Abstract
Background: Breast cancer bone metastasis (BCBM)-specific genes have been reported without considering biological differences based on estrogen receptor (ER) status. The aims of this study were to identify BCBM-specific genes using our patient dataset and validate previously reported BCBM-specific genes, and to determine whether ER-status-related biological differences matter in identification of BCBM-specific genes.
Methods: We used Affymetrix GeneChips to analyze 365 primary human epidermal growth factor receptor 2 (HER2)-negative invasive breast cancer specimens. Genes that were differentially expressed between patients who developed bone metastasis and those who developed non-bone metastasis were identified using Cox proportional hazards model, and differential expression of gene sets was assessed using gene set analysis. We performed gene set analysis to determine whether biological function associated with bone metastasis were different by ER status using 2,246 functionally annotated gene sets assembled from Gene Ontology data base.
Results: Among 16,712 probe sets, 592 were overexpressed in the bone metastasis cohort compared to the non-bone-metastasis cohort (false discovery rate ≤ 0.05). However, no BCBM-specific genes met our significance tests when the cancers were stratified by ER status. In ER-positive and ER-negative breast cancers, 151 and 125 gene sets, respectively, were overexpressed for BCBM and the majority of BCBM-related pathways were different. Of significant gene sets, only 13 gene sets were overlapped between ER-positive and -negative cohorts.
Conclusion: ER-positive and ER-negative breast cancers have different biological pathways in BCBM development. We have yet to explore BCBM-related biomarkers and targets considering the biological features associated with BCBM depending on the ER status.
Keywords: Breast cancer, bone metastasis, gene expression, gene sets, estrogen receptor status.
Introduction
Bone is the first site of metastasis in 26% to 50% of breast cancer patients 1-4. However, the mechanism of breast cancer bone metastasis (BCBM) has not been clarified. In the past decade, microarray analysis has enabled successful prediction of patients who have a high risk of recurrence 5, 6, but the sites of metastasis are not predictable. If we could identify patients likely to develop BCBM in the future, we might be able to assess the effect of a bisphosphonate in the adjuvant setting to reduce the incidence of bone metastases for these high risk population.
Smid et al. 7 have studied genes associated with breast cancer metastatic to bone. Using 107 primary breast cancer samples, they found 69 genes significantly overexpressed in patients whose relapses manifested in bone compared to those whose relapses manifested in other sites (non-bone). In that study, there was a significant difference in estrogen receptor (ER) positivity between patients with bone metastasis and those with non-bone metastasis (82.6% vs 60.5%, P = 0.019). This difference indicates that the significant difference in gene expression between the patient groups may have been strongly affected by ER status, which is the only clinical marker associated with metastasis to bone 8.
This significant difference in ER status between bone and non-bone metastases has not been considered in previous studies identifying BCBM-specific genes. None of the previously published gene sets have demonstrated that there are different biological features in ER-positive and ER-negative breast cancers that develop BCBM. Therefore, we hypothesized that ER-positive and ER-negative breast cancers have a different biological pathway in development of BCBM.
The first aim of this study was to identify BCBM-specific genes using our patient dataset and validate previously reported BCBM-specific genes. The second aim was to determine whether the biological function associated with bone metastasis were different by ER status in identification of BCBM-specific genes by gene set analysis.
Materials and methods
Patient data and tumor samples
Tumor samples were collected prospectively from 365 patients with primary invasive breast cancer who underwent surgery from November 1999 to August 2008 at The University of Texas MD Anderson Cancer Center. We excluded patients who were diagnosed with ductal carcinoma in situ, metaplastic carcinoma, sarcoma, or clinical stage IV breast cancer. Patients with human epidermal growth factor receptor 2 (HER2)-positive breast cancer were also excluded because whether they received trastuzumab may have been affected occurrence of metastasis. Participation was voluntary, and all patients gave written informed consent. The study was performed under institutional review board approval.
Tumor samples were obtained from fine-needle aspiration (FNA) of the primary tumor before initiation of any systemic therapy. The following clinical data were collected from MD Anderson's Breast Cancer Management System database.
Metastases were confirmed by imaging and/or bone biopsy. The time to metastasis and site of first metastasis were recorded. We classified the samples into three cohorts: bone as the first metastatic site, non-bone as the first metastatic site, and no metastasis (no mets). The bone cohort consisted of patients whose first metastases manifested in bone or in bone and other sites at the same time. The non-bone cohort consisted of patients whose first metastases manifested only in other sites.
Statistical analysis
Patient characteristics by type of first metastasis were summarized with medians and ranges for age and follow-up time and with frequency and percentage for all other characteristics. Bone metastasis-free survival was defined as the time interval from the diagnosis of primary breast cancer until the diagnosis of bone metastasis or the last follow-up date. Non-bone metastasis-free survival was defined as the time interval from the diagnosis of primary breast cancer until the diagnosis of non-bone metastasis or the last follow-up date. We compared the bone and non-bone cohorts according to clinical and tumor characteristics of patients using Fisher's exact test. All statistical tests were two-sided.
Gene expression data
These datasets are referred to as the MicroArray Quality Control Phase II (MAQC-II) Project (GSE16716) that has been published. We extracted a part of the dataset which included HER2-negative patients treated with weekly paclitaxel (T) (80 mg/m2) for 12 treatments, followed by 5-fluorouracil (500 mg/m2), doxorubicin (50 mg/m2), and cyclophosphamide (500 mg/m2) for four treatments, given once every 21 days (FAC), with available clinical data. Samples were placed in RNAlater (Ambion, Austin, TX) storage reagent and stored at -80°C until gene expression analysis. As described by Hess et al., all gene expression data were generated using Affymetrix U133A gene chips (Affymetrix, Inc, Santa Clara, CA) 9. Gene expression data were normalized with the MAS5 algorithm, mean centered to 600, and log 2 transformed before further analysis. Probe sets with the lowest 25% mean expression value were removed from all higher level analyses to reduce noise from low expressed probe sets. This left 16,712 probe sets for analysis. Tumors with normalized ESR1 mRNA expression (probe set 205225_at) >10.18 were considered ER-positive 10. Tumors with normalized HER2 mRNA expression (probe set 216836_s_at) >12.54 were considered HER2-positive 10. All statistical analysis was performed with BRB-ArrayTools version 3.8.1 α software and R software version 2.10.1.
Identification of differentially expressed genes and gene sets
We compared gene expression data among the bone, non-bone, and no mets cohorts without any further stratification by clinical or molecular class using the Cox proportional hazards model to test whether expression of a particular gene significantly influences development of metastasis. We also performed similar class comparisons separately for ER-positive and ER-negative cancers. False discovery rates (FDRs) were calculated to adjust for the multiple comparisons using BRB-ArrayTools. The FDR was calculated with the significance analysis of microarrays (SAM) tool as the median number of false-positive genes from permutation testing divided by the number of nominally significant genes defined from the unperturbed data 11. We mapped significantly differentially expressed genes to functional pathways using Ingenuity pathway analysis (IPA) software (http://www.ingenuity.com) 12, 13.
We next examined differential expression of prior defined gene sets using gene set analysis (GSA) 12, 13 to assess whether members of a set of genes that corresponded to a particular biological pathway tended to occur toward the top or the bottom of a rank-ordered gene list (rank ordered by differential expression among the bone, non-bone, and no-Mets cohorts) 14. We tested 2,246 different, functionally annotated gene sets from the Gene Ontology (GO) database that collectively represent most known biological pathways in eukaryotic cells. Gene sets with a minimum of 10 genes and maximum of 100 genes were selected for inclusion in this analysis. We used the Efron and Tibshirani GSA method that employs “maxmean” statistics and is implemented by BRB-ArrayTools 15, 16. We determined whether gene sets were differentially expressed among cohorts, with statistical significance being determined by a permutation test (n = 1,000). The null hypothesis was that the average degree of differential expression of members of a given gene set among the bone, non-bone, and no mets cohorts is the same as expected from a random set of genes of similar size.
Results
Patient characteristics
Clinicopathological characteristics of the 365 patients are summarized in Table 1. The bone metastasis cohort consisted of 34 patients (9.3%); the non-bone metastasis cohort, 32 patients (8.8%); and the no-mets cohort, 299 patients (81.9%).
Table 1.
Clinical and tumor characteristics of patients
| Characteristics | All patients | No mets | Bone | Non-bone | P value*(bone vs non-bone) | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | ||
| No. of patients | 365 | 100% | 299 | 100% | 34 | 100% | 32 | 100% | |
| Median age (years) | 51 | 51 | 52 | 51 | |||||
| Range (years) | 24-85 | 25-82 | 24-85 | 29-73 | |||||
| Clinical stage | |||||||||
| 0 | 1 | 0.3% | 0 | 0.0% | 1 | 2.9% | 0 | 0.0% | 1 |
| I | 18 | 4.9% | 18 | 6.0% | 0 | 0.0% | 0 | 0.0% | |
| II | 204 | 55.9% | 180 | 60.2% | 12 | 35.3% | 12 | 37.5% | |
| III | 142 | 38.9% | 101 | 33.8% | 21 | 61.8% | 20 | 62.5% | |
| Tumor grade | |||||||||
| Poor | 230 | 63.0% | 183 | 61.2% | 21 | 61.8% | 26 | 81.3% | 0.057 |
| Good to moderate | 134 | 36.7% | 116 | 38.8% | 13 | 38.2% | 5 | 15.6% | |
| Unknown | 1 | 0.3% | 0 | 0.0% | 0 | 0.0% | 1 | 3.1% | |
| ER status | |||||||||
| Positive | 214 | 58.6% | 185 | 61.9% | 19 | 55.9% | 10 | 31.3% | 0.052 |
| Negative | 151 | 41.4% | 114 | 38.1% | 15 | 44.1% | 22 | 68.8% | |
| Neoadjuvant chemotherapy | |||||||||
| Received | 338 | 92.6% | 277 | 92.6% | 30 | 88.2% | 31 | 96.9% | 0.357 |
| Not received | 27 | 7.4% | 22 | 7.4% | 4 | 11.8% | 1 | 3.1% | |
| Adjuvant chemotherapy | |||||||||
| Received | 56 | 15.3% | 45 | 15.1% | 5 | 14.7% | 6 | 18.8% | 0.748 |
| Not received | 309 | 84.7% | 254 | 84.9% | 29 | 85.3% | 26 | 81.3% | |
ER = estrogen receptor. *Fisher's exact test.
We compared the bone and non-bone cohorts according to ER status. One hundred eight seven of the 214 with ER-positive tumor (87.3%) received hormonal therapy. There was a trend of high occurrence of ER-positive breast cancer among patients with bone metastasis compared to those with non-bone metastasis (55.9% vs 31.3%, P = 0.052). This trend was similar to that shown in Smid et al.'s study 7. Other characteristics of the patients did not differ significantly between patients with bone metastasis and those with non-bone metastasis.
Site-specific relapse-free survival according to ER status
Next, we examined the relapse-free survival of patients according to bone metastasis or non-bone metastasis as a first metastatic site in ER-positive and ER-negative breast cancers using the Cox proportional hazards model. When we evaluated bone-metastasis-free survival, there was no significant difference in survival times between patients with ER-positive and ER-negative breast cancers. However, when we evaluated non-bone-metastasis-free survival, patients with ER-negative breast cancer had shorter metastasis-free survival than did patients with ER-positive breast cancer. These results indicated that bone and non-bone metastasis have a distinct clinical course relative to ER status, which might come from distinct biological differences related to ER status. Therefore, we sought to assess the gene expression associated with BCBM in ER-positive and ER-negative breast cancers separately.
Gene expression associated with BCBM
To determine the differences between bone metastasis and non-bone metastasis, individual gene (i.e., probe set)-level analysis was performed using the Cox proportional hazards model for 14 genes that have been reported to have an important role in development of BCBM 17-32. In 66 patients with metastasis, without any stratification for ER status, 6 probe sets—BMP1, BMP10, TNFRSF11A, TGFB2, CXCR4, and ESR1—were significantly overexpressed in the bone metastasis cohort compared to the non-bone metastasis cohort, with FDR ≤ 0.05. However, when this analysis was done in ER-positive and ER-negative breast cancers separately, no gene (FDR ≤ 0.05) was found to be significantly overexpressed in both patient populations. This result may suggest that previously reported BCBM-specific genes might be capturing gene differences based on ER status, not BCBM status.
Next, to examine possible differences between BCBM and non-bone metastasis, we performed the same analysis using all 16,712 probe sets. In 66 patients with metastasis without any stratification by ER status, 592 probe sets were overexpressed in the bone metastasis cohort compared to the non-bone metastasis cohort with FDR ≤ 0.05 (Table 2). However, no BCBM-specific genes were found when ER-positive and ER-negative breast cancers were analyzed separately.
Table 2.
Gene sets overexpressed in the bone-metastasis cohort compared with the non-bone-metastasis cohort
| FDR | All (n=66) | ER positive (n=29) | ER negative (n=37) |
|---|---|---|---|
| ≤ 0.3 | 6,427 | 3,617 | 0 |
| ≤ 0.1 | 2,785 | 0 | 0 |
| ≤ 0.05 | 592 | 0 | 0 |
| ≤ 0.01 | 2 | 0 | 0 |
FDR = false discovery rate; ER = estrogen receptor.
Gene set analysis
Validation of previously published BCBM-specific gene sets
Because no statistical differences in any individual gene expression level between bone and non-bone metastases were shown among ER-positive or -negative patients, we determined to, using gene set enrichment analysis, detect consistent but relatively small-scale differences in the expression of gene sets that belong to functional pathways 12, 13. We tested five gene sets that were previously reported to be associated with BCBM from breast cancer 7, 23, 32-34. Four of the five BCBM-specific gene sets were not validated in our patients for their predictive value for occurrence of BCBM. Only the BCBM-specific gene set from the study by Kang et al. 33, which was detected using the breast cancer cell line MDA-MB-231, was significantly overexpressed in the bone cohort (Table 3). Also, when we compared the bone and non-bone cohorts by ER status separately, in ER-positive patients, only that gene set was significantly overexpressed in the bone cohort. None of the five sets were overexpressed in ER-negative patients.
Table 3.
Comparison of overexpression of previously reported BCBM-associated gene sets between the bone-metastasis and non-bone-metastasis cohorts when stratified by ER status
| Gene sets | Number of genes | All | ER positive | ER negative |
|---|---|---|---|---|
| P value (direction) | P value (direction) | P value (direction) | ||
| Kang et al [33] | 15 | 0.013 (Bone) | 0.028 (Bone) | 0.259 (Bone) |
| Wilson et al [34] | 31 | 0.164 (Others) | 0.238 (Bone) | 0.397 (Others) |
| Nannuru et al [32] | 23 | 0.448 (Bone) | 0.378 (Bone) | 0.13 (Bone) |
| Zhang et al [23] | 36 | 0.193 (Bone) | 0.225 (Bone) | 0.391 (Others) |
| Smid et al [7] | 86 | 0.001 (Others) | 0.029 (Others) | 0.166 (Others) |
*Efron-Tibshirani's GSA test.
Identification of biological difference between ER status
The outcome that we could not identify BCBM-specific genes when patients were stratified by ER status indicated the possibility of the presence of a biological difference between ER status in development of BCBM. Therefore, for the second aim of our study, we sought to detect, using GSA, differences in biological features related to developing BCBM between ER-positive and ER-negative breast cancers. We also determined 2,246 functionally annotated gene sets assembled from Gene Ontology (http://www.geneontology.org/) to examine possible differences between bone and non-bone cohorts over a broad range of biological processes (Table 4). At a P value of ≤0.05 by Efron-Tibshirani test, 151 gene sets in ER-positive breast cancer and 125 gene sets in ER-negative breast cancer were overexpressed in the bone metastasis cohort compared to the non-bone metastasis cohort. Only 13 gene sets were common to both ER-positive and ER-negative breast cancers. These results demonstrated that the majority of bone-metastasis-related pathways were differently expressed in ER-positive (138/151, 91.4%) and ER-negative breast cancers (112/125, 89.6%).
Table 4.
Number of gene sets overexpressed in the bone metastasis cohort when stratified by ER status
| P value* | ER positive | ER negative | Common |
|---|---|---|---|
| 0.1 | 313 | 261 | 49 |
| 0.05 | 151 | 125 | 13 |
| 0.01 | 22 | 21 | 0 |
ER = estrogen receptor.
*bone metastasis vs non-bone metastasis
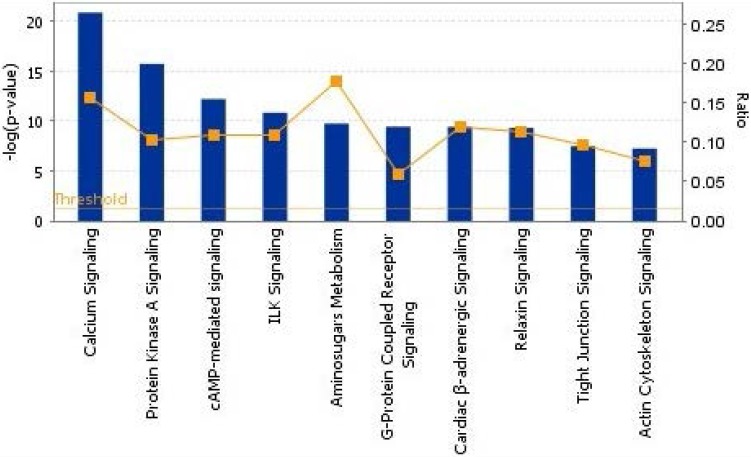
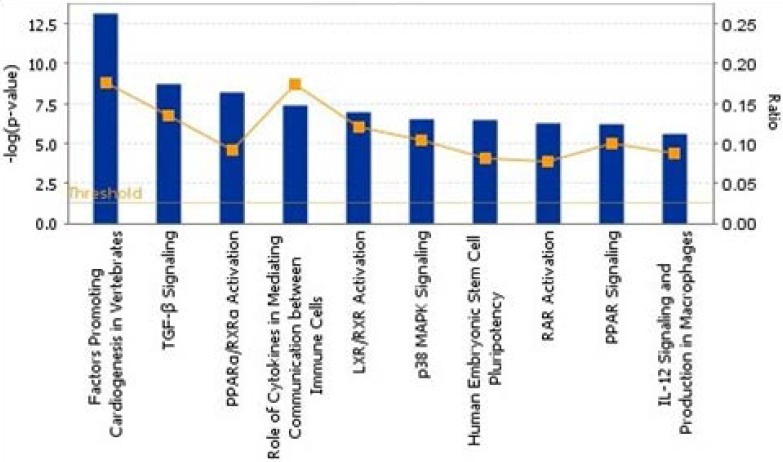
To show the biological process, we mapped the top 10 canonical pathways significantly overexpressed for the bone metastasis cohort in ER-positive breast cancers and in ER-negative breast cancers using the top 20 gene sets overexpressed in ER-positive breast cancers and ER-negative breast cancers by Ingenuity pathway analysis, separately (Figure 1 and 2). Cellular growth and proliferation, as well as intracellular and second messenger signaling pathways were overexpressed for the bone cohort in ER-positive breast cancer (Figure 1). Nuclear receptor signaling and cytokine signaling-related pathways were overexpressed in ER-negative breast cancer (Figure 2). Kang et al. 33 have reported that transforming growth factor β (TGF-β) signaling plays an important role for BCBM; in our study, the TGF-β signaling pathway was the 2nd most highly overexpressed pathway for the bone cohort in ER-negative breast cancer.
Figure 1.
The top 10 canonical pathways by significantly overexpressed genes associated with bone metastasis in ER-positive breast cancers assessed by Ingenuity pathway analysis software results. The bars show logarithms of P values that were calculated by Fisher's exact test. The line indicates the ratio for the percentage of genes in a pathway that were found in our uploaded list. The threshold indicates the line at P = 0.05.
Figure 2.
The top 10 canonical pathways by significantly overexpressed genes associated with bone metastasis in ER-negative breast cancers assessed by Ingenuity pathway analysis software results.
Discussion
We demonstrated that there are no unique genes that predict BCBM when we stratified the ER status. This is partly due to the major biological difference between ER-positive and ER-negative groups in development of BCBM, as we hypothesized. Notably, many genes were overexpressed in the bone metastasis cohort as a whole, without stratification by ER positivity, as described in previous studies. However, when gene-expression-level analysis was performed with ER status stratification, there were no bone-metastasis-specific genes. Therefore, these results suggest that ER-positive and ER-negative breast cancers have a biological difference in their mechanism to develop BCBM.
When we validated five previously reported bone-associated gene sets, one showed significant association with BCBM in ER-positive breast cancer in our patients. Kang et al. identified that set of 122 bone-metastasis-specific genes from tumor cells of BCBM of mice injected with MDA-MB-231 cells 33. The top 11 genes overexpressed in BCBM in the study were validated as significant in ER-positive but not in ER-negative breast cancer in our validation analysis.
From 107 primary breast tumor samples, Smid et al. 7 found 69 genes significantly differentially expressed between patients who experienced relapse to bone and those who experienced relapse to non-bone sites. Combining their 69-gene list with that of Kang et al. 7, 23, 32-34 revealed that the fibroblast growth factor receptor-p42/44 mitogen-activated protein kinase (FGFR-MAP) K signaling pathway was in preference of tumor cells that metastasize to bone in an animal model system. They developed a classifier of 31 genes, which in an independent validation set correctly predicted all tumors spreading to bone with a specificity of 50%. They suggested that this model could predict BCBM more accurately than ER status. However, we could not identify a BCBM-specific gene signature by gene-expression-level analysis using our dataset in either ER-positive or ER-negative breast cancers because of the very subtle differences in the expression of genes.
Kang et al. 33 also reported that the TGF-β signaling pathway is associated with occurrence of BCBM, inducing overexpression of interleukin-11 (IL-11) and CTGF, which were the most highly overexpressed genes in osteolytic metastasis. TGF-β not only facilitates tumorigenesis and producing osteoclasts 35, 36 but also causes osteolytic change by inducing expression of PTHrP 31 and IL-11 37, RANKL 27, and matrix metalloproteinases 25, 29. Furthermore, TGF-β stimulates angiogenesis 38 and induces epithelial-to-mesenchymal transition 39, 40. Similar to Kang et al.'s description 33, the TGF-β signaling pathway was significantly overexpressed for our bone cohort in ER-negative breast cancer compared to our non-bone cohort (P < 0.05).
There are some limitations of exploratory research using microarray analysis for prediction of BCBM. Only a few genes that were detected to be associated with BCBM overlapped among previous studies 7, 23, 32-34. In addition, many genes relating to proteins such as PTHrP, IL-8, insulin-like growth factor (IGF-1), and platelet-derived growth factor (PDGF), which have been reported as important factors in development of BCBM, were not identified as bone-metastasis-specific genes in any studies 35, 36. The possible reasons for the differences in gene signatures among the studies may be due to the various backgrounds of the experiments 41. For example, tumor samples in various studies were taken from a breast cancer cell line 33, mouse model 32, 34, or human samples of primary tumor (including stage IV cancers) 7 or of metastatic sites 23. No previous studies analyzed the gene expression in ER-positive and -negative breast cancers separately. Samples were collected by FNA or dissection 7, 23, analyzed for late-onset BCBM 23 or BCBM occurring at any time course, and compared between the tumor-bone interface and tumor itself 32, or among bone metastasis, non-bone metastasis, and no metastasis. These various experimental backgrounds may have induced the different results. Reproducibility of microarray analyses may be another issue 42.
We demonstrated no bone-metastasis-specific genes for early-onset BCBM. But because of the short median follow-up time and small number of events, we still need to determine whether such a specific gene can predict for late onset of BCBM. Because the number of patients with ER-positive breast cancer who develop metastasis will continue to increase 5 years after treatment 43, 44, we need a definitive study with a longer follow-up time and a larger number of events to assess the biological difference in developing BCBM between ER-positive and ER-negative breast cancers.
Our results indicate that the biological differences between ER-positive and ER-negative breast cancers affect development of BCBM. Therefore, we need to assess the biological features associated with BCBM depending on the ER status.
Acknowledgments
We thank Sunita Patterson, from MD Anderson's Department of Scientific Publications, for editorial review.
A part of this study was presented in part at the 34th San Antonio Breast Cancer Symposium, December 6-10, 2011, San Antonio, TX
Financial support
This work was supported by the National Institutes of Health through MD Anderson's Cancer Center Support Grant (CA016672) and an R01 grant (CA123318-01); the Kleberg Center for Molecular Markers at MD Anderson Cancer Center; a donation from Mr. and Mrs. Sidney J. Jansma, Jr.; a grant from the I.C.N. Foundation; and the Nellie B. Connally Breast Cancer Research Fund (to N.T.U.).
References
- 1.Coleman RE, Rubens RD. The clinical course of bone metastases from breast cancer. Br J Cancer. 1987;55:61–6. doi: 10.1038/bjc.1987.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hortobagyi GN. Bone metastases in breast cancer patients. Semin Oncol. 1991;18:11–5. [PubMed] [Google Scholar]
- 3.Solomayer EF, Diel IJ, Meyberg GC, Gollan C, Bastert G. Metastatic breast cancer: clinical course, prognosis and therapy related to the first site of metastasis. Breast Cancer Res Treat. 2000;59:271–8. doi: 10.1023/a:1006308619659. [DOI] [PubMed] [Google Scholar]
- 4.Hortobagyi GN. Novel approaches to the management of bone metastases in patients with breast cancer. Semin Oncol. 2002;29:134–44. doi: 10.1053/sonc.2002.34066. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D. et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24:2261–7. doi: 10.1200/JCO.2005.03.8802. [DOI] [PubMed] [Google Scholar]
- 8.James JJ, Evans AJ, Pinder SE, Gutteridge E, Cheung KL, Chan S. et al. Bone metastases from breast carcinoma: histopathological - radiological correlations and prognostic features. Br J Cancer. 2003;89:660–5. doi: 10.1038/sj.bjc.6601198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA. et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–44. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 10.Gong Y, Yan K, Lin F, Anderson K, Sotiriou C, Andre F. et al. Determination of oestrogen-receptor status and ERBB2 status of breast carcinoma: a gene-expression profiling study. Lancet Oncol. 2007;8:203–11. doi: 10.1016/S1470-2045(07)70042-6. [DOI] [PubMed] [Google Scholar]
- 11.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwamoto T, Bianchini G, Booser D, Qi Y, Coutant C, Shiang CY. et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J Natl Cancer Inst. 2010;103:264–72. doi: 10.1093/jnci/djq524. [DOI] [PubMed] [Google Scholar]
- 13.Iwamoto T, Bianchini G, Qi Y, Cristofanilli M, Lucci A, Woodward WA. et al. Different gene expressions are associated with the different molecular subtypes of inflammatory breast cancer. Breast Cancer Res Treat. 2011;125:785–95. doi: 10.1007/s10549-010-1280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efron B, Tishirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1:107–29. [Google Scholar]
- 16.Jiang Z, Gentleman R. Extensions to gene set enrichment. Bioinformatics. 2007;23:306–13. doi: 10.1093/bioinformatics/btl599. [DOI] [PubMed] [Google Scholar]
- 17.Dai KR, Xu XL, Tang TT, Zhu ZA, Yu CF, Lou JR. et al. Repairing of goat tibial bone defects with BMP-2 gene-modified tissue-engineered bone. Calcif Tissue Int. 2005;77:55–61. doi: 10.1007/s00223-004-0095-z. [DOI] [PubMed] [Google Scholar]
- 18.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol. 2006;175:111–20. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye L, Bokobza S, Li J, Moazzam M, Chen J, Mansel RE. et al. Bone morphogenetic protein-10 (BMP-10) inhibits aggressiveness of breast cancer cells and correlates with poor prognosis in breast cancer. Cancer Sci. 2010;101:2137–44. doi: 10.1111/j.1349-7006.2010.01648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casimiro S, Guise TA, Chirgwin J. The critical role of the bone microenvironment in cancer metastases. Mol Cell Endocrinol. 2009;310:71–81. doi: 10.1016/j.mce.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Onishi T, Hayashi N, Theriault RL, Hortobagyi GN, Ueno NT. Future directions of bone-targeted therapy for metastatic breast cancer. Nat Rev Clin Oncol. 2010;7:641–51. doi: 10.1038/nrclinonc.2010.134. [DOI] [PubMed] [Google Scholar]
- 22.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD. et al. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M. et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabioglu N, Sahin AA, Morandi P, Meric-Bernstam F, Islam R, Lin HY. et al. Chemokine receptors in advanced breast cancer: differential expression in metastatic disease sites with diagnostic and therapeutic implications. Ann Oncol. 2009;20:1013–9. doi: 10.1093/annonc/mdn740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duivenvoorden WC, Hirte HW, Singh G. Transforming growth factor beta1 acts as an inducer of matrix metalloproteinase expression and activity in human bone-metastasizing cancer cells. Clin Exp Metastasis. 1999;17:27–34. doi: 10.1023/a:1026404227624. [DOI] [PubMed] [Google Scholar]
- 26.Garnero P, Borel O, Byrjalsen I, Ferreras M, Drake FH, McQueney MS. et al. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J Biol Chem. 1998;273:32347–52. doi: 10.1074/jbc.273.48.32347. [DOI] [PubMed] [Google Scholar]
- 27.Ishida A, Fujita N, Kitazawa R, Tsuruo T. Transforming growth factor-beta induces expression of receptor activator of NF-kappa B ligand in vascular endothelial cells derived from bone. J Biol Chem. 2002;277:26217–24. doi: 10.1074/jbc.M111093200. [DOI] [PubMed] [Google Scholar]
- 28.Myoui A, Nishimura R, Williams PJ, Hiraga T, Tamura D, Michigami T. et al. C-SRC tyrosine kinase activity is associated with tumor colonization in bone and lung in an animal model of human breast cancer metastasis. Cancer Res. 2003;63:5028–33. [PubMed] [Google Scholar]
- 29.Sanchez-Sweatman OH, Lee J, Orr FW, Singh G. Direct osteolysis induced by metastatic murine melanoma cells: role of matrix metalloproteinases. Eur J Cancer. 1997;33:918–25. doi: 10.1016/s0959-8049(97)00513-3. [DOI] [PubMed] [Google Scholar]
- 30.Silver IA, Murrills RJ, Etherington DJ. Microelectrode studies on the acid microenvironment beneath adherent macrophages and osteoclasts. Exp Cell Res. 1988;175:266–76. doi: 10.1016/0014-4827(88)90191-7. [DOI] [PubMed] [Google Scholar]
- 31.Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R. et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nannuru KC, Futakuchi M, Varney ML, Vincent TM, Marcusson EG, Singh RK. Matrix metalloproteinase (MMP)-13 regulates mammary tumor-induced osteolysis by activating MMP9 and transforming growth factor-beta signaling at the tumor-bone interface. Cancer Res. 2010;70:3494–504. doi: 10.1158/0008-5472.CAN-09-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 34.Wilson TJ, Nannuru KC, Futakuchi M, Sadanandam A, Singh RK. Cathepsin G enhances mammary tumor-induced osteolysis by generating soluble receptor activator of nuclear factor-kappaB ligand. Cancer Res. 2008;68:5803–11. doi: 10.1158/0008-5472.CAN-07-5889. [DOI] [PubMed] [Google Scholar]
- 35.Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–64. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Karsenty G. Transcription factors in bone: developmental and pathological aspects. Trends Mol Med. 2002;8:340–5. doi: 10.1016/s1471-4914(02)02340-7. [DOI] [PubMed] [Google Scholar]
- 37.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR. et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–14. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth-inhibiting factors, and their receptors in invasive breast cancer. II: Correlations with proliferation and angiogenesis. J Pathol. 1998;184:53–7. doi: 10.1002/(SICI)1096-9896(199801)184:1<53::AID-PATH6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Beck C, Schreiber H, Rowley D. Role of TGF-beta in immune-evasion of cancer. Microsc Res Tech. 2001;52:387–95. doi: 10.1002/1097-0029(20010215)52:4<387::AID-JEMT1023>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 40.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127:2021–36. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YC, Sosnoski DM, Mastro AM. Breast cancer metastasis to the bone: mechanisms of bone loss. Breast Cancer Res. 2010;12:215. doi: 10.1186/bcr2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pusztai L, Hess KR. Clinical trial design for microarray predictive marker discovery and assessment. Ann Oncol. 2004;15:1731–7. doi: 10.1093/annonc/mdh466. [DOI] [PubMed] [Google Scholar]
- 43.Demicheli R, Abbattista A, Miceli R, Valagussa P, Bonadonna G. Time distribution of the recurrence risk for breast cancer patients undergoing mastectomy: further support about the concept of tumor dormancy. Breast Cancer Res Treat. 1996;41:177–85. doi: 10.1007/BF01807163. [DOI] [PubMed] [Google Scholar]
- 44.Karrison TG, Ober C. Recurrent miscarriage (REMIS) study: how should data from women who do not become pregnant be handled? Control Clin Trials. 1998;19:430–9. doi: 10.1016/s0197-2456(98)00019-1. [DOI] [PubMed] [Google Scholar]