20 (e.g. 17 nt in this study), Cpf1 still partially cleaves the nearby bases besides of the 14th base on the non-target strand (e.g. the 15th base in Figure 1B). To solve this problem, a careful design of the spacer sequence targeting the inserted sequence (e.g. the ermP sequence in this study) might be needed. For example, in this study, spacer sequences for targeting the ermP fragment were designed to be the same as those targeting the vector, And with the same spacer sequences, Cpf1 may have the same preference in the choice of the cleavage sites. In theory, the inaccurately cleaved inserts can be ligated with those inaccurately cleaved vectors, generating correct clones and thus the influence of partial inaccurate cleavage can be neglected.
To perform the DNA ligation experiment, T4 DNA ligase was firstly used, but positive rates were extremely low (<25% as determined by the size of the clones, data not shown). In consideration of the long sticky ends, Taq DNA ligase was then used to join the complementary ends under a higher temperature. In addition, because Taq DNA ligase only recognizes accurately paired ends (32), mismatched ligation would be excluded, which could be reflected by the high positive rates and 100% accuracy in this study.
Future perspectives for the CCTL method
In our previous work, Cpf1 was employed in the establishment of the C-Brick assembly standard (13). Based on the knowledge that Cpf1 cleaves after the 18th and 23rd bases on the two strands, respectively, which generates 5-nt cohesive ends, 5-bp overlaps are designed between DNA parts. While based on the findings in this study, with full length of spacer, both 5- and 8-nt cohesive ends can be produced, which could have been the reason for the limited accuracy in C-Brick method (13). In future, the C-Brick standard can be re-designed (e.g. with the use of crRNAs of shorter spacers and Taq DNA ligase), which would improve both ligation efficiency and ligation accuracy but would leave a longer linker (i.e. no less than 8 nt).
Besides, although the Cpf1 PAM sites are widespread, the application of CCTL method still relies on the existence of PAM sequence and is not absolutely sequence-independent. Therefore, a PAM-independent Cpf1 would be a prerequisite for the development of an absolutely sequence-independent method. Such Cpf1s can be either isolated from other origins or obtained through artificial engineering. Actually, with the availability of protein structures, change of the PAM recognition has been successfully achieved in Cas9 (33,34) and should be workable in Cpf1. Meanwhile, Cpf1s originated from different species have already shown different preference on the recognition PAM sites (6), which may provide experimental clues for further investigation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tolo Biotech. (Shanghai, China) for their help in gene synthesis, plasmid construction and protein expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB19040200]; Youth Innovation Promotion Association CAS; the China National Basic Research Program [973 Program No. 2012CB721102]; National Natural Science Foundation of China [31421061, 31430004; 31300031]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDB19040200]; Youth Innovation Promotion Association CAS; the China National Basic Research Program [973 Program No. 2012CB721102]; National Natural Science Foundation of China [31421061, 31430004; 31300031].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015; 526:55–61. [DOI] [PubMed] [Google Scholar]
- 3. Wang H., La Russa M., Qi L.S.. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016; 85:227–264. [DOI] [PubMed] [Google Scholar]
- 4. Kleinstiver B.P., Tsai S.Q., Prew M.S., Nguyen N.T., Welch M.M., Lopez J.M., McCaw Z.R., Aryee M.J., Joung J.K.. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016; 34:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim D., Kim J., Hur J.K., Been K.W., Yoon S.H., Kim J.S.. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016; 34:863–868. [DOI] [PubMed] [Google Scholar]
- 6. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong D., Ren K., Qiu X., Zheng J., Guo M., Guan X., Liu H., Li N., Zhang B., Yang D. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016; 532:522–526. [DOI] [PubMed] [Google Scholar]
- 10. Yamano T., Nishimasu H., Zetsche B., Hirano H., Slaymaker I.M., Li Y., Fedorova I., Nakane T., Makarova K.S., Koonin E.V. et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell. 2016; 165:949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fonfara I., Richter H., Bratovic M., Le Rhun A., Charpentier E.. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016; 532:517–521. [DOI] [PubMed] [Google Scholar]
- 12. Gao P., Yang H., Rajashankar K.R., Huang Z., Patel D.J.. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016; 26:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S.Y., Zhao G.P., Wang J.. C-Brick: a new standard for assembly of biological parts using Cpf1. ACS Synth. Biol. 2016; 5:1383–1388. [DOI] [PubMed] [Google Scholar]
- 14. Engler C., Marillonnet S.. Golden Gate cloning. Methods Mol. Biol. 2014; 1116:119–131. [DOI] [PubMed] [Google Scholar]
- 15. Chen W.H., Qin Z.J., Wang J., Zhao G.P.. The MASTER (methylation-assisted tailorable ends rational) ligation method for seamless DNA assembly. Nucleic Acids Res. 2013; 41:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geu-Flores F., Nour-Eldin H.H., Nielsen M.T., Halkier B.A.. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007; 35:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:U343–U341. [DOI] [PubMed] [Google Scholar]
- 18. Gibson D.G., Benders G.A., Andrews-Pfannkoch C., Denisova E.A., Baden-Tillson H., Zaveri J., Stockwell T.B., Brownley A., Thomas D.W., Algire M.A. et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008; 319:1215–1220. [DOI] [PubMed] [Google Scholar]
- 19. Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010; 329:52–56. [DOI] [PubMed] [Google Scholar]
- 20. Dong D., Ren K., Qiu X.L., Zheng J.L., Guo M.H., Guan X.Y., Liu H.N., Li N.N., Zhang B.L., Yang D.J. et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016; 532:522–526. [DOI] [PubMed] [Google Scholar]
- 21. Carneiro F.R., Silva T.C., Alves A.C., Haline-Vaz T., Gozzo F.C., Zanchin N.I.. Spectroscopic characterization of the tumor antigen NY-REN-21 and identification of heterodimer formation with SCAND1. Biochem. Biophys. Res. Commun. 2006; 343:260–268. [DOI] [PubMed] [Google Scholar]
- 22. Flett F., Mersinias V., Smith C.P.. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997; 155:223–229. [DOI] [PubMed] [Google Scholar]
- 23. Chen W.H., Qin Z.J.. Development of a gene cloning system in a fast-growing and moderately thermophilic Streptomyces species and heterologous expression of Streptomyces antibiotic biosynthetic gene clusters. BMC Microbiol. 2011; 11:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kieser T.B.M., Buttner M.J., Chater K.F., Hopwood D.A.. Practical Streptomyces Genetics. 2000; Norwich: The John Innes Foundation [Google Scholar]
- 25. Borodina I., Siebring J., Zhang J., Smith C.P., van Keulen G., Dijkhuizen L., Nielsen J.. Antibiotic overproduction in Streptomyces coelicolor A32 mediated by phosphofructokinase deletion. J. Biol. Chem. 2008; 283:25186–25199. [DOI] [PubMed] [Google Scholar]
- 26. Shao Z.H., Ren S.X., Liu X.Q., Xu J., Yan H., Zhao G.P., Wang J.. A preliminary study of the mechanism of nitrate-stimulated remarkable increase of rifamycin production in Amycolatopsis mediterranei U32 by RNA-seq. Microb. Cell Fact. 2015; 14:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujii T., Gramajo H.C., Takano E., Bibb M.J.. redD and actII-ORF4 pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing sigma(hrdD). J. Bacteriol. 1996; 178:3402–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 29. Zhang L., Zhao G., Ding X.. Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci. Rep. 2011; 1:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang W., Zhao X., Gabrieli T., Lou C., Ebenstein Y., Zhu T.F.. Cas9-assisted targeting of chromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 2015; 6:8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li L., Zhao Y., Ruan L., Yang S., Ge M., Jiang W., Lu Y.. A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab. Eng. 2015; 29:12–25. [DOI] [PubMed] [Google Scholar]
- 32. Barany F. Genetic-disease detection and DNA amplification using cloned Thermostable ligase. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K.. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015; 33:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.