Abstract
Pemphigus is an autoimmune disease that causes blisters and erosions in the skin and mucous membranes. The development of pemphigus is associated with the imbalance of T-cell and humoral responses. MicroRNAs (miRNAs) can regulate many cell functions. However, whether miRNA expression is altered in peripheral blood mononuclear cells (PBMCs) during the pathogenesis of pemphigus has not been clarified. The aim of the present study was to examine the miRNA expression profiles of PBMCs from patients with pemphigus. The expression profiles of miRNAs in PBMCs from patients with active pemphigus (n=3) and healthy subjects (n=3) were analyzed by microarray. The relative levels of miR-424-5p expression in PBMCs from 9 patients and controls were validated by RT-qPCR. The functional and biological processes of the differentially expressed miRNAs were analyzed by bioinformatics. There were 124 differentially expressed miRNAs in PBMCs from the patients with pemphigus, compared with healthy controls, including 71 that were upregulated (P<0.05, fold change >2), and 53 that were downregulated (P<0.05, fold change <0.5). miR-424-5p was highly expressed in patients with pemphigus. Bioinformatics analysis indicated that the genes targeted by miR-424-5p were involved in intracellular signaling cascades, phosphate metabolism and regulation of kinase activity. The predicted target genes were associated with the T-cell receptor and mitogen-activated protein kinase signaling pathways as well as others. In conclusion, the results have demonstrated the miRNA expression profile, and verified that miR-424-5p was upregulated in PBMCs from patients with pemphigus. The biological function and potential pathways of miR-424-5p in pemphigus were predicted. Thus, miR-424-5p may contribute to the pathogenesis of pemphigus.
Keywords: pemphigus, microarray, miR-424-5p, bioinformatics analysis
Introduction
Pemphigus is a group of blistering autoimmune diseases affecting the skin and mucous membranes (1). Although the cause of pemphigus remains unknown, it is thought that humoral responses to desmogleins are associated with the development of bullation. Desmoglein 1 and 3 are the main components of desmosomes, involved in connecting keratinocytes. Autoantibodies against desmogleins promote the hydrolysis of plasminogen into plasmin in keratinocytes, leading to the loss of cell-cell adhesion and the formation of blisters. It is well known that CD4+ T cells can regulate humoral responses and may contribute to the pathogenesis of pemphigus. Pro-inflammatory T cells enhance humoral responses while regulatory T cells (Tregs) inhibit B-cell activation and antibody production (1–4). However, T-cell activation and functional regulation of the humoral response during the pathogenesis of pemphigus have not been clarified.
MicroRNAs (miRNAs) are small non-coding RNAs that regulate growth, development, aging, apoptosis and other important processes in vivo (5). Currently, thousands of miRNAs have been identified in humans and many of them are expressed in immune cells. They are involved in regulating maturation, differentiation and signal transduction (6–8). Some miRNAs can regulate the development and progression of autoimmune diseases by modulating T- and B-cell function (9,10). However, to the best of our knowledge, there is no information on how miRNA expression profiles change in human peripheral blood mononuclear cells (PBMCs) during the pathogenesis of pemphigus.
To explore the potential role of miRNAs in the pathogenesis of pemphigus, we characterized the expression profiles of miRNAs in PBMCs from patients with pemphigus and age- and gender-matched healthy subjects by microarray analysis. Furthermore, we validated the high levels of miR-424-5p expression in PBMCs from patients with pemphigus. The potential gene targets of miR-424-5p and their possible functional networks were predicted by bioinformatics.
Patients and methods
Human subjects
Patients with pemphigus diagnosed according to their clinical, histopathological, and immunological parameters were recruited at the Nanfang Hospital of Southern Medical University (Guangzhou, China) (11). They had new blisters and had not been treated with immunosuppressive drugs. The exclusion criteria were serious systemic disease, infection, tumors or any other autoimmune disease. The age- and gender-matched healthy subjects were recruited from the Physical Examination Center of the same hospital. Written informed consent was obtained from individual participants and the experimental protocol was approved by the Institute Review Board of Nanfang Hospital of Southern Medical University.
Blood sample collection and pretreatment
A volume of 5 ml of venous blood was obtained from the patients and controls, and plasma samples were prepared. PBMCs were isolated by Ficoll-Hypaque (TBD Science, Tianjin, China) density gradient centrifugation, lysed in Mix RNAiso blood buffer (Takara Bio, Inc., Otsu, Japan) and stored at −80°C until use.
Isolation and quality control of RNA
Total RNA was extracted from individual PBMC samples and purified using the miRNeasy Mini kit (cat. no. 217004; Qiagen GmbH, Hilden, Germany), according to the manufacturer's instructions. The integration of the purified RNA was characterized in the Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA).
miRNA microarray analysis
The total RNAs were dephosphorylated by phosphatase and incubated with the Labeling Spike-In kit (Agilent Technologies Inc.) at 37°C for 30 min. The dephosphorylated RNAs were denatured by dimethyl sulfoxide (DMSO) and subsequently incubated at 16°C in a circulating water-bath or cool block for 2 h. The labeled RNAs were purified with spin columns to remove DMSO in the samples, dried in vacuum concentrators at 45–55°C for 1 h and dissolved in nuclease-free water. The dissolved RNAs were mixed with Hyb Spike-In solution (Agilent Technologies Inc.) to assemble the hybridization mixture. The mixture was hybridized to the Agilent Human miRNA array V19.0 (Agilent Technologies Inc.), which covers 2,006 human miRNAs, at 55°C for 20 h. The arrays were washed with the Gene Expression Wash Buffer kit and subsequently scanned by the Agilent Microarray Scanner (both from Agilent Technologies Inc.). Data on miRNA microarray images were extracted by Feature Extraction software 10.7.1.1 and normalized by Gene Spring software 12.6 (both from Agilent Technologies, Inc.). The similarity between the samples was analyzed by the principal component analysis (PCA) and correlation plot.
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was used to validate the expression level of differentially expressed miRNAs in 9 patients and 9 healthy controls. Total RNAs were isolated from PBMCs and subsequently reverse transcribed to cDNAs using miScript II RT kit (Qiagen, Valencia, CA, USA). Briefly, 1 µg total RNA was used as template in the reaction, and reverse transcribed in a 20-µl reaction mixture containing 4 µl 5X miScript HiFlex Buffer, 2 µl 10X miScript Nucleics Mix, 2 µl miScript Reverse Transcriptase Mix and 12 µl H2O. The thermal profile conditions for reverse transcription were 37°C for 60 min, 95°C for 5 min and 4°C indefinitely. The prepared cDNAs were stored at −20°C until use.
The relative levels of miRNA transcripts normalized to the internal control (U6) were determined by qPCR using the miRcute miRNA qPCR Detection kit containing a universal 3′ primer (SYBR-Green; Tiangen Biotech Co., Ltd., Beijing, China) on a 7900 HT Sequence Detection system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The 5′ sequences of primers for U6, 5′-GCTCGCTTCGGCAGCACAT-3′ and for miR-424-5p, 5′-CAGCAGCAATTCATGTTTTAAAA-3′ (Invitrogen Life Technologies). The reaction mixture contained 5 µl 2X miRcute miRNA Premix, 0.8 µl 50X ROX Reference Dye, 0.2 µl forward primer, 0.2 µl reverse primer, 1 µl prepared RT products and 2.8 µl H2O. All reactions were ran in triplicate at 95°C for 2 min, and subjected to 40 cycles at 94°C for 15 sec and 60°C for 1 min. The data were first normalized to the levels of U6 and analyzed by 2−ΔΔCq.
Statistical analysis
The differences between groups were analyzed using the Student's t-test. Data from the miRNA microarrays were considered differentially expressed when there was a P-value of <0.05 and fold changes were >2-fold or <0.5-fold. The target genes of differentially expressed miRNA were predicted with miRanda, miRWalk and TargeScan. The biological processes and signaling pathways of the differentially expressed miRNAs were analyzed by the Gene Ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Microarray analysis
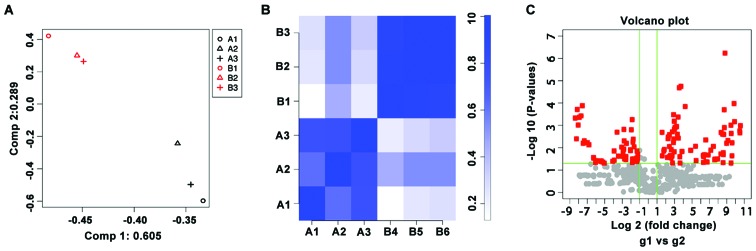
To analyze the miRNA expression profiles, 3 patients with pemphigus and 3 healthy subjects were recruited and RNA from PBMCs was extracted for microarray analysis. The similarity and correlation of miRNA expression profiles between the patients and healthy subjects were analyzed by the PCA and correlation plot (Fig. 1). There was clear separation in the miRNA profiles between the 2 groups and the intra-group miRNA profiles had high correlation coefficients. Therefore, the samples in patients were different from healthy controls according to the biological characteristics, indicating that the samples were well selected. There were 71 upregulated (P<0.05, fold change >2) and 53 downregulated (P<0.05, fold change <0.5) miRNAs on the Agilent Human miRNA array V19.0. Of these, miR-424-5p was one of the miRNAs that was upregulated by >1,000-fold in the patients with pemphigus (Table I).
Figure 1.
(A) The PCA analysis of miRNA microarray data. Group A represents samples from patients, while group B represents samples from healthy controls. (B) Correlation plot analysis of miRNA microarray data. The correlation coefficients between the samples are expressed by the shades of color. (C) Volcano plot analysis of miRNA microarray data. The dots represent the differentially expressed miRNAs selected from 2,006 miRNA probes. PCA, principal component analysis; microRNA, miRNA.
Table I.
Ten most upregulated or downregulated miRNAs in PBMC from patients with pemphigus vs. healthy controls.
| miRNAs | Fold change | P-value | miRNA | Fold change | P-value |
|---|---|---|---|---|---|
| miR-424-5p | 1507.524 | 0.0010 | miR-595 | 0.007866 | 0.0060 |
| miR-338-3p | 1451.776 | 0.0021 | miR-557 | 0.006176 | 0.0049 |
| miR-340-5p | 1430.471 | 0.0018 | miR-4726-5p | 0.005665 | 0.0062 |
| miR-30e-3p | 942.3967 | 0.0007 | miR-4472 | 0.005267 | 0.0001 |
| miR-145-5p | 908.3626 | 0.0001 | miR-4632-5p | 0.004766 | 0.0004 |
| miR-130b-3p | 781.2204 | 0.0060 | miR-5088 | 0.004145 | 0.0004 |
| miR-199b-5p | 772.3043 | 0.0230 | miR-3648 | 0.003781 | 0.0010 |
| miR-128 | 732.1749 | 0.0023 | miR-4430 | 0.00352 | 0.0002 |
| miR-590-5p | 722.1934 | 0.0479 | miR-4767 | 0.003312 | 0.0040 |
| miR-324-5p | 714.0275 | 0.0010 | miR-1180 | 0.003013 | 0.0005 |
MicroRNAs, miRNAs; PBMC, peripheral blood mononuclear cell.
RT-qPCR analysis of miR-424-5p
To validate the higher expression of miR-424-5p, another 9 patients and age- and gender-matched healthy subjects were recruited and the levels of miR-424-5p in their PBMCs were determined by RT-qPCR. The relative levels of miR-424-5p normalized to U6 in the PBMCs from the patients were significantly higher than in the healthy subjects (Fig. 2).
Figure 2.
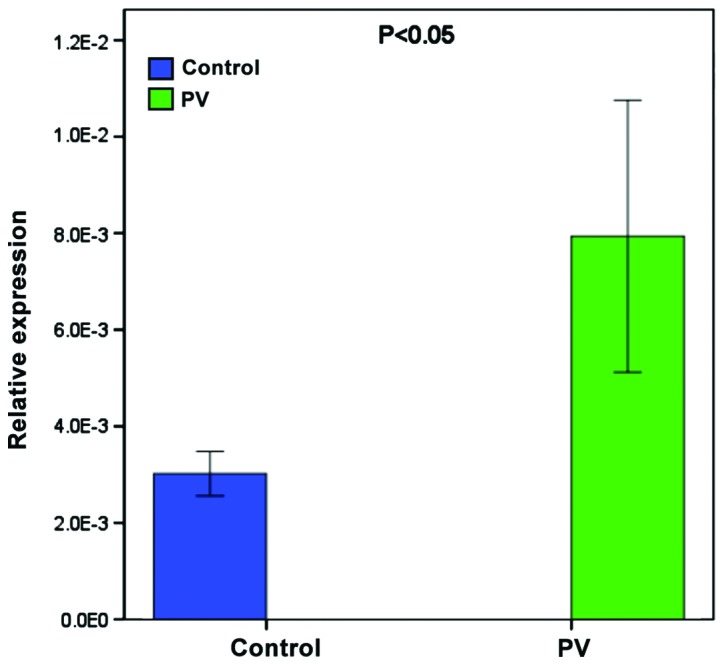
Validation of higher levels of miR-424-5p expression in patients with pemphigus. The relative levels of miR-424-5p normalized to U6 expression in PBMC from 9 patients with pemphigus and 9 healthy subjects were determined by qRT-PCR. Data are expressed as mean ± SD of each group. PBMC, peripheral blood mononuclear cell; SD, standard deviation.
Bioinformatics analysis
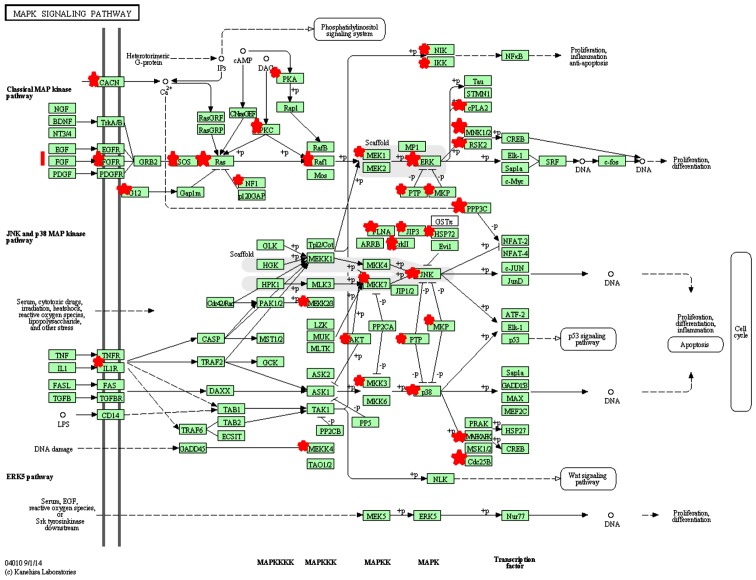
The target genes of miR-424-5p were predicted using miRanda, miRWalk and TargeScan. As predicted, an intersection of 3,539 miRNAs were regulated by miR-424-5p. Of these, 2,430 miRNAs were expressed in humans. GO analysis indicated the functional categorization of predicted target genes. There were 20 biological processes (P<0.05, Benjamini <0.05) (12), including intracellular signaling cascades, phosphate metabolism and regulation of kinase activity (Table II). There were 242 potential target genes involved in the process of intracellular signal transduction. Functional annotation of miR-424-5p was analyzed by KEGG pathway analysis. The target genes were enriched in 10 signaling pathways (P<0.05, Benjamini <0.05), including the T-cell receptor signaling pathway and mitogen-activated protein kinase (MAPK) signaling pathway, suggesting that miR-424-5p may influence these signaling pathways through target genes (Table III and Fig. 3).
Table II.
Biological process and functional annotation of hsa-miR-424-5p analyzed by GO.
| GO_ID | Term | Count of genes | P-value | Benjamini |
|---|---|---|---|---|
| GO:003556 | Intracellular signal transduction | 242 | 7.9E-10 | 3.3E-6 |
| GO:0051174 | Regulation of phosphorus metabolic process | 110 | 1.7E-8 | 3.5E-5 |
| GO:0019220 | Regulation of phosphate metabolic process | 110 | 1.7E-8 | 3.5E-5 |
| GO:0042325 | Regulation of phosphorylation | 106 | 2.7E-8 | 3.7E-5 |
| GO:0043549 | Regulation of kinase activity | 85 | 9.4E-8 | 9.7E-5 |
| GO:0045859 | Regulation of protein kinase activity | 82 | 1.8E-7 | 1.4E-4 |
| GO:0051338 | Regulation of transferase activity | 85 | 6.1E-7 | 4.2E-4 |
| GO:0006796 | Phosphate metabolic process | 183 | 7.4E-7 | 4.4E-4 |
| GO:0006793 | Phosphorus metabolic process | 183 | 7.4E-7 | 4.4E-4 |
| GO:0006468 | Protein amino acid phosphorylation | 133 | 1.4E-6 | 7.0E-4 |
| GO:0007264 | Small GTPase mediated signal transduction | 72 | 1.4E-6 | 6.4E-4 |
| GO:0035556 | Protein kinase cascade | 81 | 6.5E-6 | 2.7E-3 |
| GO:0016055 | Wnt receptor signaling pathway | 36 | 4.8E-5 | 1.8E-2 |
| GO:0006812 | Cation transport | 107 | 6.0E-5 | 2.1E-2 |
| GO:0001932 | Regulation of protein amino acid phosphorylation | 43 | 7.0E-5 | 2.2E-2 |
| GO:0016310 | Phosphorylation | 145 | 8.1E-5 | 2.4E-2 |
| GO:0033674 | Positive regulation of kinase activity | 53 | 8.9E-5 | 2.4E-2 |
| GO:0006811 | Ion transport | 139 | 1.2E-4 | 3.1E-2 |
| GO:0000165 | MAPK cascade | 44 | 1.5E-4 | 3.5E-2 |
| GO:0030001 | Metal ion transport | 91 | 1.5E-4 | 3.4E-2 |
P-value <0.05, Benjamini <0.05. GO, Gene Ontology; MAPK, mitogen-activated protein kinase.
Table III.
KEGG pathway analysis of miR-424-5p.
| KEGG pathway term | Count of genes | P-value | Benjamini |
|---|---|---|---|
| Insulin signaling pathway | 43 | 5.4E-8 | 1.0E-5 |
| Pathways in cancer | 74 | 4.1E-6 | 3.8E-4 |
| Neurotrophin signaling pathway | 35 | 2.3E-5 | 8.5E-4 |
| Wnt signaling pathway | 39 | 6.2E-5 | 1.9E-3 |
| mTOR signaling pathway | 19 | 8.0E-5 | 2.1E-3 |
| T cell receptor signaling pathway | 29 | 3.4E-4 | 4.8E-3 |
| GnRH signaling pathway | 27 | 3.7E-4 | 4.9E-3 |
| Fc epsilon RI signaling pathway | 23 | 4.1E-4 | 5.1E-3 |
| ErbB signaling pathway | 24 | 8.3E-4 | 8.6E-3 |
| MAPK signaling pathway | 52 | 4.8E-3 | 3.8E-2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase.
Figure 3.
The MAPK signaling pathway. Genes with red stars are targeted by miR-424-5p. MAPK, mitogen-activated protein kinase.
Discussion
The present study examined the miRNA expression profiles in PBMCs from patients with pemphigus and healthy subjects by miRNA microarray analysis. We identified 124 differentially expressed miRNAs, 71 of which were upregulated and 53 of which were downregulated. Interestingly, we found higher levels of miR-424-5p expression in PBMCs from patients with pemphigus. We also described the biological function and regulation network of miR-424-5p, which may help explore the regulatory role of miR-424-5p in the pathogenesis of pemphigus.
Previous studies reported that miR-424-5p expression was dysregulated and correlated with age-related macular degeneration (13), endometriosis (14), cancer (15,16) and Mycobacterium tuberculosis infection (17). The difference is likely due to the different biological processes regulated by miR-424-5p in different tissues and organs. However, our data are in disagreement with a previous study that miR-424-5p expression was significantly decreased specifically in psoriasis skin. Downregulation of miR-424-5p expression leads to overexpression of the target genes MEK1 and cyclin E1, causing the hyper-proliferation of keratinocytes (18). Furthermore, miR-424 promotes monocyte differentiation, together with miR-155, miR-222 and miR-503 (19). The difference may stem from the different types of cells examined.
We analyzed the potential function of miR-424-5p by bioinformatics and found many potential target genes in functionally related groups. GO and KEGG analysis indicated that the biological function of the potential genes targeted by miR-424-5p were enriched in intracellular signaling cascades, phosphate metabolism and regulation of kinase activity. These potential target genes may regulate a wide range of pathways, such as the p38 and other MAPK signaling pathways. The MAPK signaling pathway is dysregulated in the pathogenesis of pemphigus (20). miR-424-5p may regulate the pathogenesis of pemphigus by targeting the MAPK signaling pathway. A previous study showed that p38 MAPK is important for regulating autoimmune responses, cell survival, proliferation, differentiation and apoptosis (21). Activation of p38 MAPK was associated with collapse of the cytoskeleton, disassembly of desmosomes, and keratinocyte apoptosis (22). Pemphigus-specific autoantibodies can induce p38 MAPK activation in human keratinocytes and mouse keratinocytes, and inhibition of p38 MAPK activity can prevent pemphigus-specific, autoantibody-induced cytoskeletal reorganization, and heat shock protein (HSP) 27 phosphorylation in human keratinocytes (23). Furthermore, treatment with p38 inhibitors prevented skin blistering by inhibiting pemphigus IgG-activated signaling in epidermal cells in a mouse model of pemphigus induced by adoptive transfer of pemphigus-specific antibodies (24,25). Thus, the p38 MAPK signaling pathway is crucial for the pathogenesis of pemphigus. In addition, HSP27 participates in the pathogenesis of pemphigus and phosphorylated HSP27 is observed in skin biopsies from patients with pemphigus, mouse pemphigus models and cultured keratinocytes. HSP27 phosphorylation has been considered to be involved in regulation of the cytoskeletal assembly of actin filaments and keratin intermediate filaments. Dysregulated cytoskeletal arrangements are associated with the loss of cell-cell adhesion, and it is possible that miR-424-5p regulates p38 MAPK activation and HSP27 phosphorylation, and blister formation during the pathogenesis of pemphigus (23,25,26).
We predicted that there were 52 target genes of miR-424-5p enriched in the MAPK signaling pathway. This indicates that miR-424-5p may regulate p38 MAPK activation and HSP27 phosphorylation in human PBMCs. miR-424-5p may regulate HSP27 phosphorylation by targeting the upstream MAPKAPK3 and CDC25B. MAPKAPK3 is a member of the Ser/Thr protein kinase family. This kinase can be activated by growth factors and stress stimuli, and is involved in cell responses and gene regulation (27). CDC25B is a member of the CDC25 phosphatase family and is regulated by the p38 MAPK and/or MAPKAP kinase-2 pathways (28,29). The dysregulated expression of MAPKAPK3 is associated with the development of various types of cancer, but its specific role in tumor formation remains to be determined (30–32). Further investigations are required focusing on the exact roles of MAPKAPK3 and CDC25B in the pathogenesis of pemphigus.
In conclusion, we examined the miRNA expression profile of PBMCs from patients with pemphigus by miRNA microarray analysis. To the best of our knowledge, we are the first to identify differentially expressed miRNAs in PBMCs from patients with pemphigus and demonstrated higher levels of miR-424-5p in PBMCs from these patients. A bioinformatics approach predicted the potential target genes, biological functions, and pathways of miR-424-5p, indicating that miR-424-5p may contribute to the pathogenesis of pemphigus by regulating the p38 MAPK signaling pathway. The influence of miR-424-5p on HSP27 phosphorylation by targeting MAPKAPK3 and CDC25B may also regulate the pathogenesis of pemphigus.
References
- 1.Rizzo C, Fotino M, Zhang Y, Chow S, Spizuoco A, Sinha AA. Direct characterization of human T cells in pemphigus vulgaris reveals elevated autoantigen-specific Th2 activity in association with active disease. Clin Exp Dermatol. 2005;30:535–540. doi: 10.1111/j.1365-2230.2005.01836.x. [DOI] [PubMed] [Google Scholar]
- 2.Sugiyama H, Matsue H, Nagasaka A, Nakamura Y, Tsukamoto K, Shibagaki N, Kawamura T, Kitamura R, Ando N, Shimada S. CD4+CD25high regulatory T cells are markedly decreased in blood of patients with pemphigus vulgaris. Dermatology. 2007;214:210–220. doi: 10.1159/000099585. [DOI] [PubMed] [Google Scholar]
- 3.von Bubnoff D, Andrès E, Hentges F, Bieber T, Michel T, Zimmer J. Natural killer cells in atopic and autoimmune diseases of the skin. J Allergy Clin Immunol. 2010;125:60–68. doi: 10.1016/j.jaci.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 4.Xu RC, Zhu HQ, Li WP, Zhao XQ, Yuan HJ, Zheng J, Pan M. The imbalance of Th17 and regulatory T cells in pemphigus patients. Eur J Dermatol. 2013;23:795–802. doi: 10.1684/ejd.2013.2177. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 7.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 8.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. 2015;125:2242–2249. doi: 10.1172/JCI78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heegaard NH, Carlsen AL, Skovgaard K, Heegaard PM. Circulating extracellular microRNA in systemic autoimmunity. EXS. 2015;106:171–195. doi: 10.1007/978-3-0348-0955-9_8. [DOI] [PubMed] [Google Scholar]
- 11.Amagai M, Tanikawa A, Shimizu T, Hashimoto T, Ikeda S, Kurosawa M, Niizeki H, Aoyama Y, Iwatsuki K, Kitajima Y. Committee for Guidelines for the Management of Pemphigus Disease: Japanese guidelines for the management of pemphigus. J Dermatol. 2014;41:471–486. doi: 10.1111/1346-8138.12486. [DOI] [PubMed] [Google Scholar]
- 12.Noble WS. How does multiple testing correction work? Nat Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassmann F, Schoenberger PG, Brandl C, Schick T, Hasler D, Meister G, Fleckenstein M, Lindner M, Helbig H, Fauser S, et al. A circulating microRNA profile is associated with late-stage neovascular age-related macular degeneration. PLoS One. 2014;9:e107461. doi: 10.1371/journal.pone.0107461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braza-Boïls A, Salloum-Asfar S, Marí-Alexandre J, Arroyo AB, González-Conejero R, Barceló-Molina M, García-Oms J, Vicente V, Estellés A, Gilabert-Estellés J, et al. Peritoneal fluid modifies the microRNA expression profile in endometrial and endometriotic cells from women with endometriosis. Hum Reprod. 2015;30:2292–2302. doi: 10.1093/humrep/dev204. [DOI] [PubMed] [Google Scholar]
- 15.Wu K, Hu G, He X, Zhou P, Li J, He B, Sun W. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19:739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia X, Zhao W, Huai W, Qiu Y, Sun L, et al. miR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. doi: 10.1038/srep06248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng QL, Liu F, Yang XY, Liu XM, Zhang X, Zhang C, Zhang ZD. Identification of latent tuberculosis infection-related microRNAs in human U937 macrophages expressing Mycobacterium tuberculosis Hsp16.3. BMC Microbiol. 2014;14:37. doi: 10.1186/1471-2180-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinnin M. Recent progress in studies of miRNA and skin diseases. J Dermatol. 2015;42:551–558. doi: 10.1111/1346-8138.12904. [DOI] [PubMed] [Google Scholar]
- 19.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Ishii N, Ohata C, Furumura M, Hashimoto T. Signalling pathways in pemphigus vulgaris. Exp Dermatol. 2014;23:155–156. doi: 10.1111/exd.12317. [DOI] [PubMed] [Google Scholar]
- 21.Coulthard LR, White DE, Jones DL, McDermott MF, Burchill SA. p38(MAPK): stress responses from molecular mechanisms to therapeutics. Trends Mol Med. 2009;15:369–379. doi: 10.1016/j.molmed.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chernyavsky AI, Arredondo J, Kitajima Y, Sato-Nagai M, Grando SA. Desmoglein versus non-desmoglein signaling in pemphigus acantholysis: characterization of novel signaling pathways downstream of pemphigus vulgaris antigens. J Biol Chem. 2007;282:13804–13812. doi: 10.1074/jbc.M611365200. [DOI] [PubMed] [Google Scholar]
- 23.Berkowitz P, Hu P, Liu Z, Diaz LA, Enghild JJ, Chua MP, Rubenstein DS. Desmosome signaling. Inhibition of p38MAPK prevents pemphigus vulgaris IgG-induced cytoskeleton reorganization. J Biol Chem. 2005;280:23778–23784. doi: 10.1074/jbc.M501365200. [DOI] [PubMed] [Google Scholar]
- 24.Lee HE, Berkowitz P, Jolly PS, Diaz LA, Chua MP, Rubenstein DS. Biphasic activation of p38MAPK suggests that apoptosis is a downstream event in pemphigus acantholysis. J Biol Chem. 2009;284:12524–12532. doi: 10.1074/jbc.M808204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkowitz P, Hu P, Warren S, Liu Z, Diaz LA, Rubenstein DS. p38MAPK inhibition prevents disease in pemphigus vulgaris mice; Proc Natl Acad Sci USA; 2006; pp. 12855–12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkowitz P, Diaz LA, Hall RP, Rubenstein DS. Induction of p38MAPK and HSP27 phosphorylation in pemphigus patient skin. J Invest Dermatol. 2008;128:738–740. doi: 10.1038/sj.jid.5701080. [DOI] [PubMed] [Google Scholar]
- 27.Meunier I, Lenaers G, Bocquet B, Baudoin C, Piro-Megy C, Cubizolle A, Quilès M, Jean-Charles A, Cohen SY, Merle H, et al. A dominant mutation in MAPKAPK3, an actor of p38 signaling pathway, causes a new retinal dystrophy involving Bruch's membrane and retinal pigment epithelium. Hum Mol Genet. 2016;25:916–926. doi: 10.1093/hmg/ddv624. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire M, Ducommun B, Nebreda AR. UV-induced downregulation of the CDC25B protein in human cells. FEBS Lett. 2010;584:1199–1204. doi: 10.1016/j.febslet.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 29.Lemaire M, Froment C, Boutros R, Mondesert O, Nebreda AR, Monsarrat B, Ducommun B. CDC25B phosphorylation by p38 and MK-2. Cell Cycle. 2006;5:1649–1653. doi: 10.4161/cc.5.15.3006. [DOI] [PubMed] [Google Scholar]
- 30.Song GQ, Zhao Y. MicroRNA-211, a direct negative regulator of CDC25B expression, inhibits triple-negative breast cancer cells' growth and migration. Tumour Biol. 2015;36:5001–5009. doi: 10.1007/s13277-015-3151-6. [DOI] [PubMed] [Google Scholar]
- 31.Kar S, Wang M, Ham SW, Carr BI. Fluorinated Cpd 5, a pure arylating K-vitamin derivative, inhibits human hepatoma cell growth by inhibiting Cdc25 and activating MAPK. Biochem Pharmacol. 2006;72:1217–1227. doi: 10.1016/j.bcp.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Wang M, Zhu XY, Wang L, Lin Y. Expression and signi-ficance of CDC25B, PED/PEA-15 in esophageal carcinoma. Cancer Biother Radiopharm. 2015;30:139–145. doi: 10.1089/cbr.2014.1701. [DOI] [PubMed] [Google Scholar]