Abstract
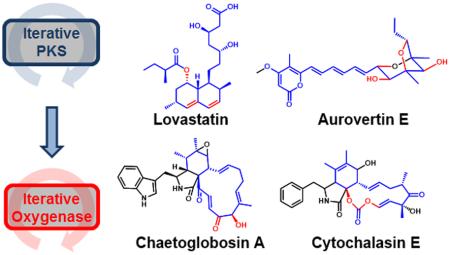
Fungal polyketides are natural products with great chemical diversity that exhibit a wide range of biological activity. This chemical diversity stems from specialized enzymes encoded in the biosynthetic gene cluster responsible for the natural product biosynthesis. Fungal polyketide synthases (PKS) are the megasynthases that produce the carbon scaffolds for the molecules. Subsequent downstream tailoring enzymes such as oxygenases will then further modify the organic framework. In fungi, many of these enzymes have been found to work iteratively—catalyzing multiple reactions on different sites of the substrate. This perspective will analyze several examples of fungal polyketides that are assembled from a scaffold-building iterative PKS and an accompanying iterative tailoring oxygenase. In these examples, the PKS product is designed for downstream iterative oxygenations to generate additional complexity. Together, these iterative enzymes orchestrate the efficient biosynthesis of elaborate natural products such as lovastatin, chaetoglobosin A, cytochalasin E, and aurovertin E.
Keywords: polyketide synthase, monooxygenase, natural products, biosynthesis, catalysis
Perspective
Polyketides play a vital role in modern medicine as some of the most important therapeutic drugs in the world.1 The concise biosynthetic pathways of several fungal polyketide natural products feature the combination of iterative polyketide synthases (PKSs) and multifunctional and iterative oxygenases.2–4 An iterative enzyme can be defined as a protein that reuses its catalytic unit to sequentially catalyze distinct modifications on a substrate. Typical examples of iterative enzymes include ubiquitous proteins such as nucleases, proteases and glycosidases. In the context of secondary metabolism, we define iterative enzymes as those that contain active sites that can catalyze multiple rounds of structural modifications during the biosynthesis of a natural product.
Fatty acid synthases (FASs) serve as a well-studied example of iterative enzymes.5 FASs catalyze the homo-polymerization of malonyl-CoA and complete β-reduction to afford a saturated lipid chain. Although iterative PKSs share many similarities with FASs including domain architecture and catalytic mechanisms, they are programmed in a more sophisticated fashion to carry out combinatorial catalysis. The β-reduction domains are differentially used during each iteration of chain elongation to generate the structural diversity observed in many fungal natural products.2,3 The diverse polyketide backbones are then further functionalized by tailoring enzymes such as oxygenases. Some of these tailoring oxygenases function iteratively and catalyze multiple rounds of oxidative modifications to generate the mature and bioactive polyketide natural product.
Together, the synergistic PKS and oxidative enzymes have enabled fungi to adeptly introduce chemical complexity to efficiently biosynthesize many chemically diverse and biologically active molecules such as lovastatin (anti-hypercholesterolemia),6,7 chaetoglobosin A (apoptosis-inducer),8 cytochalasin E (anti-angiogenesis),9,10 and aurovertins (ATP synthase inhibitor).11,12 Inspired by their complex chemical structures and useful properties, these fungal polyketides are attractive targets for total chemical synthesis.13–16 Some of these total syntheses draw strategies from Nature and employ biomimetic methodologies.16,17 Yet, the elegance, brevity and synchronization of many enzymatic transformations often lack an equivalent chemical parallel. This viewpoint will highlight several succinct biosyntheses of fungal polyketides that are collaboratively forged by these iterative enzymes.
Overview of Type I Iterative Fungal Reducing Polyketide Synthase (PKS)
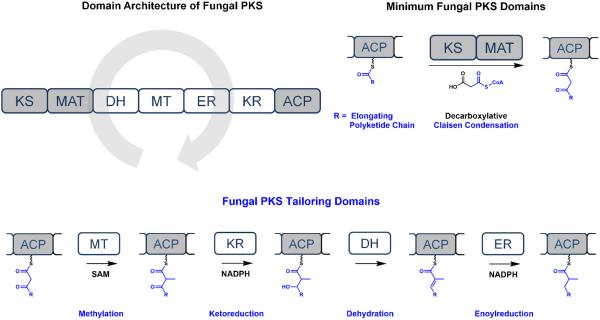
Type I PKSs are multi-domain megasynthases that possess the catalytic domains required for polyketide biosynthesis.18 In most type I bacterial PKSs,19 multiple sets of domains are typically compiled into modules and their biosynthesis proceeds in an assembly-line fashion. In contrast, type I fungal reducing PKSs use a single set of domains in a highly programmed and permutative fashion.2,3 The architecture of the fungal reducing PKSs consist of the minimal fungal PKS components and the auxiliary tailoring domains (Figure 1). The β-ketoacyl synthase (KS),20 malonyl-CoA: ACP transacylase (MAT) and acyl carrier protein (ACP)21 form the minimal fungal PKS components—the basis for the chain-extending iterations through decarboxylative Claisen condensations (Figure 1). During each iteration, the minimal fungal PKS components catalyze the decarboxylative polymerization of malonyl-CoA to elongate the polyketide chain by a ketide (two carbons).
Figure 1.
Illustration of the domain architecture and the catalysis of the minimum fungal PKS domains and the fungal PKS tailoring domains in fungal polyketide synthases. In fungal PKSs, a single set of domains is used iteratively and permutatively to generate backbone structural diversity.
Following each chain extension step, the ACP-bound, β-ketothioester intermediate may undergo a series of modifications from the tailoring domains such as α-methylation by the methyltransferase (MT), β-ketoreduction by the ketoreductase (KR),22 dehydration by the dehydratase (DH),23 and enoylreduction by the enoylreductase (ER) domains (Figure 1).24 The MT domain utilizes S-adenosylmethionine (SAM) as the methylating agent while the reductive domains use nicotinamide adenine dinucleotide phosphate hydride (NADPH) as the reducing agent. The α and β position of each ketide unit will differ depending on the extent of methylation and reduction during each cycle. Through different permutative tailoring modifications following each chain extension, the same set of tailoring domains can install structural diversity into the α- and β- positions of polyketide backbones.2,3 The elongation-tailoring events proceed iteratively until the polyketide chain extension is terminated through product off-loading such as hydrolysis or reductive release. While these permutative programming steps may seem randomly coded at first glance, the examples shown in this perspective show that the complete polyketide products are exquisitely and precisely functionalized to facilitate the downstream modifications—many of which are oxidative. Currently, underlying programming rules for the iterative catalysis of both bacterial and fungal polyketide synthases remain an active area of research.
Overview of Post-PKS Iterative Oxygenases
After the biosynthesis of the polyketide scaffold, further modifications are catalyzed by post-PKS tailoring enzymes. Oxygenases are common tailoring enzymes responsible for much of the chemical diversity of natural products.25 These enzymes often carry out a diverse range of redox reactions including hydroxylations, epoxidations, dehydrogenations, cyclizations, and various rearrangements—often decreasing the lipophilicity of polyketide metabolites.26 Recent discoveries have found several multifunctional oxygenases that can act iteratively on multiple sites of their substrates.4,27 Thus, iterative oxygenases are enzymes that can introduce multiple oxygen atoms from molecular oxygen at different sites on a single substrate.
There are several major classes of oxygenases including cytochrome P450 monooxygenases (P450s), flavin-containing monooxygenases (FMOs), and non-heme, iron- and α-ketoglutarate-dependent dioxygenases. Examples of iterative catalysis are found in each of these classes in fungal polyketide biosynthesis. Monooxygenases incorporate one oxygen atom from molecular oxygen (O2) while dioxygenases can incorporate both oxygen atoms. A notable example of an iterative, multifunctional α-ketoglutarate-dependent dioxygenase is the AusE enzyme, which catalyzes several interesting oxidations on the terpene portion of the fungal polyketide hybrid molecule austinol.28 However, this perspective will focus on examples which feature iterative oxidations from P450s and FMOs that work together with iterative PKSs.
P450s are heme-dependent enzymes
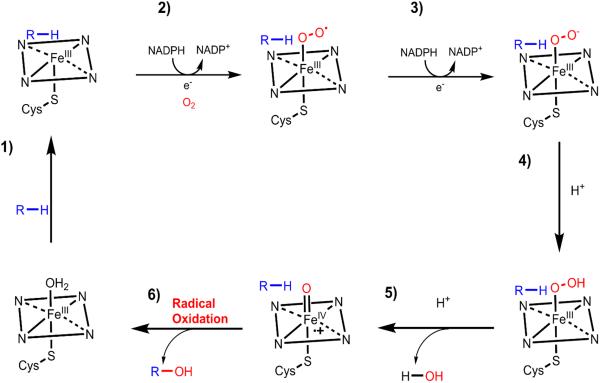
P450s are ubiquitous oxidative enzymes that span across all organisms, from bacteria to humans. P450s are heme-binding enzymes and share a highly conserved protein fold.29 These oxygenases use heme complexes to oxidize a vast multitude of different substrates using molecular oxygen.30 Despite their substantial range of substrate diversity, P450 oxidations can be highly stereoselective and regioselective. Furthermore, P450s can be organized into Class I and Class II subgroups, differing in their associated reductive partner. Fungal biosynthetic pathways typically use Class II P450s—microsomal transmembrane enzymes often associated with a transmembrane cytochrome P450 reductase (CPR) partnering enzymes which together moderate electron transfer from NADPH.29 P450 enzymes in natural product biosynthesis use a single-electron manifold to produce radical intermediates. These reactive radical intermediates can lead to a variety of modifications including hydroxylation, epoxidation, dehydrogenation, radical homo-coupling, etc. A well-known example of an iterative P450-catalyzed reaction is the repeated oxidation of methyl groups to corresponding carboxylic acids via alcohol and aldehyde intermediates.31–33 This set of transformations is widely found in plant and fungal biosynthetic pathways and is also prominently featured in the conversion of lanosterol into cholesterol.34
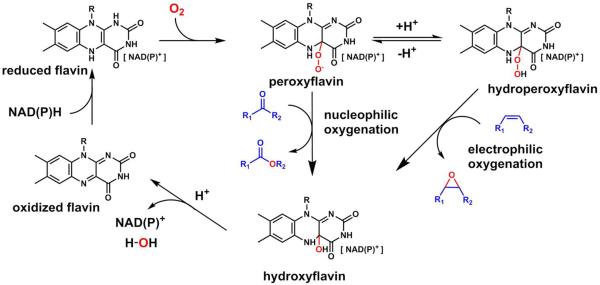
FMOs are versatile oxidases and oxygenases
FMOs are widespread enzymes that catalyze a large variety of substrate oxidations such as dehydrogenation, hydroxylations, epoxidations, Baeyer-Villiger oxidations, and sulfoxidations.35,36 Unlike P450s which use a heme prosthetic group to activate molecular oxygen, FMOs instead use a flavin cofactor such as flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN), to generate reactive peroxyl species that serve as nucleophiles (peroxyflavin, Fl-OO−) or electrophiles (hydroperoxyflavin, Fl-OOH).36 The flavin coenzymes are often tightly or covalently bound to the FMO.36 After each round of catalysis, the flavin cofactor can be reduced in the presence of NAD(P)H to repeat the catalytic cycle.
Lovastatin
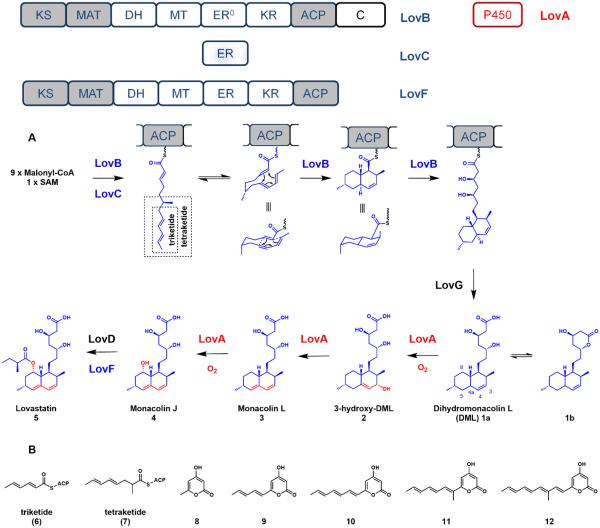
Lovastatin (5) is one of the most pharmaceutically relevant fungal natural products in use today. Lovastatin and its synthetic analogue (simvastatin) exhibit potent cholesterol-lowering activity by inhibiting the HMG-CoA reductase (HMGR) enzyme in cholesterol biosynthesis.37 Lovastatin represents a classical example of a fungal natural product that is synthesized by highly-reducing iterative PKSs. Produced by the filamentous fungus Aspergillus terreus, 5 is the esterified product of two highly-reduced polyketide chains catalyzed by LovB and LovF, respectively.
Polyketide Core Assembly (LovB/LovC and LovF)
LovB (nonaketide synthase) and LovC (trans-acting ER partner) catalyze ~35 highly programmed reactions to produce the ACP-bound dihydromonacolin L (DML, 1a), the nonaketide precursor to 5.38 The multiple reductions are catalyzed by the KR and DH domains within LovB in concert with LovC. LovB also catalyzes a Diels-Alder reaction to form the decalin core, a yet unresolved step proposed to occur at the hexaketide stage. The polyketide chain is then offloaded from LovB by the thioesterase LovG to yield 1a, which can undergo iterative, oxidative tailoring around the decalin core to yield monacolin J (4). The acyltransferase LovD can then transesterify an α-methylbutyrate diketide from LovF onto the newly installed 8-hydroxyl group of 4 to afford 5 (Figure 4A).
Figure 4.
A) Biosynthesis of lovastatin. The polyketide products produced from an iterative PKS are highlighted in blue and oxidative tailoring produced from an iterative oxygenase are highlighted in red. The portions of the polyketide chain reflected by intermediates 6 and 7 are shown in the dashed boxes. B) The natural triketide and tetraketide intermediates and the offloaded polyketide pyrone shunt products.
LovB exhibits extraordinary specificity and precision in catalyzing the formation of 1a. Each structural portion of lovastatin is important for its activity as a HMGR inhibitor. The 3,5-dihydroxy acid portion is the warhead that binds to the active site of HMGR, while the decalin ring provides the hydrophobic anchor for high affinity binding. To accomplish this, LovB shows an impressive ability to control the timing of reductions and methylation steps during polyketide chain elongation, with an ability to correct its mistakes.38 In the first two rounds of polyketide chain elongation, only the KR and DH tailoring domains are active to afford the triketide (6). In the next round, the MT of LovB and the LovC will also function to produce the methylated tetraketide (7). As a method of proofreading, aberrantly tailored acyl intermediates are offloaded through α-pyrone formation. In vitro studies with LovB showed that the absence of the reductive NADPH cofactor led to polyketide chain offloading by lactonization to produce the triketide α-pyrone (8).38 In the absence of either of SAM or LovC, offloading would also proceed to form other α-pyrones (Figure 4B). These findings illustrate that if the PKS cannot correctly proceed to later stages of biosynthesis, the PKS is programmed to catalyze two additional condensation cycles (KS/MAT) to offload the shunt polyketide products.38 This proofreading mechanism prevents stalling of the incorrectly modified acyl chain on LovB.
Furthermore, mechanistic studies of LovB provided valuable insight into the programming of iterative PKSs. For example at the tetraketide stage, the methylation step is a prerequisite for ER activity as LovC fails to recognize substrate that is not methylated.38 Recent kinetic studies also compared the substrate specificity of the KR and MT domains.39 While the KR domain displays broader substrate promiscuity, the MT domain exhibits high specificity only for the tetraketide substrate. The experiments also illustrated that the kinetic competition between the KR and MT domains prevents methylation at incorrect positions. These experiments suggest that the substrate specificity of the catalytic domains may decide the sequence of reactions in the polyketide biosynthesis. Studies on LovB have unveiled key features of PKSs, including the strict checks and balances of individual tailoring domains on the fidelity of previous iterations.38,39 Subtle mistakes are recognized and rejected in the later iterations. Collectively, LovB is an incredibly accurate enzyme. This accuracy is clearly exemplified in the linear hexaketide triene intermediate. The presence of the three double bonds is a result of precise tailoring to set up the intramolecular Diels-Alder reaction that forges the decalin core (Figure 4A). Clearly, any mistakes prior to that would not lead to formation of the decalin core.
Additional functionalization by post-PKS iterative oxygenase (LovA)
After formation of 1a, several oxidation steps are required to generate the penultimate intermediate 4, which is acylated at the C8-hydroxyl group by the LovF product to yield 5. Although the lovastatin biosynthetic gene cluster encodes two P450 genes (LovA and Orf17), gene disruption of LovA in Aspergillus terreus confirmed its role as the only oxidative enzyme required in lovastatin biosynthesis.7 Furthermore, heterologous reconstitution of LovA and its cytochrome P450 oxidoreductase (CPR) partner in Saccharomyces cerevisiae confirmed the iterative oxygenase activity of LovA.40 It was demonstrated that LovA first converted 1a to monacolin L (3), and then to 4 via sequential radical hydroxylation reactions (Figure 4A). The conversion of 1a to 3-hydroxymonacolin L (2) proceeds through allylic hydrogen abstraction followed by an oxygen rebound to introduce the hydroxyl group at the C3α position (Figure 2 and 4A). Dehydration of the hydroxyl group yields the diene in the decalin core of 3. LovA then performs the second oxidation on the C8α position of the decalin to introduce the hydroxyl group.
Figure 2.
The well-accepted mechanism of P450 catalyzed, radical-mediated hydroxylation of a substrate (R).
Intriguingly, the second olefin in the decalin core of 3 cannot be produced by a PKS alone. However, LovB shrewdly placed the first unsaturation between C3-C4 in DML to activate the decalin ring for an allylic hydroxylation-elimination sequence to yield the second unsaturation between C4a and C5. The C3-C4 π-bond has been experimentally demonstrated to be vital for substrate recognition by LovA since the saturated DML derivative cannot be taken as a substrate.40 Furthermore, the lack of hydroxylation at the C8α position prior to the second unsaturation possibly also suggests the order of oxidation events catalyzed by LovA is exact (see contrast by chaetoglobosin P450 below). It is possible that the conformational change (chair to half-chair) upon diene formation in the decalin substrate may be required for the second hydroxylation by LovA to occur.
Chaetoglobosin A
Cytochalasans are a family of fungal polyketide-amino acid hybrid natural products that have interesting biological activies.41 Many cytochalasans exhibit antibiotic, anti-inflammatory, anti-angiogenic and anti-cytokinesis activity.41 Furthermore, the representative members of the cytochalasan family all share an isoindolone moiety fused to a macrocycle in its tricyclic core. Among the cytochalasans, the highly oxygenated chaetoglobosin A (20) inhibits actin polymerization and was the first member to have its biosynthetic gene cluster elucidated.42
Polyketide core assembly (CheA/CheB and CHGG_01239/CHGG_01240)
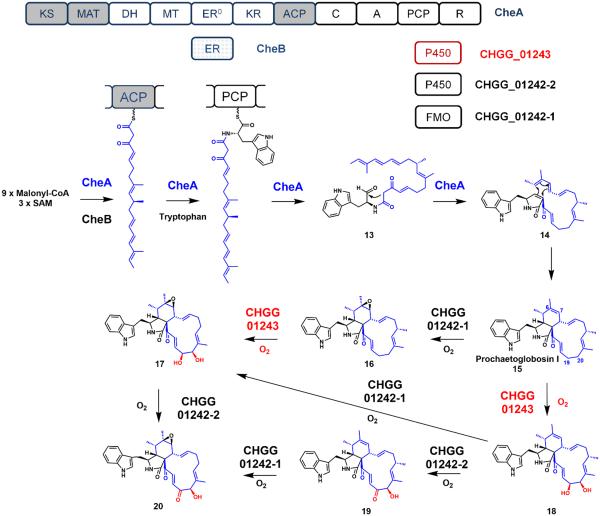
The backbone of chaetoglobosin A is assembled through the polyketide synthase-nonribosomal synthetase (PKS-NRPS) hybrid enzyme, CheA (Figure 5). This finding was first confirmed by RNA-mediated gene silencing in Penicillium expansum.42 A functionally equivalent CheA homolog was also discovered from another chaetoglobosin A producer, Chaetomium globosum.43 Similar to the LovB PKS, the PKS portion of CheA also lacks a functional ER domain (Figure 5), and is complemented with a trans-acting ER partnering enzyme (CheB) in the construction of the nonaketide portion of the molecule. Differing from the lovastatin nonaketide precursor, the CheA PKS product is then condensed with a tryptophanyl thioester attached to the fused NRPS module of CheA. The nonaketide-tryptophanyl thioester intermediate 13 is then reductively offloaded from CheA by the reduction (R) domain on the NRPS module (Figure 5).42,43
Figure 5.
Biosynthetic pathway of chaetoglobosin A. The polyketide products produced from an iterative PKS are highlighted in blue and oxidative tailoring produced from an iterative oxygenase are highlighted in red.
An intramolecular Knoevenagel condensation between the β-ketoamide and the aldehyde produces the pyrrolinone 14 that can serve as a dienophile in a subsequent intramolecular Diels-Alder reaction. The resulting Diels-Alder reaction precisely and stereoselectively forges the perhydro-isoindolone core of prochaetoglobosin I 15.44 The cyclized 15 requires a series of downstream oxidations to produce the final chaetoglobosin A product.
Comparing the reducing PKS domain architecture of LovB and CheA, the main difference lies in the complete NRPS module appended at the C-terminus of CheA (Figure 4 and 5). Condensation of PKS product with an aminoacyl building block serves several purposes: 1) this represents one method of chain termination which is likely controlled through the substrate specificity of the corresponding C domain in the NRPS module; 2) the amino acid unit introduces structural variation, especially a basic nitrogen that is absent from the PKS alone—providing a convenient entry point for equipping polyketide products with the versatile nitrogen atom; and 3) formation of the pyrrolinone through Knoevenagel condensation generates a dienophile for a Diels-Alder reaction. Again, the diene is formed from the permutative activities of the PKS module. Interestingly, the diene is part of a triene moiety and the Diels-Alderase is able to regioselectively and stereoselectively perform the cycloaddition to yield 15. The functional differences between LovB and CheA therefore result in vastly different polyketide structures and biological activities. The contrast highlights the importance of understanding the programming rules of PKSs from seemingly similar PKS architecture.
Additional functionalization by post-PKS iterative oxygenase (CHGG_01243)
After assembly of the tricyclic core of 15, downstream oxidative tailoring by three oxidative enzymes CHGG_01242-2 (FMO), CHGG_01242-1 (P450), and CHGG_01243 (P450) decorate the polyketide portion of the molecule with different oxygenated functional groups (alcohol, ketone and epoxide) to form 20.43 Different combinations of targeted gene deletions of these three oxygenases were constructed to generate single and double gene deletion mutants. Product analysis from these mutants indicated that P450 CHGG_01243 performs the iterative hydroxylations on C19 and C20; both are allylic carbons within the macrocycle scaffold (Figure 5). Subsequently, the FMO CHGG_01242-2 catalyzes oxidation of the newly introduced C19 hydroxyl to a ketone. Finally, the P450 CHGG_01242-1 performs the epoxidation across the C6–C7 π-bond to yield 20.
When analyzing the product profile from the various mutants, it was discovered that the order of the downstream tailoring oxidations is not strictly linear (Figure 5). The ketone formation must occur after CHGG_01243 dihydroxylation, but the epoxidation by CHGG_01242-1 can occur either before or after these two reactions. This shows a relatively broad substrate specificity of the iterative oxygenase CHGG_01243 towards the remote segment of the molecule, as it can utilize either 15 or 16 as substrates. This contrasts with the highly selective nature of the LovA iterative oxygenase in the lovastatin biosynthetic pathway.
Cytochalasin E and Cytochalasin K
Despite the similar name, the cytochalasins are actually specific metabolites of the general cytochalasan family.41 The cytochalasins are fungal polyketide-phenylalanine hybrids45,46 that inhibit several cellular processes such as the polymerization of actin and cell division.47,48 For example, cytochalasin E (26) has garnered significant attention due to its strong anti-angiogenic properties.49 The ccs gene cluster in Aspergillus clavatus responsible for cytochalasin E and cytochalasin K (cytochalasin E/K, 26 and 27) biosynthesis was discovered for members of this family.9
Polyketide core assembly (CcsA/CcsC)
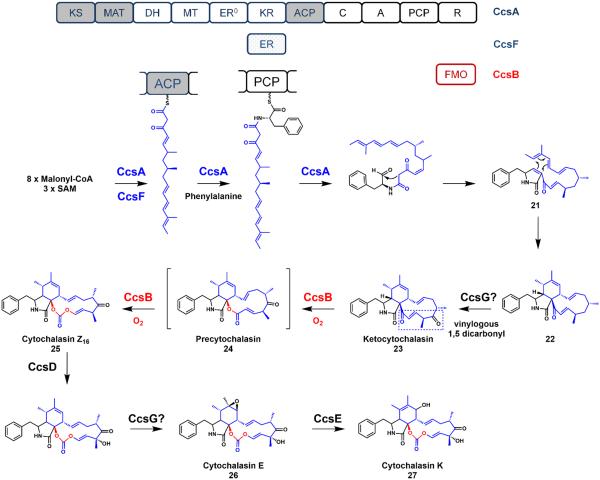
Considering the structural similarities between all cytochalasans, the reducing PKSs responsible for the biosynthesis of chaetoglobosin A and cytochalasin E/K expectedly share similar domain architectures. When comparing the reducing PKSs for chaetoglobosin A (CheA/B) and cytochalasin E/K (CcsA/C), both multidomain enzymes possess the same set of tailoring domains including the MT, all of the reductive domains, a partnering trans-acting ER, and a fused NRPS module.9 Furthermore, the biosyntheses of their linear polyketide precursor are strikingly similar—differing only by a double bond and an ethylene unit (Figure 5 and 6).
Figure 6.
Biosynthesis of cytochalasin E and cytochalasin K. The polyketide products produced from an iterative PKS are highlighted in blue and oxidative tailoring produced from an iterative oxygenase are highlighted in red.
The distinguishing feature of the cytochalasins from other members of the cytochalasan family is the incorporated amino acid, phenylalanine.46 The ACP-bound linear polyketide precursor is condensed with the phenylalanyl thioester by the NRPS module to produce the β-ketoamide that can be cyclized by a Knoevenagel condensation to yield the pyrrolinone 21. Operating under similar programming rules as CheA, the essential structural features required for the cycloaddition are found in the acyclic precursor, such as the diene in the triene tail and the dienophile. An intramolecular Diels-Alder reaction similarly furnishes the perhydro-isoindolone core and the large carbocycle 2250—the hallmark of the cytochalasan family (Figure 6).41 The Diels-Alder adduct 22 then undergoes an iterative oxidation at C17 to produce the essential vinylogous, 1,5-dicarbonyl system in ketocytochalasin 23 (Figure 6 and 7).
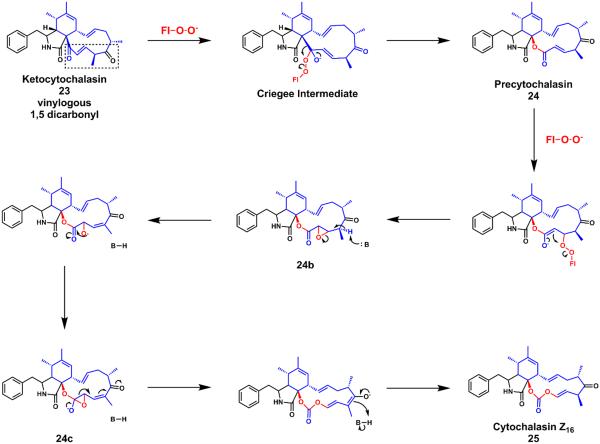
Figure 7.
Proposed mechanism of the CcsB catalyzed conversion of ketocytochalasin to cytochalasin Z16. Alternatively, a direct attack of precytochalasin (24), followed by a vinyl migration on the Criegee intermediate can also form the same epoxy-alkoxide system (24c) that can collapse to form 25; this would make CcsB a true, iterative BVMO.
The subtle differences between the iterative reducing PKSs for chaetoglobosin A and cytochalasin E/K further highlights the precise calibration of iterative catalysis by the reducing PKS. In the chaetoglobosin A biosynthesis, the absent ethylene unit is flanked by π bonds which forms the vicinal allylic carbons. These activated carbons are exactly the targets of the iterative allylic hydroxylation by the P450 enzyme, CHGG_01243, in the cheA pathway.43 Although cytochalasin E/K lacks the diol derived from the vicinal allylic carbons, the vinylogous 1,5-dicarbonyl sets up a most fascinating set of iterative oxygenation to yield the carbonate group found in the final natural products.
Additional functionalization by post-PKS iterative oxygenase (CcsB)
Among the cytochalasans, cytochalasin E/K are particularly interesting due to the unique vinyl carbonate moiety incorporated in the macrocyclic ring.51 The exceptionally rare carbonate group in natural products is essential for the cytotoxic properties of cytochalasin E/K, as the corresponding ester is ~500 fold lower in activity.52,53 Isotopic labeling experiments suggested that a Baeyer-Villiger monooxygenase (BVMO)54,55 can convert deoxaphomin (carbocycle) to cytochalasin B (lactone) via an oxygen derived from molecular oxygen.10,56,57 Consequently, the carbonate moiety in cytochalasin E/K has been postulated to result from two consecutive oxygen insertions via a single BVMO enzyme. This is an unprecedented reaction in both synthetic and biosynthetic chemistry.
Deletion of the BVMO, CcsB, in the A. clavatus strain abolished the production of cytochalasin E/K and resulted in the accumulation of 23, the ketone containing substrate.10 In vitro assays with purified CcsB enzyme demonstrated the conversion of 23 to the carbonate cytochalasin Z16 25 via two oxygen atom insertions and a vinyl ketone ester shunt product (not shown).10 The shunt product is isomerized from the ester product precytochalasin 24, which was not isolated under assay conditions. A point mutation on the FMO enzyme's catalytic arginine to alanine also abolished the formation of 23 and the lactone shunt product (Figure 7). Together, these findings illustrate the first known example of an FMO enzyme iteratively catalyzing two oxygen insertions to convert a ketone functional group to a carbonate moiety.
While the first oxygen insertion to form 24 is expected to follow the classical BVMO mechanism with regards to migratory aptitude, the unusual second oxygen insertion is expected to be inserted via a completely different mechanism. Based on experimental data and survey of all known cytochalasins, it appears that the vinylogous-1,5-dicarbonyl system is a prerequisite for carbonate formation.10 The energetically-favored mechanism for the second oxygen insertion could proceed by an initial Michael addition 24 by the peroxyflavin anion to furnish the α-β epoxide on the lactone (24b) (Figure 7). A subsequent ring-opening elimination can then produce an epoxy-alkoxide product (24c), which upon on ring opening across the enone can then yield the carbonate moiety in cytochalasin Z16 (25). Formation of the carbonate product requires cleavage of the C-C bond in the epoxide and the remote vinyl ketone serving as an electron sink. Based on this mechanism, the iterative PKS CcsA precisely orchestrates the construction of the polyketide precursor that can be oxidized to yield the essential vinylogous-1,5-dicarbonyl 23. CcsB can utilize the unique electronic properties of the polyketide system to convert a ketone into the carbonate found in cytochalasin E/K. While sequence alignment comparisons suggest that CcsB is a typical BVMO, its pairing with a unique substrate yields a remarkably rare functional group among natural products.
Aurovertin E
Aurovertin E (34) represents the biosynthetic precursor and the representative structure of the fungal polyketide family of aurovertins. The acetylated derivative, aurovertin B, exhibits potent and non-competitive inhibition of F1 ATPase, as well as induction of apoptosis and cell arrest in breast cancer cells.12,58 The defining structural hallmark of the aurovertins includes a methylated α-pyrone attached to a 2,6-dioxabicyclo[3.2.1]octane (DBO) ring system via a triene linker. Unlike the lovastatin, chaetoglobosin A and cytochalasin E examples, the heterocyclic portion of aurovertin E (34) is not derived from a Diels-Alder reaction, but rather through the iterative epoxidation of a polyene precursor and a cascade of concerted epoxide openings that result in bridged, cyclic ether formations.
Polyketide core assembly (AurA)
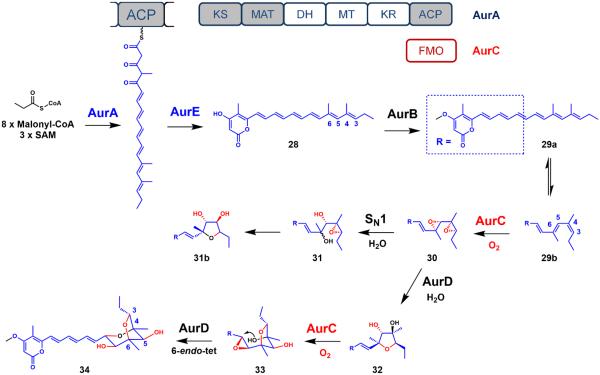
Another key differentiating feature of aurovertin E is the conjugated polyene structure deficient in fully reduced carbons. The only methylene unit within the entire structure has been shown to be derived from a propionate starter unit through isotope labeling.59 This structural feature is manifested in the domain architecture of the PKS. The AurA enzyme from Calcarisporium arbuscula is a reducing PKS that possesses the minimal PKS unit along with the MT, KR and DH tailoring domains. However, it lacks a cis-acting or trans-acting ER domain. Consequently, the PKS can only reduce each β-carbonyl to the enoyl functional group, consistent with the observed polyolefin structure. The programming rules of AurA are remarkably precise, as the PKS employs all of its reductive domains for six iterations to generate a hexaene thioester intermediate. However, in the final two iterations, the PKS is able to shut down the reductive domains to generate a tricarbonyl tail that is required for subsequent chain release steps. Furthermore, the PKS precisely uses its MT domain to introduce two α-methyl groups at C4 and C6 only. These methyl groups may be later involved in the post-PKS oxidation that forges the DBO ring system.
The tricarbonyl hexaene linear precursor is offloaded by enzymatic or spontaneous intramolecular cyclization (Figure 8). The offloading proceeds by enolization of the δ-carbonyl, followed by an additional-elimination on the thioester carbonyl to produce the α-pyrone-polyene conjugated intermediate 28. Curiously, this α-pyrone-polyene motif is also produced by LovB in the absence of its ER partner (LovC) (Figure 4). This scaffold is also present in other related fungal pyrone-polyene metabolites such as citreoviridin,60 asteltoxin,61 and asteltoxin B.62 Pausing of the KR and DH activities in the final two extension cycles is essential for the enol-induced lactonization to produce the α-pyrone substructure in 28.
Figure 8.
Proposed biosynthesis of aurovertin E. The polyketide products produced from an iterative PKS are highlighted in blue and oxidative tailoring produced from an iterative oxygenase are highlighted in red.
Additional functionalization by post-PKS iterative oxygenases (AurC)
Isotope 18O-labeling studies shed valuable insight on the origin of the oxygen atoms in the DBO ring system. Using 18O2, the oxygen atom within the C3-O-C6 ether linkage as well as the equatorial hydroxyl groups on C5 and C7 were shown to be derived from molecular oxygen. The lack of labeling of the oxygen within the C4-O-C8 ether linkage implies that the oxygen atom is most certainly derived from water.63 Despite having three oxygen atoms derived from O2 in the DBO ring system, the identified aur gene cluster revealed only a single FMO, AurC. Taken together, these observations hinted the utilization of an iterative FMO enzyme in selective epoxidations of the double bonds in 28.64
Considering that the pyrone-polyene intermediate 28 possesses six consecutive alkenes, an oxygenase must exhibit high regioselectivity to oxidize the distal alkenes but not the proximal triene linker. Interestingly, these distal olefins are substituted with methyl groups installed by the MT domain. Density functional theory (DFT) calculations on the pyrone-polyene intermediate 28 revealed the non-planarity on C3-C4-C5-C6 in its preferred conformation. Presumably, this conformation avoids the possible A1,3-allylic strain between the α-methyl groups. The induced asymmetry may therefore play a role in the specific epoxidation by the FMO enzyme.
Based on the absolute stereochemistry, isotope labeling experiments and detailed biochemical studies, formation of the DBO system was shown to be constructed using an epoxide-opening, cyclic ether ring-closing cascade.63,64 Gene inactivation of the AurC oxygenase enzyme in Calcarisporium arbuscula abolished the production of aurovertin E and led to the accumulation of a methylated pyrone-polyene intermediate 29a. The reactive 29b pyrone-polyene resulted from the trans-cis isomerization of 29a.
Furthermore, coexpression of the AurA (PKS), AurC (FMO) and AurB (O-MT) in yeast led to the production of tetrahydrofuranyl isomers 32 and 31b. Chemical complementation with 32 to the ΔaurA (PKS-knockout) strain restored the production of aurovertin E. The formation of 32 is consistent with the biomimetic total synthesis16 and the structures of related natural products.60–62 An epoxide hydrolase, AurD, was identified to control the regioselectivity of the epoxide openings (Figure 8). Overall, the distal olefins were converted to the DBO substructure by an iterative FMO enzyme and an assisting epoxide hydrolase. The MT domain may have subtly introduced an asymmetry in the polyene for FMO substrate recognition. The collaboration between two iterative enzymes produced the complex framework of aurovertin E.
Conclusion
As we have shown above in four examples, the synchronization between an iterative PKS and an iterative oxygenase can succinctly construct complex fungal polyketide natural products. The examples discussed in this perspective underscore how the iterative PKS produces precisely chiseled intermediates specifically for the iterative oxygenase partner. Many hypotheses may emerge to account for the evolutionary origin to develop these highly efficient tandem enzymatic systems. Naturally, one also may speculate on the evolutionary advantages of iterative enzymes as a whole. Considering the ubiquitous nature of many iterative enzymes such as non-specific proteases, a rational assumption could be the metabolic efficiency of multifunctional catalysis. Recognizing that polyketides constitute the most abundant secondary metabolites in fungi,65 it is also tempting to consider the selective advantages that these iterative enzymes partners offer. Fungi are well known as rich producers of secondary metabolites, yet they are constrained by their microbial genome size. Iterative enzymes in natural product biosynthesis likely have evolved to help circumvent these intrinsic limitations.
Figure 3.
The mechanism of FMO mediated nucleophilic oxygenation (see cytochalasin E/K) and electrophilic oxygenation (see aurovertin E).
ACKNOWLEDGMENT
This work was supported by the by NIH (1R01GM085128 and 1DP1GM106413) YT.
References
- (1).Weissman KJ. Philos. T. Roy. Soc. A. 2004;362:2671. doi: 10.1098/rsta.2004.1470. [DOI] [PubMed] [Google Scholar]
- (2).Chooi YH, Tang Y. J. Org. Chem. 2012;77:9933. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cox RJ, Simpson TJ. Method. Enzymol. 2009;459:49. doi: 10.1016/S0076-6879(09)04603-5. [DOI] [PubMed] [Google Scholar]
- (4).Cochrane RV, Vederas JC. Acc. Chem. Res. 2014;47:3148. doi: 10.1021/ar500242c. [DOI] [PubMed] [Google Scholar]
- (5).Jenni S, Leibundgut M, Boehringer D, Frick C, Mikolasek B, Ban N. Science. 2007;316:254. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- (6).Hendrickson L, Ray Davis C, Roach C, Kim Nguyen D, Aldrich T, McAda PC, Reeves CD. Chem. & Biol. 1999;6:429. doi: 10.1016/s1074-5521(99)80061-1. [DOI] [PubMed] [Google Scholar]
- (7).Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Science. 1999;284:1368. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- (8).Oikawa H, Murakami Y, Ichihara A. J. Chem. Soc. Perk. T. 1992;1:2955. [Google Scholar]
- (9).Qiao KJ, Chooi YH, Tang Y. Metab. Eng. 2011;13:723. doi: 10.1016/j.ymben.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hu YC, Dietrich D, Xu W, Patel A, Thuss JAJ, Wang JJ, Yin WB, Qiao KJ, Houk KN, Vederas JC, Tang Y. Nat. Chem. Biol. 2014;10:552. doi: 10.1038/nchembio.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang F, Luo DQ, Liu JK. J. Antibiot. 2005;58:412. doi: 10.1038/ja.2005.53. [DOI] [PubMed] [Google Scholar]
- (12).vanRaaij MJ, Abrahams JP, Leslie AGW, Walker JE. P. Natl. Acad. Sci. U. S. A. 1996;93:6913. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hirama M, Iwashita M. Tetrahedron Lett. 1983;24:1811. [Google Scholar]
- (14).Haidle AM, Myers AG. P. Natl. Acad. Sci. U. S. A. 2004;101:12048. doi: 10.1073/pnas.0402111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Nishiyama S, Toshima H, Kanai H, Yamamura S. Tetrahedron. 1988;44:6315. [Google Scholar]
- (16).Forbes JE, Pattenden G. J. Chem. Soc. Perk. T. 1991;1:1959. [Google Scholar]
- (17).Stork G, Nakahara Y, Nakahara Y, Greenlee WJ. J. Am. Chem. Soc. 1978;100:7775. [Google Scholar]
- (18).Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- (19).Dutta S, Whicher JR, Hansen DA, Hale WA, Chemler JA, Congdon GR, Narayan ARH, Hakansson K, Sherman DH, Smith JL, Skiniotis G. Nature. 2014;510:512. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Tang YY, Kim CY, Mathews II, Cane DE, Khosla C. P. Natl. Acad. Sci. U. S. A. 2006;103:11124. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lim J, Kong R, Murugan E, Ho CL, Liang ZX, Yang D. Plos One. 2011;6:e20549. doi: 10.1371/journal.pone.0020549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zheng JT, Keatinge-Clay AT. MedChemComm. 2013;4:34. [Google Scholar]
- (23).Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Structure. 2010;18:94. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ames BD, Nguyen C, Bruegger J, Smith P, Xu W, Ma S, Wong E, Wong S, Xie XK, Li JWH, Vederas JC, Tang Y, Tsai SC. P. Natl. Acad. Sci. U. S. A. 2012;109:11144. doi: 10.1073/pnas.1113029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Walsh CT. Nat. Chem. Biol. 2015;11:620. doi: 10.1038/nchembio.1894. [DOI] [PubMed] [Google Scholar]
- (26).Cashman JR. Biochem. Bioph. Res. Co. 2005;338:599. doi: 10.1016/j.bbrc.2005.08.009. [DOI] [PubMed] [Google Scholar]
- (27).Wu LF, Meng S, Tang GL. BBA-Proteins Proteom. 2016;1864:453. doi: 10.1016/j.bbapap.2016.01.012. [DOI] [PubMed] [Google Scholar]
- (28).Matsuda Y, Awakawa T, Wakimoto T, Abe I. J. Am. Chem. Soc. 2013;135:10962. doi: 10.1021/ja405518u. [DOI] [PubMed] [Google Scholar]
- (29).Dawson J. Science. 1996;271:1507. [Google Scholar]
- (30).Coon MJ. Annu. Rev. Pharmacol. 2005;45:1. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- (31).Abe S, Sado A, Tanaka K, Kisugi T, Asami K, Ota S, Il Kim H, Yoneyama K, Xie X, Ohnishi T, Seto Y, Yamaguchi S, Akiyama K, Yoneyama K, Nomura T. P. Natl. Acad. Sci. U. S. A. 2014;111:18084. doi: 10.1073/pnas.1410801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. P. Natl. Acad. Sci. U. S. A. 2001;98:2065. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MCY, Withers ST, Shiba Y, Sarpong R, Keasling JD. Nature. 2006;440:940. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- (34).Risley JM. J. Chem. Educ. 2002;79:377. [Google Scholar]
- (35).van Berkel WJ, Kamerbeek NM, Fraaije MW. J. Biotechnol. 2006;124:670. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- (36).Walsh CT, Wencewicz TA. Nat. Prod. Rep. 2013;30:175. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tobert JA. Nat. Rev. Drug Discov. 2003;2:517. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- (38).Ma SM, Li JWH, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie XK, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Science. 2009;326:589. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cacho RA, Thuss J, Xu W, Sanichar R, Gao ZZ, Nguyen A, Vederas JC, Tang Y. J. Am. Chem. Soc. 2015;137:15688. doi: 10.1021/jacs.5b11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Barriuso J, Nguyen DT, Li JWH, Roberts JN, MacNevin G, Chaytor JL, Marcus SL, Vederas JC, Ro D-K. J. Am. Chem. Soc. 2011;133:8078. doi: 10.1021/ja201138v. [DOI] [PubMed] [Google Scholar]
- (41).Scherlach K, Boettger D, Remme N, Hertweck C. Nat. Prod. Rep. 2010;27:869. doi: 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- (42).Schumann J, Hertweck C. J. Am. Chem. Soc. 2007;129:9564. doi: 10.1021/ja072884t. [DOI] [PubMed] [Google Scholar]
- (43).Ishiuchi K, Nakazawa T, Yagishita F, Mino T, Noguchi H, Hotta K, Watanabe K. J. Am. Chem. Soc. 2013;135:7371. doi: 10.1021/ja402828w. [DOI] [PubMed] [Google Scholar]
- (44).Sato M, Yagishita F, Mino T, Uchiyama N, Patel A, Chooi YH, Goda Y, Xu W, Noguchi H, Yamamoto T, Hotta K, Houk KN, Tang Y, Watanabe K. ChemBioChem. 2015;16:2294. doi: 10.1002/cbic.201500386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Binder M, Kiechel JR, Tamm C. Helvetica Chimica Acta. 1970;53:1797. doi: 10.1002/hlca.19700530728. [DOI] [PubMed] [Google Scholar]
- (46).Vederas JC, Tamm C. Helvetica Chimica Acta. 1976;59:558. doi: 10.1002/hlca.19760590221. [DOI] [PubMed] [Google Scholar]
- (47).Cooper JA. J. Cell. Biol. 1987;105:1473. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sampath P, Pollard TD. Biochemistry. 1991;30:1973. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- (49).Udagawa T, Yuan J, Panigrahy D, Chang YH, Shah J, D'Amato RJ. J. Pharmacol. Exp. Ther. 2000;294:421. [PubMed] [Google Scholar]
- (50).Vedejs E, Campbell JB, Gadwood RC, Rodgers JD, Spear KL, Watanabe Y. J. Org. Chem. 1982;47:1534. [Google Scholar]
- (51).Buchi G, Kitaura Y, Yuan SS, Wright HE, Clardy J, Demain AL, Glinsuko T, Hunt N, Wogan GN. J. Am. Chem. Soc. 1973;95:5423. doi: 10.1021/ja00797a060. [DOI] [PubMed] [Google Scholar]
- (52).Zhang H, Liu HB, Yue JM. Chem. Rev. 2014;114:883. doi: 10.1021/cr300430e. [DOI] [PubMed] [Google Scholar]
- (53).Wang FZ, Wei HJ, Zhu TJ, Li DH, Lin ZJ, Gu QQ. Chem. Biodivers. 2011;8:887. doi: 10.1002/cbdv.201000133. [DOI] [PubMed] [Google Scholar]
- (54).de Gonzalo G, Mihovilovic MD, Fraaije MW. ChemBioChem. 2010;11:2208. doi: 10.1002/cbic.201000395. [DOI] [PubMed] [Google Scholar]
- (55).Leisch H, Morley K, Lau PCK. Chem. Rev. 2011;111:4165. doi: 10.1021/cr1003437. [DOI] [PubMed] [Google Scholar]
- (56).Robert JL, Tamm C. Helvetica Chimica Acta. 1975;58:2501. doi: 10.1002/hlca.19750580830. [DOI] [PubMed] [Google Scholar]
- (57).Vederas JC, Nakashima TT, Diakur J. Planta Med. 1980;39:201. [Google Scholar]
- (58).Huang TC, Chang HY, Hsu CH, Kuo WH, Chang KJ, Juan HF. J. Proteome. Res. 2008;7:1433. doi: 10.1021/pr700742h. [DOI] [PubMed] [Google Scholar]
- (59).Steyn PS, Vleggaar R, Wessels PL. J. Chem. Soc. Perk. T. 1981;1:1298. [Google Scholar]
- (60).Ueno Y, Ueno I. Jpn J. Exp. Med. 1972;42:91. [PubMed] [Google Scholar]
- (61).Kawai K, Fukushima H, Nozawa Y. Toxicol. Lett. 1985;28:73. doi: 10.1016/0378-4274(85)90012-8. [DOI] [PubMed] [Google Scholar]
- (62).Adachi H, Doi H, Kasahara Y, Sawa R, Nakajima K, Kubota Y, Hosokawa N, Tateishi K, Nomoto A. J. Nat. Prod. 2015;78:1730. doi: 10.1021/np500676j. [DOI] [PubMed] [Google Scholar]
- (63).Steyn PS, Vleggaar R. Chem. Commun. 1985:1796. [Google Scholar]
- (64).Mao XM, Zhan ZJ, Grayson MN, Tang MC, Xu W, Li YQ, Yin WB, Lin HC, Chooi YH, Houk KN, Tang Y. J. Am. Chem. Soc. 2015;137:11904. doi: 10.1021/jacs.5b07816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Keller NP. Nat. Chem. Biol. 2015;11:671. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]