Abstract
The motion of polarizable particles in a non-uniform electric field, i.e., dielectrophoresis, has been extensively used for concentration, separation, sorting, and transport of biological particles, from cancer cells and viruses to biomolecules such as DNAs and proteins. However, current approaches to dielectrophoretic manipulation are not sensitive enough to selectively target individual molecular species. Here we describe the application of the dielectrophoretic principle for selective detection of DNA and RNA molecules using an engineered biological nanopore. The key element of our approach is a synthetic polycationic nanocarrier that selectively binds to the target biomolecules, dramatically increasing their dielectrophoretic response to the electric field gradient generated by the nanopore. The dielectrophoretic capture of the nanocarrier-target complexes is detected as a transient blockade of the nanopore ionic current while any non-target nucleic acids are repelled from the nanopore by electrophoresis and thus do not interfere with the signal produced by the target’s capture. Strikingly, we show that even modestly charged nanocarriers can be used to capture DNA or RNA molecules of any length or secondary structure and simultaneously detect several molecular targets. Such selective, multiplex molecular detection technology would be highly desirable for real-time analysis of complex clinical samples.
Keywords: Single-molecule detection, molecular dynamics simulation, HIV-TAT, nucleic acids, microRNA, single nucleotide polymorphism
INTRODUCTION
Dielectrophoresis (DEP) is an electrokinetic technology for collection, enrichment, and sorting of pathogenic cells and viruses.1–4 Subject to a non-uniform electric field, the induced dipoles of such large polarizables particles produces electrostatic force that moves the particles along or against the field gradient.5, 6 Recently, DEP has been developed for protein and DNA manipulation,7–13 enabling applications of DEP at the molecular level, for example, enrichment of circulating nucleic acids in cancer detection,13 concentration of charged proteins in a nanopipet,14 and ultra-sensitive DNA detection in a metallic nanopore.15 In spite of these advances, however, the DEP technology still cannot precisely discriminate target molecules, such as a nucleic acid biomarker, from a sample containing non-target species. The problem is that individual biomolecules have very similar and very small induced dipole moments, requiring high field gradient in order to generate a substantial DEP force.11 Hence, although current DEP technologies are capable of detecting a molecular induced dipole, they lack the sensitivity to discriminate induced dipoles of different molecular species.
Nanopores have been developed as next-generation sensors of genetic, epigenetic and proteomic biomarkers, and drug compounds16–27 with applications ranging from gene sequencing28–32 to molecular diagnostics.33 Nanopore detection of biomarkers relies on ionic current signatures produced by the translocation of biomarkers in the nanopore, which is driven by electrophoresis and electroosmosis.34–36 However, non-target molecules, which are typically present in clinical samples, can enter the nanopore as well, producing ionic current signatures that interfere with the target signal, severely lowering the target detection accuracy. An ideal nanopore sensor would thus have high sensitivity to the target biomarkers and minimal interactions with all non-target species.
In this report, we describe a carrier-guided nanopore dielectrophoresis, a single molecule detection method that overcomes the selectivity challenges of conventional dielectrophoresis and nanopore sensing. Rather than relying on the target’s native polarizability, we engineered a small polycationic nanocarrier to impose a prescribed dipole moment onto the target nucleic acid, granting both sensitivity and selectivity to the nanopore detection method. Subject to the electric field gradient at the nanopore entrance, any target molecule bound to a nanocarrier is driven into the nanopore by the DEP force. All non-target molecules, having no bound carrier and hence no enhanced dipole moment, are instead electrophoretically repelled from the nanopore and thus do not generate any interfering signals. As we show below, this approach is effective for either single or double-stranded DNA or RNA of any length and permits simultaneous detection of several target biomarkers.
RESULTS
Nanocarrier-enabled anti-field capture of target nucleic acids
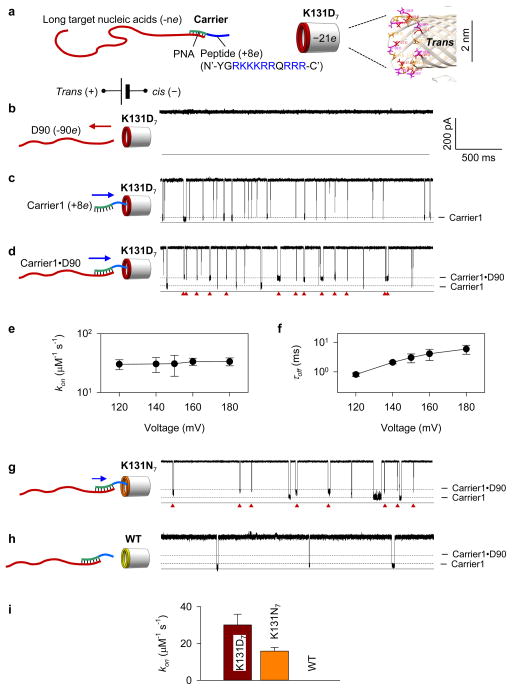
For the nanopore experiments (Supplementary Methods 1), we used the K131D7 variant of α-hemolysin, which has a negatively charged ring (−21e, where e is the charge of a proton) formed by seven copies of D128, D129 and D131 at the trans entrance (Fig. 1a right). The K131D7 pore was previously used by the Movileanu group to trap proteins containing positively charged tags37, 38. The nanocarrier used for driving the target nucleic acid into the nanopore contained a polycationic peptide tag (charge +8e, Fig. 1a left) covalently attached to a fragment of neutral target-specific peptide nucleic acid (PNA). When the trans compartment was positively biased with respect to the grounded cis compartment, no nucleic acids were observed to enter the nanopore from the trans solution and thus change the nanopore conductance (Fig. 1b). In contrast, the cationic nanocarriers could be electrically attracted and captured by the nanopore, reducing the nanopore current I to I/I0=10.0±0.4% of the open pore current I0 (Fig. 1c, Fig. S1). Thus, both nucleic acids and nanocarries were observed to move in the direction prescribed by the transmembrane bias.
Figure 1.
Anti-field capture of long DNA molecules. a. Nanocarrier (Carrier1) consisting of a polycationic peptide (+8e, blue) linked to a PNA (green) that is hybridized with a long target DNA (red). The complex can be anti-field captured by the K131D7 α-hemolysin pore containing seven copies of D127, D128, and D131 residues (−21e) at the trans entrance. b–d. Representative nanopore current traces recorded at +180 mV applied from the trans side and in the presence of various analytes in trans solution: (b) a mixture of DNA constructs including D10 to D90 (500 nM each), (c) free Carrier1 (100 nM), (d) a mixture of 100 nM Carrier1 and 100 nM D90. The traces collectively show that free DNA molecules are pushed away from the K131D7 pore without altering the pore conductance (b); free nanocarriers are attracted to and pass through the pore, producing characteristic blockades (c); when bound to the nanocarrier, DNA can be anti-field captured by the pore, producing distinct blockades (d, marked by red triangles). Here and in all subsequent figures, the horizontal arrows schematically show the expected direction of the effective force acting on the analytes. e–f, Variation of kon (e) and τoff (f) with the voltage for the DNA construct D90 (90 nts). g–h, Current traces recorded at +180 mV showing the Carrier1•D90 capture blockades in the K131N7 pore (–14e, e) and WT pore (–7e, f). i, Carrier1•D90 capture rate (kon) in the K131D7, K131N7 and WT pores. Compared to the K131D7 pore, the Carrier1•DNA complex generates fewer signature blockades in the K131N7 pore; no blockade signatures indicative of Carrier1•DNA capture was observed for the WT pore.
In comparison to the nanocarrier, its target DNA molecules carry a much greater (by magnitude) negative charge (Table S1 for sequences). Clearly, when these DNA molecules are hybridized with the nanocarrier, their complex remains highly negatively charged, and should, at first look, be electrostatically pushed away from the K131D7 pore. Our experimental observations, however, suggest the opposite. As shown in Fig. 1d (for D90) and Fig. S2 (for all DNA targets), the carrier•DNA complexes transiently reduce the nanopore current to a level (I/I0=27–29%) that is distinctly higher than the blockades produced by the nanocarrier alone (I/I0=9.5%); the hybridization of the nanocarriers with DNA considerably reduces the frequency of ionic current blockades produced by the free nanocarriers. Thus, in spite of carrying an overall negative charge, the carrier•DNA complex can be attracted to the nanopore and captured rather than repelled. The capture rate (Fig. 1e) exhibits a very weak dependence on the voltage, which is discussed in more details in the subsequent sections. The increase of the carrier•DNA blockade duration (τoff) with voltage (Fig. 1f and Figs. S3–S4) suggests that a captured carrier•DNA complex does not translocate through the pore. As the DNA•PNA duplex domain is wider than the trans entrance of the nanopore, the duplex is likely to remain outside the nanopore (Fig. 1a right) and only the peptide domain of the complexes to thread into the nanopore stem. This configuration was supported by our observation that the magnitude of the blockade current produced by the carrier•DNA complex (27~29%) is close to the blockade current produced by the peptide tag of the nanocarrier alone (I/I0=21%, Fig. S5).
According to the above observation, a small cationic carrier can reverse the effective force of the transmembrane bias on long target nucleic acids, making the latter move along the direction of the applied field (opposite to that prescribed by its overall electrical charge) and toward a negatively charged nanopore. Since both the carrier•DNA complex and free DNA (without carrier binding) carry large negative charge, the reversal of their effective driving force cannot be explained by electrophoresis. Indeed, the experiment shows that increasing the voltage was seen to very slightly increase the capture efficiency (Fig. 1e and Fig. S6). For example, the capture rate kon for the Carrier1•D90 complex was 29±4 μM−1s−1 at +120 mV and 33±6 μM−1s−1 at +180 mV (Fig. 2e). At the same time, reversal of the driving force cannot be explained by the electro-osmotic effect as it would apply equally to both the carrier•DNA complex and the free DNA. Furthermore, although the K131D7 pore is slightly cation selective, its low K+/Cl− permeability ratio (P+/P−=1.6, estimated from the reverse potential value of +10 mV measured for a cis/trans=1 M/0.2 M KCl asymmetric solution) is not expected to generate a sufficiently large ion and water flux. However, in contrast to voltage, we observed a dramatic modulation of the capture efficiency when altering the charge of the nanopore entrance from −21e of K131D7 to −14e of K131N7 or −7e of wild type (WT) variants, respectively. We found that kon for Carrier1•D90 was reduced from 33±6 μM−1·s−1 in the K131D7 pore (Fig. 1d) to 16±2 μM−1·s−1 in the K131N7 pore (Fig. 1g and i). The Carrier1•D90 blockades were not observed for the WT pore (Fig. 1h and i). A similar trend was observed for all DNA targets (Fig. S7). As the presence of the charge at the nanopore entrance produces a strong and non-uniform electrical field extending from the nanopore entrance to the bulk solution, we hypothesized that this non-uniform electric field is responsible for the capture of the carrier•DNA complexes.
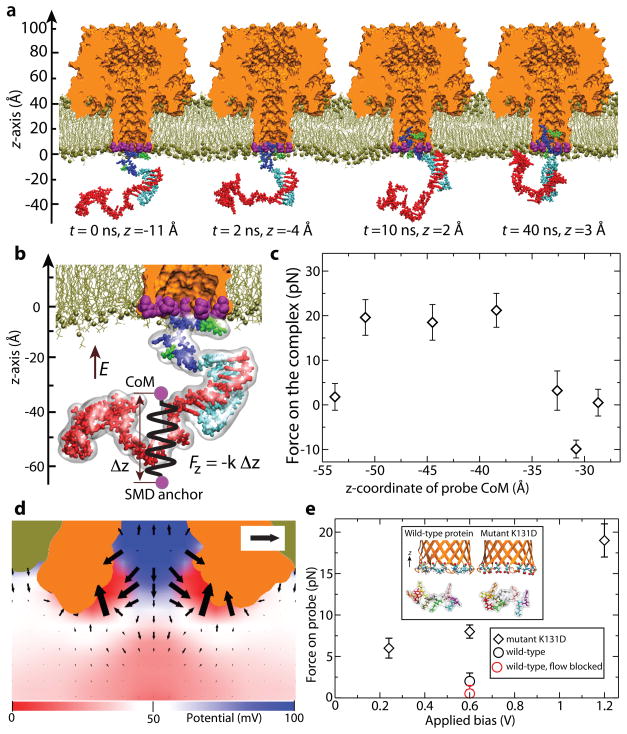
Figure 2.
MD simulations of nanopore dielectrophoresis. a, MD simulation of carrier•DNA capture. The K131D7 variant of αHL is shown as a cut-away orange molecular surface and the seven Asp131 residues as purple vdW spheres; the lipid bilayer membrane is shown as lines and spheres (tan); water and ions are not shown. The carrier•DNA complex consists of a 28-nt ssDNA strand (red) hybridized to an 8-residue fragment of PNA (cyan) that is covalently connected to a tag peptide of 11 amino acids (positive residues shown in blue, polar residues shown in green). A transmembrane bias of 1.2 V is applied along the z axis. The z-coordinate under each snapshot indicates the location of the peptide tag’s CoM. b, Schematic illustration of the SMD method for determining the effective force on the probe. A semi-transparent molecular surface indicates the approximate physical volume occupied by the carrier•DNA complex. The CoM of the carrier•DNA complex is restrained to the SMD anchor by a harmonic potential of spring constant k. The distance between the CoM of the complex and the SMD anchor is exaggerated for clarity. c, The effective force acting on the carrier•DNA complex under a +1.2 V bias versus the z-coordinate of the complex’ CoM. The z-axis is defined in panel b. A positive force is directed along the z-axis. Each data point was obtained from a ~40 ns MD trajectory. d, The difference of the K131D7 and WT electrostatic potential and local force maps. The force scale bar indicates a 3pN force. e, Mean force on the peptide probe (charge +8e) vs. applied bias for the K131D7 and WT simulation systems. In these simulations, the backbone atoms of the peptide were restrained to maintain their initial z coordinates and remain within 17 Å of the pore axis. Each data point represents a 200 ns trajectory average of instantaneous forces sampled every 200 fs. In one simulation of the WT system, water flow through the pore was blocked by harmonically restraining all water molecules to their initial coordinates within a 10-Å segment of the pore’s stem. The inset illustrates the simulation systems used to measure the effect of the nanopore charge on the effective force.
Microscopic simulations of the anti-field capture
To understand the force on and movement mechanism of the carrier•DNA complex, we conducted molecular dynamics (MD) simulation of the atomic-scale models of the experimental systems39 (Fig. 2, Supplementary Methods 2 and Figs. S8–S11). The simulation systems contained one copy of the K131D7 or WT pore embedded in a lipid bilayer membrane, a probe molecule placed in front of the trans entrance of the pore, and water and ions corresponding to 1 M KCl solution (Fig. 2a). The model of the carrier•DNA complex consisted of a +8e polycationic peptide appended to a neutral PNA that was hybridized to a 28-nt ssDNA. The total charge of this carrier•DNA complex was −19e. In agreement with our interpretation of experimental results, we observed capture of the complex by the K131D7 pore after 20 ns of MD simulations at a 1.2V bias, despite the overall negative charge of the complex (Fig. 2a and Fig. S8). The complex’ capture was also observed in the case of the WT pore, but the complex did not permeate deep inside the pore stem and remained close to the rim of the trans entrance (Fig. S8).
To determine the direction and magnitude of the effective force on the complex, we utilized the steered molecular dynamics (SMD) method to harmonically restrain the center of mass (CoM) of the complex40 (Fig. 2b). The displacement of the complex produced by the electric field reported on the restraint force, which was averaged over the MD trajectory to calculate the effective force on the complex as a function of the complex’ position along the pore axis (Fig. 2c). The results of these simulations show that the effective force on the complex is directed toward the pore when the complex is located 5.5 nm away from the pore entrance. The force reaches 20 pN (at +1.2V bias) as the CoM of the complex moves closer to the pore but drops to zero about 3 nm away from the pore entrance. In the latter configuration, the peptide domain of the complex resides inside the nanopore whereas the duplex domain is anchored at the pore entrance; the effective force of the electric field is balanced by the reaction force from the pore rim.
To explore the effect of the pore’s local electric field on the complex capture, we computed the distributions of the electrostatic potential in the WT and K131D7 pores in the absence of the complex (Fig. S9). Point-by-point subtraction of the two electrostatic maps (Fig. 2d) indicates that a positive unit charge near the K131D7 lumen experiences up to 3 pN more force toward the lumen than the same charge in the WT pore (at +600 mV bias). Therefore, placement of more negative charges at the pore entrance considerably increases the effective capture force. The simulated effective force was also affected by the transmembrane voltage (Fig 2e). The force on the peptide tag of the complex significantly increased from 8±1 pN at +600 mV to 19±3 pN at +1.2 V. At lower biases, however, the simulated force increased only slightly, from 6±2 pN at +240 mV to 8±1 pN at +600 mV, which is consistent with the weak voltage-dependence of the carrier•DNA capture rate observed experimentally in the 100 to 200 mV range (Fig. S6). Similar simulations performed using a DNA fragment as a probe revealed that DNA is repulsed from the pore roughly as strongly as the peptide is attracted (Fig. S10). Additional simulations ruled out electro-osmosis as a cause of carrier•DNA capture (Fig. 2e and Fig. S11).
Dielectrophoretic mechanism of nanocarrier-facilitated nanopore capture
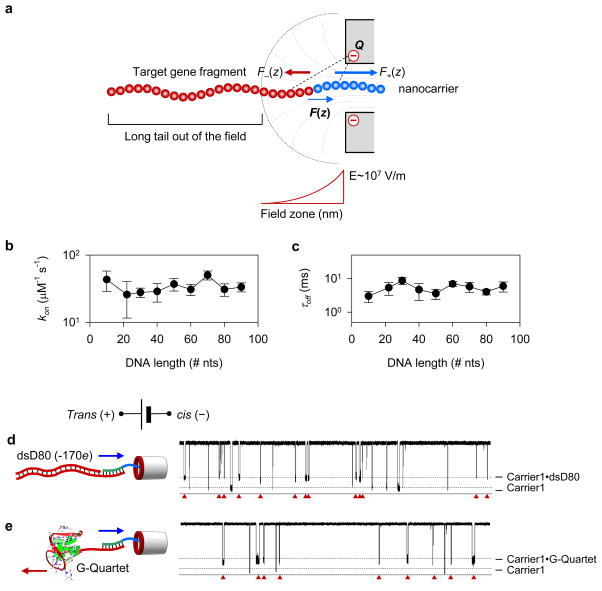
Our experimental and simulation findings both suggest that the force driving the anti-field motion of the carrier•DNA complex originates from a short-ranged, non-uniform electric field contributed by the negative charge at the nanopore trans entrance. According to our theoretical model (Fig. 3a, Figs. S12–13, and Supplementary Methods 3), the field strength rapidly increases near the nanopore, producing an extremely high field gradient of 107 V·m−1 per nanometer within several nanometers of the nanopore entrance (Fig. S12). The carrier•DNA complex placed in that field is acted on by both an attractive force on the nanocarrier and a repulsive force on the nucleic acid. However, simple addition of the two opposite forces located the same distance away from the nanopore does not produce a net attractive force (Fig. S10). Hence, the carrier•DNA complex is to be considered as a large synthetic dipole, with the counter charges of the peptide and nucleic acid being separated by a physical distance. Placed in a non-uniform electric field, the counter charges of the dipole experience different field strength. The peptide, which is closer to the nanopore, experiences higher field strength than the DNA tail, resulting in a net attractive force driving the carrier•DNA dipole toward the pore. The magnitude of the effective force would then depend on both the field gradient and the spatial separation of charge along the carrier•DNA dipole, a signature feature of dielectrophoresis.
Figure 3.
Dielectrophoretic mechanism of carrier-guided nanopore capture. a. Schematics of a theoretical model describing the capture of a nanocarrier•DNA complex (Fig. S12–13, Supplementary Methods 3). The nanopore contains a ring of negative charge (Q) at the pore entrance, which forms a non-uniform electric field (Fig. S12). Color arrows indicate the direction and relative amplitude (arrow length) of the forces acting on the cationic peptide (blue arrows) and anionic DNA (red arrows) as the carrier•DNA complex moves toward the pore. Because of the sharp field gradient and the separation of charge in the carrier•DNA complex, the attractive force on the peptide can be larger than the repulsive force on the DNA, generating a net attractive force that drives the complex toward the nanopore (Fig. S12–13). Because of its narrow range, the field does not affect the long DNA tail and, therefore, no matter how long the DNA is, it can be anti-field captured by the nanopore when bound to the nanocarrier. b–c, The rate of the carrier•DNA capture (kon, b) and the blockade duration (τoff, c) as a function of the DNA length. The data shown correspond to +180 mV voltage. For other voltages, see Fig. S6 for kon and Fig. S3 for τoff . The independence of the effective force on the DNA length is also predicted by theoretical considerations, Fig. S12. d–e, Current traces at +180 mV for a mixture of 100 nM Carrier1 and 100 nM 80-bp dsD80 (–170e, d), and the tertiary G-quadruplex (thrombin-binding aptamer, e). The traces collectively show that when bound to the nanocarrier, dsDNA (e) and DNA of complex tertiary structure (f) can be anti-field captured by the pore, producing distinct blockades (marked by red triangles).
The dielectrophoretic mechanism of carrier-DNA capture implies that the capture efficiency should not depend on the length of the DNA target. Indeed, the range of the electric field outside the nanopore is only several nanometers. Such a short-range, rapidly decaying field acts on only those nucleotides that are located near the nanopore entrance (8 nucleotides for a 5-nm-wide field zone), whereas the rest of the nucleotides experience a negligible force. Thus, regardless of their length, target nucleic acids would experience a similar force from the short-range field and captured by the nanopore with the same frequency at the same concentration. Our experimental observations confirm this conjecture. The average capture rate (kon) for DNA targets varying in length from 10 (D10) to 90 (D90) nucleotides varies only slightly between 23 and 43 μM−1·s−1 at +180 mV (Fig. 3b) and exhibits no systematic dependence on the DNA length, while the duration of the blockades (τoff) fluctuates between 3.0 and 8.7 ms (Fig. 3c). Similar capture rates and blockade durations were also observed at other voltages (Fig. S6 for kon and Fig. S3 for τoff). As the DNA part of the constructs does not participate in the force generation by the DEP mechanism, the DEP force should not depend on the DNA conformation. This can be seen in Fig. 3d and e, in which the same nanocarrier can also be used to capture long fragments of double-stranded DNA, such as the 80-basepair dsD80 construct (−170e charge, Fig. 3d), and nucleic acids with tertiary structures, such as the G-quadruplex (Fig. 3e). The capture efficiencies for the latter two constructs were similar to that of ssDNA: kon=44±7 μM−1·s−1 for dsD80 (Fig. 3d) and 22±6 μM−1·s−1 for the G-quadruplex (Fig. 3e). Once captured, the cationic peptide of the complex is trapped in the pore stem, in which the peptide is held by an electric field generated by the voltage. As suggested by the model (Fig. S13), increasing the voltage will enhance this in-the-pore field strength, thus providing a larger holding force. This explains the observation that the capturing duration (τoff) for each carrier•DNA complexes is prolonged as the voltage increases (Fig. 1f).
Interference-free detection of nucleic acid targets
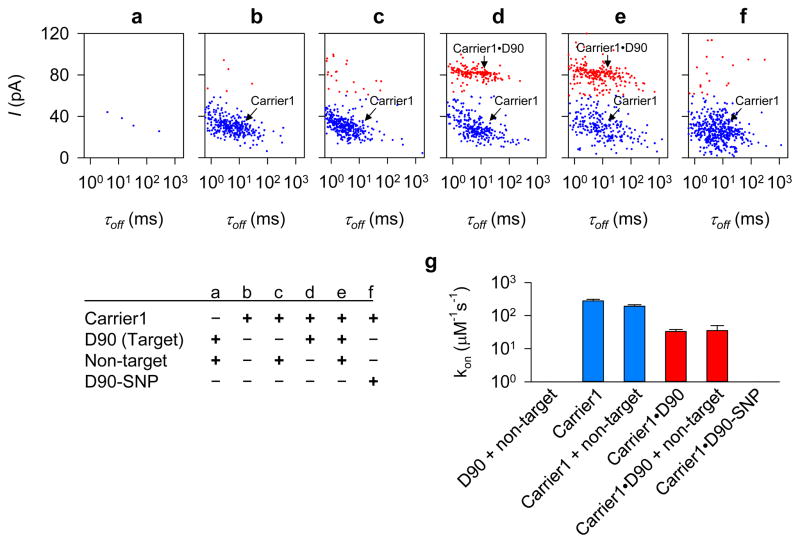
The dielectrophoretic mechanism for carrier-assisted nanopore capture immediately suggests an interference-free approach to selective detection of genetic biomarkers from contaminated samples. As only the target nucleic acids are selectively attracted and captured by the nanopore, all signals produced by non-target species are eliminated (Fig. 4). Indeed, we find that the presence of non-target DNA molecules in the sample influences neither the capture efficiency of the free nanocarriers (Fig. 4b, c and g) nor that of the carrier•DNA complexes (Fig. 4d, e and g), indicating that the nanocarrier’s binding is specific to the target and that formation of the carrier•DNA complex is not influenced by the presence of non-target DNA. Furthermore, the synthetic dipole-assisted nanopore capture is sensitive enough to enable single nucleotide discrimination. When a mismatched base pair is introduced in the carrier•DNA complex (Carrier1•D90-SNP), the frequency of the carrier•DNA blockades is greatly reduced (Fig. 4f, g, Fig. S14), suggesting that the carrier•DNA formation is sensitive to single-nucleotide variation in a target gene fragment. The results of our experiments demonstrate selective, interference-free detection of target nucleic acid in the presence of non-target species, which is a typical situation encountered in the analysis of clinical samples.
Figure 4.
Selective and interference-free detection of target gene fragment in the presence of non-target species. a–f, Scatter plots of blockade current amplitude (I) and duration (τoff) for the mixture of D90 (target) and non-target DNA (a), free Carrier1 (b), Carrier1 in the presence of non-target DNA (c), the mixture of Carrier1 and D90 in the absence (d) and presence (e) of non-target DNA, and the mixture of Carrier1 and D90-SNP. Blue and red dots indicate the blockades associated with the capture of free carrier and carrier•DNA complexes, respectively. Non-target DNA mixture included pT7 plasmid DNA (10.2 ng·μL−1) and a 225-bp BRAF gene fragment PCR reaction mixture (37.2 ng·μL−1). D90-SNP is a variant of D90, which forms a single mismatched basepair with Carrier1 (Table S1, Fig. S14 for traces). g, Comparison of the capture rate (kon) for the carrier•DNA complex for conditions shown in a–f. The presence of non-target DNA did not influence the capture efficiencies for both free nanocarrier and the carrier•DNA complex, suggesting that the nanocarrier is specific to the target and does not bind to non-target DNA, and that the formation of the carrier•DNA complex is not influenced by non-target DNA.
Applications to RNA and multi-target detection
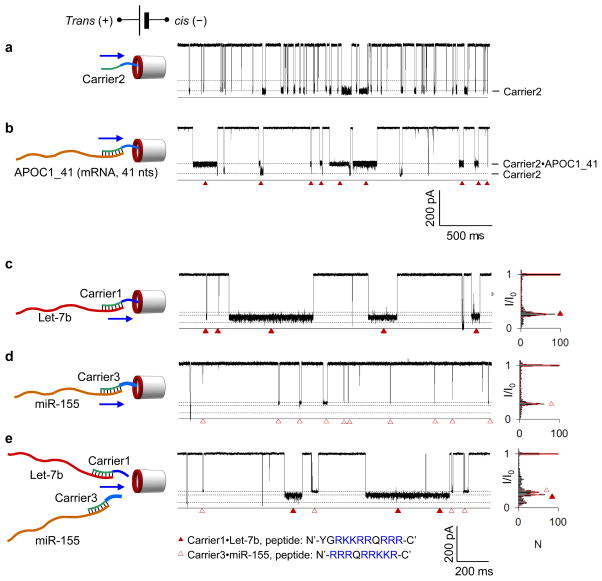
To demonstrate the generality of our approach, we applied it to RNA detection. RNA, such as mRNA and various non-coding RNAs, are important not only because they carry templates for protein expression and gene regulators, but also because they are biomarkers for disease development and diagnosis. We have previously demonstrated detection of short microRNAs (18–22 bases) using a peptide-PNA design.41 Here we demonstrate that the nanocarrier can selectively guide the capture of long RNA. Our target was a 41-nt fragment of Apolipoprotein C1 (APOC1) mRNA (APOC1_41), a biomarker associated with the occurrence of Alzheimer’s disease.42–44 We designed a second nanocarrier (referred to as Carrier2) to bind a fragment of APOC1_41 with its complementary PNA fragment (Table S1) and have the same polycationic peptide tag as Carrier1. In our nanopore experiments, we found the Carrier2•APOC1_41 complex to block the ionic current much more effectively (I/I0=31.3±1.5%, Fig. 5b) than Carrier2 alone (I/I0=12.1±1.3%, Fig. 5a); the blockades occurred with a high capture rate of kon=44 μM−1·s−1. Interestingly, some of the Carrier2•APOC1-41 blockade signatures feature a current transition, which can be interpreted as an indication of unzipping of the Carrier2•APOC1_41 complex. Upon dehybridization, the RNA molecule returns to the trans solution, while the cationic nanocarrier translocates through the pore, transiently lowering the blockade level. Note that blockades signatures indicating Carrier1•DNA dehybridization under the same experimental conditions are rare (Fig. 1d and Fig. S2).
Figure 5.
Dielectrophoretic detection of RNA targets. a–b, Current traces showing blockades produced by the translocation of Carrier2 (Table S1, 100 nM) alone (a) and when bound to a 40-nt fragment of Apolipoprotein C1 mRNA, APOC1_41 (100 nM, b), from the trans solution. c–e, Simultaneous capture and characterization of two types of carrier•RNA complexes by a nanopore. The targets are microRNA Let-7b and miR-155 (Table S1). Their specific carriers are Carrier1 and Carrier3, which differ from one another by both the peptide and PNA sequences (Table S1). Carrier1•Let-7b (solid triangles in panel c) and Carrier3•Let-7b (empty triangles in panel d) produced blockades of different conductance levels. In panel e, the mixture of two targets and two carriers were presented in the trans solution (100 nM each species). The difference in the peptide sequence of the two nanocarriers causes the two distinct blockade levels: the lower conductance blockade is generated by Carrier1•Let-7b and the higher conductance blockade by Carrier3•Let-7b.
To demonstrate multi-target detection, we designed two nanocarriers, Carrier1 and Carrier 3, to specifically bind two different microRNA targets (see Table S1). The peptide tags of the two carriers had identical amino acid compositions but different (reversed with respect to one another) amino acid sequences. We found that such peptide tag design can not only produce distinct blockade signatures to signal the capture of each target, but also preserve the overall high capture rate. When the two nanocarries were simultaneously introduced into the trans solution containing a mixture of Let-7b and miR-155 microRNA, two distinct populations of ionic current blockades emerged, one at a blockade level of I/I0=29.0±1.5% and another at I/I0=33.7±1.0% (Fig. 5e). Knowing the blockade levels produced by each type of the carrier•target complex (Fig. 5c and d), we could identify the former and the latter blockade events as being produced by Carrier1•Let-7b and Carrier3•miR-155 complexes, respectively. The ~14 pA current difference (at +180 mV) was sufficient to accurately discriminate the two types of capture events. As expected, the capture rates for the Carrier1•Let-7b (kon=24±7 M−1·s−1) and Carrier3•miR-155 (18±9 M−1·s−1) complexes were similar. Thus, altering the chemical structure of the nanocarrier’s peptide tag can produce distinct nanopore signatures, enabling simultaneous and selective dielectrophoretic detection of multiple biomarkers.
CONCLUSIONS
We have demonstrated a carrier-guided dielectrophoretic approach for interference-free nanopore detection of genetic biomarkers. Unlike induced dipole-based dielectrophoresis for manipulation of large biological particles such as cells and viruses, our approach is enabled by an engineered cationic nanocarrier that forms a non-covalent synthetic dipole with its target nucleic acid upon hybridization. Placing such a synthetic dipole in the large field gradient near the charge-terminated nanopore produces a substantial dielectrophoretic force on the target molecule, which enables anti-field capture of the dipole by the nanopore. The use of such synthetic dipoles considerably increases the strength of the dielectrophoretic effect and makes the dielectroporetic effect specific to the target species. For DNA and RNA detection, such specificity allows for detection of molecular sequences differing by a single nucleotide. Furthermore, minor modifications of the nanocarrier structure enable simultaneous detection of multiple targets via characteristic ionic current signatures. Our approach can be, in principle, used to selectively and sensitively detect any type of nucleic acids, DNA or RNA, long or short, and of any complex shape. This work suggests a nanosensor design for possible applications not only in cancer detection and other human disease molecular diagnostics, but also more broadly in fields such as plant science and foodborne detection where rapid genetic testing is required.
METHODS
Chemicals and materials
Chemical and materials including DNA constructs, peptide-PNA, and proteins are described in Supplementary Methods 1.
Electrophysiology recordings from single protein pores
Nanopore electrical recording was conducted according to previously reported protocols,45 and is described in Supplementary Methods 1.
RNA in vitro transcription, purification and hybridization with carrier
RNA target APOC1-41 (Table S1) was synthesized by in vitro transcription.46 The procedure is described in Supplementary Methods 1.
Summary of MD methods
All MD simulations were performed using the NAMD2 program,47 CHARMM27 force field,48 custom NBFIX corrections to describe ion-DNA interactions49 and CHARMM-compatible force field for PNA.50 The systems were constructed using the previously described models51 of alpha-hemolysin and ssDNA.52 A transmembrane bias was induced by applying an external electric field.53 The effective force was measured as equilibrium displacement of the complex subject to harmonic restraint.40 Detailed description of the simulation protocols are provided in Supplementary Methods 2.
Theoretical model of carrier-guided nanopore dielectrophoresis
The modeling method is described in Supplementary Methods 3.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01-GM079613 and R01-GM114204 to L.-Q.G. and A.A.). The authors acknowledge supercomputer time at the Blue Waters Sustained Petascale Facility (University of Illinois) and at the Texas Advanced Computing Center (Stampede, allocation award MCA05S028).
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: The Supporting Information is available free of charge on the ACS Publications website http://pubs.acs.org/, and includes sequences of nanocarriers and all nucleic acid targets, examples of nanopore blockades produced by free nanoarrier and all carrier•DNA complexes, variation of the carrier•DNA capture rate and capture duration with DNA length and voltage, MD simulation of the carrier•DNA capture by the WT and K131D7 variants of αHL, distribution of the electrostatic potential and magnitude and direction of local electrostatic force on one proton charge in the open-pore K131D7 and WT systems, SMD simulations of the effective force on the DNA and peptide domains of the carrier•DNA complex, electro-osmotic flow as a function of the applied bias for αHL systems with and without a peptide tag added, detection of SNP using nanopore dielectrophoresis, MD protocol, and mechanistic model of nanopore dielectrophoresis for capturing carrier•DNA complex in a non-uniform electric field.
References
- 1.Braff WA, Willner D, Hugenholtz P, Rabaey K, Buie CR. Dielectrophoresis-Based Discrimination of Bacteria at the Strain Level Based on Their Surface Properties. PLoS ONE. 2013;8:e76751. doi: 10.1371/journal.pone.0076751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grom F, Kentsch J, Müller T, Schnelle T, Stelzle M. Accumulation and Trapping of Hepatitis A Virus Particles by Electrohydrodynamic Flow and Dielectrophoresis. Electrophoresis. 2006;27:1386–1393. doi: 10.1002/elps.200500416. [DOI] [PubMed] [Google Scholar]
- 3.Lapizco-Encinas BH, Davalos RV, Simmons BA, Cummings EB, Fintschenko Y. An Insulator-Based (Electrodeless) Dielectrophoretic Concentrator for Microbes in Water. J Microbiol Methods. 2005;62:317–326. doi: 10.1016/j.mimet.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 4.Lapizco-Encinas BH, Simmons BA, Cummings EB, Fintschenko Y. Dielectrophoretic Concentration and Separation of Live and Dead Bacteria in an Array of Insulators. Anal Chem. 2004;76:1571–1579. doi: 10.1021/ac034804j. [DOI] [PubMed] [Google Scholar]
- 5.Pohl HA. The Motion and Precipitation of Suspensoids in Divergent Electric Fields. J App Phys. 1951;22:869–871. [Google Scholar]
- 6.Pohl HA. Dielectrophoresis: The Behavior of Neutral Matter in Nonuniform Electric Fields. Cambridge University Press; Cambridge; New York: 1978. [Google Scholar]
- 7.Chou CF, Tegenfeldt JO, Bakajin O, Chan SS, Cox EC, Darnton N, Duke T, Austin RH. Electrodeless Dielectrophoresis of Single- and Double-Stranded DNA. Biophys J. 2002;83:2170–2179. doi: 10.1016/S0006-3495(02)73977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng L, Brody JP, Burke PJ. Electronic Manipulation of DNA, Proteins, and Nanoparticles for Potential Circuit Assembly. Biosens & Bioelectron. 2004;20:606–619. doi: 10.1016/j.bios.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 9.Clarke RW, White SS, Zhou D, Ying L, Klenerman D. Trapping of Proteins under Physiological Conditions in a Nanopipette. Angew Chem. 2005;117:3813–3816. doi: 10.1002/anie.200500196. [DOI] [PubMed] [Google Scholar]
- 10.Regtmeier J, Duong TT, Eichhorn R, Anselmetti D, Ros A. Dielectrophoretic Manipulation of DNA: Separation and Polarizability. Anal Chem. 2007;79:3925–32. doi: 10.1021/ac062431r. [DOI] [PubMed] [Google Scholar]
- 11.Parikesit GOF, Markesteijn AP, Piciu OM, Bossche A, Westerweel J, Young IT, Garini Y. Size-Dependent Trajectories of DNA Macromolecules Due to Insulative Dielectrophoresis in Submicrometer-Deep Fluidic Channels. Biomicrofluidics. 2008;2:024103. doi: 10.1063/1.2930817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalir H, Yanagida Y, Hatsuzawa T. Probing DNA Mechanical Characteristics by Dielectrophoresis. Sens Actuators, B: Chemical. 2009;136:472–478. [Google Scholar]
- 13.Sonnenberg A, Marciniak JY, Rassenti L, Ghia EM, Skowronski EA, Manouchehri S, McCanna J, Widhopf GF, Kipps TJ, Heller MJ. Rapid Electrokinetic Isolation of Cancer-Related Circulating Cell-Free DNA Directly from Blood. Clin Chem. 2014;60:500–509. doi: 10.1373/clinchem.2013.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke RW, White SS, Zhou D, Ying L, Klenerman D. Trapping of Proteins Under Physiological Conditions in a Nanopipette. Angew Chem Int Ed Engl. 2005;44:3747–50. doi: 10.1002/anie.200500196. [DOI] [PubMed] [Google Scholar]
- 15.Freedman KJ, Otto LM, Ivanov AP, Barik A, Oh SH, Edel JB. Nanopore Sensing at Ultra-Low Concentrations Using Single-molecule Dielectrophoretic Trapping. Nat Commun. 2016;7:10217. doi: 10.1038/ncomms10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of Epigenetic DNA Modifications with a Protein Nanopore. Chem Commun. 2010;46:8195–7. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid Electronic Detection of Probe-Specific MicroRNAs Using Thin Nanopore Sensors. Nat Nanotechnol. 2010 doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 18.Clamer M, Hofler L, Mikhailova E, Viero G, Bayley H. Detection of 3′-End RNA Uridylation with a Protein Nanopore. ACS Nano. 2013 doi: 10.1021/nn4050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Enhanced Translocation of Single DNA Molecules through Alpha-Hemolysin Nanopores by Manipulation of Internal Charge. Proc Natl Acad Sci US A. 2008;105:19720–5. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc Natl Acad Sci US A. 2009;106:7702–7. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, Bartlett JJ, Pavlenok M, Gundlach JH. Detection and Mapping of 5-Methylcytosine and 5-Hydroxymethylcytosine with Nanopore MspA. Proc Natl Acad Sci. 2013;110:18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Larrea D, Bayley H. Protein Co-Translocational Unfolding Depends on the Direction of Pulling. Nat Commun. 2014;5:4841. doi: 10.1038/ncomms5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nir I, Huttner D, Meller A. Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores. Biophys J. 2015;108:2340–2349. doi: 10.1016/j.bpj.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammad MM, Movileanu L. Impact of Distant Charge Reversals within a Robust Beta-Barrel Protein Pore. J Phys Chem B. 2010;114:8750–8759. doi: 10.1021/jp101311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RP, Fleming AM, Burrows CJ, White HS. Effect of an Electrolyte Cation on Detecting DNA Damage with the Latch Constriction of Alpha-Hemolysin. J Phys Chem Lett. 2014;5:3781–3786. doi: 10.1021/jz502030e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghale G, Lanctot AG, Kreissl HT, Jacob MH, Weingart H, Winterhalter M, Nau WM. Chemosensing Ensembles for Monitoring Biomembrane Transport in Real Time. Angew Chem Int Ed Engl. 2014;53:2762–5. doi: 10.1002/anie.201309583. [DOI] [PubMed] [Google Scholar]
- 28.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, et al. The Potential and Challenges of Nanopore Sequencing. Nat Biotechnol. 2008;26:1146–53. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated Forward and Reverse Ratcheting of DNA in a Nanopore at 5-A Precision. Nat Biotech. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual Polynucleotide Molecules Using a Membrane Channel. Proc Natl Acad Sci. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R, Doering K, Shendure J, Gundlach JH. Decoding Long Nanopore Sequencing Reads of Natural DNA. Nat Biotechnol. 2014;32:829–33. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Reading DNA at Single-Nucleotide Resolution with a Mutant MspA Nanopore and Phi29 DNA Polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu LQ, Cheley S, Bayley H. Electroosmotic Enhancement of the Binding of a Neutral Molecule to a Transmembrane Pore. Proc Natl Acad Sci. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howorka S, Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 36.van Dorp S, Keyser UF, Dekker NH, Dekker C, Lemay SG. Origin of the Electrophoretic Force on DNA in Solid-State nanopores. Nat Phys. 2009;5:347–351. [Google Scholar]
- 37.Mohammad MM, Movileanu L. Excursion of a Single Polypeptide into a Protein Pore: Simple Physics, but Complicated Biology. Eur Biophys J. 2008;37:913–25. doi: 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- 38.Bikwemu R, Wolfe AJ, Xing X, Movileanu L. Facilitated Translocation of Polypeptides through a Single Nanopore. J Phys : Condens Matter. 2010;22:454117. doi: 10.1088/0953-8984/22/45/454117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aksimentiev A. Deciphering Ionic Current Signatures of DNA Transport through a Nanopore. Nanoscale. 2010;2:468–83. doi: 10.1039/b9nr00275h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luan B, Aksimentiev A. Electro-Osmotic Screening of the DNA Charge in a Nanopore. Phys Rev E. 2008;78:021912. doi: 10.1103/PhysRevE.78.021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian K, He Z, Wang Y, Chen SJ, Gu LQ. Designing a Polycationic Probe for Simultaneous Enrichment and Detection of MicroRNAs in a Nanopore. ACS Nano. 2013 doi: 10.1021/nn305789z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Zhao F, Lv ZP, Zheng CG, Zheng WD, Sun L, Wang NN, Pang S, de Andrade FM, Fu M, He XH, Hui J, Jiang W, Yang CY, Shi XH, Zhu XQ, Pang GF, Yang YG, Xie HQ, Zhang WD, et al. Association between APOC1 Polymorphism and Alzheimer’s Disease: A Case-Control Study and Meta-Analysis. PLoS One. 2014;9:e87017. doi: 10.1371/journal.pone.0087017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, et al. Candidate Single-Nucleotide Polymorphisms from a Genomewide Association Study of Alzheimer Disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 44.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, et al. Genome-Wide Association Analysis Reveals Putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–32. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. Designed Protein Pores as Components for Biosensors. Chem Biol. 1997;4:497–505. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 46.Cazenave C, Uhlenbeck OC. RNA Template-Directed RNA Synthesis by T7 RNA Polymerase. Proc Natl Acad Sci US A. 1994;91:6972–6. doi: 10.1073/pnas.91.15.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 49.Yoo J, Aksimentiev A. Improved Parametrization of Li+, Na+, K+, and Mg2+ Ions for All-Atom Molecular Dynamics Simulations of Nucleic Acid Systems. J Phys Chem Lett. 2012;3:45–50. [Google Scholar]
- 50.Weroński P, Jiang Y, Rasmussen S. Molecular Dynamics Study of Small PNA Molecules in Lipid-Water System. Biophys J. 92:3081–3091. doi: 10.1529/biophysj.106.097352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhattacharya S, Muzard J, Payet L, Mathé J, Bockelmann U, Aksimentiev A, Viasnoff V. Rectification of the Current in α-Hemolysin Pore Depends on the Cation Type: The Alkali Series Probed by Molecular Dynamics Simulations and Experiments. J Phys Chem C. 2011;115:4255–4264. doi: 10.1021/jp111441p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattacharya S, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH, Aksimentiev A. Molecular Dynamics Study of MspA Arginine Mutants Predicts Slow DNA Translocations and Ion Current Blockades Indicative of DNA Sequence. ACS Nano. 2012;6:6960–8. doi: 10.1021/nn3019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aksimentiev A, Schulten K. Imaging Alpha-Hemolysin with Molecular Dynamics: Ionic Conductance, Osmotic Permeability, and the Electrostatic Potential Map. Biophys J. 2005;88:3745–61. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.