Abstract
Prasinophytes occur in all oceans but rarely dominate phytoplankton populations. In contrast, a single ecotype of the prasinophyte Micromonas is frequently the most abundant photosynthetic taxon reported in the Arctic from summer through autumn. However, seasonal dynamics of prasinophytes outside of this period are little known. To address this, we analyzed high-throughput V4 18S rRNA amplicon data collected from November to July in the Amundsen Gulf Region, Beaufort Sea, Arctic. Surprisingly during polar sunset in November and December, we found a high proportion of reads from both DNA and RNA belonging to another prasinophyte, Bathycoccus. We then analyzed a metagenome from a December sample and the resulting Bathycoccus metagenome assembled genome (MAG) covered ~90% of the Bathycoccus Ban7 reference genome. In contrast, only ~20% of a reference Micromonas genome was found in the metagenome. Our phylogenetic analysis of marker genes placed the Arctic Bathycoccus in the B1 coastal clade. In addition, substitution rates of 129 coding DNA sequences were ~1.6% divergent between the Arctic MAG and coastal Chilean upwelling MAGs and 17.3% between it and a South East Atlantic open ocean MAG in the B2 Clade. The metagenomic analysis also revealed a winter viral community highly skewed toward viruses targeting Micromonas, with a much lower diversity of viruses targeting Bathycoccus. Overall a combination of Micromonas being relatively less able to maintain activity under dark winter conditions and viral suppression of Micromonas may have contributed to the success of Bathycoccus in the Amundsen Gulf during winter.
Introduction
Over half of global photosynthetic production occurs in the oceans, with picophytoeukaryotes (0.8–3 μm diameter) frequently accounting for much of this production (Jardiller et al., 2010; Forest et al., 2011), and phytoplankton biomass (Li, 1994; Worden et al., 2004; Dasilva et al., 2013). Picophytoeukaryotes are phylogenetically diverse and include heterokonts, haptophytes, cryptophytes and about a third are in the chlorophyte chlorophyll b lineages. Among these are the Mamiellophyceae within the polyphyletic marine prasinophytes (Vaulot et al., 2008). The Arctic Ocean and shelf seas are unusual compared with more temperate seas, in that outside of spring blooms, phytoplankton pigment studies have highlighted a surprisingly high proportion of chlorophyll b phototrophs (Vidussi et al., 2004; Coupel et al., 2015). While the heterokont Phaeocystis is reported in the picophytoeukaryote fraction of Atlantic influenced waters of Fram Straight in the European Arctic (Kilias et al., 2014), over the summer and autumn throughout much of the Arctic, including at the North Pole, a single mamiellophyte represented by the cultured strain Micromonas sp. CCMP2099 (Lovejoy et al., 2007; Balzano et al., 2012; Zhang et al., 2015) dominates picophytoeukaryotes. A second mamiellophyte, Bathycoccus prasinos, is also consistently reported from Arctic marine waters but as a minor community constituent (Lovejoy and Potvin, 2011; Balzano et al., 2012). Ostreococcus, which is the smallest known photosynthetic microbial eukaryote is also in the Mamiellophyceae, but has never been reported from the Arctic.
Species occurrences are also influenced by differential loss processes, for example, grazability (Apple et al., 2011), and viral burden (Mojica et al., 2016). In particular, prasinoviruses (Phycodnaviridae) are thought to exert control over Mamiellophyceae (Clerissi et al., 2014). Given the predominance of Mamiellophyceae in the Arctic, prasinoviruses could contribute to species sorting in Arctic waters, and the prevalence of a single Micromonas ecotype represent populations with resistance to prasinoviruses as described by Thomas et al. (2011).
Micromonas and Bathycoccus are often reported from other oceanic regions (Monier et al., 2016; Simmons et al., 2016) and the persistent co-occurrence of the two genera could be enigmatic. They are similar in size (1 to 2 μm), with a single mitochondrion and a single chloroplast. However, observations of live cells and electron microscopy reveal striking morphological differences, with Micromonas having a single distinct flagellum and no scales, whereas Bathycoccus has no flagella, but is covered in organic ‘spider web' scales (Eikrem and Throndsen, 1990). There are also marked differences in the known phylogenetic diversity of the two genera; Bathycoccus is much less diverse than Micromonas, which could be an evidence of alternative evolutionary adaptive strategies. Bathycoccus consists of two clades (Simmons et al., 2016), based on the internal transcribed spacer 2 region (ITS2) with clade B1 proposed to be adapted to coastal environments and B2 to the open ocean (Vaulot et al., 2012: Monier et al., 2013). The two clades could be separate species, as ITS2 variation can indicate sexual incompatibility (Amato et al., 2007; Coleman, 2007; Kaczmarska et al., 2009) and potential speciation (Isaka et al., 2012). However, the genus remains monospecific with B. prasinos the sole valid species to date. Until recently Micromonas was also a monospecific genus, however phylogenies of 18S rRNA and other genes convincingly show that Micromonas consists of at least five well-supported clades and several subclades (Šlapeta et al., 2006; Worden et al., 2009). Recently, van Baren et al. (2016) compared the two Micromonas strains with complete genomes available RCC299 in Clade A and CCMP1545 in Clade D (Šlapeta et al., 2006) and elevated clade A to species level, described as M. commoda. The remaining clades of M. pusilla, including the Arctic Micromonas in clade E2 (Simmons et al., 2015) are in need of similar taxonomic treatment.
In the Arctic Ocean, winter darkness and early spring sea-ice cover precludes photosynthesis, but small algae persist in surface waters over winter even during the Polar Night (Marquardt et al., 2016). In a study in Franklin Bay (Amundsen Gulf, Canada), Micromonas-like cells were detected via epifluorescence microscopy throughout the winter and began exponential growth under ice in late winter (Lovejoy et al., 2007). Yet, it is often challenging to separate Bathycoccus and Micromonas using microscopy alone and the contribution of Bathycoccus to the Arctic winter phytoplankton remains unknown. Here we mined high-throughput (HTS) amplicon tag libraries targeting the V4 region of the 18S rRNA gene (rDNA) and 18S rRNA (rRNA) to identify prasinophytes collected from November through July in Amundsen Gulf. Prasinophyte reads were identified to at least the level of genus, with Micromonas classified to the clade level using a curated 18S rRNA reference database (Lovejoy et al., 2016). To gain additional insight into the winter prevalence of Bathycoccus and Micromonas, we interrogated a metagenome from a sample collected in the same region on 13 December 2007. To have a clearer perception of the genetic variability of this globally distributed genus, we compared Bathycoccus from the Arctic metagenome to the published Bathycoccus genome (strain Ban7, RCC1105; Moreau et al., 2012) and to other available Bathycoccus metagenome assembled genomes (MAGs). To evaluate the viral community that could potentially influence the survival of Mamiellophyceae, we searched for prasinoviruses in the Arctic metagenome.
Materials and methods
Field sampling
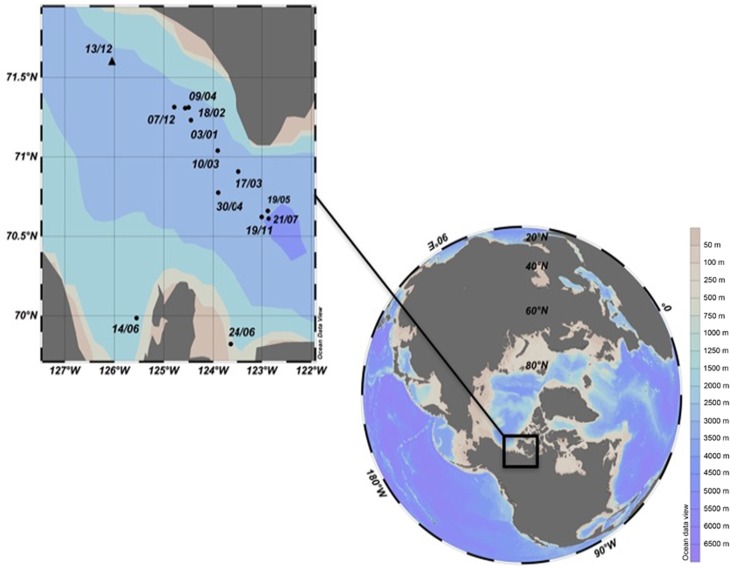
Sampling was carried out during the International Polar Year Circumpolar Flaw Lead study in Amundsen Gulf, Canadian Beaufort Sea, from November 2007 to July 2008 (Figure 1). The samples were collected every 2–4 weeks as described in Terrado et al. (2011) from the surface Polar Mixed Layer (~10 m depth) and from the top of the halocline that separates the Polar Mixed Layer from Pacific Water (Supplementary Table S1), which is where the subsurface chlorophyll maximum layer forms in summer (Monier et al., 2015). The samples for amplicon tag HTS were collected and preserved as in Terrado et al. (2011). For the metagenome, 7 liters of water from 10 m was collected on the 13 December 2007 (Supplementary Table S1).
Figure 1.
Map of the sampling sites of the overwintering study. The samples were collected from surface waters and halocline from 11 November to 21 July. The metagenomic sample was collected on 13 December. All the stations were located in the main basin of Amundsen Gulf with the exception of samples collected on 14 June from Darnley Bay and 24 June from Franklin Bay.
Extraction, library preparation and sequencing
DNA and RNA were extracted from two size fractions (3−50 μm and 0.2−3 μm) as in Terrado et al. (2011). The V4 region of the 18S rRNA was targeted for HTS as in Comeau et al. (2011) using previously reported primers E572F (CYG CGG TAA TTC CAG CTC) and E1009R (CRA AGA YGA TYA GAT ACC RT). With an aim to retrieve the entire microbial eukaryotic community, small and large fractions were mixed based on the size-fractionated chlorophyll (Chl a) concentrations for a given date (Supplementary Table S2) and sequenced at the Université Laval Plate-forme d'Analyses Génomiques using the 454-GS-FLX (Roche, Branford, CT, USA). The resulting reads were processed using packages implemented in Quantitative Insights into Microbial Ecology (QIIME, Caporaso et al., 2010a). The samples were demultiplexed, primers trimmed, short reads removed using mothur (Schloss et al., 2009), then denoised following Reeder and Knight (2010). Chimeras were checked using UCHIME (Edgar, 2004; Edgar et al., 2011) by comparing de novo and against the SILVA (release 102) reference database (Pruesse et al., 2007), and chimeric sequences removed. Operational Taxonomic Units (>98% similarity level using mothur) and representative sequences were picked by comparing with the same reference database using USEARCH (Edgar, 2010). Representative sequences were aligned in PyNast (Caporaso et al., 2010b), manually curated in BioEdit v7.2.5 (Hall, 1999) and a phylogenetic tree was generated by FastTree version 2.1.3 (Price et al., 2010). Taxonomic assignment was carried out using a curated 18S rRNA gene reference database (Comeau et al., 2016; Lovejoy et al., 2016). As our focus was on single-celled microbial eukaryotes, fungi and metazoans were removed from the data set. The raw reads are available at NCBI under Bioproject PRJNA283142. Tables of relative abundances of taxa are available on Zenodo (http://doi.org/10.5281/zenodo.163540; http://doi.org/10.5281/zenodo.163541).
For the winter metagenome, DNA from the 0.2 to 3 μm fraction was extracted using a phenol–chloroform protocol (Diez et al., 2001). The extracted DNA was fragmented with a Rapid Library Nebulizer (Roche), followed by TruSeq shotgun library preparation for Illumina sequencing. Library quality was checked using a DNA High Sensitivity chip on a BioAnalyzer 2100 (Agilent, Santa Clara, CA, USA) and paired-end sequenced using Illumina HiSeq 2000 system by the McGill University and Genome Quebec Innovation Center (Montreal, QC, Canada). Raw reads are available at NCBI with Biosample accession number SAMN05514161.
Metagenomic bioinformatics
The reads were assembled using Megahit (Li et al., 2015), via succinct de Bruijn graphs and a multiple k-mer size strategy. Assembled contigs are available at NCBI under Biosample SAMN05514161. Assembly quality and statistical analysis were carried out using the quality assessment tool for genome assemblies (QUAST) metaquast option (Gurevich et al., 2013).
A BLASTn (Altschul et al., 1990) of all Arctic metagenomic reads was carried out against the genomes of Bathycoccus Ban7 and Micromonas RCC299. The contigs and reads (paired-end mode) with BLASTn matches to the reference genomes were aligned against the two reference genomes using BWA mem algorithm (Li, 2013). Genome coverage was calculated using BEDtools (Quinlan, 2014) and statistics were performed using samtools flagstat (Li H et al.,2009). In addition, contigs were aligned to other available genomes (Micromonas CCMP1545) and transcriptomes (Bathycoccus RCC716 and Micromonas CCMP2099; Supplementary Table S3).
The differences between Arctic Bathycoccus and Ban7 genomes were examined using a variant call approach in Platypus (Rimmer et al., 2014). Variants with a minimum coverage of five reads were further examined for effects at the DNA level using SnpEff (Cingolani et al., 2012). The genes with software-called high effects were mapped to KEGG pathways (Kanehisa and Goto, 2000; Kanehisa et al., 2014; http://www.genome.jp/kegg/tool/map_pathway1.html).
To identify the dominant microbes in the Arctic metagenome, Metawatt (Strouss et al., 2012) was used to bin and taxonomically classify all contigs using a curated metawatt reference database that included 2843 genomes including 58 microbial eukaryote reference genomes (see http://doi.org/10.5281/zenodo.164419). In Metawatt, each contig was fragmented (500 bp) and every fragment BLASTn searched (e-value <1.10−6) separately against the database. Then, for each contig the BLAST results were analyzed and the contig classified to the taxa most frequently encountered as the best BLAST hit. A new BLASTn was then carried out between Coding DNA Sequences (CDS) of Ban7 against the Arctic Bathycoccus-like contigs. The matching regions were aligned using a Smith–Waterman local alignment implemented in EMBOSS (Rice et al., 2000) to determine the similarity between the two genomes.
Two MAGs from the Chilean coast, one South East (SE) Atlantic (Vaulot et al., 2012; Monier et al., 2013; Supplementary Table S4) and our Arctic Bathycoccus were blasted against CDS of Ban7 (Moreau et al., 2012) to identify homologous sequences. CDS with 90% minimum query coverage in the five genomes were retained for similarity comparisons. The selected CDS were aligned using MUSCLE (Edgar, 2004) and distance matrices between each pair of sequences were generated using ‘distmat' implemented in EMBOSS (Rice et al., 2000), with Kimura correction (Kimura, 1980) determined after model selection in jModelTest v.2 (Darriba et al., 2012). The substitution rates per 100 bp were summed and divided by the number of genes for a global overview of the similarity of the 129 CDS among the five data sets.
Gene phylogenies
The 18S rRNA gene and ITS2 sequences were recovered from the Arctic metagenome with a BLASTn search using a 10−6 e-value and 99% identity cut-offs. Reads and contigs were aligned de novo to reference sequences from Ban7, MAGs from the Chilean Coast and the MAG from the SE Atlantic (Supplementary Table S4) using MAFFT v7 (Katoh et al., 2002). Phylogenetic trees were constructed using randomized accelerated maximum likelihood (RAxML v8, GTR+G model, 100 RAxML bootstrap replicates) in Geneious (Kearse et al., 2012).
The presence of the functional gene-processing factor 8 protein (PRP8), which encodes the largest protein of the spliceosomal machinery, was determined first by predicting open-reading frames (ORF) on Arctic Bathycoccus-like contigs with a minimum ORF length of 60 amino acids. These ORFs were then searched for PRP8 sequences using hmmsearch (Eddy, 1998), in HMMer v3, with the Pfam (Punta et al., 2012) PRP8 models (PF09092 and PF12134). Only two PRP8-like putative PRP8 ORFs originating from metagenomic contigs longer than 1000 nt were retained. Additional Mamiellophyceae PRP8 protein sequences from GenBank and the Marine Microbial Eukaryotic Transcriptome Sequencing Project (Keeling et al., 2014) database, were identified using hmmsearch. PRP8 protein sequences were aligned using MAFFT and the multiple sequence alignment was then converted to nucleotides (back translated to a codon alignment based on gene information). ML inferences and nonparametric bootstrapping were carried out using RAxML v8 (Liu et al., 2011) with the GTR+CAT model.
Families of genes implicated in meiosis were targeted based on Ostreococcus (Derelle et al., 2006) and Micromonas (Worden et al., 2009) genomes. These meiosis-related genes were searched for in the Bathycoccus-like contigs using BLASTn.
Viruses in the metagenome
Diversity of prasinoviruses in the Arctic metagenome was assessed from DNA polymerase B gene (polB) sequences. Environmental reads or contigs coding for polB were identified using HMM searches (after a six-frame translation to ORFs) with hmmer v3 (http://hmmer.org/; Eddy, 2011) and the corresponding Pfam model (http://PFAM.sanger.ac.uk/; Punta et al., 2012), PF00136. ORFs that passed the HMM search gathering threshold and that were most similar to polB sequences of eukaryotic viruses after a BLASTp against UniProtKB were selected for subsequent phylogenetic analysis. Viral reference sequences and metagenomic ORFs with >200 amino-acid residues were aligned using MAFFT v7 (in local pair, iterative refinement mode), and sites with ⩾50% gaps were discarded using trimAl v1.4 (Capella-Gutiérrez et al., 2009). The best ML tree was retrieved from 100 ML topological searches and bootstrap support was determined from 100 nonparametric replications using RAxML v8 with the LG+G+I+F model. The latter model was selected based on the Akaike Information Criterion using ProtTest v3 (Darriba et al., 2011). The tree was rooted using other non-prasinovirus as outgroups.
Results
Environmental conditions and temporal patterns
The physical oceanography over the 9-month sampling period have been reported elsewhere (Forest et al., 2011; Barber et al., 2012). The environmental conditions corresponding to the samples reported here are given in Supplementary Table S1. In situ chlorophyll a fluorescence was negligible throughout winter, and increased on 9 April, with a maximum in our samples on 19 May.
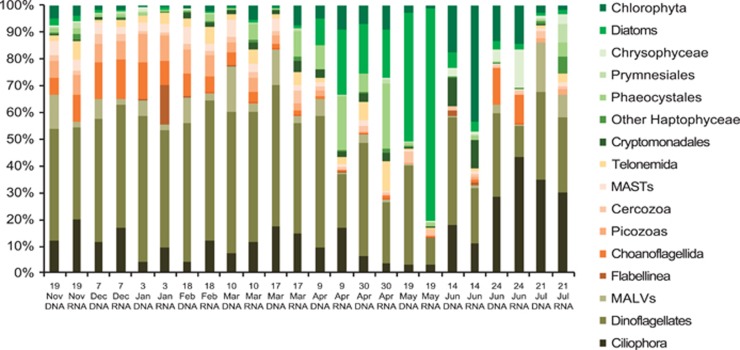
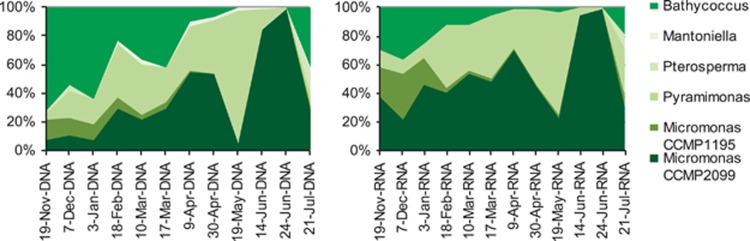
Overall, heterotrophs dominated the microbial eukaryotic reads for all samples over most of the sampling period, except during the diatom maxima in May (Figure 2). Prasinophytes accounted for 0.2 to 17% of whole community rDNA reads, and 0.8 to 42% of the rRNA reads over 9 months, with Arctic Micromonas (CCMP2099) and Bathycoccus always present. Pyramimonadales were also detected, with Pyramimonas prevalent in spring and summer and Pterosperma in July and December (Figure 3). A second Micromonas in Clade C (Šlapeta et al., 2006) accounted for 12% of the prasinophyte reads in December. Another Mamiellales, Mantoniella, occurred later in winter but was not detected in November or December (Supplementary Table S5B). Among prasinophytes, Bathycoccus dominated in winter with over half (54–72%) of prasinophyte rDNA reads at the surface from November to January and a third of the rRNA reads in November and December. From mid-February, the Arctic Micromonas predominated with highest proportions in June, with up to 90% of prasinophyte rDNA and rRNA reads, while Bathycoccus accounted for 1%, except for the July surface with 41% of the rDNA reads (Figure 3, Supplementary Table S5B).
Figure 2.
Relative abundance of the major taxonomic groups as percent (%) of the total reads from surface microbial eukaryotic. Dates of collection and template are given for 18S rRNA (RNA) and the 18S rRNA gene or rDNA (DNA). Taxonomic names follow NCBI, the phylogenetically diverse marine stramenopiles (MASTs) are grouped as are the marine alveolates (MALVs).
Figure 3.
Proportion of prasinophyte taxa out of total prasinophyte reads (see above as Chlorophyta) from DNA (left) and RNA (right) templates.
Winter metagenome
Approximately 157 million reads from the Arctic metagenome were assembled into 17 million contigs (Supplementary Table S6). After binning, taxonomic assignment indicated that although a marine bacterium Pelagibacter (12.9 Mb corresponding to 17 666 contigs binned) had high representation in the metagenome, the second largest bin was Bathycoccaceae (12.5 Mb corresponding to 10 194 contigs binned). Because of differences in genome size, the 12.9 Mb of contigs assigned to ‘Candidatus Pelagibacter ubique' potentially covered the genome 10 × compared with closer to 1 × coverage of the Bathycoccus genome. The remaining bins were <4 Mb and matched other Pelagibacter, Flavobacteria, Acidobacteria and Verrucomirobia (Table 1).
Table 1. Metagenomic contigs taxonomically binned using a BLAST and contig fragmentation approach in Metawatt. Mb refers to the combined length of all contigs that were taxonomically classified to the corresponding species.
| Species | Mb |
|---|---|
| Candidatus Pelagibacter ubique HTCC1062 | 12.9 |
| Bathycoccus (Bathycoccaceae) | 12.5 |
| Polaribacter sp. MED152 (Flavobacteriaceae) | 3.6 |
| Candidatus Pelagibacter sp. IMMCC9063 | 3.1 |
| Lacinutric sp. 5H-3-7-4 (Flavobacteriaceae) | 3.1 |
| Ilumatobacter (Acdimicrobiaceae) | 3.1 |
| Opitus terrae PB90-1 (Verrucomicrobia) | 2.1 |
| Alpha proteobacterium HIMB5 (Pelagibacteraceae) | 1.6 |
A much higher percentage of metagenomics contigs were aligned to the reference genome of Bathycoccus Ban7 (88.8%) compared with Micromonas RCC299 (21.2%). Using reads from the metagenome increased the percent alignment to 95.4% of Ban7, but was insignificant for Micromonas RCC299. Similarly when aligning our metagenomics contigs against Bathycoccus RCC716 and Micromonas CCMP2099 from the Marine Microbial Eukaryotic Transcriptome Sequencing Project database, metagenome contigs aligned with 53% of RCC716 transcripts but <4% of the CCMP2099 transcripts (Supplementary Table S3). At the level of individual Bathycoccus chromosomes; metagenomic reads covered 91.3 to 100% of the total length of 18 chromosomes, although less than half of chromosome 19 was covered. The GC content of the majority of individual Arctic chromosomes (47.7 to 49.3% Table 2) was comparable to Ban7, with an estimated GC content of 48% (Moreau et al., 2012).
Table 2. Assignment of the Arctic metagenomic reads to individual chromosomes of B. prasinos Ban7 and associated statistics.
| Ref names | Ref length (bp) | # Reads | Alignment (%) | Depth cover | % ID | % GC |
|---|---|---|---|---|---|---|
| chrom_01 | 1 352 724 | 155 626 | 97.6 | 10.7 | 92.9 | 49.3 |
| chrom_02 | 1 122 692 | 127 118 | 97.2 | 10.5 | 92.8 | 49 |
| chrom_03 | 1 091 008 | 123 431 | 96.3 | 10.6 | 92.9 | 49.2 |
| chrom_04 | 1 037 991 | 122 919 | 97 | 11 | 92.5 | 48.5 |
| chrom_05 | 1 019 276 | 114 827 | 96.5 | 10.5 | 92.8 | 48.9 |
| chrom_06 | 989 707 | 110 283 | 96.2 | 10.4 | 92.8 | 48.8 |
| chrom_07 | 955 652 | 108 599 | 96.3 | 10.6 | 92.9 | 48.9 |
| chrom_08 | 937 610 | 104 766 | 95.7 | 10.4 | 92.9 | 49 |
| chrom_09 | 895 536 | 101 602 | 96.7 | 10.6 | 92.9 | 49 |
| chrom_10 | 794 368 | 86 786 | 95.1 | 10.2 | 92.8 | 48.9 |
| chrom_11 | 741 603 | 108 169 | 96.5 | 13.1 | 93.2 | 48.7 |
| chrom_12 | 712 459 | 79 697 | 96.3 | 10.4 | 92.8 | 48.9 |
| chrom_13 | 708 035 | 83 561 | 96.8 | 11 | 92.6 | 49 |
| chrom_14 | 663 424 | 59 280 | 96.5 | 8.4 | 93 | 43.6 |
| chrom_15 | 519 835 | 58 731 | 96.6 | 10.5 | 92.8 | 48.8 |
| chrom_16 | 494 108 | 53 034 | 93.2 | 9.9 | 92.8 | 48.7 |
| chrom_17 | 465 570 | 46 946 | 91.3 | 9.3 | 92.7 | 48.4 |
| chrom_18 | 310 170 | 32 728 | 95.4 | 9.6 | 92.6 | 47.7 |
| chrom_19 | 146 238 | 3740 | 41.9 | 2.2 | 92.1 | 42.4 |
| Mito | 43 614 | 21 546 | 100 | 39.6 | 95.5 | 45.5 |
| Chloro | 72 700 | 40 406 | 69.2 | 45.8 | 96.6 | 46.3 |
Reference names (Ref Names) lists the individual chromosomes (1 to 19), the mitochondria (Mito) and chloroplast (Chloro). Reference length (Ref Length) is the length of the reference sequences for the chromosomes in base pair (bp). Number of reads (# Reads) mapped to the reference sequence. Alignment is the percentage of the chromosome length with aligned reads. Depth of coverage (Depth Cover) is average number of reads aligning to a given position of the reference chromosome. Percent identity (% ID) is average pairwise identity over the alignment and the percent GC (%GC) is GC content of mapped Arctic reads.
Bathycoccus genes
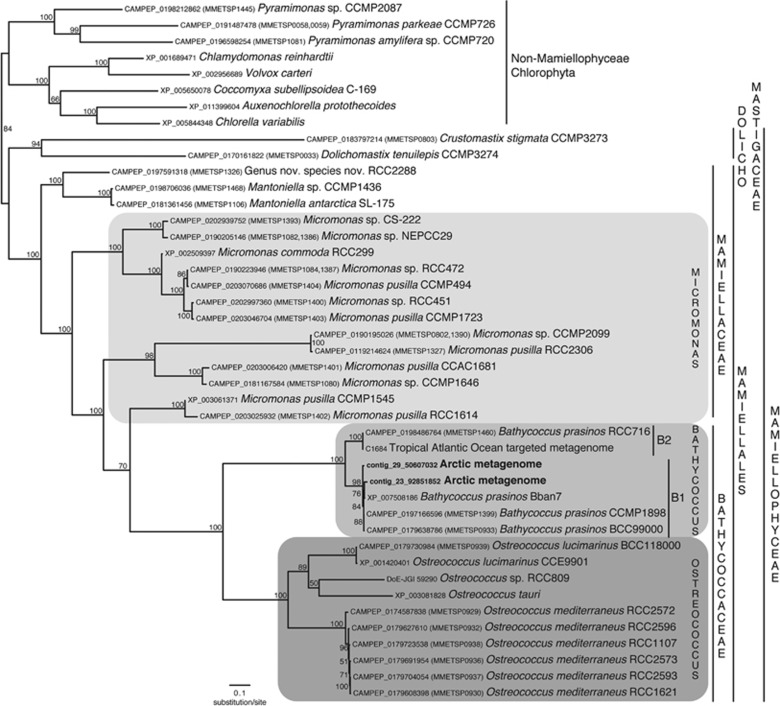
A single ~331 bp fragment of the 18S rRNA gene was recovered from a global search of the Arctic metgenome contigs. The 18S rRNA gene region had 100% sequence similarity with Ban7. Another contig containing the complete ITS2 sequence of Bathycoccus placed the Arctic Bathycoccus into the B1 coastal clade (Supplementary Figure S1). The phylogenetic analysis of the PRP8 gene also placed the Arctic Bathycoccus into the B1 clade (Figure 4) along with most other available Bathycoccus PRP8 genes. The SE Atlantic MAG and one isolate from the Indian Ocean (Bathycoccus RCC716) made up clade B2. Both the B2 clade PRP8 genes contained inteins, but at a different insert locations. Because sexual stages have been suggested as a survival strategy in Mamiellophyceae, we also targeted meiosis-related genes in the contigs binned as Bathycoccaceae. All the targeted meiosis-related genes were found in the Arctic Bathycoccus (Supplementary Table S7).
Figure 4.
Phylogenetic tree of PRP8 protein marker gene (see text). Maximum-likelihood (ML) reconstruction using 100 nonparametric bootstrap and gtr+cat model (marked at nodes). The two Bathycoccus clades are indicated as B1 and B2.
A similarity search of Ban7 CDS against the Arctic Bathycoccus-like contigs identified 7255 putative genes with an average of 95% similarity in the two genomes. Among these were 129 CDS that were present in Ban7 and all of Bathycoccus MAGs (ours from the Arctic, two Pacific and one Atlantic). Based on this conserved set of genes, the Arctic Bathycoccus was 82.7% similar to the SE Atlantic Bathycoccus and 98% similar to the two Pacific Bathycoccus, which were highly similar to each other (Table 3).
Table 3. Summary of the substitution rate per 100 bp of the 129 coding genes common to the five data sets.
| Atlantic | Pacific 1 | Pacific 2 | Arctic | Mediterranean | |
|---|---|---|---|---|---|
| Atlantic | 0 | 17.34 | 17.36 | 17.33 | 17.36 |
| Pacific 1 | 0 | 1.12 | 1.97 | 1.33 | |
| Pacific 2 | 0 | 1.84 | 1.32 | ||
| Arctic | 0 | 1.85 | |||
| Mediterranean | 0 |
The Atlantic refers to Bathycoccus metagenomic contigs from the South East (SE) Atlantic (Monier et al., 2013). Pacific 1 and 2 refer to the two metagenomes from the upwelling zone of Pacific water off the Chilean coast (Vaulot et al., 2012). Mediterranean refers to the reference genome of Bathycoccus Ban7 (Moreau et al., 2012) and Arctic refers to Bathycoccus-like contigs from the metagenomic sample from the present study.
Bathycoccus variants
Global differences between Arctic and Ban7 Bathycoccus genomes were identified by variant call analysis, where a variant represents a Ban7 nucleotide position that differed from the sequence of the Arctic reads mapped to the same position. In total 141 795 variants were detected, with over 99% of the variants predicted to have little impact on the final protein. About 0.73% of the variants were CDS frame shift insertion/deletion (indels), splice acceptor/donor sites, non-start/stop codons substituted for start/stop codons and other modifications affecting final putative protein product. Within the pool of these higher impact variants, 200 had ⩾5 occurrences (reads) and could be matched to KEGG (KO) numbers (Supplementary Table S8).
Viral diversity in the Arctic metagenome
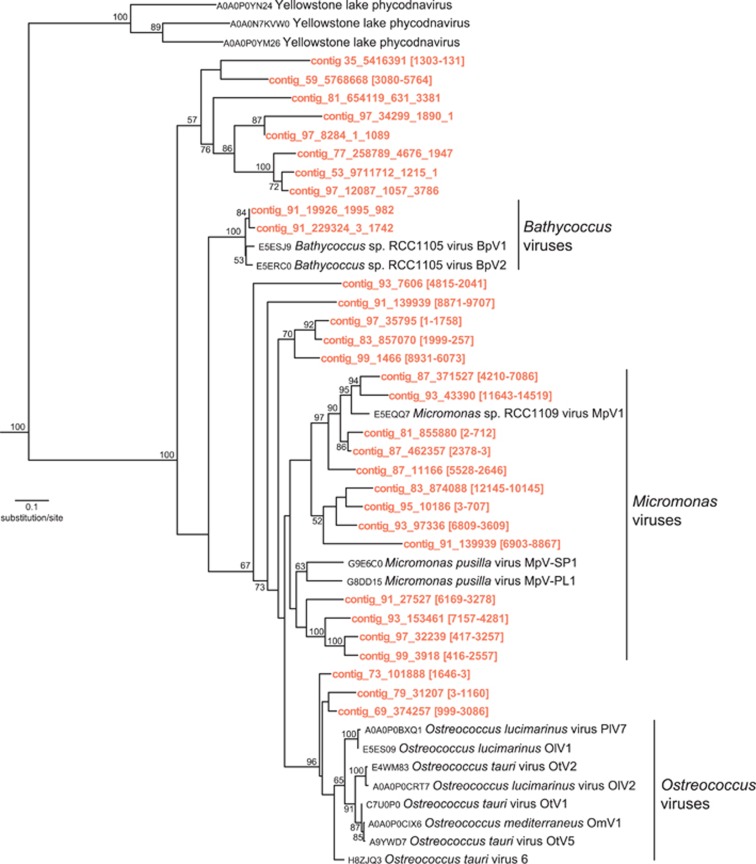
Prasinovirus infection is common in Mamiellophyceae and for this reason, we looked for sequences of polB, which is a prasinovirus marker gene. We found that in the winter metagenome Micromonas polB gene was much more diverse than those associated with Bathycoccus (Figure 5). The polB phylogeny also showed a novel environmental cluster at the base of the Phycodnaviridae.
Figure 5.
Phylogenetic tree of DNA polymerase B (polB) gene. ML reconstruction using model GTR+G based on a multiple sequence alignment of nucleotides (Boobstrap replicate=100). Only nodes with bootstrap values >50% are displayed. Contigs from the Arctic Bathycoccus MAG are in the lighter bold font.
Discussion
Bathycoccus and Micromonas
In the summer Arctic, a single ecotype of Micromonas (CCMP2099), dominates picoeukaryote 18S rRNA gene surveys (Lovejoy and Potvin 2011; Terrado et al., 2011; Balzano et al., 2012). Micromonas (ecotype CCMP2099) has also been reported from DNA collected from Isfjorden, West Spitbergan during the Polar night (Vader et al., 2015; Marguardt et al., 2016). However, data on the taxonomic makeup of Arctic winter and spring microbial eukaryote communities is rare. Here we expected to find Micromonas over the winter and the high representation of Bathycoccus 18S rRNA gene and 18S rRNA reads from November to December was surprising. Close investigation of a metagenome from early December also indicated that Bathycoccus was a major component of the microbial community at that time. As Bacteria usually dominate shotgun metagenomes (Piganeau et al., 2008), the high proportion of Bathycoccus reads and contigs in the whole community metagenome was also unexpected, Micromonas contigs and reads were also found in the metagenome but at much lower levels, with only 20% of reads and contigs aligning to the Micromonas reference genome, compared with >90% for Bathycoccus. These results raise questions as to why Bathycoccus, which is usually relatively rare in the Arctic compared with the Arctic ecotype of Micromonas, was so abundant in early December in Amundsen Gulf.
Globally, Micromonas and Bathycoccus often co-occur (Not et al., 2004, 2005). Outside the Arctic, Bathycoccus concentrations are reported to be less variable than Micromonas, for example, a seasonal study based on 16S rRNA chloroplast genes at the Bermuda Biological Time Series (BATS) station reported more variability in Micromonas compared with Bathycoccus (Treusch et al., 2012). In the Monterey Bay Drift Study, Bathycoccus was found in all samples with Micromonas more variable (Simmons et al., 2016). Finally, in a seasonal study off the coast of the Mediterranean, Micromonas became rare following deep winter mixing, while Bathycoccus persisted (Zhu et al., 2005). These examples suggest that Bathycoccus could have an advantage under low-light conditions in more temperate waters. However, this explanation does not fit well with the continuous presence of the Micromonas arctic ecotype over winter, both in our samples and around Svalbard (Vader et al., 2015), during the Polar Night.
One mechanism for surviving during prolonged dark periods is the capacity to form cysts, which is associated with a sexual stage in many algae (Doucette and Fryxell, 1983; French and Hargraves, 1985). Similar to other Mamiellophyceae (Derelle et al., 2006; Worden et al., 2009; Grimsley et al., 2010), the arctic Bathycoccus MAG contained meiosis-implicated gene families and low GC regions, consistent with potential for a sexual stage. We also noted the presence of a full intein inside the PRP8 protein in the Atlantic MAG that could also indicate sexuality (Monier et al., 2013). However, no inteins were found in the Arctic clade. The genomic evidence now points to at least ancestral sexual reproduction in the Mamiellophyceae and the possibility of Micromonas and Bathycoccus having resistant resting stages, enabling survival when conditions are not suitable for active growth. As both share this trait, any advantage would need to be linked to environmental triggers.
Another survival strategy during prolonged darkness, could be related to alternative carbon and energy acquisition strategies. Many phytoflagellates are mixotrophic and survive in the dark when prey are available (Bell and Laybourn-Parry, 2003; Hartman et al., 2012). Burns et al. (2015), using comparative genomics, found several genes associated with phagotrophic heterotrophs in the marine prasinophyte Cymbomonas tetramitiformis. However, that alga grows best in the presence of bacteria under low light and short day lengths (Maruyama and Kim, 2013). In laboratory studies, bacterial ingestion rates by Micromonas CCMP2099 are greatest in the light and under low-nutrient conditions (McKie-Krisberg and Sanders, 2014). Similarly, in the Arctic, Micromonas is reported to actively take up labeled beads or bacteria in summer when light is available and nitrogen limiting (Gonzalez et al., 1993; Sherr et al., 2003). These reports suggests that the Arctic Micromonas uses prey as a source of nutrients rather than for energy acquisition (Mitra et al., 2016). During our study, the winter (November–January) bacterial concentrations and production rates were low (Nguyen et al., 2012) and Micromonas may have been unable to compete with true heterotrophs for bacterial prey. Monier et al. (2015) also noted that Micromonas did not sustain high relative populations under light-limiting conditions caused by surface shading from the McKenzie river plume, consistent with light as a primary factor limiting Micromonas. However, bacterial production was also relatively low in the subsurface chlorophyll maximum during that study (Ortega-Retuerta et al., 2012) and bacterial prey may also have been limiting. Although phagotrophy has not been reported in Bathycoccus, the possibility of using osmotrophy in the dark cannot be ruled out.
The predominance of Bathycoccus compared with Micromonas in November and December could also be consistent with different loss rates. In summer and autumn, Bathycoccus is normally rare in the Arctic, and would have lower encounter rates with potential predators (Cram et al., 2016). However, in winter, both genera remain potential prey for microzooplankton and smaller zooplankton, which are active during the Polar Night (Berge et al., 2015). Interestingly, the organic scales on the surface of Bathycoccus could act as a deterrent to grazers (Moreau et al., 2012). Having scales could also decrease viral attachment efficiency, making Bathycoccus a defense specialist.
Being rare would also reduce encounter rates with viruses (Winter et al., 2010; Becket and Williams, 2013). Micromonas, which is normally more abundant compared with Bathycoccus in summer and autumn (Data from Comeau et al., 2011; Genbank SRA029114), would have been more exposed to viral infection before November. Overall, there was an anomaly in the ratio of Bathycoccus reads to Bathycoccus polB genes compared with the ratio of Micromonas reads to Micromonas polB genes, which would not be consistent with all the polB genes being predominantly intercellular. Given that DNA viruses persist following infection, this leads to the prediction that a higher proportion of DNA viruses targeting Micromonas compared with those targeting Bathycoccus would be evidence for past viral activity. The high number of Micromonas-specific prasinovirus marker genes in the metagenome is consistent with such a scenario (c.f. Moreau et al., 2010). In which case, this resembles a ‘kill the winner' outcome (Thingstad, 2000) with Micromonas succumbing to viral infection, leaving Bathycoccus temporarily more abundant. A single polB gene phylotype of the Bathycoccus virus was also found (Figure 5) indicating ongoing dynamics between the viruses and their hosts.
Arctic-Boreal prasinophytes and polB diversity
Using HTS, we detected a second Micromonas phylotype with closest affinities to CCMP1195 (Clade C; Šlapeta et al., 2006; Simmons et al., 2015) in winter and in the July surface sample. Lovejoy and Potvin (2011) reported the same phylotype from an Amundsen Gulf Pacific Halocline clone library, and speculated that it was from Pacific waters flowing along the shelf break. CCMP1195 was originally isolated from a winter Gulf of Maine sample, and could represent a Boreal-Arctic cold-water form, analogous to Thalassiosira species with both Arctic and North Atlantic distributions (Luddington et al., 2016). The phylotype may have also been a source for some of the prasinoviruses genes found in the winter metagenome. Other potential hosts for the non-Micromonas, non-Bathycoccus prasinoviruses, could include Pterosperma, another Arctic-Boreal species detected in December and July. In addition, Pyramimonas spp. are common in the Arctic (Lovejoy et al., 2002; Niemi et al., 2011; Monier et al., 2015) and although the maximum occurrence during our study was in March and April, Pyramimonas were present in winter. The diversity of prasinoviruses even when hosts may have been rare, is consistent with reports of persistence of phycodnaviruses, which could infect susceptible hosts when host populations reach a threshold level (Short et al., 2011).
Seasonal recovery of the status quo
In our study, Micromonas re-established rapidly once surface irradiances reached ~5 mol phot m−2 per day (Nguyen et al., 2015), which is equivalent to ~60 μmol phot m−2 per day or 8–10 μmol phot m−2 per day at the depth of collection for our surface samples (assuming 1% light levels at 40 m). The relative increase in Micromonas in March and April despite low irradiance levels in the water column, due to low sun angles and ice cover, is consistent with the Arctic ecotype having the capacity to carry out photosynthesis at low temperatures under a wide range of light levels (Lovejoy et al., 2007; Ni et al., 2016). The Amundsen Gulf phytoplankton community abruptly changed in May, with longer days and higher surface temperatures promoting a diatom dominated spring phytoplankton bloom (Figure 2,Forest et al., 2011; Terrado et al., 2011). Following the bloom, the Arctic Micromonas again dominated phytoplankton reads, consistent with smaller cells with higher cell surface to volume ratios adapted to low surface nitrate concentrations (Raven, 1998; Li WKW et al., 2009). The sudden higher proportion of Bathycoccus in July surface samples was enigmatic but may have been associated with offshore upwelling and spreading of winter waters in the Polar Mixed Layer (Garneau et al., 2006), or more speculatively, a viral attack on Micromonas triggered by specific oceanographic conditions.
Ecotypes and phylogenetic placement of Bathycoccus from the Arctic
Compared to Micromonas and Ostreococcus, diversity of the 18S rRNA gene in Bathycoccus is genuinely low, which was consistent with the low rate of variance between the Arctic and Ban7 Bathycoccus. We examined potential variation in the hypervariable internal transcribed spacer 2 (ITS2), which is useful for separating populations or species (Kaczmarska et al., 2009). Previous phylogeny of ITS2 and PRP8 sequences suggests two ecotypes of Bathycoccus (Vaulot et al., 2012; Monier et al., 2013), one adapted to coastal waters or more nutrient-rich environments (Clade B1) and the other to open ocean more oligotrophic environments (Clade B2). The MAGs from the upwelling zone of the Pacific (Vaulot et al., 2012) and coastal Mediterranean belong to B1, and the SE Atlantic MAG belongs to B2 (Monier et al., 2013). Occurrence of B1 in the Arctic is consistent with the Amundsen Gulf region dominated by cross shelf processes (Williams and Carmack, 2015). However, the distribution and identity of microbial eukaryotes in deeper Arctic waters is practically unknown (Pedrós-Alió et al., 2015). The deeper Arctic Basins remain undersampled and ITS2 or PRP8 gene surveys are needed to resolve the distribution of the two clades in the Arctic. Finally, at the genome level, the high similarity of the Arctic Bathycoccus to others in clade B1 contrasts with Micromonas and other eukaryotic phytoplankton that have representative species restricted to the Arctic. Ecophysiological studies and complete reference genomes, starting with cultured Arctic Bathycoccus are needed to verify whether Bathycoccus is truly cosmopolitan across oceans.
Acknowledgments
This study was part of the Circumpolar Flaw Lead—International Polar Year (CFL-IPY) study supported by the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Network of Centers of Excellence ArcticNet. NJ received scholarships from Université Laval and the Canadian Excellence Research Chair—Remote Sensing of Canada's New Arctic Frontier (CERC) grant to Marcel Babin. Additional support came from an NSERC Discovery and Northern Supplement grants to CL and the Fonds de recherche du Québec Nature et Technologies (FRQNT) to Quebéc-Océan. We are also grateful to Jean-Éric Tremblay for nutrient data, C. Evans for April samples and Ramon Terrado who supervised the sampling over the deployment, Emmanuel Medrinal, Marianne Potvin and Brian Boyle for laboratory work and Jerome Laroche for intensive bioinformatic support.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Amato A, Kooistra WHCF, Hee J, Ghiron L, Mann DG. (2007). Reproductive isolation among sympatric cryptic species in marine diatoms. Protist 158: 193–207. [DOI] [PubMed] [Google Scholar]
- Apple JK, Strom SL, Palenik B, Brahamsha B. (2011). Variability in protist grazing and growth on different marine Synechococcus isolates. Appl Environ Microbiol 77: 3074–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzano S, Marie D, Gourvil P, Vaulot D. (2012). Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J 6: 1480–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber DG, Asplin MG, Papakyriakou TN, Miller L, Else BGT, Asselin NC et al. (2012). Consequences of change and variability in sea ice on marine ecosystem and biogeochemical processes during the 2007–2008 Canadian International Polar Year program. Clim Change 115: 135–159. [Google Scholar]
- Barber DG, Asplin MG, Raddatz RL, Candlish LM, Nickels S, Prinsenberg SJ. (2012). Change and variability in sea ice during the 2007–2008 Canadian International Polar Year program. Clim Change 115(S1): 115–113. [Google Scholar]
- Beckett SJ, Williams HTP, Beckett SJ. (2013). Coevolutionary diversification creates nested-modular structure in phage – bacteria interaction networks. Interface Focus 3: 20130033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EM, Laybourn-Parry J. (2003). Mixotrophy in the antarctic phytoflagellate, Pyramimonas gelidicola (Chlorophyta: Prasinophyceae). J Phycol 39: 644–649. [Google Scholar]
- Berge J, Daase M, Renaud PE, Ambrose WG, Darnis G, Last KS et al. (2015). Unexpected levels of biological activity during the polar night offer new perspectives on a warming arctic. Current Biology 25: 2555–2561. [DOI] [PubMed] [Google Scholar]
- Burns JA, Paasch A, Narechania A, Kim E. (2015). Comparative genomics of a bacterivorous green alga. Genome Biol Evol 7: 3047–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. a). Correspondence QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nature 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w 1118; iso-2; iso-3. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerissi C, Grimsley N, Subirana L, Maria E, Oriol L, Ogata H et al. (2014). Prasinovirus distribution in the Northwest Mediterranean Sea is affected by the environment and particularly by phosphate availability. Virology 466: 146–157. [DOI] [PubMed] [Google Scholar]
- Coleman AW. (2007). Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res 35: 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Li WKW, Tremblay J-É, Carmack EC, Lovejoy C. (2011). Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6: e27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Vincent WF, Bernier L, Lovejoy C. (2016). Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci Rep 6: 30120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupel P, Matsuoka A, Gosselin M, Marie D, Tremblay J, Babin M et al. (2015). Pigment signatures of phytoplankton communities in the Beaufort Sea. Biogeosciences 12: 991–1006. [Google Scholar]
- Cram JA, Parada AE, Fuhrman JA. (2016). Dilution reveals how viral lysis and grazing shape microbial communities. Limnol Oceanogr 61: 889–905. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2011). Protest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012). Europe PMC Funders Group jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasilva CR, Li WKW, Lovejoy C. (2013). Phylogenetic diversity of eukaryotic marine microbial plankton on the Scotian Shelf Northwestern Atlantic Ocean. J Plank Res 36: 344–363. [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S et al. (2006). Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez B, Pedrós-Alió C, Massana R. (2001). Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by Small-Subunit rRNA gene cloning and sequencing. Appl Environ Microbiol 67: 2932–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucette GJ, Fryxell GA. (1983). Thalassiosira antarctica: vegetative and resting stage chemical composition of an ice-related marine diatom. Marine Biol 78: 1–6. [Google Scholar]
- Eddy S. (1998). HMMER: profile HMMs for protein sequence analysis. Bioinformatics 14: 755–763.9918945 [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Eikrem W, Throndsen J. (1990). The ultrastructure of Bathycoccus gen. nov. and B. prasinos sp. nov., a non-motile picoplanktonic alga (Chlorophyta, Prasinophyceae) from the Mediterranean and Atlantic. Phycologia 29: 344–350. [Google Scholar]
- Forest A, Tremblay J-Éric, Gratton Y, Martin J, Gagnon J, Darnis G et al. (2011). Progress in Oceanography Biogenic carbon flows through the planktonic food web of the Amundsen Gulf (Arctic Ocean): a synthesis of field measurements and inverse modeling analyses. Prog Oceanogr 91: 410–436. [Google Scholar]
- French FW, Hargraves PE. (1985). Spore formation in the life cycles of the diatoms Chaetoceros diadema and Leptocylindrus danicus. J Phycol 21: 477–483. [Google Scholar]
- Garneau MÈ, Vincent WF, Alonso-Sáez L, Gratton Y, Lovejoy C. (2006). Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat Microb Ecol 42: 27–40. [Google Scholar]
- Gonzalez JM, Sherr BF, Sherr EB. (1993). Digestive enzyme activity as a quantitative measure of protistan grazing: the acid lysozyme assay for bacterivory. Mar Ecol Prog Ser 100: 197–206. [Google Scholar]
- Grimsley N, Pequin B, Bachy C, Moreau H, Piganeau G. (2010). Cryptic sex in the smallest eukaryotic marine green alga. Mol Biol Evol 27: 47–54. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ et al. (2012). Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Natl Acad Sci USA 109: 5756–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka N, Kawai-Toyooka H, Matsuzaki R, Nakada T, Nozaki H. (2012). Description of two new monoecious species of Volvox sect Volox (Volvocaceae, Chlorophyceae), based on comparative morphology and molecular phylogeny of cultured material. J Phycol 48: 759–767. [DOI] [PubMed] [Google Scholar]
- Jardillier L, Zubkov M V, Pearman J, Scanlan DJ. (2010). Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J 4: 1180–1192. [DOI] [PubMed] [Google Scholar]
- Kaczmarska I, Lovejoy C, Potvin M, Macgillivary M. (2009). Morphological and molecular characteristics of selected species of Minidiscus (Bacillariophyta, Thalassiosiraceae). Eur J Phycol 44: 461–475. [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilias ES, Nothig E, Wolf C, Metfies K. (2014). Picoeukaryote plankton composition off West Spitsbergen at the entrance to the Arctic Ocean. J Eukaryot Microbiol 61: 569–579. [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, Lam TW. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31: 1674–1676. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303: 3997. [Google Scholar]
- Li WKW, McLaughlin FA, Lovejoy C, Carmack EC. (2009). Smallest algae thrive as the Arctic Ocean freshens. Science 326: 539. [DOI] [PubMed] [Google Scholar]
- Li WKW. (1994). Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: Measurements from flow cytometric sorting. Limnol Oceanogr 39: 169–175. [Google Scholar]
- Liu K, Linder CR, Warnow T. (2011). RAxML and FastTree: Comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One 6: e27731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C, Comeau A, Thaler M. (2016) Curated reference database of SSU rRNA for northern marine and freshwater communities of Archaea, Bacteria and microbial eukaryotes, v. 1.0. Nordicana D23, doi:10.5885/45409XD-79A199B76BCC4110.
- Lovejoy C, Legendre L, Martineau M. (2002). Distribution of phytoplankton and other protists in the North Water. Deep Sea Res 49: 5027–5047. [Google Scholar]
- Lovejoy C, Potvin M. (2011). Microbial eukaryotic distribution in a dynamic Beaufort Sea and the Arctic Ocean. J Plank Res 33: 431–444. [Google Scholar]
- Lovejoy C, Vincent WF, Bonilla S, Roy S, Martineau M-J, Terrado R et al. (2007). Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in Arctic Seas. J Phycol 43: 78–89. [Google Scholar]
- Luddington IA, Lovejoy C, Kaczmarska I. (2016). Species-rich meta-communities of the diatom order Thalassiosirales in the Arctic and northern Atlantic Ocean. J Plank Res 38: 781–797. [Google Scholar]
- Marquardt M, Vader A, Stübner EI, Reigstad M, Gabrielsen TM. (2016). Strong seasonality of marine microbial eukaryotes in a high-arctic fjord (Isfjorden, in West Spitsbergen, Norway). Appl Environ Microbiol 82: 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Kim E. (2013). A modern descendant of early green algal phagotrophs. Curr Biol 23: 1081–1084. [DOI] [PubMed] [Google Scholar]
- McKie-Krisberg ZM, Sanders RW. (2014). Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J 10: 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Flynn KJ, Tillmann U, Raven JA, Caron D, Stoecker DK et al. (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167: 106–120. [DOI] [PubMed] [Google Scholar]
- Mojica KDA, Huisman J, Wilhelm SW, Brussaard CPD. (2016). Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean. ISME J 10: 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Comte J, Babin M, Forest A, Matsuoka A, Lovejoy C. (2015). Oceanographic structure drives the assembly processes of microbial eukaryotic communities. ISME J 9: 990–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Sudek S, Fast NM, Worden AZ. (2013). Gene invasion in distant eukaryotic lineages: discovery of mutually exclusive genetic elements reveals marine biodiversity. ISME J 7: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Worden AZ, Richards TA. (2016). Phylogenetic diversity and biogeography of the Mamiellophyceae lineage of eukaryotic phytoplankton across the oceans. Environ Microbiol Rep 8: 461–469. [DOI] [PubMed] [Google Scholar]
- Moreau H, Piganeau G, Desdevises Y, Cooke R, Derelle E, Grimsley N. (2010). Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J Virol 84: 12555–12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau H, Verhelst B, Couloux A, Derelle E, Rombauts S, Grimsley N et al. (2012). Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol 13: R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Maranger R, Balague V, Lovejoy C, Pedro C. (2015). Winter diversity and expression of proteorhodopsin genes in a polar ocean. ISME J 9: 1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Maranger R, Tremblay J-Éric, Gosselin M. (2012). Respiration and bacterial carbon dynamics in the Amundsen Gulf, Western Canadian Arctic. J Geophys Res 117: C00G16. [Google Scholar]
- Ni G, Zimbalatti G, Murphy CD, Barnett AB, Arsenault CM, Li G et al. (2016). Arctic Micromonas uses protein pools and non-photochemical quenching to cope with temperature restrictions on Photosystem II protein turnover. Photosynth Res 131: 203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi A, Michel C, Hille K, Poulin M. (2011). Protist assemblages in winter sea ice: setting the stage for the spring ice algal bloom. Polar Biol 34: 1803–1817. [Google Scholar]
- Not F, Latasa M, Marie D, Cariou T, Vaulot D, Simon N. (2004). A single species, Micromonas pusilla (Prasinophyceae), dominates the eukaryotic picoplankton in the Western English Channel. Appl Environ Microbiol 70: 4064–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Massana R, Latasa M, Marie D, Colson C, Eikrem W et al. (2005). Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol Oceanogr 50: 1677–1686. [Google Scholar]
- Ortega-Retuerta E, Jeffrey WH, Babin M, Bélanger S, Benner R, Marie D et al. (2012). Carbon fluxes in the Canadian Arctic: Patterns and drivers of bacterial abundance, production and respiration on the Beaufort Sea margin. Biogeosciences 9: 3679–3692. [Google Scholar]
- Pedrós-Alió C, Potvin M, Lovejoy C. (2015). Diversity of planktonic microorganisms in the Arctic Ocean. Prog Oceanogr 139: 233–243. [Google Scholar]
- Piganeau G, Desdevises Y, Derelle E, Moreau H. (2008). Picoeukaryotic sequences in the Sargasso Sea metagenome. Genome Biol 9: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill P, Eberhardt R, Mistry J, Tate J, Boursnell C et al. (2012). The Pfam protein families' databases. Nucleic Acids Res 40: D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. (2014). BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. (1998). The twelfth Tansley Lecture. Small is beautiful: the picophytoplankton. Functional Ecol 12: 503–513. [Google Scholar]
- Reeder J, Knight R. (2010). Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7: 668–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM et al. (2014). Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46: 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF, Wheeler PA, Thompson K. (2003). Temporal and spatial variation in stocks of autotrophic and heterotrophic microbes in the upper water column of the central Arctic Ocean. Deep Sea Res I 50: 557–571. [Google Scholar]
- Short CM, Rusanova O, Short SM. (2011). Quantification of virus genes provides evidence for seed-bank populations of phycodnaviruses in Lake Ontario, Canada. ISME J 5: 810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Bachy C, Sudek S, van Baren MJ, Sudek L, Ares M Jr et al. (2015). Intron invasions trace algal speciation and reveal nearly identical Arctic and Antarctic Micromonas populations. Mol Biol Evol 32: 2219–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Sudek S, Monier A, Limardo AJ, Jimenez V, Perle CR et al. (2016). Abundance and biogeography of picoprasinophyte ecotypes and other phytoplankton in the Eastern North Pacific Ocean. Appl Environ Microbiol 82: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šlapeta J, López-García P, Moreira D. (2006). Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol 23: 23–29. [DOI] [PubMed] [Google Scholar]
- Strous M, Kraft B, Bisdorf R, Tegetmeyer HE. (2012). The binning of metagenomic contigs for microbial physiology of mixed cultures. Front Microbiol 3: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrado R, Medrinal E, Dasilva C, Thaler M, Vincent WF, Lovejoy C. (2011). Protist community composition during spring in an Arctic flaw lead polynya. Polar Biol 34: 1901–1914. [Google Scholar]
- Thingstad TF. (2000). Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr 45: 1320–1328. [Google Scholar]
- Thomas R, Grimsley N, Escande M, Subirana L, Derelle E, Moreau H. (2011). Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ Microbiol 13: 1412–1420. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Demir-Hilton E, Vergin KL, Worden AZ, Carlson CA, Donatz MG et al. (2012). Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids. ISME J 6: 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader A, Marquardt M, Meshram AR, Gabrielsen TM. (2015). Key Arctic phototrophs are widespread in the polar night. Polar Biol 38: 13–21. [Google Scholar]
- Van Baren MJ, Bachy C, Reistetter EN, Purvine SO, Grimwood J, Sudek S et al. (2015). Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. BMC Genomics 17: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, Eikrem W, Viprey M, Moreau H. (2008). The diversity of small eukaryotic phytoplankton in marine ecosystems. FEMS Microbiol Rev 32: 795–820. [DOI] [PubMed] [Google Scholar]
- Vaulot D, Lepère C, Toulza E, De la Iglesia R, Poulain J, Gaboyer F et al. (2012). Metagenomes of the picoalga Bathycoccus from the Chile coastal upwelling. PLoS One 7: e39648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidussi F, Roy S, Lovejoy C, Gammelgaard M, Thomsen HA, Booth B et al. (2004). Spatial and temporal variability of the phytoplankton community structure in the North Water Polynya, investigated using pigment biomarkers. Can J Fish Aquat Sci 2052: 2038–2052. [Google Scholar]
- Williams WJ, Carmack EC. (2015). The ‘interior'shelves of the Arctic Ocean: physical oceanographic setting, climatology and effects of sea-ice retreat on cross-shelf exchange. Prog Oceanogr 139: 24–41. [Google Scholar]
- Winter C, Bouvier T, Weinbauer MG, Thingstad TF. (2010). Trade-offs between competition and defense specialists among unicellular planktonic organisms: the 'Killing the Winner' hypothesis revisited. Microbiol Mol Biol Rev 74: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden A, Nolan J, Palenik B. (2004). Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr 49: 168–179. [Google Scholar]
- Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272. [DOI] [PubMed] [Google Scholar]
- Zhang F, He J, Lin L. (2015). Dominance of picophytoplankton in the newly open surface water of the central Arctic Ocean. Polar Biol 38: 1081–1089. [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. (2005). Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52: 79–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.