Abstract
Background: Alginate lyases belonging to polysaccharide lyase family-7 (PL-7) are the most well studied on their structures and functions among whole alginate lyases. However, all characterized PL-7 alginate lyases are from prokaryotic bacteria cells. Here we report the first identification of eukaryotic PL-7 alginate lyase from marine red alga Pyropia yezoensis.
Methods: The cDNA encoding an alginate lyase PyAly was cloned and was used for the construction of recombinant PyAly (rPyAly) expression system in Escherichia coli. Purified rPyAly was assayed to identify its enzymatic properties. Its expression pattern in P. yessoensis was also investigated.
Results: PyAly is likely a secreted protein consisting of an N-terminal signal peptide of 25 residues and a catalytic domain of 216 residues. The amino-acid sequence of the catalytic domain showed 19-29% identities to those of bacterial characterized alginate lyases classified into family PL-7. Recombinant PyAly protein, rPyAly, which was produced with E. coli BL21(DE3) by cold-inducible expression system, drastically decreased the viscosity of alginate solution in the early stage of reaction. The most preferable substrate for rPyAly was the poly(M) of alginate with an optimal temperature and pH at 35oC and 8.0, respectively. After reaction, unsaturated tri- and tetra-saccharides were produced from poly(M) as major end products. These enzymatic properties indicated that PyAly is an endolytic alginate lyase belonging to PL-7. Moreover, we found that the PyAly gene is split into 4 exons with 3 introns. PyAly was also specifically expressed in the gametophytic haplopid stage.
Conclusion: This study demonstrates that PyAly in marine red alga P. yezoensis is a novel PL-7 alginate lyase with an endolytic manner. PyAly is a gametophyte-specifically expressed protein and its structural gene is composed of four exons and three introns. Thus, PyAly is the first enzymatically characterized eukaryotic PL-7 alginate lyase.
Keywords: Alginate, Alginate lyase, poly(M) lyase, Family PL-7, Pyropia yezoensis, Red algae
INTRODUCTION
Life cycle of red alga laver such as Pyropia and Porphyra consists of two generations, leafy gametophyte and filamentous sphorophyte differences in the composition of cell-wall [1]. It has been demonstrated that gametophytic cell wall contains a large amount of β-1,3-xylan, mannan, and sulfated galactan such as porphyran [2], while cellulose is the major polysaccharide in sporophytic cell wall [3]. In addition, cell wall of sporophytes also contains sulfated galactan as a minor polysaccharide; however, its chemical properties are distinctly different from that in gametophytes [4]. These differences in cell-wall constituents between gametophyte and sporophyte suggest that the biosynthesis and degradation pathways for each polysaccharide are significantly different between the two generations. Nevertheless, little is known about the enzymes involved in the synthesis and degradation of cell-wall polysaccharides in Pyropia yezoensis.
In order to elucidate the regulation of cell wall synthesis in P. yezoensis, we have been attempting to identify the genes encoding polysaccharide-degrading enzymes such as xylanase, mannanase, and agarase in the EST database of P. yezoensis ( http://est.kazusa.or.jp/en/plant/porphyra/EST/index.html ) [5,6]. Although we have not yet identified the EST clones encoding these enzymes, EST clones encoding a putative alginate lyase were unexpectedly found. Alginate lyase is an enzyme that depolymerizes alginate, an acidic polysaccharide comprising β-D-mannuroic acid (M) and α-L-guluronic acid (G) [7,8]. Alginate is known as a structural polysaccharide of brown algae [9] and certain bacteria [10-14], but not in red algae. Thus, the presence of the EST-clones encoding alginate lyase-like-protein in the EST database of P. yezoensis seemed to be quite curious for us. If an alginate lyase is expressed by itself in P. yezoensis, this may become a clue to discover a novel function of alginate lyases in marine multicellular algae.
Amino acid sequences of the alginate lyase-like-protein deduced from P. yezoensis EST clones showed the high homology with those of bacterial alginate lyases belonging to family PL-7. Alginate lyases in this family have been well characterized in some bacteria such as Flavobacterium sp. [15,16], Vibrio sp. [17-21], Sphingomonas sp. [22,23], Stre-ptomyces sp. [24,25], Corynebacterium sp. [26], Pseudoalte-romonas sp. [27], Azotobacter vinelandii [28], Pseudomonas aeruginosa [29], Photobacterium sp. [30], Agarivorans sp. [18,31], Saccharophagus degradans [32], Klebsiella pneumonia [33], and Zobellia galactanivorans [34]. Of these, five protein’s three-dimensional structures were determined [23,29,34,35]. According to CAZy database ( http://www.cazy.org/ ), five proteins from eukaryota, P. yezoensis (present study), Achlya hypogyna, Aspergillus oryzae, Botryotinia fuckeliana, Thielavia terrestris, are found in family PL-7 to date, but there are no information on their enzyme properties.
In the present study, we determined enzymatic properties of the alginate lyase PyAly encoded by EST clones of P. yezoensis by using the recombinant protein produced with an Escherichia coli cold-inducible expression system. Moreover, to exclude a possibility of bacterial contamination in the EST database, the presence of introns in the gene for this protein and the expression of this gene in gametophytic and sphorophytic generations of P. yezoensis were examined. As far as we know, this is the first report of a PL-7 alginate-lyase gene in red algae.
MATERIALS AND METHODS
Materials
Alginate from Macrocystis pyrifera was purchased from Sigma (St. Louis, MO, USA) and poly(M), poly(G), and poly(MG) were prepared according to the method of Gacesa and Wusteman [36]. Restriction and DNA-modifying enzymes used were purchased from New England Biolabs (Ipswich, MA, USA), Takara (Shiga, Japan), and Toyobo (Osaka, Japan). A set of E. coli DH5α and plasmid vector pTac-1 and the other set of E. coli BL21(DE3) and cold-shock vector pCold I were purchased from Biodynamics (Tokyo, Japan) and Takara, respectively.
Gametophytes of P. yezoensis
Pyropia yezoensis Ueda (strain TU-1) [37] was cultured in ESL medium containing 3.5% Sealife powder (Marinetech, Tokyo, Japan) and 1% ESS2 stock solution [38].
cDNA Cloning of PyAly from P. yezoensis
Protoplasts were prepared from gametophytes of P. yezoensis (0.01 g) by the method of Araki et al. [39] and total RNA was extracted from the protoplasts with an RNeasy mini kit (Qiagen, Hilden, Germany). First-stranded cDNA was prepared from the total RNA with an oligo-dT primer and PrimeScript reverse transcriptase (Takara), and then subjected to RT-PCR. The RT-PCR was performed with TaKaRa Ex Taq DNA polymerase (Takara). Forward and reverse primers, PA-F1 and PA-R1 (Table 1), were determined by the basis of the nucleotide sequences of the P. yezoensis EST clones accession numbers AV434206 and AV436451, respectively. The PCR was carried out with a successive incubation at 94oC for 2 min, 30 cycles of 94 oC for 30 sec, 55 oC for 30 sec, and at 72 oC for 1 min. Amplified DNA was cloned with a pTac-1 vector, and the nucleotide sequence of the cloned DNA was determined with a BigDye Terminator v3.1 Cycle Sequencing Kit and a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequence data is available from the DDBJ, EMBL and GenBank with an accession number AB512413.
Table 1.
Primers used in this study.
| Primers | Sequences |
|---|---|
| PA-F1 | 5’-GGGCTCGGCCCCTTCGTTTCGGCATAGG-3’ |
| PA-R1 | 5’-GGGCTTGCCCTTCTCGGACACCATCATC-3’ |
| PA-R2 | 5’-CCCTCTTTCCCTCGCGCTTGCCTCTCTG-3’ |
| PyElf1-F | 5’-CACCATGGGCAAGCGCAAA-3’ |
| PyElf1-R | 5’-CTCCTCCCCCCCATTCACCT-3’ |
| PyAly-XhoF | 5’-GCTCGAGGCCCGCAGGCCGCCATGGGAC-3’ |
| PyAly-HinR | 5’-GAAGCTTCTACGACACAGTCAGACCATG-3’ |
Xho I and Hind III sites are underlined for PyAly-XhoF and PyAly-HinR, repectively.
Computational Analysis of Signal Peptide
The location of signal peptide of PyAly was predicted using the SignalP 3.0 server ( http://www.cbs.dtu.dk/services/SignalP/ ) [40].
Production of Recombinant Protein
The open reading frame of PyAly-cDNA was subcloned into pCold I expression vector (Takara) as follows. Restriction sites Xho I and Hind III were introduced by PCR to 5’- and 3’-end of the cDNA, respectively, using primers PyAly-XhoF and PyAly-HinR (Table 1). The amplified cDNA was then digested with Xho I and Hind III, and inserted into pCold I already digested with the same restriction enzymes. Thus, a 6 x His-tag was added to the recombinant PyAly (rPyAly) at the N-terminus. BL21(DE3) transformed with the resultant plasmid was cultured overnight at 37 oC. Expression of the recombinant protein was induced by decreasing the temperature to 15 oC along with the addition of 0.1 mM IPTG. After the incubation for 12 h, cells were harvested by centrifugation at 5,000 x g for 15 min, and sonicated in a buffer containing 10 mM imidazole-HCl (pH 8.0), 0.5 M NaCl, 1% Triton X-100, and 0.01 mg/ml lysozyme. After centrifugation at 10,000 x g for 20 min, the supernatant was subjected to a column packed with Ni-NTA agarose (1 cm x 3 cm, Qiagen). The matrix was washed with 10-bed volumes of 30 mM imidazole-HCl (pH 8.0) containing 0.5 M NaCl and the recombinant protein was eluted with 150 mM imidazole-HCl buffer (pH 8.0) containing 0.5 M NaCl. Purified protein was dialyzed against 10 mM sodium phosphate buffer (pH 7.5) containing 0.5 M NaCl and stored on ice until use. Protein concentration was determined by the Bradford’s method [41] using bovine serum albumin fraction V as a standard protein. Molecular mass of the purified protein was estimated by SDS-polyactylamide gel electrophoresis (SDS-PAGE) according to the procedure described by Porzio and Pearson [42] using a 10% polyacrylamide slab gel.
Assay for Alginate Lyase Activity
Decrease in viscosity of alginate solution by the enzyme reaction was determined at 30oC with an Ostwald-type viscometer and the reaction mixture containing 10 mM sodium phosphate (pH 8.0), 0.1 M NaCl, 0.3% sodium alginate, and 0.05 mg/ml rPyAly.
Alginate lyase activity was measured by the increase in absorbance at 235 nm derived from the formation of double bond between C4 and C5 at the non-reducing end by cleavage of alginate. Standard assay for alginate lyase activity was carried out at 30oC in a reaction mixture containing 10 mM sodium phosphate (pH 8.0), 0.1 M NaCl, 0.05 mg/ml rPyAly, and 0.15% (w/v) substrate (sodium alginate, poly(M), poly(MG), or poly(G)). Degradation of substrates was monitored by measuring absorbance at 235 nm with a spectrophotometer Hitachi model U-3010 (Tokyo, Japan) equipped by a thermal controlling apparatus SP-12R (Taitec, Tokyo, Japan). One unit of alginate lyase was defined as the amount of enzyme that increases absorbance at 235 nm to 0.01 for 1 min. Assays were done three times and the data were shown as mean ± SD.
Degradation products of alginate produced by the enzyme reaction were analyzed by thin-layer chromatography (TLC) with silica gel 60 plate (Merck, Darmstadt, Germany) and a developing solvent consisting of ethyl acetate, acetic acid, and water (2:2:1 (v:v:v)). Sugars developed on the TLC plate were detected by spraying 10% (v/v) sulfuric acid in ethanol followed by heating at 130oC for 10 min.
Cloning of the Genomic DNA Encoding PyAly from P. yezoensis
Gametophytic blades of P. yezoensis were quickly frozen in liquid nitrogen and immediately ground into powder. Genomic DNA was extracted from the powder with the DNeasy Plant kit (Qiagen) according to the manufacturer’s instruction. Using the genomic DNA as a template, PCR was carried out under the following conditions: an incubation at 94 oC for 2 min followed by 30 cycles of 94 oC for 30 sec, 60 oC for 30 sec, and 72 oC for 2 min using the primer set, PA-F1 and PA-R2 (Table 1) with KOD-Plus-DNA polymerase (Toyobo). Final extension was carried out at 72 oC for 7 min. The amplified DNA was subcloned and sequenced as mentioned above. The sequence data is available from the DDBJ, EMBL and GenBank with an accession number AB512414.
Expression Analysis for the PyAly Transcripts
Total RNAs were prepared separately from gametophytes and sporophytes of P. yezoensis as mentioned above. RT-PCR using the PyAly-specific primers (PA-F1 and PA-R2, Table 1) and PrimeSTAR HS DNA polymerase (Takara) was performed as follows: initial incubation at 94 oC for 1 min, and 30 cyles of 94 oC for 30 sec, 55 oC for 30 sec, and 72 oC for 1 min then final incubation at 72oC for 7 min. As an internal control, RT-PCR was performed with a specific primer pair PyElf1-F and PyElf1-R (Table 1) that can amplify a transcription elongation factor, PyElf1, of P. yezoensis [43].
RESULTS
cDNA Cloning of Alginate Lyase PyAly from P. yezoensis
A nucleotide sequence of the full open reading frame of alginate lyase-like protein termed as PyAly was predicted by overlapping two EST clones, accession numbers AV434206 and AV436451 (Fig. 1). On the basis of the sequences around 5’- and 3’-untranslated regions of these clones, we synthesized specific forward and reverse primers, PA-F1 and PA-R1, respectively (Suppl. Fig. S1 (420.7KB, pdf) and Table 1), and amplified a PyAly-cDNA as a single DNA fragment. Its cDNA comprises 876 bp and the single open reading frame is spanning from 64th to 789th nucleotide positions encoding an amino-acid sequence of 241 residues (Fig. S1 (420.7KB, pdf) ). Since the N-terminal region of 25 residues was predicted as a signal peptide, a mature PyAly appeared to be a secretory protein of 216 residues with the calculated molecular mass of 24,747.
Fig. (1).
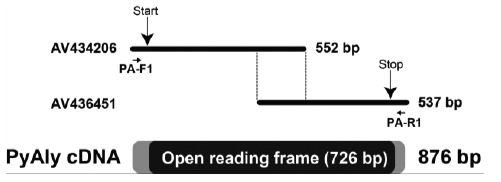
Schematic drawing of P. yezoensis EST clones for the alginate lyase-like protein, PyAly, and an amplified PyAly cDNA. The 151 bp of the 3'-terminal and 5'-terminal of P. yezoensis EST clones AV434206 and AV436451, respectively, can overlap entirely. The annealing positions of specific primers for PCR amplification are represented by side arrows.
The amino acid sequence of PyAly showed the significant identity to those of bacterial characterized PL-7 alginate lyases; for instance, 29, 22, 20, 21, and 19% identities with Vibrio sp. AlyVOB [20], Flavobacterium sp. FlAlyA [15], Z. galactanivorans AlyA1 [34], Sphingomonas sp. A1-II’ [23], P. aeruginosa PA1167 [29], respectively (Fig. 2).
Fig. (2).
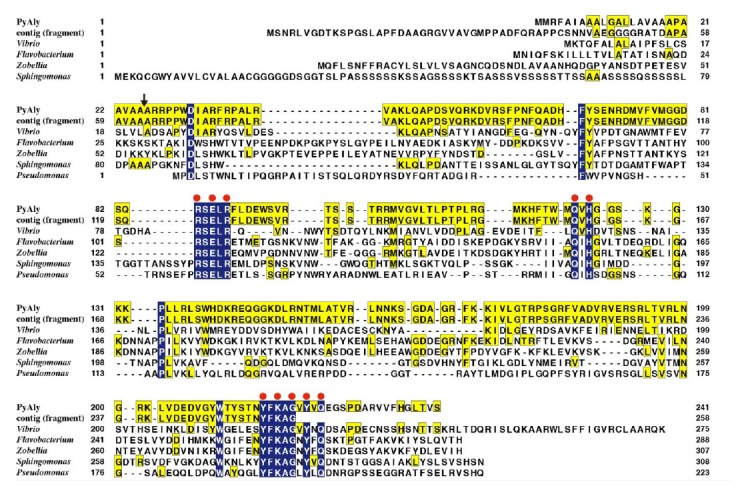
Comparison of the amino acid sequences of PyAly, its homolog protein deduced from genome sequence of P. yezoensis, and characterized PL-7 family enzymes. PyAly., P. yezoensis PyAly in present study; contig (fragment), PyAly homolog protein sequence derived from P. yezoensis genome database (contig_11106_g2658,) [44]; Vibrio, Vibrio sp. O2 alginate lyase AlyVOB [20]; Flavobacterium, Flavobacterium sp. UMI-01 alginate lyase FlAlyA [15]; Zobellia, Zobellia galactanivorans alginate lyase AlyA1 [34]; Sphingomonas, Sphingomonas A1 alginate lyase A1-II’ [23]; Pseudomonas., Pseudomonas aeruginosa PAO1 alginate lyase PA1167 [29]. Residues identical between PyAly and other enzymes are indicated with yellow boxes. Outline characters on a blue background show conserved residues in all listed sequences. Red circles represent positions for catalytic residues known in the PL-7 family enzymes. An arrow shows the location of the predicted signal peptide cleavage site.
Interestingly, a PyAly homolog protein encoding by contig_11106_g2658, was found in the annotated protein database derived from genome sequence of symbiont-free P. yezoensis strain U-51 ( http://nrifs.fra.affrc.go.jp/ResearchCenter/5_AG/genomes/nori/ ) [44]. Although homolog sequence in this database does not cover the entire protein due to partially lacking of the C-terminal region, overlapped sequences with PyAly were completely matched except N-terminal predicted signal sequence of PyAly (Fig. 2).
When these amino acid sequences were aligned, we found that residues Asp32, Phe67, Arg84-Arg89, Gln123, His125, Pro133, Trp211, Tyr217-Gly221, Tyr223, and Gln225 were completely conserved among these proteins (Fig. 2). Of these, since Arg84, Glu86, Arg88, Gln123, His125, Tyr217, Lys219, Gly221, Tyr223, and Gln225 in
PyAly are known to take part in the catalytic site of PL-7 enzymes [29]. These conserved amino acids strongly support that PyAly is a member of family PL-7.
Enzymatic Properties of PyAly
In order to investigate whether PyAly possesses alginate lyase activity, we expressed and purified the His-tagged recombinant PyAly (rPyAly, Fig. 3A) and subjected to the activity assay. As shown in Fig. (3B), that the purified rPyAly showed a single band on SDS-PAGE with approx. 29 kDa, which was comparable to the calculated molecular weight (Fig. 3B). The yield was about 1.2 mg from 1 L culture medium.
Fig. (3).
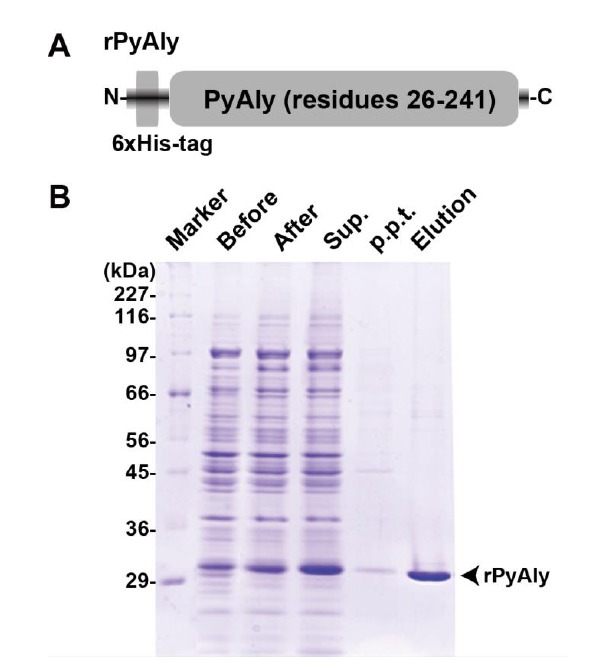
Bacterial expression and purification of rPyAly. A, schematic drawing of rPyAly. B, SDS-PAGE of cell extracts before (Before) and after (After) cold-induction, supernatant (Sup.) and precipitation (p.p.t.) after centrifugation of the induced cell extracts after, and purified rPyAly (Elution). Marker is a molecular mass protein marker.
We first examined the changes in viscosity of alginate solution by rPyAly. As shown in Fig. (4), the viscosity was drastically decreased within 30 min after addition of rPyAly, and slowly in the subsequent reaction. These results indicate that rPyAly degraded alginate substrate with an endolytic manner.
Fig. (4).
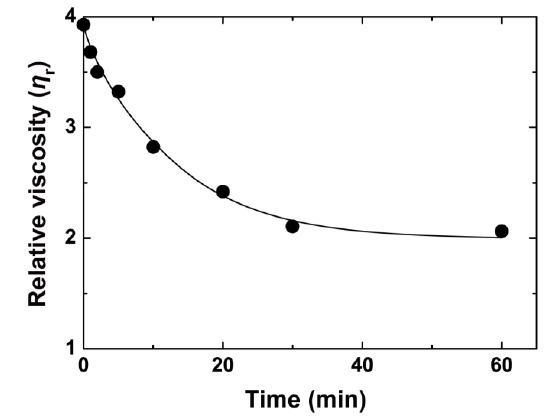
Decreasing viscosity of an alginate solution brought about by rPyAly. The time course of alginate degradation by rPyAly was measured in terms of decreasing viscosity. The reactions were conducted at 30 oC using 0.3% sodium alginate in a buffer containing 50 mM sodium phosphate (pH 7.5) and 0.1 M NaCl.
Next, substrate specificity of rPyAly was examined. Among alginate, poly(M), poly(G), and poly(MG), rPyAly preferably degraded poly(M) substrate with the activity of 1,460 U/mg (Fig. 5). This activity level is 3- and 5-times higher than those for poly(G)- and poly(MG)-block substrates, respectively (Fig. 5).
Fig. (5).
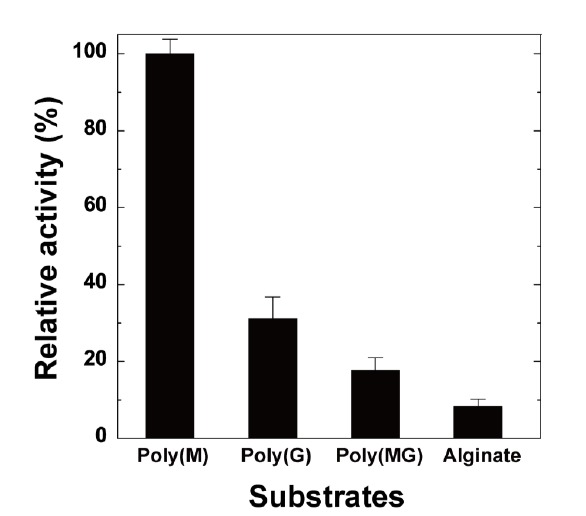
Substrate specificity of rPyAly. The enzyme reaction was performed in a buffer containing 10 mM sodium phosphate (pH 8.0), 0.1 M NaCl, and 0.15% of each substrate at 30 oC. A relative activity of 100% is equivalent to 1,320 U/mg. Error bars show SD for triplicate samples.
Then, we measured pH and temperature dependence of rPyAly activity. As shown in Fig. (6A, B), the optimum pH and temperature were observed at 8.0 and 35oC, respectively (Fig. 6A, B). Thermostability of rPyAly was assessed by measuring the activity remaining after the incubation at various temperatures for 30 min (Fig. 6C). By the incubation, the activity of rPyAly was significantly decreased above 30oC showing 50% activity at 32.5 oC. Complete loss of activity was observed above 50oC.
Fig. (6).
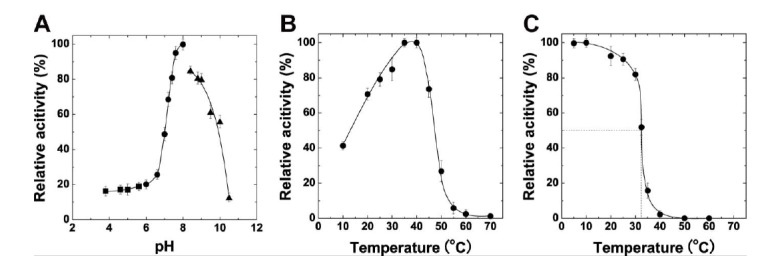
General enzymatic properties of rPyAly. A, effect of pH on rPyAly. The assay was conducted using 0.15% poly(M) as a substrate. 50 mM sodium acetate buffer (square, pH 3.8-5.6), 50 mM sodium phosphate buffer (circle, pH 6.0-8.0), and 50 mM glycine-NaOH buffer (triangle, pH 8.6-10.5) were used at the indicated pH. A relative activity of 100% is equivalent to 1,380 U/mg. B, effect of temperature on rPyAly. Lyase activity was measured at the indicated temperature using 0.15% poly(M). A relative activity of 100% is equivalent to 1,740 U/mg. C, heat stability of rPyAly. The enzyme was incubated at the indicated temperature for 30 min, and then lyase activity was measured using 0.15% poly(M). A relative activity of 100% is equivalent to 1,410 U/mg. Error bars show SD for triplicate samples.
Moreover, when the degradation products of poly(M)-block substrate produced by rPyAly was analyzed by TLC, tri-, tetra-, penta-, and hexa-saccharides were observed in the early phase of reaction, of which tri- and tetrasaccharides as the major end products (Fig. 7). These results indicate that rPyAly is an endolytic poly(M) lyase.
Fig. (7).
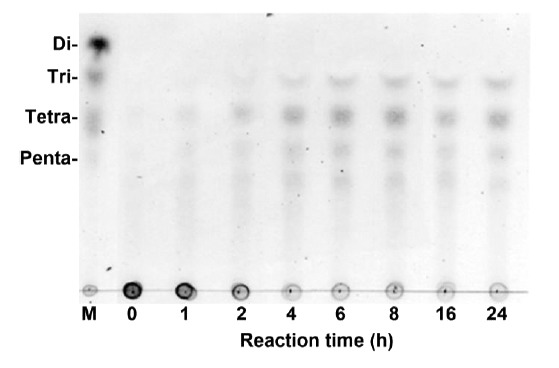
TLC analysis of degradation products by rPyAly. Poly(M) (0.5%) was incubated with rPyAly in a buffer containing 50 mM sodium phosphate (pH 8.0) and 0.1 M NaCl at 25 oC for several hours. Di, Tri, Tetra, and Penta indicate di-, tri-, tetra-, and pentasaccharides yielded from the enzymatic action on alginate, respectively. Lane M is the marker oligoalginates.
Structural Organization of the PyAly Gene
Since alginate substrate was considered to be absent in cell wall of P. yezoensis, the physiological significance of this alginate-lyase was quite obscure. Thus, it is possible that EST clones for PyAly is derived from some bacteria contaminated in algal materials. Therefore, we attempted to clone the structural gene of PyAly from the genome of P. yezoensis and obtained about 1.7 kb-DNA fragment as the PyAly gene (Fig. 8). Nucleotide sequence analysis indicated that the PyAly gene is comprised four exons intervened by three introns with the GT-AG splicing rule (Suppl. Fig. S2 (420.7KB, pdf) ). Taken together with the presence of introns in the structural gene and the finding of the homolog protein in P. yezoensis genomic database [44], our results unambiguously indicate that P. yezoensis has the PyAly gene in its genome and the identified EST clones are not contamination of bacterial genomes.
Fig. (8).
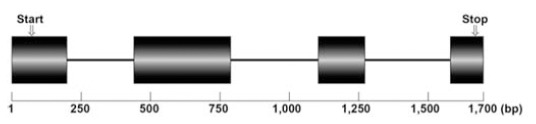
Schematic representation of the structure of the PyAly gene. Exons and introns are represented with boxes and lines, respectively.
Gametophyte-Specific Expression of the PyAly Gene
Besides the two EST clones encoding PyAly, i.e., AV434206 and AV436451, we identified additional 19 clones encoding partial sequences of PyAly in the EST database (accession numbers AV430057, AV430116, AV432029, AV432230, AV433187, AV433686, AV434264, AV434276, AV434521, AV434731, AV434833, AV435288, AV435984, AV436727, AV439370, AV432965, AV434062, AV434314, and AV435145). Since these clones were isolated only from the gametophyte EST library, it is possible to consider that the PyAly gene is expressed specifically in gametophytic cells. To examine this possibility, expression levels of the PyAly gene were compared between gametophytic and sporophytic cells by RT-PCR with specific primers, PA-F1 and PA-R2 (Table 1). As shown in Fig. (9), DNA fragment of approx. 850 bp was amplified only from gametophytic cDNA, but not sporophytic cDNA. The amplification level was not significantly affected by differences in cultivation temperature for gametophytes (Fig. 9). Accordingly, we consider that expression of the PyAly gene is restricted in gametophytic cells of P. yezoensis.
Fig. (9).
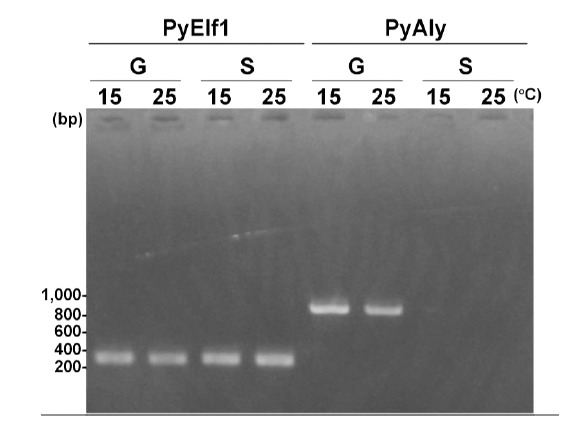
RT-PCR analysis of mRNA expression of PyAly. “G” and “S” represent the gametophyte and sporophyte of P. yezoensis, respectively. “15” and “25” show the culture-temperatures, respectively. A primer set of P. yezoensis transcription elongation factor (PyElf1) was used as a control.
DISCUSSION
In the present study, we enzymatically characterized the gametophyte-specific expressed PL-7-class alginate lyase, PyAly, from P. yezoensis. The activity assay for the recombinant enzyme, rPyAly, indicated that this protein is an endolytic alginate lyase.
To date, alginate lyases have been found in a virus, bacteria, fungi, brown algae, and marine invertebrates, and they have been classified under seven families, PL-5, 6, 7, 14, 15, 17, and 18 on the basis of the hydrophobic cluster analysis of their primary structures [45]. Bacterial alginate lyases are found in all families; however, eukaryotic enzymes are involved in only PL-7 and PL-14 families. Eukaryotic PL-14 enzymes were well characterized in marine gastropods such as abalone [46,47], turban shell [48,49], sea hare [50-52]. Although eukaryotic PL-7 enzymes have been suggested at the gene level in the A. hypogyna, A. oryzae, B. fuckeliana, and T. terrestris, their enzymatic properties have not been investigated yet. In algae, the alginate lyase activity was detected only in brown algae to date [53,54]; however, no alginate lyase and its gene has been isolated. Against this general consensus, we here represented the presence of alginate lyase, PyAly, in the marine red alga P. yezoensis. Thus, PyAly seemed to be the only algal family PL-7 alginate lyase whose properties were characterized.
PyAly was regarded as a poly(M) lyase; however, the substrate preference of this enzyme was considerably different from those of known poly(M) lyases. For example, the abalone HdAly, a typical poly(M) lyase from mollusk, can degrade both poly(M)-block and alginate substrate in high rates [46], while PyAly degraded only poly(M)-block with high rate (Fig. 6). This may indicate that PyAly has been specialized to degrade short alginate chains rich in β-D-mannuronate residues. The actual alginate substrate of PyAly in nature may be such short alginate materials contained in P. yezoensis.
Crystal structures of PL-7 alginate lyases provided deep insights for the mechanisms of substrate binding [23,29,34,35]. Based on the information, Uchimura et al. [18] proposed two characteristic sequences involving substrate specificity by comparison of several PL-7 alginate lyases. In Poly(G) or poly(MG) preferable lyases, RSELREM and QIH positioned at the region to form the cavity involving substrate binding are highly conserved. However, in poly(M) preferable lyases, some amino acids of the former sequence are replaced and the latter sequence is conserved as QVH. Corresponding sequences of PyAly are 84RSELRFL90 and 123QVH125, respectively, therefore, these residues are thought to be responsible for poly(M) preferability.
Recently, much attention is paid to alginate as a source of biofuel. In 2014, bioethanol production from 4-deoxy-L-erythro-5-hexoseulose uronate (DEH) by an engineered yeast was reported [55]. In this experiment, DEH was prepared beforehand by the degradation of alginate by alginate lyases in vitro and was added to medium. Although alginate-metabolizing yeast has not been found yet in nature, the utilization of an yeast having alginate degradation activity would be key to improve this system. So far, the yeast Yarrowia lipolytica displaying an alginate lyase AlyV1 from Vibrio sp. on the cell surface was constructed, and recombinant yeast showed the alginate degradation activity [56]. However, substrates (alginate, polyM, or polyG) were not completely depolymerized and oligosaccharides were poorly produced. If this low activity is ascribed to the incompatibility between yeast cells and introduced prokaryotic alginate lyase gene, eukaryotic alginate lyase gene, such as PyAly, would support to develop the better alginate-degrading yeast.
The physiological significance of PyAly in P. yezoensis is still obscure because alginate substrate has not been found in this alga. Bacterially expressed rPyAly exhibited 40-70% of the maximal activity at 10-20 oC, which that is consistent with the normal habitat temperature of P. yezoensis (Fig. 6A). In addition, rPyAly showed significant activity at a pH range from 7 to 10 (Fig. 6B). These results suggest that PyAly may function in natural sea-water conditions.
Previously, Okazaki et al. [57] reported that alginate or alginate-like polysaccharides were contained in cell wall of red alga Serraticardia maxima (Corallinaceae). This may indicate the presence of some biosynthetic pathways for alginate in this alga. However, to date, there has been no report dealing with the presence of alginate in red algae other than the above Corallinaceae. One possible role of alginate in Corallinaceae seems to strengthen the cell wall matrices and/or allow the alga to adhere to solid bases through the formation of elastic gel. If this explanation is appropriate, alginate-degrading enzymes such as alginate lyases are important for Corallinaceae to sever alginate and make normal life.
In case of P. yezoensis, an alginate lyase, PyAly, may have a similar role to those in Corallinaceae although alginate substrate has not been identified yet in this alga. Another possibility is that PyAly may serve as part of a cellular defense system against pathogenic bacteria that produce alginate as a biofilm. To clarify these hypotheses, further study focusing especially on the presence/absence of alginate, the expression and localization of PyAly, and the identification of a natural substrate for PyAly in vivo are essential.
CONCLUSION
This study demonstrates that PyAly in marine red alga P. yezoensis is a novel PL-7 alginate lyase with an endolytic manner. PyAly is a gametophyte-specifically expressed protein and its structural gene is composed of four exons and three introns. Thus, PyAly is the first enzymatically characterized eukaryotic PL-7 alginate lyase.
ACKNOWLEDGEMENTS
This study was supported in part by the the Program for Constructing “Tohoku Marine Science Bases” supported by Ministry of Education, Culture, Sports, Science and Technology, Japan.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Conway E., Cole K. Studies in the Bangiaceae: structure and reproduction of the conchocelis of Porphyra and Bangia in culture (Bangiales, Rhodophyceae). Phycologia. 1977;16:205–216. [Google Scholar]
- 2.Usov A.I. Polysaccharides of the red algae. Adv. Carbohydr. Chem. Biochem. 2011;65:115–217. doi: 10.1016/B978-0-12-385520-6.00004-2. [DOI] [PubMed] [Google Scholar]
- 3.Mukai L.S., Craigie J.S., Brown R.G. Chemical composition and structure of the cell walls of the conchocelis and thallus phases of Porphyra tenera (Rhodophyceae). J. Phycol. 1981;17:192–198. [Google Scholar]
- 4.Gretz M.R., Mccandless E.L., Aronson J.M., Sommerfeld M.R. The galactan sulphates of the conchocelis phases of Porphyra leucostricta and Bangia atropurpurea (Rhodophyta). J. Exp. Bot. 1983;34:705–711. [Google Scholar]
- 5.Nikaido I., Asamizu E., Nakajima M., Nakamura Y., Saga N., Tabata S. Generation of 10,154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- 6.Asamizu E., Nakajima M., Kitade Y., Saga N., Nakamura Y., Tabata S. Comparison of RNA expression profiles between the two generations of Porphyra yezoensis (Rhodophyta), based on expressed sequence tag frequency analysis. J. Phycol. 2003;39:923–930. [Google Scholar]
- 7.Haug A., Larsen B., Smidsrod O. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 1967;21:691–704. [Google Scholar]
- 8.Alginates G.P. Carbohydr. Polym. 1988;8:161–182. [Google Scholar]
- 9.Draget K.I. Handbook of Hydrocolloids. Elsevier; 2009. [Google Scholar]
- 10.Linker A., Jones R.S. A polysaccharide resembling alginic acid from a Pseudomonas micro-organism. Nature. 1964;204:187–188. doi: 10.1038/204187a0. [DOI] [PubMed] [Google Scholar]
- 11.Gorin P.A., Spencer J.F. Exocellular alginic acid from Azotobacter vinelandii. Can. J. Chem. 1966;44:993–998. [Google Scholar]
- 12.Govan J.R., Fyfe J.A. JARMAN TR. Isolation of alginate-producing mutants of Pseudomonas fluorescens, Pseudomonas putida and Pseudomonas mendocina. J. Gen. Microbiol. 1981;125:217–220. doi: 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- 13.Costerton J.W., Cheng K.J., Geesey G.G., Ladd T.I., Nickel J.C., Dasgupta M., et al. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 14.Cote G.L., Krull L.H. Characterization of the exocellular polysaccharides from Azotobacter chroococcum. Carbohydr. Res. 1988;181:143–152. [Google Scholar]
- 15.Inoue A., Takadono K., Nishiyama R., Tajima K., Kobayashi T., Ojima T. Characterization of an alginate lyase, FlAlyA, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs. 2014;12:4693–4712. doi: 10.3390/md12084693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang L., Zhou J., Li X., Peng Q., Lu H., Du Y. Characterization of a new alginate lyase from newly isolated Flavobacterium sp. S20. J. Ind. Microbiol. Biotechnol. 2013;40:113–122. doi: 10.1007/s10295-012-1210-1. [DOI] [PubMed] [Google Scholar]
- 17.Sugimura I., Sawabe T., Ezura Y. Cloning and sequence analysis of Vibrio halioticoli genes encoding three types of polyguluronate lyase. Mar. Biotechnol. (NY) 2000;2:65–73. doi: 10.1007/s101269900010. [DOI] [PubMed] [Google Scholar]
- 18.Uchimura K., Miyazaki M., Nogi Y., Kobayashi T., Horikoshi K. Cloning and sequencing of alginate lyase genes from deep-sea strains of Vibrio and Agarivorans and characterization of a new Vibrio enzyme. Mar. Biotechnol. (NY) 2010;12:526–533. doi: 10.1007/s10126-009-9237-7. [DOI] [PubMed] [Google Scholar]
- 19.Badur A.H., Jagtap S.S., Yalamanchili G., Lee J-K., Zhao H., Rao C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol. 2015;81:1865–1873. doi: 10.1128/AEM.03460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamoto H., Horibe A., Miki Y., Kimura T., Tanaka K., Nakagawa T., et al. Cloning and sequencing analysis of alginate lyase genes from the marine bacterium Vibrio sp. O2. Mar. Biotechnol. (NY) 2006;8:481–490. doi: 10.1007/s10126-005-6157-z. [DOI] [PubMed] [Google Scholar]
- 21.Han F., Gong Q-H., Song K., Li J-B., Yu W-G. Cloning, sequence analysis and expression of gene alyVI encoding alginate lyase from marine bacterium Vibrio sp. QY101. DNA Seq. 2004;15:344–350. doi: 10.1080/10425170400019300. [DOI] [PubMed] [Google Scholar]
- 22.Yoon H.J., Hashimoto W., Miyake O., Okamoto M., Mikami B., Murata K. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr. Purif. 2000;19:84–90. doi: 10.1006/prep.2000.1226. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki M., Ogura K., Hashimoto W., Mikami B., Murata K. A structural basis for depolymerization of alginate by polysaccharide lyase family-7. J. Mol. Biol. 2005;352:11–21. doi: 10.1016/j.jmb.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 24.Kim D.E., Lee E.Y., Kim H.S. Cloning and characterization of alginate lyase from a marine bacterium Streptomyces sp. ALG-5. Mar. Biotechnol. (NY) 2008;11:10–16. doi: 10.1007/s10126-008-9114-9. [DOI] [PubMed] [Google Scholar]
- 25.Kim H.S. Characterization of recombinant polyg-specific lyase from a marine bacterium, Streptomyces sp. M3. J. Life Sci. 2010;20:1582–1588. [Google Scholar]
- 26.Matsubara Y., Kawada R., Iwasaki K., Kimura Y., Oda T., Muramatsu T. Cloning and sequence analysis of a gene (aly PG) encoding poly(alpha-L-guluronate)lyase from Corynebacterium sp. strain ALY-1. J. Biosci. Bioeng. 2000;89:199–202. doi: 10.1016/s1389-1723(00)88738-1. [DOI] [PubMed] [Google Scholar]
- 27.Duan G., Han F., Yu W. Cloning, sequence analysis, and expression of gene alyPI encoding an alginate lyase from marine bacterium Pseudoalteromonas sp. CY24. Can. J. Microbiol. 2009;55:1113–1118. doi: 10.1139/w09-051. [DOI] [PubMed] [Google Scholar]
- 28.Gimmestad M., Ertesvåg H., Heggeset T.M., Aarstad O., Svanem B.I., Valla S. Characterization of three new Azotobacter vinelandii alginate lyases, one of which is involved in cyst germination. J. Bacteriol. 2009;191:4845–4853. doi: 10.1128/JB.00455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasaki M., Moriwaki S., Miyake O., Hashimoto W., Murata K., Mikami B. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold. J. Biol. Chem. 2004;279:31863–31872. doi: 10.1074/jbc.M402466200. [DOI] [PubMed] [Google Scholar]
- 30.Malissard M., Duez C., Guinand M., Vacheron M-J., Michel G., Marty N., et al. Sequence of a gene encoding a (poly ManA) alginate lyase active on Pseudomonas aeruginosaalginate. FEMS Microbiol. Lett. 1993;110:101–106. doi: 10.1111/j.1574-6968.1993.tb06302.x. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Yang X., Zhang L., Yu W., Han F. Cloning, expression and characterization of a cold-adapted and surfactant-stable alginate lyase from marine bacterium Agarivorans sp. L11. J. Microbiol. Biotechnol. 2015;25:681–686. doi: 10.4014/jmb.1409.09031. [DOI] [PubMed] [Google Scholar]
- 32.Kim H.T., Ko H-J., Kim N., Kim D., Lee D., Choi I-G., et al. Characterization of a recombinant endo-type alginate lyase (Alg7D) from Saccharophagus degradans. Biotechnol. Lett. 2012;34:1087–1092. doi: 10.1007/s10529-012-0876-9. [DOI] [PubMed] [Google Scholar]
- 33.Baron A.J., Wong T.Y., Hicks S.J., Gacesa P., Willcock D., McPherson M.J. Alginate lyase from Klebsiella pneumoniae, subsp. aerogenes: gene cloning, sequence analysis and high-level production in Escherichia coli. Gene. 1994;143:61–66. doi: 10.1016/0378-1119(94)90605-x. [DOI] [PubMed] [Google Scholar]
- 34.Thomas F., Lundqvist L.C., Jam M., Jeudy A., Barbeyron T., Sandström C., et al. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J. Biol. Chem. 2013;288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osawa T., Matsubara Y., Muramatsu T., Kimura M., Kakuta Y. Crystal structure of the alginate (poly α-l-guluronate) lyase from Corynebacterium sp. at 1.2Å resolution. J. Mol. Biol. 2005;345:1111–1118. doi: 10.1016/j.jmb.2004.10.081. [DOI] [PubMed] [Google Scholar]
- 36.Gacesa P., Wusteman F.S. Plate assay for simultaneous detection of alginate lyases and determination of substrate specificity. Appl. Environ. Microbiol. 1990;56:2265–2267. doi: 10.1128/aem.56.7.2265-2267.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwano K., Aruga Y., Saga N. Cryopreservation of clonal gametophytic thalli of Porphyra (Rhodophyta). Plant Sci. 1996;116:117–124. [Google Scholar]
- 38.Kitade Y., Fukuda S., Nakajima M., Watanabe T., Saga N. Isolation of a cDNA encoding a homologue of actin from Porphyra yezoensis (Rhodophyta). J. Appl. Phycol. 2002;14:135–141. [Google Scholar]
- 39.Araki T., Hayakawa M., Tamaru Y., Yoshimatsu K., Morishita T. Isolation and regeneration of haploid protoplasts from Bangia atropurpurea (Rhodophyta) with marine bacterial enzymes. J. Phycol. 1994;30:1040–1046. [Google Scholar]
- 40.Bendtsen J.D., Nielsen H. Heijne von G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 41.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Porzio M.A., Pearson A.M. Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim. Biophys. Acta. 1977;490:27–34. doi: 10.1016/0005-2795(77)90102-7. [DOI] [PubMed] [Google Scholar]
- 43.Uji T., Takahashi M., Saga N., Mikami K. Visualization of nuclear localization of transcription factors with cyan and green fluorescent proteins in the red alga Porphyra yezoensis. Mar. Biotechnol. (NY) 2010;12:150–159. doi: 10.1007/s10126-009-9210-5. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura Y., Sasaki N., Kobayashi M., Ojima N., Yasuike M., Shigenobu Y., et al. The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS One. 2013;8:e57122. doi: 10.1371/journal.pone.0057122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong T.Y., Preston L.A., Schiller N.L. ALGINATE LYASE: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000;54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu E., Ojima T., Nishita K. cDNA cloning of an alginate lyase from abalone, Haliotis discus hannai. Carbohydr. Res. 2003;338:2841–2852. doi: 10.1016/j.carres.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki H., Suzuki K-I., Inoue A., Ojima T. A novel oligoalginate lyase from abalone, Haliotis discus hannai, that releases disaccharide from alginate polymer in an exolytic manner. Carbohydr. Res. 2006;341:1809–1819. doi: 10.1016/j.carres.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 48.Muramatsu T., Yamada K., Date M., Yoshioka S. Action of poly(β-D-mannuronate)lyase from Turbo cornutus on oligomeric substrates. Biosci. Biotechnol. Biochem. 1993;57:1990–1994. [Google Scholar]
- 49.Muramatsu T., Komori K., Sakurai N., Yamada K., Awasaki Y., Fukuda K., et al. Primary structure of mannuronate lyases SP1 and SP2 from Turbo cornutus and involvement of the hydrophobic C-terminal residues in the protein stability. J. Protein Chem. 1996;15:709–719. doi: 10.1007/BF01887144. [DOI] [PubMed] [Google Scholar]
- 50.Rahman M.M., Inoue A., Tanaka H., Ojima T. Isolation and characterization of two alginate lyase isozymes, AkAly28 and AkAly33, from the common sea hare Aplysia kurodai. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010;157:317–325. doi: 10.1016/j.cbpb.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Rahman M.M., Inoue A., Tanaka H., Ojima T. cDNA cloning of an alginate lyase from a marine gastropod Aplysia kurodai and assessment of catalytically important residues of this enzyme. Biochimie. 2011;93:1720–1730. doi: 10.1016/j.biochi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Rahman M.M., Wang L., Inoue A., Ojima T. cDNA cloning and bacterial expression of a PL-14 alginate lyase from a herbivorous marine snail Littorina brevicula. Carbohydr. Res. 2012;360:69–77. doi: 10.1016/j.carres.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 53.Madgwick J., Haug A., Larsen B. Alginate lyase in the brown alga Laminaria digitata (Huds.) Lamour. Acta Chem. Scand. 1973;27:711–712. doi: 10.3891/acta.chem.scand.27-0711. [DOI] [PubMed] [Google Scholar]
- 54.Shiraiwa Y., Abe K., Sasaki S.F., Ikawa T., Nisizawa K. Alginate Lyase Activities in the Extracts from Several Brown Algae. Bot. Mar. 1975:18. [Google Scholar]
- 55.Enquist-Newman M., Faust A.M., Bravo D.D., Santos C.N., Raisner R.M., Hanel A., et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature. 2014;505:239–243. doi: 10.1038/nature12771. [DOI] [PubMed] [Google Scholar]
- 56.Liu G., Yue L., Chi Z., Yu W., Chi Z., Madzak C. The surface display of the alginate lyase on the cells of Yarrowia lipolytica for hydrolysis of alginate. Mar. Biotechnol. (NY) 2009;11:619–626. doi: 10.1007/s10126-009-9178-1. [DOI] [PubMed] [Google Scholar]
- 57.Okazaki M., Furuya K., Tsukayama K., Nisizawa K. Isolation and identification of alginic acid from a calcareous red alga Serraticardia maxima. Bot. Mar. 1982;25:123–132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.


