Abstract
Marine Euryarchaeota remain among the least understood major components of marine microbial communities. Marine group II Euryarchaeota (MG-II) are more abundant in surface waters (4–20% of the total prokaryotic community), whereas marine group III Euryarchaeota (MG-III) are generally considered low-abundance members of deep mesopelagic and bathypelagic communities. Using genome assembly from direct metagenome reads and metagenomic fosmid clones, we have identified six novel MG-III genome sequence bins from the photic zone (Epi1–6) and two novel bins from deep-sea samples (Bathy1–2). Genome completeness in those genome bins varies from 44% to 85%. Photic-zone MG-III bins corresponded to novel groups with no similarity, and significantly lower GC content, when compared with previously described deep-MG-III genome bins. As found in many other epipelagic microorganisms, photic-zone MG-III bins contained numerous photolyase and rhodopsin genes, as well as genes for peptide and lipid uptake and degradation, suggesting a photoheterotrophic lifestyle. Phylogenetic analysis of these photolyases and rhodopsins as well as their genomic context suggests that these genes are of bacterial origin, supporting the hypothesis of an MG-III ancestor that lived in the dark ocean. Epipelagic MG-III occur sporadically and in relatively small proportions in marine plankton, representing only up to 0.6% of the total microbial community reads in metagenomes. None of the reconstructed epipelagic MG-III genomes were present in metagenomes from aphotic zone depths or from high latitude regions. Most low-GC bins were highly enriched at the deep chlorophyll maximum zones, with the exception of Epi1, which appeared evenly distributed throughout the photic zone worldwide.
Introduction
Marine archaea are important marine microbes in terms of their metabolic activity and abundance (Karner et al., 2001; Li et al., 2015). Ammonia-oxidizing Thaumarchaeota (Brochier-Armanet et al., 2008) are the most abundant archaeal phylum in the oceans and have a key role in the marine nitrogen cycle (Konneke et al., 2005; Qin et al., 2014). Studies have also identified three major groups of marine Euryarchaeota: (i) group II (MG-II) (DeLong, 1992; Fuhrman et al., 1992; Fuhrman and Davis, 1997; Massana et al., 2000), (ii) group III (MG-III) (Fuhrman and Davis, 1997; Lopez-Garcia et al., 2001a), and (iii) group IV (MG-IV) (Lopez-Garcia et al., 2001b). So far, there are no cultured representatives of marine Euryarchaeota and little is known about their physiology and ecological role in the oceans. MG-II are widely distributed within the euphotic zone of temperate waters. MG-II are the dominant archaeal community not only in the surface and in the deep chlorophyll maximum (DCM) (Massana et al., 2000; Karner et al., 2001; Herndl et al., 2005; DeLong et al., 2006; Galand et al., 2010; Belmar et al., 2011; Martin-Cuadrado et al., 2015) but have also been found in deep-sea waters (Lopez-Garcia et al., 2001a, Martin-Cuadrado et al., 2008; Li et al., 2015). The other two marine Euryarchaeota groups, MG-III and MG-IV, are considered to be rare components of deep-sea communities (Lopez-Garcia et al., 2001a,b; Galand et al., 2009).
MG-III were first described by Fuhrman and Davis, 1997 from deep marine plankton samples and have subsequently been found in 16S-rRNA gene surveys from most deep oceanic regions, albeit at very low abundance (Massana et al., 2000; Lopez-Garcia et al., 2001a,b), and by metagenomics throughout the water column in the central Pacific gyre (DeLong et al., 2006). However, occasionally, they have been identified at much higher proportions. For instance, 16S-rRNA sequences from MG-III represented one of the largest archaeal groups in the deep Arctic Ocean (>40% of tag sequences) (Galand et al., 2009) and between 30% and 50% of the archaeal sequences from a deep (500–1250 m) Marmara Sea metagenome (Quaiser et al., 2011). They were also relatively abundant (ca.18% of the total archaeal population) in the oxygen minimum zone (50–400 m) in the Eastern tropical South Pacific (Belmar et al., 2011). Only a few studies report the presence of MG-III in the photic zone. They represented 0.4% of all the archaeal sequences obtained in surface Arctic waters (Galand et al., 2009) and up to 10% in samples recovered along 4.5 years in the Mediterranean DCM (Galand et al., 2010).
The initial analysis of three MG-III fosmids from deep-sea metagenomic libraries allowed a first glance at their metabolic potential (Martin-Cuadrado et al., 2008). The presence of some fermentation-related genes led to the hypothesis that they could be facultative anaerobes. In a recent study, up to 3% of the single amplified genomes of archaea recovered from mesopelagic waters from South-Atlantic and North-Pacific gyres belonged to MG-III (Swan et al., 2014). However, only two single amplified genomes classified as MG-III, SCGC-AAA-007-O11 (isolated at 800 m in the South-Atlantic sub-tropical gyre) and SCGC-AAA-288-E19 (from 770 m in the North-Pacific sub-tropical gyre), have been deposited in GenBank. Only the SCGC-AAA-288-E19 partial genome had ribosomal RNA genes that corresponded to MG-III, but contig annotation showed contamination with Chloroflexi (32 genome fragments out of the 102). Complete archaeal fosmids (452 adding up to 16 Mb of sequence) from deep Mediterranean samples belonging to MG-II/III have been published (Deschamps et al., 2014) and five MG-III partial genomes (31–65% completeness) were assembled from metagenomes from the Guaymas basin (1993 m, Gulf of California) and the Mid-Cayman Rise (2040–2238 m and 4869–4946 m, Caribbean Sea) (Li et al., 2015). Based on the genes present in these genomes, it was proposed that the microbes they represented are motile heterotrophs with different mechanisms for scavenging organic matter.
Binning the assembled fragments by oligonucleotide frequencies, GC content and differential recruitment in metagenomes is a successful strategy for the discovery of novel microbial lineages (Tyson et al., 2004; Ghai et al., 2012; Iverson et al., 2012; Narasingarao et al., 2012; Martin-Cuadrado et al., 2015; Li et al., 2015; Vavourakis et al., 2016). We applied this approach to recover MG-III sequences using several metagenomic fosmid libraries from the Mediterranean Sea (collections KM3, AD1000 (Martin-Cuadrado et al., 2008) and MedDCM-OCT2007 (Ghai et al., 2010)) and from the assemblies of 16 metagenomes (four collections from the Mediterranean: MedDCM-JUL2012 (Martin-Cuadrado et al., 2015), MedDCM-SEP2014 (this work), Med-Io7–77mDCM and Med-Ae2–600mDeep (Mizuno et al., 2016) and 12 from TARA microbiomes (Sunagawa et al., 2015)). We obtained a total of eight different MG-III genome bins. Six of them belong to novel surface MG-III lineages distantly related to the previously described deep MG-III sequence bins (Li et al., 2015). They are the first near-complete genomes of MG-III living in the photic zone. Some of them appear to be widespread in the ocean; their distribution in different water masses has been analyzed.
Materials and methods
Sampling and sequencing
A fosmid metagenomic library of ca. 13 000 clones was constructed with biomass recovered in October 2007 (50 m deep) at the Mediterranean DCM (38°4′6.64″N 0°13′55.18″W). Partial results of almost 7000 fosmid sequences have been described previously in Ghai et al. (2010) and Martin-Cuadrado et al. (2015). Metagenomes were also sequenced from samples recovered at the same location and at a similar depth the following years (MedDCM-JUL2012 (Martin-Cuadrado et al., 2015) and MedDCM-SEP2014) from one sample recovered at the DCM from the Ionian Sea (Med-Io7–77mDCM) and from a sample collected from the deep Aegean Sea (Med-Ae2–600mDeep) (Mizuno et al., 2016). For these metagenomes, sea water was collected and sequentially filtered on-board using a positive pressure system through a 20 μm pore filter followed by a 5 μm pore size polycarbonate filter and, finally, 0.22 μm pore size Sterivex filters (Durapore; Millipore, Billerica, MA, USA). Filters were frozen on dry ice and kept at −80 °C until processed in the laboratory. Filters were thawed on ice and then treated with 1 mg ml−1 lysozyme and 0.2 mg ml−1 proteinase K (final concentrations). Nucleic acids were extracted with phenol–chloroform–isoamyl alcohol and chloroform–isoamyl alcohol. Sequencing was carried out using Illumina HiSeq2000 (PE, 100 bp) (Macrogen, Seoul, Korea and BGI, Hong Kong).
‘De novo’ assembly, gene annotation and binning of the MG-III sequences
A schematic of the assembly pipeline is shown in Supplementary Figure S1. The assembly of the fosmids from the MedDCM-OCT2007, KM3 and AD1000 metagenomic fosmid libraries has been previously described (Ghai et al., 2010; Deschamps et al., 2014; Martin-Cuadrado et al., 2015). Sequences from metagenomes MedDCM-JUL2012, MedDCM-SEP2014, Med-Io7–77mDCM and Med-Ae2–600mDeep were quality trimmed and assembled independently using IBDA-UD (Peng et al., 2012) with the following parameters: –mink 70, –maxk 100, –step 10, –pre_correction. Gene predictions on the assembled sequences were carried out using Prodigal (Hyatt et al., 2010). Ribosomal genes were identified using ssu-align (Nawrocki, 2009) and meta_rna (Huang et al., 2009). Functional annotation was performed by comparing predicted protein sequences against the NCBI-nr database, Pfam (Bateman et al., 2004), arCOGS (Makarova et al., 2015) and TIGRfams (Haft et al., 2001) (cutoff E-value 10–5). Based on sequence similarity against the non-redundant NCBI database, the best hit for each gene was determined and used to bin to top-level taxa. Bona fide Euryarchaeota genome fragments were defined as having >50% of the predicted open reading frames with best hits to other Euryarchaeota genes. The resulting sequences were used to screen for their presence in several metagenomes (in subsets of 20 million reads, where applicable): the TARA data sets (Sunagawa et al., 2015), the GOS collection (Rusch et al., 2007), the depth profiles collections from the subtropical gyres of North Atlantic (Bermuda Atlantic Time Series, BATS) and North Pacific (Hawaii Ocean Time-Series, HOT) (DeLong, 2006; Coleman and Chisholm, 2010), several Mediterranean Sea metagenomes at different depths (Ghai et al., 2010; Quaiser et al., 2011; Smedile et al., 2012; Martin-Cuadrado et al., 2015), and a number of deep ocean and cold waters metagenomes (Alonso-Saez et al., 2012; Larsson et al., 2014). The collections coming from the surroundings of hydrothermal vents published in Li et al. (2015) were also included. The screening was performed using Usearch6 (Edgar, 2010), with a cutoff of 95% identity over an alignment length of at least 50 bp (approximately species-level divergence, Konstantinidis and Tiedje, 2005). To compare the results among different data sets, the number of reads was normalized to the metagenome size and the sequence length. The final coverage results were expressed as the number of reads per kilobase of the fragment per gigabase of metagenome collection (rpkg). Only metagenomes in which any of the MG-III sequences recruited reads at over 3 rpkg, a total of 33 metagenomes, were used for genome assembly (Supplementary Table S1).
All the sequences obtained from these assemblies were binned together in order to cluster them by their tetranucleotide frequencies, GC content and coverage values (Supplementary Figure S2 and Supplementary Table S1). Tetranucleotide frequencies were computed using the ‘wordfreq’ program from the EMBOSS package (Rice et al., 2000) and the coverage values were calculated as rpkg as described before. Only those clusters with >10 sequences and containing at least one gene marker with a clear affiliation to MG-III were retained. The phylogenetic assignment to MG-III was determined by the presence of at least one housekeeping gene in the same bin (see below). Following this method, a total of 375 genomic fragments >10 Kb could be classified into 10 different MG-III bins of sequences, Epi1, Epi2A, Epi2B, Epi2C, Epi3, Epi4, Epi5, Epi6, Bathy1 and Bathy2. We also considered 16 MG-III sequences that contained a ribosomal or a housekeeping gene but that could not be included in any of the bins by the criteria used (Supplementary Table S2).
In order to improve the completeness and remove the redundancy present in the initial MG-III bins, a second assembly was performed combining the sequences >10 Kb with the short paired-end Illumina reads of the metagenomes from where they were assembled (Tables 1 and 2 and Supplementary Figure S3). For each of the MG-III sequence bins, we used the BWA aligner (Li and Durbin, 2009; default parameters) to recover the short pair-reads that mapped onto the >10 Kb contigs. For each bin, these reads were then pooled and assembled together with the large DNA contigs previously assembled using SPAdes (Bankevich et al., 2012). The final assemblies were termed ‘composite genomes’ (CGs), as they belong to similar MG-III cellular lineages (defined by the MG-III bins) but from different samples (Supplementary Table S3). The completeness of the reconstructed archaeal genomes was estimated by three different criteria and based on the presence of essential/core genes using HMMER (35, 112 and 53 genes (Raes et al., 2007; Narasingarao et al., 2012; Albertsen et al., 2013)). An E-value <10–5 and an alignment coverage >65% were used as cutoffs to define homologs of the essential/core genes. Analysis of the contamination within the CGs was performed using CheckM (Parks et al., 2014) (Table 1). Average nucleotide identity (ANI) and conserved DNA fraction between reconstructed and/or reference genomes were calculated based on the whole-genome sequence as in Goris et al. (2007) (Supplementary Figure S4). GC content was calculated using the ‘geecee’ tool from the emboss package (Rice et al., 2000).
Table 1. General features of the MG-III bins and the composite genomes CG-MGIII.
| MG-III bin | No. of contigs | %GC±s.d. | No. of Mb | % Genome (1) (2) (3)a | No. of genomes | Largest contig (Kb) | No. of CDS/nr CDSb | Intergenic region (bp) | Median gene size (bp) | CheckMc % contamination |
|---|---|---|---|---|---|---|---|---|---|---|
| Epi1 | 136 | 36.6±0.9 | 2.95 | 80.0/35.1/83.0 | 2 | 120.8 | 3058/1307 | 42 | 752 | — |
| CG-Epi1 | 25 | 36.6±0.8 | 1.18 | 85.7/34.2/84.9 | 1 | 135.9 | —/1106 | 40 | 792 | 2 |
| Epi2A | 34 | 36.0±0.9 | 0.54 | 54.3/16.2/45.3 | 1 | 27.4 | 519/— | 41 | 765 | — |
| Epi2B | 26 | 36.2±0.9 | 0.56 | 57.1/18.0/54.7 | 1 | 79.1 | 525/— | 40 | 768 | — |
| Epi2C | 10 | 36.1±0.9 | 0.31 | 5.7/4.5/5.7 | 1 | 92.8 | 282/— | 40 | 807 | — |
| CG-Epi2 | 43 | 36.1±1.1 | 1.22 | 80.0/30.6/75.5 | 1 | 101.9 | —/1182 | 42 | 777 | 25 |
| Epi3 | 35 | 35.9±1.2 | 0.71 | 48.6/19.8/45.3 | 1 | 55.8 | 675/673 | 37 | 798 | — |
| CG-Epi3 | 30 | 35.9±1.2 | 0.79 | 62.9/24.3/62.3 | 1 | 71.7 | —/741 | 40 | 786 | 0.8 |
| Epi4 | 31 | 36.5±0.8 | 0.66 | 34.3/15.3/39.6 | 1 | 47.2 | 612/610 | 37 | 794 | — |
| CG-Epi4 | 24 | 36.4±0.7 | 0.71 | 34.3/16.2/43.4 | 1 | 80 | —/666 | 37 | 786 | 0 |
| Epi5 | 18 | 36.1±0.8 | 0.26 | 2.9/3.6/7.6 | 1 | 23.4 | 242/221 | 36 | 767 | — |
| CG-Epi5 | 22 | 36.3±0.8 | 0.39 | 2.9/4.5/9.4 | 1 | 47.6 | —/370 | 38 | 804 | 0 |
| Epi6 | 26 | 36.6±0.9 | 0.88 | 51.4/15.3/54.7 | 1 | 111.1 | 854/594 | 47 | 705 | — |
| CG-Epi6 | 26 | 36.3±1.1 | 0.54 | 57.1/21.6/60.4 | 1 | 50.3 | —/542 | 47 | 714 | 1.6 |
| Bathy1 | 39 | 36.9±0.9 | 1.2 | 54.3/23.4/60.4 | 1 | 94.7 | 1143/1007 | 46 | 807 | — |
| CG-Bathy1 | 22 | 37.5±1.8 | 1.04 | 60.0/26.1/64.2 | 1 | 211.7 | —/988 | 46 | 792 | 4.8 |
| Bathy2 | 37 | 64.6±1.8 | 1.06 | 68.6/18.9/58.5 | 1 | 41.7 | 1023/654 | 40 | 771 | — |
| CG-Bathy2 | 18 | 64.4±1.5 | 0.77 | 68.6/21.6/58.5 | 1 | 130.1 | —/704 | 41 | 816 | 2.4 |
Abbreviations: CDS, Coding DNA sequence; nr-CDS, non-redundant CDS.
(1) Raes et al. (2007); (2) Albertsen et al. (2013); (3) Narasingarao et al. (2012).
nr-CDS: non-redundant CDS clustered at 80% similarity and 70% coverage.
Table 2. Environmental collections from where MG-III sequences were assembled.
| Depth (m) | Fraction size (μm) | Epi1 | Epi3 | Epi4 | Epi5 | Epi6 | Epi2A | Epi2B | Epi2C | Bathy1 | Bathy2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total, Kb | 2950.4 | 707.0 | 631.3 | 259.7 | 848.7 | 542.7 | 564.7 | 305.0 | 1196.5 | 1061.4 | ||
| ERR598993 (TARA_18)a | 5 | 0.22–1.6 | 658.3 | |||||||||
| ERR599073 (TARA_18)a | 60 | 0.22–1.6 | 54.6 | |||||||||
| ERR315859 (TARA_023)a | 55 | 0.22–01.6 | 11.7 | |||||||||
| ERR594297 (TARA_068)a | 5 | 0.45–0.8 | 25.3 | |||||||||
| ERR594294 (TARA_068)a | 50 | 0.22–0.45 | 367.2 | 47.4 | ||||||||
| ERR594348 (TARA_068)a | 50 | 0.45–0.8 | 159.3 | |||||||||
| ERR594335 (TARA_070)a | 5 | 0.45–0.8 | 41.9 | |||||||||
| ERR598942 (TARA_133)a | 45 | 0.22–3 | 707.0 | 60.9 | ||||||||
| ERR598983 (TARA_145)a | 5 | 0.22–3 | 198.8 | 422.4 | 305.0 | |||||||
| ERR598996 (TARA_150)a | 40 | 0.22–3 | 128.0 | |||||||||
| ERR598976 (TARA_151)a | 5 | 0.22–3 | 264.7 | |||||||||
| ERR598986 (TARA_151)a | 80 | 0.22–3 | 216.5 | |||||||||
| MedDCM-OCT2007b | 60 | 0.22–5 | 1034.7 | 34.5 | 733.8 | |||||||
| MedDCM-JUL2012c | 75 | 0.22–5 | 542.7 | 142.3 | ||||||||
| MedDCM-SEP2014d | 60 | 0.22–5 | 596.7 | |||||||||
| AD1000e | 1000 | 0.22–5 | 38.7 | |||||||||
| Med-Ae2–600mDeepf | 600 | 0.22–5 | 1017.6 | |||||||||
| Med-Io7–77mDCMf | 77 | 0.22–5 | 55.8 | |||||||||
| KM3e | 3000 | 0.22–5 | 140.1 | 1059.6 |
Phylogenetic analysis
16S-rRNA and 23S-rRNA gene sequences detected in the MG-III genomic fragments were used to retrieve rRNA gene sequences from the most closely related euryarchaeal genomes and selected genome fragments in GenBank using BLAST (Altschul et al., 1990). 16S-rRNA sequences from metagenome collections were screened and trimmed using ssu-align (Nawrocki, 2009). Archaeal 16S-rRNA and 23S-rRNA gene sequences were then aligned using MUSCLE (Edgar, 2004). Phylogenetic reconstructions were conducted by maximum likelihood using MEGA6-v.0.6 (Tamura-Nei model, 100 bootstraps, gamma distribution with (five discrete categories), all positions with <80% site coverage were eliminated) (Tamura et al., 2013) (Supplementary Figure S5). For the protein trees of RecA, RpoB, SecY, geranylgeranylglyceryl phosphate synthase, DnaK, GyrA, GyrB, photolyase and rhodopsin (Supplementary Figures S6–S14), sequences were selected based on existing literature. Sequences were aligned using MUSCLE (Edgar, 2004) and a maximum likelihood tree was constructed using MEGA6-v.0.6 (Jones-Taylor-Thornton model, 100 bootstraps, gamma distribution with five discrete categories, positions with <80% site coverage were eliminated). Taxonomic affiliation of the selected bins was also determined by a phylogenomic tree based on concatenates of several ribosomal proteins (L13, S9, L5, S8, L6, S5, S12, S7, L11, L3, L4, L2, L22, S3, L14, S17, L15 and L18). A balanced taxonomic representation of other archaeal genomes was included as reference. Shared proteins were concatenated and aligned using Kalign (Lassmann and Sonnhammer, 2005) and a maximum likelihood tree was made using MEGA6-v.0.6.
Genome comparisons
Synteny among the CG-MGIII was examined with CIRCOS (Krzywinski et al., 2009) and defined as arrays of contiguous genes in tracts of DNA >5 Kb and having >70% of identity. For each of the MG-III bins, non-redundant protein databases were constructed clustering the coding DNA sequences with UCLUST (Edgar, 2010) (cutoff: 80% similarity in 70% of their length). These subsets of proteins were compared among themselves using a reciprocal best-hit analysis of putative homologs by BLASTP. Reciprocal relations were plotted using CYTOSCAPE (Shannon et al., 2003). In order to identify the unique proteins of each of the bins, UCLUST was used with a cutoff of 30% similarity along 70% of their length.
Accession numbers
Mediterranean metagenomes used for recruitment are available at NCBI-BioProjects: PRJNA257723 (MedDCM-SEP2014, MedDCM-JUL2012 and MedDCM-OCT2007), PRJNA305355 (Med-Io7–77mDCM, Med-Io16–70mDCM, Med-Io17–3500mDeep, Med-Ae1–75mDCM and Med-Ae2–600mDeep). Sequences >10 Kb and the reconstructed CGs genomes have been deposited in Bio-Project number: PRJNA335308. TARA metagenomes were downloaded from the European-Bioinformatics-Institute (http://www.ebi.ac.uk/services/tara-oceans-data).
Results and Discussion
General features of MG-III archaeal genomes
Following assembly and binning, we obtained 375 genomic fragments that clustered into 8 MG-III bins (Supplementary Figure S1). Six bins, Epi1–Epi6, were from epipelagic origin (photic zone) and contained a total of 386 genomic fragments with a total of 8.3 Mb. Two bins, Bathy1 and Bathy2, were from deep marine samples (aphotic zone) and contained 76 fragments for a total of 2.3 Mb. Manual inspection of the differential coverage of the sequences in each bin identified three subsets of Epi2, referred to as Epi2A, Epi2B and Epi2C. Further genomic comparisons indicated that these bins were very similar to each other (93–96% ANI, Supplementary Figure S4) and represent genomes from related species, likely within the same genus.
Remarkably, seven genome bins were formed by sequences primarily from a single sampling site (Table 2). The exception was Epi1, which includes sequences retrieved from nine different sites in the Mediterranean Sea, Atlantic and North-Pacific oceans. These findings suggest that the organisms represented by Epi1 are cosmopolitan in temperate epipelagic waters, whereas the other groups are only abundant enough to assemble from metagenomes at specific sites (endemic) or under transient environmental conditions causing significant growth (for example, blooms; see below).
To improve the analysis of each genome bin, a second assembly was performed and CGs were reconstructed using sequences from different samples and geographic origins (Supplementary Figure S1). These CGs are non-redundant and consist of genomic fragments from similar lineages of MG-III cells but not necessarily from the same sample. In this further assembly, subsets Epi2A, 2B and 2C were condensed into a single bin, CG-Epi2. Genomic features of the genome bins can be found in Tables 1 and 2 and the complete list of the MG-III contigs and the CGs are given in Supplementary Tables S2 and S3. Using the criteria of Narasingarao et al. (2012), the genome bins with highest degree of completeness were CG-Epi1 (85%), followed by CG-Epi2 (75%) and the mesopelagic CG-Bathy1 (64%). Based on the number of different variants of single copy genes in each bin, all our CGs contained a single microbial species each (Supplementary Table S4).
All MG-III bins had low GC content (36–36.8%) with the exception of Bathy2 (64.2%). Previously described MG-III sequences from different bathypelagic samples were all high GC (62.8%–65.4%) except for Guaymas32 (36.8%) (Li et al., 2015). It has been noted that GC content tends to increase with depth (Romero et al., 2009; Mizuno et al., 2016). Selection for less nitrogen demand has been proposed as the main drive toward low genomic GC content in free-living marine bacterioplankton. In epipelagic waters, nitrogen is more likely to be the limiting nutrient, in contrast to the dark, energy-limited but relatively nitrogen-rich, deep ocean (Dufresne et al., 2005; Swan et al., 2013; Batut et al., 2014; Giovannoni and Nemergut, 2014). Nevertheless, Bathy1 and Guaymas32 have similar low GC content to surface MG-III bins, suggesting that other factors might be also important.
In general, epipelagic MG-III bins were more genetically heterogeneous. Among the low GC-MGIII bins, the ANI varied from 68% to 85.4%, whereas the high GC-MGIII bins (Bathy2 is 90.8% similar to Cayman92) showed higher degrees of conservation, with ANIs ranging 89.5% to 96.2% (Supplementary Figure S4). This apparently higher diversity of the epipelagic groups may reflect the chemical and physical heterogeneity of surface water layers, which are submitted to stronger hydrodynamic, seasonal and geographical variations (Bryant et al., 2015). In contrast, MG-III representatives from the deep ocean inhabit a more stable environment and might consequently be less diverse, with more homogenous genomes.
Phylogenetic affiliation of the genomic bins
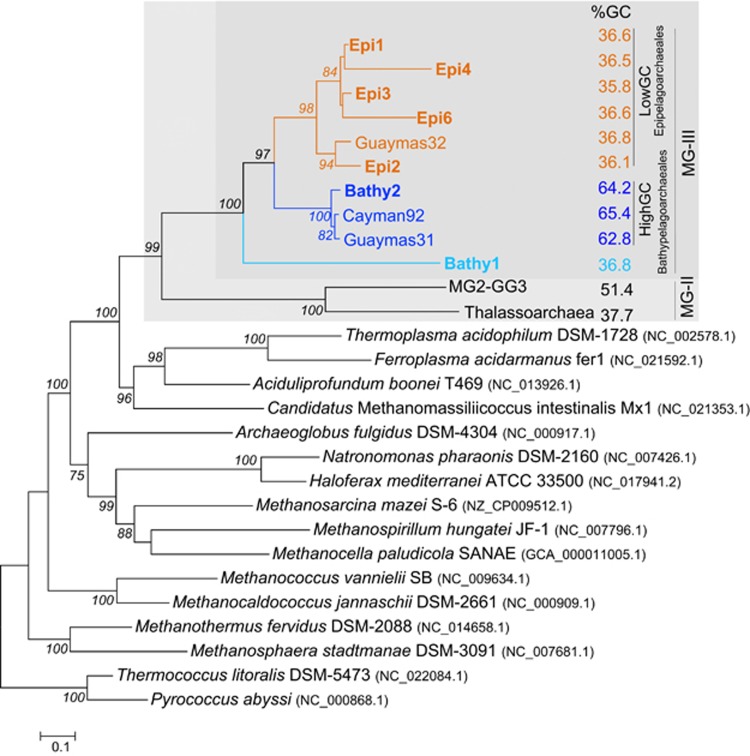
Genes coding for rRNA are difficult to bin because (i) rRNA genes assemble poorly due to their conservation and duplication in genomes and (ii) they recruit metagenomic reads at much higher levels making coverage-based approaches impractical. Most of the rRNA sequences came from fosmid-libraries (Km3 and AD1000) and did not cluster within any of the bins described here. The only assigned 16S-rRNA sequence (372 bp) belonged to Bathy1 and it appears distantly related to the previously described OTU-D (Galand et al., 2009) and DH148-W24 clusters (Lopez-Garcia et al., 2001a,b) (Supplementary Figure S5a). A similar result was obtained with the 23S-rRNA gene identified in Bathy1 (Supplementary Figure S5b). Therefore, we looked for other housekeeping genes that might be helpful to define the phylogenetic relationships of the novel MG-III with other archaea. We identified and constructed phylogenetic trees for RecA, RpoB, SecY, the geranylgeranylglyceryl phosphate synthase, DnaK and the two gyrase subunits, GyrA and GyrB (Supplementary Figures S6–S12). Although DnaK, GyrA and GyrB have a complex history of horizontal gene transfer (HGT) (Gribaldo et al., 1999; Petitjean et al., 2012; Raymann et al., 2014), their phylogenetic analysis clearly showed the split between MG-II and MG-III sequences. The MG-III housekeeping genes retrieved from epipelagic waters clustered into two groups, one represented only by Epi2 and the other including Epi1, 3, 4, 5 and 6. Bathy2 appeared as a separate cluster from the epipelagic MG-III, and Bathy1 sequences appeared as the most divergent and basal branch. The phylogenomic analysis of the concatenated ribosomal proteins revealed a similar topology (Figure 1). The two epipelagic clusters shared similar GC content. Accordingly, they were named LowGC-MGIII (comprising two subclades: LowGC1-MGIII (Epi1, 3, 4 and 6) and LowGC2-MGIII (Epi2 and Guaymas32)), and a separate clade, containing bins exclusively of bathypelagic origin (Bathy2, Cayman92 and Guaymas31), was named HighGC-MGIII. Bin Epi5 lacks the ribosomal operon, but it was included into the LowGC1-MGIII based on the phylogenetic analysis of the other housekeeping genes (Supplementary Figures S11 and S12). Bathy1 consistently appeared as a separate basal branch, which might reflect the intermediate depth (600 m), location (Aegean Sea) and physicochemical conditions (highly saline, relatively warm and extremely oligotrophic) of the samples contributing sequences to this genomic bin. The position of Guaymas32 (retrieved from 1993 m), which clusters with Epi2 (5–75 m), might be explained by the presence of two different microbial species in the Guaymas32 bin (Li et al., 2015). One appears to be most similar to the surface Epi2 sequences (80.8% ANI), while the other is closer to the deeper Bathy1 sequences (72.9% ANI) (also observed in the synteny plot of Figure 2a) (see below). Another plausible explanation is that Guaymas32 might be a surface organism dragged to the bottom by the continuous flux of surface microbes and particles into the deep. Indeed, Guaymas sediments are surprisingly enriched in surface planktonic microbes (Edgcomb et al., 2002) when compared with other deep-sea sediments (Lopez-Garcia et al., 2003). However, the lack of rhodopsins and photolyases (discussed below), together with higher recruitments from deep data sets, would suggest that Guaymas32 is a bona fide deep inhabitant.
Figure 1.
Maximum likelihood tree based on 18 ribosomal proteins concatenated present in draft MG-III archaeal genomes reconstructed from epipelagic and deep-sea metagenomes. Archaeal genomes from major orders of Euryarchaeota were included as references (accession number in brackets). Novel sequences from this work are shown in bold. Average GC content is shown on the right and colored depending on whether it is high or low GC. Only bootstrap values over >50% are shown.
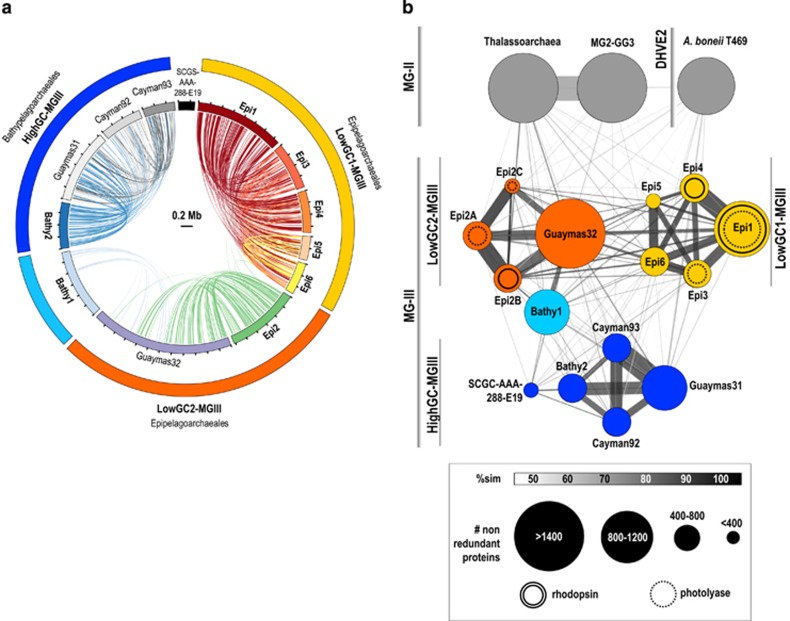
Figure 2.
(a) Overview of genomic conserved synteny among the CG-MGIII genomes. Alignments >5 Kb over 70% identity are shown. A color code is used for each MG-III bin. (b) Amino-acid comparison among the MG-III bins. Sets of non-redundant proteins (cutoff of 80% similarity over 70% of their length) were compared through reciprocal BLASTP and the average amino-acid similarity was plotted. Each circle represents a genome bin. Circles are interconnected as a function of the percentage of shared proteins and colored in accordance with their similarity. Size of the bins and width of the lines are explained in the legend. Proteins of the MG-II MG2-GG3 (Iverson et al., 2012), Thalassoarchaea (Martin-Cuadrado et al., 2015) and the deep-sea hydrothermal vent Euryarchaeota (DHVE2) Aciduliprofundum boneii T469 were included in the analysis.
Synteny and gene content
To examine the conservation of synteny across the different genome bins, we performed an all-versus-all genome comparisons with the available sequences of MG-III (Figure 2a). Within the two groups of LowGC-MGIII bins, large fragments have the same genomic context while synteny blocks are not conserved between LowGC-MGIII and HigGC-MGIII. In the case of LowGC-MGIII, the highest synteny was found between Epi1 and Epi4 (54 block alignments, 62% of Epi4 genome size). For LowGC-MGIII, only Epi2 and Guaymas32 showed a significant synteny (56 block alignments, 38% of CG-Epi2). The low level of synteny between Bathy1 and other bins confirms that the microbes represented by this bin are very distant to the other LowGC-MGIII. Among the HighGC-MGIII bins, the highest synteny was found between Bathy2 and Guaymas31 (40 block alignments, 42% of CG-Bathy2) followed closely by Cayman92 and Guaymas31 (42 block alignments, 40.8% of the Cayman92 genome).
Non-redundant sets of proteins were obtained for each of the bins, including MG-II relatives, and compared between bins, retaining only the best hit for each protein and using a threshhold of 80% similarity. The relationships between bins were then plotted in the similarity network showed in Figure 2b. This protein content analysis supported the clustering observed in the phylogenomic tree (Figure 1). Bathy1 and SCGC-AAA-288-E19 appeared distantly associated with Guaymas32 and Guaymas31, respectively. MG-III bins Epi1 with Epi4 had the largest percentage of shared proteins (34.8%), followed by Epi2B and Guaymas32 (24%) and then Bathy2 and Guaymas31 (25%). Only 8% of Epi1 proteins were conserved in Epi2 and 0.5% in Bathy2. Although these numbers may be biased owing to the incomplete nature of the bins, they suggest that marine Euryarchaeota are very diverse and contain very different gene pools. Similar results were obtained by Deschamps et al. (2014) who found that the core genome of the MGII/III Euryarchaeota was only 15.6% of their pangenome, while their flexible genome was almost triple that of the Thaumarchaeota.
Metabolic functional inference
Several studies have suggested that marine Euryarchaeota have a significant role in the degradation of dissolved organic matter in marine waters, for example, dissolved amino acids (Ouverney and Fuhrman, 2000) or carbohydrates (Boutrif et al., 2011). The presence of large peptidases related to protein degradation, together with enzymes for the use of fatty acids in the MG2-GG3 genome suggested that particles might be a habitat for MG-II Euryarchaeota (Iverson et al., 2012; Orsi et al., 2015). MG-II shared various features with the deep MG-III described by Li et al. (2015), suggesting that they might be aerobic heterotrophs that use proteins and polysaccharides as major energy source. In order to infer different lifestyles, the predicted open reading frames were functionally classified according the arCOG categories and their frequencies in the different genomes compared (Supplementary Tables S5 and S6 and Supplementary Figure S15).
Central carbon metabolism
MG-III genomes harbored enzymes for glycolysis, the tricarboxylic acid cycle and oxidative phosphorylation, indicating aerobic respiration (Supplementary Table S7). However, owing to the incomplete nature of these genomes, not all genes could be found, and some predictions need to be taken cautiously, especially for Bathy2. We found genes for the complete tricarboxylic acid cycle in LowGC-MGIII but three genes were absent in Bathy1. Remarkably, only the aconitase and the fumarase were found in Bathy2. As was observed in some MG-II (Martin-Cuadrado et al., 2015), MG-III appears to possess most of the enzymes of the Embden–Meyerhof–Parnas (EMP) pathway for metabolism of hexose sugars, with the exception of the first and the last enzymes of the pathway. We were unable to find any other enzyme that could serve as an alternative for the missing glucokinase. For the final step of the EMP, we propose that phosoenolpyruvate synthase, found in all of our MG-III bins, might be able to function bi-directionally and substitute for the missing pyruvate kinase, allowing the EMP to function in both directions, gluconeogenic and glycolytic. Likewise, we found typical gluconeogenesis enzymes such as phosphoenolpyruvate carboxykinases in the LowGC-MGIII and Bathy1 bins, as well as subunits of the pyruvate/oxaloacetate carboxyltransferase in all the MG-III bins. We were unable to find glucose 1-dehydrogenase, gluconolactonase and 2-keto-3-deoxy gluconate aldolase homologs, suggesting that the Entner–Duodoroff hexose catabolic pathway is not present in the MG-III, unlike findings in other Euryarchaea (Makarova et al., 1999; Makarova and Koonin, 2003; Hallam et al., 2006).
Only a small number of amino-acid synthases were found in MG-III: cysteine in Bathy1 and Bathy2, glutamine in LowGC-MGIII, and for glutamate in all MG-III bins. Remarkably, many enzymes for de novo biosynthesis were missing, including those for synthesizing methionine, arginine, threonine, histidine, aromatic amino acids and branched amino acids (Supplementary Table S7). However, we observed multiple genes related with the uptake and transformation of peptides or amino acids in our MG-III bins, indicating that these organisms are capable of taking up amino acids from the environment and incorporating them into their proteins. For example, we found genes for permeases for lysine/arginine (all bins), histidine (Bathy2), glutamine (LowGC-MGIII and Bathy1), proline (LowGC-MGIII and Bathy1) and polar amino acids (Bathy2). Also, several ABC-transporter-systems were found for peptides and oligopeptides; for example, Dpp-ABC-type dipeptide/oligopeptide transporters (in all) and Liv-ABC-type branch amino-acid transporters (LowGC-MGIII and Bathy1). Several enzymes involved in the degradation of amino acids were also found, including dehydrogenases for alanine (all bins), glutamate (all bins), threonine (LowGC-MGIII and Bathy2) and proline (LowGC-MGIII), as well as several aminotransferases for branched-chain amino acids (LowGC-MGIII and Bathy1) and aspartate/tyrosine/aromatic aminotransferases (LowGC-MGIII and Bathy1). These findings suggest that there may be differences in the substrates used by the different MG-III groups. Indeed, although several subtilase-family proteases (arCOG00702 and arCOG02553) were present in all bins, some peptidases had limited distributions: dipeptidyl-aminopeptidases (LowGC-MGIII and Bathy1), C1A-peptidases (LowGC-MGIII), C25-peptidases (Bathy1), Xaa-Pro aminopeptidases (Bathy2), and several AprE-like subtilisins (arCOG06823, present in LowGC-MGIII and arCOG03610 present in Bathy1) (Supplementary Table S6).
Carbohydrates can be important carbon sources and, with the exception of Bathy1, several proteins with sugar-binding domains were found in all the bins (lectin and laminin-like). In the Epi6 bin, a cutin-like hydrolase was found (37% similar to a hydrolase from the Bacteriodetes Rufibacter sp. DG15C). Cutin is a polyester composed of hydroxyl/hydroxyepoxy fatty acids present in plants, and cutinases are produced by pathogenic fungi as extracellular degradative enzymes (Chen et al., 1997). Lipo-oligosaccharide transport systems (nodI/J-like genes) and phosphonate transporters were found exclusively in the LowGC-MGIII. As observed in MG-II Thalassoarchaea (Martin-Cuadrado et al., 2015), multidrug and antimicrobial peptide transporters (ABC-type) together with several permeases for drug/metabolites (RhaT-like family) were also abundant in all MG-III bins. Although the nature of the substrates is difficult to ascertain, these transporters may be involved in coping with high environmental concentrations of toxins such as those produced by cyanobacterial and algal blooms.
Oxygen
The presence of superoxide dismutase in all MG-III bins, together with several genes for alkyl-hydroperoxide reductases in LowGC1-MGIII and Bathy1, suggests that these microbes must cope with oxygen radicals. Complete cytochrome-C and B-B6 oxidase subunits operons were also found in LowGC1-MGIII and Bathy1 and Bathy2 bins. Copper-binding proteins and haloarchaeal-like halocyanins were found in proximity of these operons, an arrangement similar to that described for MG-II Thalassoarchaea (Martin-Cuadrado et al., 2015). It has been suggested that MG-II could be facultative anaerobes (Martin-Cuadrado et al., 2008; Belmar et al., 2011) and that sulfate could be used as terminal electron acceptor. Although no sulfate reductase-like proteins could be identified in our MG-III bins, several phosphate/sulfate permeases could be identified in Epi6 and Bathy2 and were also present in Guaymas31/32 and Cayman92. Pterin-based molybdenum enzymes (for example, sulfite oxidase, xanthine oxidase and dimethyl sulfoxide reductase) function under anaerobic conditions whereby their respective cofactors serve as terminal electron acceptors in respiratory metabolism (Schwarz et al., 2009). For Bathy2 (fosmid Km3–43-F08), a novel operon for the molybdopterin biosynthesis, was found (catalytic domains, MOCS1/S2/S3, have <55% similarities in the nr-database). However, we could not find any of the pterin-based enzymes.
Light-related genes
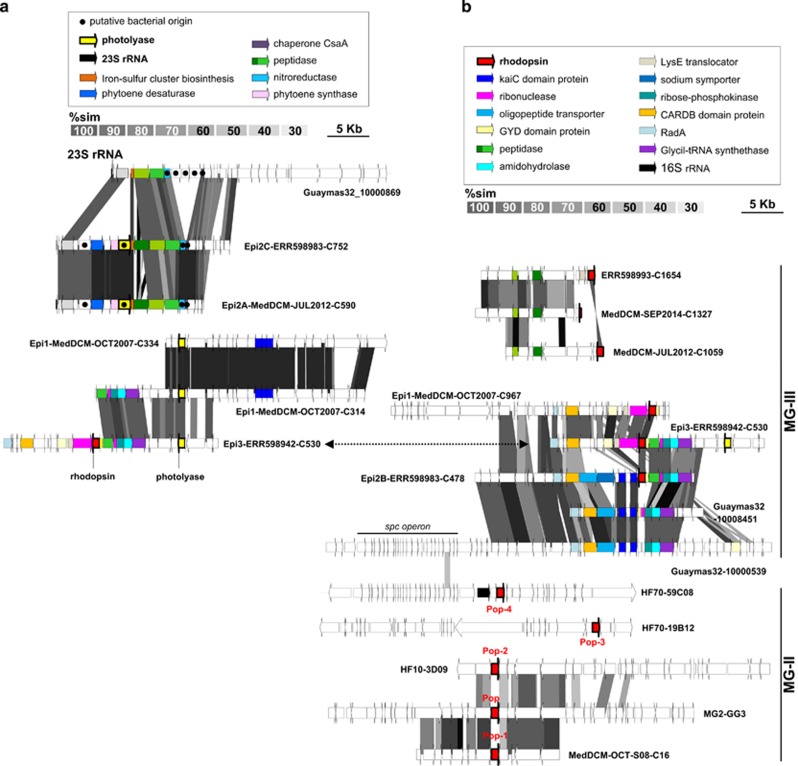
The presence of photolyases/cryptochromes among the LowGC-MGIII bins supports our hypothesis that they are bona fide epipelagic microbes (Figure 3a). Photolyases are proteins capable of photorepairing ultraviolet-induced pyrimidine dimers in the presence of light (Essen, 2006; Essen and Klar, 2006). Cryptochromes are proteins structurally similar to photolyases that act as blue light photoreceptors or regulators of the circadian rhythm (Cashmore et al., 1999) but that have lost the enzymatic photolyase activity (Chaves et al., 2011). Up to now, seven major classes of photolyase/cryptochrome families have been found (Scheerer et al., 2015). Interestingly, while the subunits found in Epi1 and Epi3 have similarity with eukaryotic cryptochromes (38–49%), the photolyases found in Epi2A and Epi2C bins have their highest similarities with Planctomycetales homologs (30–52%), suggesting potential interdomain HGT events. Five related genes, a phytoene synthase, a phytoene-desaturase, an histidine kinase, a sugar-epimerase and one hypothetical protein, were found adjacent to the photolyase gene. At the equivalent genomic position, the aphotic Guaymas32 had neither the photolyase nor the associated genes mentioned above (downstream from a 23S-rRNA gene) (Figure 3a). The phylogenetic origin of the genes flanking the photolyases was analyzed and, in several cases, were most closely related to homologs from Bacteriodetes/Planctomycetes, again suggesting instances of HGT. These included a chaperone involved in protein secretion that was 76% similar to a Rhodopirellula mairorica homolog, a nitroreductase that was 75% similar to a Gracilimonas tropica homolog and a sugar-epimerase next to the photolyase that was 58% similar to a Pirellula staleyi protein. Likewise, a hypothetical protein adjacent to the photolyase in Epi1 and Epi3 was most closely related to eukaryotic genes, suggesting that this pair of genes may have been transferred together.
Figure 3.
(a) Comparative genomic organization of MG-III sequences containing photolyases (in yellow). (b) Comparative genomic organization of MG-III sequences containing rhodopsins (in red) in context with other genomic fragments containing the MG-II Pop, Pop-1, Pop-2, Pop-3 and Pop-4 rhodopsins (bottom). Conserved genomic regions are indicated by gray shaded areas, gray intensity being a function of sequence similarity by TBLASTX. Particular open reading frames mentioned in the text are highlighted by a graphic code (see legend).
Epipelagic bins Epi1-2-3 all contained rhodopsins (Figure 2b) indicative of a photoheterotrophic lifestyle (Beja et al., 2000; Fuhrman et al., 2008; Inoue et al., 2013). In contrast, and consistent with previous reports (Deschamps et al., 2014; Li et al., 2015), Bathy1 and Bathy2 did not have rhodopsins. Phylogenetically, MG-III rhodopsins cluster with bacterial proteorhodopsins rather than with the euryarchaeal rhodopsins previously described for MG-II (Iverson et al., 2012; Martin-Cuadrado et al., 2014), suggesting that they may have been acquired by HGT from bacteria (Supplementary Figure S14). The analysis of key residues showed that all of these MG-III rhodopsins are proton pumps (Inoue et al., 2013) with a glutamine (Q) in the characteristic spectral tuning residue site indicating their ability to absorb light from the blue range (Supplementary Figure S16). In deeper waters (down to 300 m), only blue light remains available and blue rhodopsins are more suitable for generating energy. Therefore, epipelagic MG-III archaea seem to prefer low-light environments rather than the highly irradiated uppermost surface. Indeed, epipelagic MG-III bins recruited better from DCM or subsurface pelagic metagenomes (~50–70 m) than from surface (5 m) ones (see below). Genomic comparisons with MG-II rhodopsins (Martin-Cuadrado et al., 2014) revealed two new genomic contexts for this gene (Figure 3b). Interestingly, one of the clusters also contains one of the photolyase genes previously mentioned (Figure 3, contig Epi3-ERR598942-C530). Downstream from the rhodopsin genes, a gene for an unknown GYD domain protein was present. In cyanobacteria, proteins containing GYD and KaiC domains are involved in generating circadian rhythms (Chang et al., 2015). This raises the possibility that epipelagic MG-III Euryarchaeota may also have a circadian rhythm. A similar genome segment was found in two Guaymas32 sequences but, in these cases, the rhodopsin and the GYD domain-containing protein were absent.
The phylogenetic relationships of photolyases and rhodopsins, their proximity in at least one of the MG-III bins, together with the multiple putative HGT events observed in the nearby genes, leads us to hypothesize an ancestral ‘dark nature’ for MG-III. These light-related genes would have been recently transferred from epipelagic bacteria to MG-III, probably long after the massive HGT events that have been detected prior to the diversification of several mesophilic archaeal clades, including MGII/III (Deschamps et al., 2014; Lopez-Garcia et al., 2016). The acquisition of proteorhodopsins, together with ultraviolet-protection photolyases, would have promoted a better adaptation to the oligotrophic surface waters allowing MG-III clades to expand into new photic niches.
Structural components
Cell envelope
One of the advantages of generating environmental fosmid sequences is that they allow the unequivocal assembly and detection of the so-called ‘metagenomic islands’ (Coleman et al., 2006; Cuadros-Orellana et al., 2007; Rodriguez-Valera et al., 2009). These are clone-specific genome areas that, owing to their low coverage, are rarely assembled from metagenomic data sets but can be easily identified in reference-genome recruitment plots in the form of empty (or little populated) areas with virtually no environmental homologs. One example can be observed in CG-Epi1. The area of the genome shown in Figure 4b (labeled with an asterisk) is enriched in genes needed for cell wall biosynthesis and contains several glycosyl-transferases (type I/IV), together with polysaccharide synthases and genes for carbohydrate modification (acyltransferases and aminotransferases). The presence of several lipopolysaccharide biosynthesis proteins in all MG-III bins suggests a more complex cell envelope than a protein layer (S-layer). Adjacent to the CG-Epi1 island, we found a giant protein of 7258 amino acids with no similarity in sequence databases. These types of proteins have previously been observed in several bacterial and archaeal genomes (Reva and Tummler, 2008; Strom et al., 2011) and have been hypothesized to have a role in defense against predation or in cell adhesion. Although we could not predict any function for it, the presence of lectin/glucanase domains (laminin_G3), glycosyl-transferase domains (RfaB), several beta-helix repeats and copper-binding domains (NosD) suggest an extracellular function. Large proteins (>5000 amino acids) with similar domains were also detected in other bins (Epi2-3-5). The similarity found between the giant proteins present in Guaymas31 and Bathy2 (90%) was remarkable.
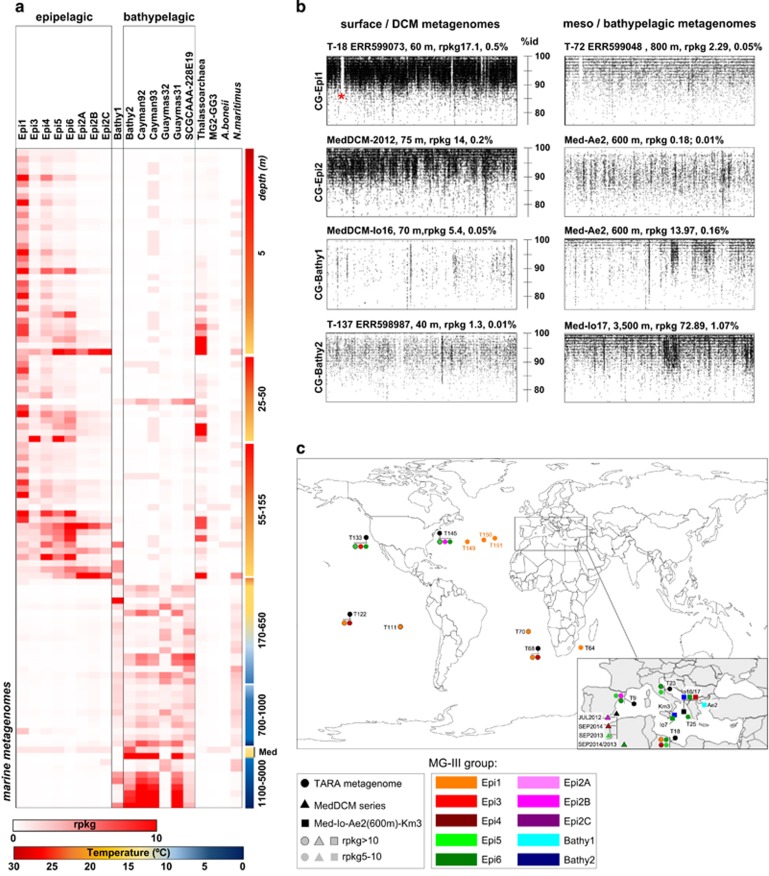
Figure 4.
(a) Heat map of the number of rpkg of each CG-MGIII of this work together with the ones of Li et al. (2015), MG-II and other archaea genomes used as references in 106 different metagenomes from different geographical points and depths. Only those collections in which any of the MG-III sequences recruited rpkg>1 were represented. (b) Recruitment plots of the CG-Epi1, CG-Epi2, CG-Bathy1 and CG-Bathy2 genomes in the metagenomes where they were better represented, from surface (<200 m) and bathypelagic (>500 m) (BLASTN-based, see Methods section). Rpkg and the percentages of the total of the reads with an identity bigger than >95% are indicated. (c) Worldwide distribution of the CG-MGIII determined by metagenomic fragment recruitment against public metagenomic databases. Only samples where the CGs recruited rpkg>5 are indicated in the map (cutoff: %identity >95% in >50 bp coverage). TARA spots are indicated by T#station.
Flagellum/Pili
Many archaeal surface structures are assembled by mechanisms related to the assembly of bacterial type IV pili (Lassak et al., 2012). With the exception of Epi5, we found several sequences containing two concatenated flaJ genes (implicated in archaeal flagellum assembly) followed by a flaI gene (a transcriptional activator). Syntenic operons were also found among deep-MG-III in Li et al. (2015). However, these gene clusters are very different from the flagellar operon found in MG2-GG3 (Iverson et al., 2012) or in any other Euryarchaeota described to date (Jarrell and McBride, 2008; Jarrell et al., 2010). Although it has been claimed that the genes found might be enough to build a functional flagellum (Li et al., 2015), the lack of a more complex gene cluster suggests that this operon might be involved in a secretion system translocating proteins rather than in cell motility.
Prevalence in the marine environment
To evaluate the relative abundance of the novel MG-III genomes, we used the non-redundant CGs to recruit reads from >200 metagenomic data sets that provide reasonably complete coverage of open-ocean waters from around the world. Among them, 106 gave values higher than one rpkg for any of the CGs tested (Figure 4 and Supplementary Table S1). Negative results are probably due to the small size of the data sets (for example, GOS) that may have poor representation of less abundant organisms. Although a considerable number of MG-III clones have been detected in cold waters such as the deep Atlantic layer of the central Arctic Ocean, (Galand et al., 2009), the MG-III bins described here were not well represented in metagenomes from cold water regions such as polar regions (Alonso-Saez et al., 2012), the Baltic (Larsson et al., 2014) or the northeast subarctic Pacific (Allers et al., 2013). This may suggest that there are other abundant MG-III groups present in high latitudes that have yet to be discovered. Even in warmer latitudes, our LowGC-MG-III bins only represent a small fraction of the total prokaryotic population of photic marine habitats. The highest abundance we found was for CG-Epi1 that accounted for 0.5% of the reads in the samples from the Mediterranean station TARA-018 (ERR599073 collection) (Figure 4b). The deep MG-III bins recruited slightly more. For instance, CG-Bathy2 recruited up to 1% of the reads in the deep sample Med-Io17 (3500 m).
Figure 4 shows a clear correlation of the two MG-III groups with depth (as already suggested by the origin of the assembled bins). Most LowGC-MGIII bins are only present in epipelagic collections, while the HighGC-MGIII plus the LowGC Bathy1 and Guaymas32 were clearly bathy or mesopelagic. CG-Epi1 seemed to be evenly distributed throughout the photic zone, but CG-Epi3, 5 and 6 increased at deeper waters (25–155 m, including the DCM) and the three CG-Epi2 showed an increase in even deeper photic zone waters. Bathy1 has its maximum at mesopelagic waters (Adriatic Sea 600 m), but it was also detected in colder bathypelagic waters (for example, the metagenomes from the Cayman-Rise and Guaymas Basin). CG-Bathy2 together with the Cayman and Guaymas bins revealed a strong correlation with deeper waters with much higher abundance in metagenomic collections <1000 m. These bins were more abundant in the warmer (13 °C) and saltier Mediterranean deep samples (KM3, 3000 m and Io17, 3500 m deep), although the temperature in most bathypelagic waters, where these microbes were detected (global ocean), typically decreases down to <5 °C. Overall, these numbers indicate that MG-III cells are relatively minor components of the archaeal communities in the photic and aphotic zones.
Using the Mediterranean DCM time series data sets, we found significant temporal variation in the abundance of the different GC bins despite a relatively constant abundance of reads attributable to euryarchaeal 16S rRNA genes (Supplementary Figure S17). For example, CG-Epi2A predominated in 2012, whereas CG-Epi6 was dominant in 2013 and CG-Epi4 in 2014. In the case of MG-II, it has been experimentally demonstrated that eukaryotic phytoplankton additions stimulate their growth in bottle incubations (Orsi et al., 2015). Also, MG-II became one of the most abundant organisms (up to 40% of prokaryotes) in a phytoplankton bloom where diatoms, small flagellates and pico-phytoplankton dominated consecutively (Needham and Fuhrman, 2016). In order to know whether MG-II and the genomes of MG-III described here respond to similar blooming patterns, we measured the recruitment of available MG-II genomes in the metagenomes from which MG-III were assembled. The results show very low numbers for MG-II genomes in these samples, close to 100 times less than for MG-III genomes (Supplementary Figure S18). These data indicate that, despite being closely related and using similar substrates, MG-II and MG-III do not bloom concurrently. Using published plankton-interactome data (Lima-Mendez et al., 2015), we constructed an interaction network for MG-III archaea (Supplementary Figure S19). The results showed that MG-III coexists mainly with Metazoa and Dinophyta, which represented 50.6% and 23.5% of the total of interactions observed. These findings may indicate that MG-III cells could be attached to other organisms and only sporadically be released to the environment.
Conclusions
The photic zone of the oligotrophic ocean, one of the largest microbial habitats on Earth, has been extensively explored by molecular and genomic approaches (DeLong, 1992; DeLong et al., 1999; Venter et al., 2004; Rusch et al., 2007; Sunagawa et al., 2015). Nevertheless, many epipelagic microbes remain to be characterized. Using metagenomics, we have uncovered eight new groups of planktonic marine Euryarchaeota that likely represent novel taxonomic orders or at least families. Based on differences in genome content and sequence identity, we propose the following nomenclature: Epipelagoarchaeales for the LowGC-MGIII and Bathypelagoarchaeales for the HighGC-MGIII. A separate and basal clade with low GC content but apparently living in the dark ocean (Bathy1) has also been uncovered. Genome comparisons between these new groups together and previously described MG-III genomes (Li et al., 2015) showed a marked differentiation between MG-III from photic and aphotic layers. Genomic analysis indicates that at least some representatives Epipelagoarchaeales (Epi1–Epi6) are planktonic photoheterotrophs. Two other groups with the Epipelagoarchaeales, Bathy1 and Guaymas32, lack genes indicating photoheterotrophy and are likely mesopelagic microbes with diverse metabolic capabilities. We hypothesize that the low GC content characteristic of the Epipelagoarchaeales may be an adaptation to the nitrogen limitation of surface waters. It is remarkable that all marine Euryarchaeota appear to possess similar metabolic profiles based on heterotrophic degradation of polymers and proteins (Iverson et al., 2012; Martin-Cuadrado et al., 2014; Li et al., 2015; Orsi et al., 2015). The broad diversity of marine microbes exploiting this habitat is likely a reflection of the enormous diversity of metabolic substrates available. Our data suggest a possible interaction of MG-III with eukaryotic cells and, more specifically, with metazoa.
Acknowledgments
We are thankful to José de la Torre for editing the manuscript. We are thankful to L Gasperini and G Bortoluzzi of the Instituto di Geologia Marina (ISMAR), CNR, Bologna (Italy) for allowing PLG to participate in the Marmara2010 R/V Urania cruise during which part of the samples analyzed in this study were collected. This work was supported by projects MEDIMAX BFPU2013–48007-P from the Spanish Ministerio de Economía y Competitividad, MaCuMBA Project 311975 of the European Commission FP7, project AQUAMET II/2014/012 from the Generalitat Valenciana and by the French Agence Nationale de la Recherche (ANR-08-GENM-024–001,EVOLDEEP). JHM was supported with a PhD fellowship from the Spanish Ministerio de Economía y Competitividad.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL et al. (2012). Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109: 17989–17994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. Isme J 7: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S et al. (2004). The Pfam protein families database. Nucleic Acids Res 32(Dataabase issue): D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut B, Knibbe C, Marais G, Daubin V. (2014). Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol 12: 841–850. [DOI] [PubMed] [Google Scholar]
- Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529. [DOI] [PubMed] [Google Scholar]
- Belmar L, Molina V, Ulloa O. (2011). Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol Ecol 78: 314–326. [DOI] [PubMed] [Google Scholar]
- Boutrif M, Garel M, Cottrell MT, Tamburini C. (2011). Assimilation of marine extracellular polymeric substances by deep-sea prokaryotes in the NW Mediterranean Sea. Environ Microbiol Rep 3: 705–709. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. (2008). Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6: 245–252. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Aylward FO, Eppley JM, Karl DM, Church MJ, DeLong EF. (2015). Wind and sunlight shape microbial diversity in surface waters of the North Pacific Subtropical Gyre. ISME J 10: 1308–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. (1999). Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. (2010). Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA 107: 18634–18639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, Delong EF et al. (2006). Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770. [DOI] [PubMed] [Google Scholar]
- Cuadros-Orellana S, Martin-Cuadrado AB, Legault B, D'Auria G, Zhaxybayeva O, Papke RT et al. (2007). Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J 1: 235–245. [DOI] [PubMed] [Google Scholar]
- Chang YG, Cohen SE, Phong C, Myers WK, Kim YI, Tseng R et al. (2015). Circadian rhythms. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science 349: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K et al. (2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364. [DOI] [PubMed] [Google Scholar]
- Chen S, Su L, Chen J, Wu J. (1997). Cutinase: characteristics, preparation, and application. Biotechnol Adv 31: 1754–1767. [DOI] [PubMed] [Google Scholar]
- DeLong EF. (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. (2006). Archaeal mysteries of the deep revealed. Proc Natl Acad Sci U S A 103: 6417–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Taylor LT, Marsh TL, Preston CM. (1999). Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol 65: 5554–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P, Zivanovic Y, Moreira D, Rodriguez-Valera F, López-García P. (2014). Pangenome evidence for extensive inter-domain horizontal transfer affecting lineage-core and shell genes in uncultured planktonic Thaumarchaeota and Euryarchaeota. Genome Biol Evol 6: 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Garczarek L, Partensky F. (2005). Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol 6: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgcomb VP, Kysela DT, Teske A, de Vera Gomez A, Sogin ML. (2002). Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc Natl Acad Sci USA 99: 7658–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO. (2006). Photolyases and cryptochromes: common mechanisms of DNA repair and light-driven signaling? Curr Opin Struct Biol 16: 51–59. [DOI] [PubMed] [Google Scholar]
- Essen LO, Klar T. (2006). Light-driven DNA repair by photolyases. Cell Mol Life Sci 63: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Davis AA. (1997). Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. March Ecol Prog Series 150: 275–285. [Google Scholar]
- Fuhrman JA, McCallum K, Davis AA. (1992). Novel major archaebacterial group from marine plankton. Nature 356: 148–149. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. (2008). Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol 6: 488–494. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. (2009). Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J 3: 860–869. [DOI] [PubMed] [Google Scholar]
- Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO. (2010). Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory). Limnol Oceanogr. 55: 2117–2125. [Google Scholar]
- Ghai R, Martin-Cuadrado A, Gonzaga A, Garcia-Heredia I, Cabrera R, Martin J et al. (2010). Metagenome of the Mediterranenan deep Chlorophyl Maximum stuidied by direct and fomid library 454 pyrosequencing. ISME J 4: 1154–1166. [DOI] [PubMed] [Google Scholar]
- Ghai R, McMahon KD, Rodriguez-Valera F. (2012). Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC. Environ Microbiol Rep 4: 29–35. [DOI] [PubMed] [Google Scholar]
- Giovannoni S, Nemergut D. (2014). Ecology. Microbes ride the current. Science 345: 1246–1247. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57: 81–91. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Lumia V, Creti R, Conway de Macario E, Sanangelantoni A, Cammarano P. (1999). Discontinuous occurrence of the hsp70 (dnaK) gene among Archaea and sequence features of HSP70 suggest a novel outlook on phylogenies inferred from this protein. J Bacteriol 181: 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT et al. (2001). TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29: 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, Sugahara J et al. (2006). Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA 103: 18296–18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A et al. (2005). Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71: 2303–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gilna P, Li W. (2009). Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25: 1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ono H, Abe-Yoshizumi R, Yoshizawa S, Ito H, Kogure K et al. (2013). A light-driven sodium ion pump in marine bacteria. Nat Commun 4: 1678. [DOI] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. (2012). Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335: 587–590. [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Jones GM, Kandiba L, Nair DB, Eichler J. (2010) S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea 2010. [DOI] [PMC free article] [PubMed]
- Jarrell KF, McBride MJ. (2008). The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6: 466–476. [DOI] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510. [DOI] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. (2005). Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102: 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B et al. (2014). Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. [DOI] [PMC free article] [PubMed]
- Lassak K, Ghosh A, Albers SV. (2012). Diversity, assembly and regulation of archaeal type IV pili-like and non-type-IV pili-like surface structures. Res Microbiol 163: 630–644. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. (2005). Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Baker BJ, Anantharaman K, Jain S, Breier JA, Dick GJ. (2015). Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun 6: 8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F et al. (2015). Ocean plankton. Determinants of community structure in the global plankton interactome. Science 348: 1262073. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Lopez-Lopez A, Moreira D, Rodriguez-Valera F. (2001. a). Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36: 193–202. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Moreira D, Lopez-Lopez A, Rodriguez-Valera F. (2001. b). A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ Microbiol 3: 72–78. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Philippe H, Gail F, Moreira D. (2003). Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci USA 100: 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Zivanovic Y, Deschamps P, Moreira D. (2016). Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol 13: 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI et al. (1999). Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res 9: 608–628. [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. (2003). Comparative genomics of Archaea: how much have we learned in six years, and what's next? Genome Biol 4: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. (2015) archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales Life (Basel) 5: 818–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Fontaine T, Esteban PF, del Dedo JE, de Medina-Redondo M, del Rey F et al. (2008). Characterization of the endo-beta-1,3-glucanase activity of S. cerevisiae Eng2 and other members of the GH81 family. Fungal Genet Biol 45: 542–553. [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Garcia-Heredia I, Molto AG, Lopez-Ubeda R, Kimes N, Lopez-Garcia P et al. (2015). A new class of marine Euryarchaeota group II from the Mediterranean deep chlorophyll maximum. ISME J 9: 1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Pasic L, Rodriguez-Valera F. (2014). Diversity of the cell-wall associated genomic island of the archaeon Haloquadratum walsbyi. BMC Genomics 16: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR et al. (2008). Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J 2: 865–886. [DOI] [PubMed] [Google Scholar]
- Massana R, DeLong EF, Pedros-Alio C. (2000). A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol 66: 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno CM, Ghai R, Saghai A, Lopez-Garcia P, Rodriguez-Valera F. (2016). Genomes of abundant and widespread viruses from the deep ocean. MBio 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP. (2009) Structural RNA homology search and alignment using covariance models, PhD thesis. Washington University in Saint Louis, School of Medicine, St. Louis, MO, USA.
- Needham DM, Fuhrman JA. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. [DOI] [PubMed]
- Orsi WD, Smith JM, Wilcox HM, Swalwell JE, Carini P, Worden AZ et al. (2015). Ecophysiology of uncultivated marine euryarchaea is linked to particulate organic matter. ISME J 9: 1747–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouverney CC, Fuhrman JA. (2000). Marine planktonic archaea take up amino acids. Appl Environ Microbiol 66: 4829–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. (2014). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Petitjean C, Moreira D, Lopez-Garcia P, Brochier-Armanet C. (2012). Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol Biol 12: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Zivanovic Y, Moreira D, Lopez-Garcia P. (2011). Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5: 285–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. (2007). Prediction of effective genome size in metagenomic samples. Genome Biol 8: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymann K, Forterre P, Brochier-Armanet C, Gribaldo S. (2014). Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol Evol 6: 192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva O, Tummler B. (2008). Think big—giant genes in bacteria. Environ Microbiol 10: 768–777. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B et al. (2009). Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7: 828–836. [DOI] [PubMed] [Google Scholar]
- Romero H, Pereira E, Naya H, Musto H. (2009). Oxygen and guanine-cytosine profiles in marine environments. J Mol Evol 69: 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S et al. (2007). The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheerer P, Zhang F, Kalms J, von Stetten D, Krauss N, Oberpichler I et al. (2015). The class III cyclobutane pyrimidine dimer photolyase structure reveals a new antenna chromophore binding site and alternative photoreduction pathways. J Biol Chem 290: 11504–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR, Ribbe MW. (2009). Molybdenum cofactors, enzymes and pathways. Nature 460: 839–847. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M et al. (2012). Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol 3: 1462–2920. [DOI] [PubMed] [Google Scholar]
- Strom SL, Brahamsha B, Fredrickson KA, Apple JK, Rodriguez AG. (2011). A giant cell surface protein in Synechococcus WH8102 inhibits feeding by a dinoflagellate predator. Environ Microbiol. [DOI] [PubMed]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Ocean plankton. Structure and function of the global ocean microbiome. Science 348: 1261359. [DOI] [PubMed] [Google Scholar]
- Swan BK, Chaffin MD, Martinez-Garcia M, Morrison HG, Field EK, Poulton NJ et al. (2014). Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One 9: e95380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, Gonzalez JM et al. (2013). Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Vavourakis CD, Ghai R, Rodriguez-Valera F, Sorokin DY, Tringe SG, Hugenholtz P et al. (2016). Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline Soda Lake Brines. Front Microbiol 7: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.