Abstract
The absent in melanoma 2 (AIM2) inflammasome plays an important role in many viral and bacterial infections, but very little is known about its role in RNA virus infection, including influenza A virus (IAV). In this study, we have designed in vivo and in vitro studies to determine the role of AIM2 in infections with lethal doses of IAVs A/PR8/34 and A/California/07/09. In wild-type mice, IAV infection enhanced AIM2 expression, induced dsDNA release, and stimulated caspase-1 activation and release of cleaved IL-1β in the lung, which was significantly reduced in AIM2-deficient mice. Interestingly, AIM2 deficiency did not affect the transcription of caspase-1 and IL-1β. In addition, AIM2-deficient mice exhibited attenuated lung injury and significantly improved survival against IAV challenges, but did not alter viral burden in the lung. However, AIM2 deficiency did not seem to affect adaptive immune response against IAV infections. Furthermore, experiments with AIM2-specific small interfering RNA–treated and AIM2-deficient human and mouse lung alveolar macrophages and type II cells indicated a macrophage-specific function of AIM2 in regulation of IAV-stimulated proinflammatory response. Collectively, our results demonstrate that influenza infection activates the AIM2 inflammasome, which plays a critical role in IAV-induced lung injury and mortality. AIM2 might serve as a therapeutic target for combating influenza-associated morbidity and mortality without compromising the host antiviral responses.
Introduction
The inflammasome is a multiprotein complex that activates caspase-1 (Casp1) and results in cleavage of IL-1β (1, 2). Different from other host defense mechanisms, the inflammasome uses intracellular pattern recognition receptors to sense pathogen and danger-associated molecular patterns to guard the host. Absent in melanoma 2 (AIM2), a member of the pyrin and HIN200 domain-containing protein family (3–5), is an intracellular pattern recognition receptor that can form an inflammasome by directly binding to dsDNA from virus, bacteria, or the host itself (5–7). The role of the AIM2 inflammasome has been reported in many viral infections including murine CMV, vaccinia, and HSV (7–11), and is critical in defense against the cytosolic bacterium Francisella and Listeria monocytogenes and Mycobacterium tuberculosis (12–14). To date, very little is known about whether the AIM2 inflammasome is activated during RNA virus infections, including influenza A virus (IAV), which can cause life-threatening diseases, especially in high-risk groups.
IAV, an ssRNA virus, is one of the most important pathogens for seasonal and pandemic respiratory illness. In severe cases of lower respiratory tract infection, IAV infects lung epithelial cells and macrophages, and causes diffuse alveolar damage and excessive inflammatory responses (12, 15, 16). By analyzing our previously published microarray data (17, 18), we found that IAV significantly increased expression of AIM2 in both human primary alveolar type II (ATII) cells and alveolar macrophages (AMs), the important targets for influenza infection (12, 15, 16). This led us to hypothesize that AIM2 might participate in regulating the influenza-induced proinflammatory response. In this study, we sought to determine the role of AIM2 in influenza-induced disease using in vitro human and mouse lung primary cells in combination with an in vivo mouse model of influenza A infections.
Our results indicate that IAVs A/PR8/34 (PR8, a widely used mouse adaptive strain) and A/California/07/09 (CA07, a clinical isolate) activate the AIM2-dependent inflammasome. AIM2 is critical for virus-induced Casp1 activation and cleavage and release of IL-1β from the lung, but not for virus-stimulated increase in transcription of these two genes. Deficiency in AIM2 leads to attenuated lung injury and inflammation, and significantly improves survival following lethal IAV infections. AIM2 deficiency appears to be dispensable for host antiviral defense and shaping the adaptive immune response. In addition, AIM2 plays a proinflammatory role specifically in human and mouse AMs, but not ATII cells. Our results suggest that the function of AIM2 is focused on the innate immune response, and AIM2 is a detrimental host factor for influenza-induced lung injury and mortality.
Materials and Methods
Human lung donors
Deidentified patient lungs that were not suitable for transplantation and donated for medical research were obtained through the International Institute for the Advancement of Medicine (Edison, NJ) and the Airway Epithelial Core at the University of Pittsburgh as we described previously (17, 19). The Committee for Oversight of Research and Clinical Training Involving Decedents and University of Pittsburgh Institutional Review Board approved use of the human tissues. The donors used in this study included eight male and eight female donors with average age of 50.2 years; there were seven current smokers, one ex-smoker, and eight nonsmokers.
Mice
AIM2 knockout (−/−), NLRP3−/−, and wild-type (WT) C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME); ASC−/− mice were from Genentech (San Francisco, CA). All mice were bred in-house, and AIM2−/− mice were further backcrossed with C57BL/6J mice for two more generations. Mice were maintained under pathogen-free conditions within the animal facilities at the Children’s Hospital of Pittsburgh of University of Pittsburgh Medical Center. All animal studies were performed on age- and sex-matched mice and conducted with approval from the University of Pittsburgh Institutional Animal Care and Use Committee.
Viruses
Influenza PR8, a laboratory-adapted H1N1 virus, was originally provided by Dr. K. Hartshorn from Boston University and CA07, a 2009 pandemic H1N1 virus, was provided by Dr. T. Ross at University of Georgia. Both viruses were propagated in MDCK cells as previously described (19); the concentration of viruses were titrated using plaque assay as previously described (17, 19). In addition, LD50 was determined according to Reed and Muench’s method (20). For PR8, 1LD50 equals 80 PFU; for CA07, 1LD50 equals 10,000 PFU.
Infection of primary human and murine ATII cells and AMs with IAVs
Human ATII cells and AMs were isolated from deidentified donor lungs and cultured as we described previously (17, 19). Mouse ATII cells from age- and sex-matched WT and AIM2−/− mice were isolated as previously described (21) and plated in DMEM with 5% rat serum in 37°C incubator with 5% CO2 for 2 d, and then cultured with 1% charcoal-stripped FBS plus 10 ng/ml keratinocyte growth factor (R&D Systems, Minneapolis, MN) for 4 d preinfection. For mouse AM isolation, lungs from WT and AIM2−/− mice were lavaged with 1 ml of sterile saline 13 times; AMs were collected by centrifuging the bronchoalveolar lavage (BAL) fluid (BALF) at 1000 rpm at 4°C for 10 min and were cultured in DMEM with 5% rat serum overnight before viral infection.
Prior to infection, human and mouse cells were washed once with DMEM and then infected with PR8 or CA07 at multiplicity of infection (moi) of 1 for 1 h at 37°C. Cells were washed postinfection and fresh media were added to the culture. At the designated time points postinfection, cells were harvested for evaluation of gene expression, viral replication, and cytokine secretion.
AIM2 small interfering RNA treatment of human ATII cells and AMs
For knockdown of AIM2 gene expression, 50 nM small interfering RNA (siRNA) against human AIM2 or nontarget control (Dharmacon, Lafayette, CO) was transfected into human ATII cells and AMs using GenomONE HVJ Envelope Vector Kit (Cosmo Bio, Carlsbad, CA) as we described previously (22). Cells were allowed to express transgene for 48 h before influenza infection, and AIM2 expression level was confirmed by real-time RT-PCR with GAPDH normalization.
Mouse influenza infection
Prior to infection, WT and age- and sex-matched AIM2−/−, NLRP3−/−, or ASC−/− mice were cohoused (for female) or exchanged bedding (for male) for >1 wk. Mice were anesthetized with isoflurane and challenged with different doses of PR8 or CA07 in 50 μl of sterile DMEM or DMEM control intranasally or intratracheally. In designated experiments, AIM2−/− and their littermate control mice were infected with PR8 or CA07. Mice were monitored daily postinfection for weight loss and signs of clinical illness and were harvested at the indicated time points.
BAL and lung tissue processing
At the indicated time points, mice were euthanized by i.p. injection of sodium pentobarbital (Henry Schein Animal Health, Lake Forest, IL). The lungs were lavaged with 1 ml of sterile saline solution, and BALF was collected and centrifuged at 4°C, 3000 rpm for 10 min. An aliquot of 100 μl of cell-free BALF was snap frozen by dry ice-ethanol bath for evaluation of viral burden by plaque assay. Another aliquot of 200 μl of cell-free BALF was treated with protease inhibitors (Fisher Scientific, Waltham, MA) and stored at −80°C for analysis of protein expression by Western blotting. The rest of the BALF was stored at −80°C for detection of albumin, lactate dehydrogenase (LDH), and cytokine by ELISA. BAL cell cytospin slides were stained with a HEMA-3 stain kit (Fisher Scientific) for inflammatory cell differential counts. Right superior, middle, and inferior lobes were collected and homogenized in 1 ml of sterile ice-cold PBS at 4°C using gentleMACS Dissociator (Miltenyi Biotec, San Diego, CA). Tissue-free lung homogenates were collected and stored at −80°C for use in plaque assay and Western blotting as described earlier. The postcaval lobe was saved for RNA assay. Left lobe was fixed with 10% neutral buffered formalin (EMD Millipore, Billerica, MA) and subjected to subsequent H&E staining.
Real-time RT-PCR
Lung tissue RNA was extracted from the postcaval lobe using TRIzol reagent (Invitrogen, Carlsbad, CA), and cellular RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany). One microgram of RNA was used as a template to generate cDNA using qScript cDNA Synthesis kit (Quanta Bioscience, Gaithersburg, MD). cDNA was then used in standard real-time PCR to measure gene expression using the Applied Biosystems 7900HT (Life Technologies, Carlsbad, CA). Reaction conditions were 95°C for 15 s and 60°C for 1 min, repeated for 40 cycles, with a 10-min hot start at 95°C. Relative mRNA level was quantified using the 2ΔCt method and standardized to the level of GAPDH. The TaqMan real-time PCR probes (Thermo Fisher Scientific, Waltham, MA) used in this study are human AIM2 (Hs00915710_m1), IL-1β (Hs00174097_m1), and GAPDH (Hs02758991_g1), as well as mouse AIM2 (Mm01295719_m1), IL-1β (Mm00434228_m1), and GAPDH (Mm99999915_g1).
Western blotting
Lung tissue and BAL proteins were quantified using BCA protein assay kit (Bio-Rad, Hercules, CA). The primary Abs used in this study were rabbit anti-Casp1 (ab108362; Abcam, Cambridge, MA), mouse anti-Casp1 (MAB6215; R&D Systems), rabbit anti-IL-1β (ab9722; Abcam), mouse anti–IL-1β (12242S; Cell Signaling, Danvers, MA), rabbit anti–β-actin (A2066; Sigma-Aldrich, St. Louis, MO), and goat anti-GAPDH (SAB2500451; Sigma). For detection of Casp1 and IL-1β abundance, 30 μg of lung homogenates or 5 μg of BAL protein per sample was loaded and separated by 4–15% Mini-PROTEAN TGX Precast Gels (Bio-Rad) and further transferred to PVDF membranes (Bio-Rad). Blots were then probed with primary Abs against IL-1β or Casp1 and subsequently probed with HRP-conjugated secondary Abs (Jackson ImmunoResearch Laboratories, West Grove, PA). In the end, membranes were developed using ECL Western blotting Substrate (Pierce, Junction City, OH), and images were captured with Fujifilm LAS-3000 (Fujifilm Life Science, Stanford, CA) and quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
Analysis of dsDNA in cell-free BALF
The amount of dsDNA in cell-free BALF from uninfected and infected mice was measured using the QuantiFluor dsDNA kit (Promega, Madison, WI) following manufacturer’s instructions. The DNA pattern was further evaluated on Agilent High Sensitivity D1000 ScreenTape (Agilent, Santa Clara, CA).
Detection of influenza-specific IgG in mouse serum
ELISA was used to detect influenza virus–specific IgG in mouse serum samples as described previously (23). In brief, formalin-inactivated PR8 was used as Ag to coat Nunc 96-well plate (eBioscience) at 4°C overnight. After washing and blocking with BSA, the plate was incubated with 1:2 serially diluted serum samples from uninfected and infected mice. Anti-influenza IgG was detected using HRP-conjugated goat anti-mouse Ab (Jackson ImmunoResearch Laboratories) and developed using TMB Substrate Set (R&D Systems). The OD was read at wavelength 450 and 470 nm (as reference). The ratio of OD from infected versus uninfected samples was calculated for each infected sample. If the ratio was >2.1, the sample was designated as “positive” for influenza-specific IgG, and the dilution factor of this sample was the IgG titer.
Flow cytometry analyses of infected lung tissues
Flow cytometric analysis was performed as previously described (24). Single-cell suspension of homogenized mouse lungs was stained with Abs against CD45 (45-0451-80), CD8 (25-0081-81), CD4 (48-0042-80) (Thermo Fisher Scientific), and TCRβ (560656; BD Biosciences, San Jose, CA). Matched isotype control Abs were used to define specific staining above autofluorescence. Data were acquired using FACS LSR II (BD Biosciences) and analyzed by FlowJo software. Percentage of blood mononuclear cells (CD45+), CD4+ T cells (CD45+TCRβ+/CD4+), and CD8+ T cells (CD45+TCRβ+/CD8+) was determined.
Statistical analysis
All data analyses were performed with Prism 6.0 (GraphPad, La Jolla, CA). Kaplan–Meier survival curve was produced and log-rank (Mantel–Cox) test was used to compare difference between two groups. Mann–Whitney U test was used for comparison of gene expression and viral replication between two groups. For other comparisons between two groups, two-tailed Student t test was used.
Results
To determine the role of AIM2 in influenza infection, we first infected primary human ATII cells and AMs from six donors with IAV PR8 and measured expression of AIM2 at 24 h postinfection (hpi) by real-time RT-PCR. The results shown in Fig. 1A confirmed the findings from our previous microarray experiments that influenza infection significantly increased expression of AIM2 in both cell types (17, 18). Compared with ATII cells, the basal expression level of AIM2 is significantly higher in human AMs (Fig. 1A), but the influenza-induced increase is less than that in ATII cells. To analyze whether the findings from human primary cells in vitro represent infection in vivo, we examined the expression of AIM2 and IL-1β in PR8-infected mouse lung tissues. Consistent with the results from human lung primary cells, viral infection significantly increased AIM2 expression in murine lungs (Fig. 1B).
FIGURE 1.
Influenza A infection increases AIM2 and stimulates dsDNA release into BAL. (A) Cultured human primary ATII cells and AMs from six donors were infected with influenza virus PR8 or MOCK control (Ctrl) at moi of 1, and RNA was extracted at 24 hpi for evaluation of AIM2 expression by real-time PCR (17, 18). Data show the relative expression of AIM2 and IL-1β mRNA normalized to the expression of human housekeeping gene GAPDH. (B–D) WT and AIM2−/− mice were challenged with 50LD50 of PR8 virus or DMEM control (Ctrl) intranasally. Lung tissues (B) were harvested at 3 dpi for evaluation of AIM2 and IL-1β expression by real-time RT-PCR. Data illustrate the relative expression of AIM2 normalized to mouse GAPDH. (C and D) Cell-free BALF was collected at 1 and 3 dpi for quantification of dsDNA (C) using QuantiFluor dsDNA kit (Promega) and analysis of DNA pattern (D) on Agilent High Sensitivity D1000 ScreenTape (Agilent). *p < 0.05, **p < 0.01, compared with the Ctrl group.
We then evaluated whether influenza infection induces dsDNA release from host because AIM2 has been demonstrated as a critical DNA sensor in host defense. We infected both WT and AIM2−/− mice with PR8, collected BALF, and determined the level of dsDNA using QuantiFluor dsDNA kit (Promega). Fig. 1C confirmed the presence of dsDNA in cell-free BALF as early as 1 day postinfection (dpi) in both WT and AIM2−/− mice, and the amount of dsDNA was further increased at 3 dpi. No significant difference in dsDNA release was detected between WT and AIM2−/− groups. We further analyzed the DNA pattern from BALF samples. As shown in representative Fig. 1D, most of the DNA fragments peaked at the size around 173 bp, which satisfies the minimum sequence length of 80 bp required for activation of the AIM2 inflammasome (11).
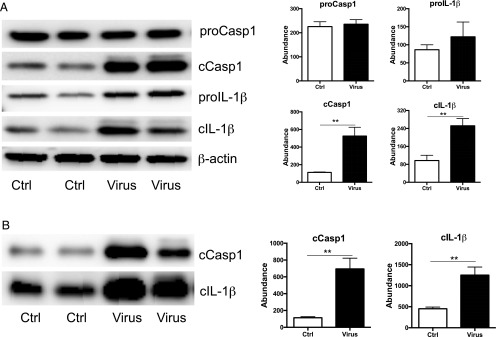
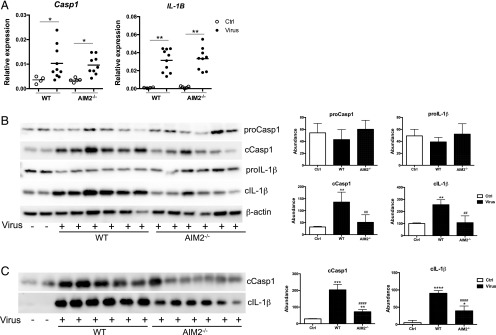
To test whether there is functional inflammasome activation in influenza-infected mouse lungs, we examined the abundance of pro- and cleaved forms of Casp1 and IL-1β in BAL and lung homogenates from uninfected and PR8-infected mice by Western blotting (Fig. 2). Both pro- and cleaved forms of Casp1 and IL-1β were present in uninfected and infected lung tissues (Fig. 2A), whereas only cleaved Casp1 (cCasp1) and IL-1β were detected in BAL samples (Fig. 2B). Viral infection significantly increased the abundance of cCasp1 and cleaved IL-1β (cIL-1β) in BAL and lung tissues but did not increase the levels of pro-Casp1 and pro–IL-1β (Fig. 2). These results indicate that influenza infection activates Casp1 and promotes processing of IL-1β. To further determine whether AIM2 is required for inflammasome activation, we performed additional experiments to compare the levels of Casp1 and IL-1β mRNA and proteins between virus-infected WT and AIM2-deficient mice (Fig. 3). AIM2 deficiency did not alter virus-stimulated increase of the transcription of Casp1 and IL-1β (Fig. 3A). However, AIM2-deficient mice displayed significantly less abundance of cCasp1 and cIL-1β in lung tissues (Fig. 3B) and BAL (Fig. 3C) than WT mice. No difference was present in proforms of these two proteins between the two groups. Further quantification of Western blot signal intensity indicated that AIM2 deficiency reduced around 60% of cIL-1β and cCasp1 (Fig. 3). We further validated these results in additional experiments with AIM2 littermate controls. As shown in Supplemental Fig. 1, PR8-infected AIM2 homozygous knockouts, heterozygous littermates, and WT littermate control mice showed similar levels of pro-Casp1 and pro–IL-1β. However, significant reduction in cCasp1 and cIL-1β was present in AIM2 homozygous knockouts compared with WT controls. The level of cCasp1 and cIL-1β in AIM2 heterozygous tended to be higher than homozygous knockouts, but lower than WT controls, although the difference was not statistically significant due to the variation (Supplemental Fig. 1), suggesting a possible gene dosage effect. These results clearly demonstrate that AIM2 is critical for influenza-stimulated activation of Casp1 and processing of IL-1β independent of transcriptional regulation.
FIGURE 2.
Influenza infection activates the inflammasome. Six-week-old C57B/6 mice were infected with PR8 (50LD50) or same volume of DMEM (Ctrl) intranasally. Mice were harvested at 3 dpi, and 5 μg cell-free BALF or 30 μg of lung tissue was analyzed for the expression of Casp1 and IL-1β by Western blotting. (A) Lung homogenate. (B) BAL. Representative images of Western blotting (left panel). Quantification of Western blotting results using ImageJ software (National Institutes of Health) (right panel). Protein abundance was shown as relative abundance to the level of β-actin in the same sample. n = 4. **p < 0.01, compared with the Ctrl group.
FIGURE 3.
AIM2 deficiency reduces PR8-induced activation of the inflammasome. Sex- and age-matched WT and AIM2−/− mice were challenged with PR8 (50LD50) or DMEM (Ctrl) intranasally. Mice were harvested at 3 dpi for evaluation of expression of Casp1 and IL-1β gene and proteins by real-time RT-PCR and Western blotting, respectively. (A) Gene expression. (B and C) Protein expression. (Left panels) Representative images of Western blotting of lung homogenates (B) and BAL samples (C). (Right panels) Quantification of Western blotting results. n = 6. *p < 0.05, **p < 0.01, compared with the Ctrl group; ##p < 0.01, ####p <0.0001, between AIM2−/− and WT groups.
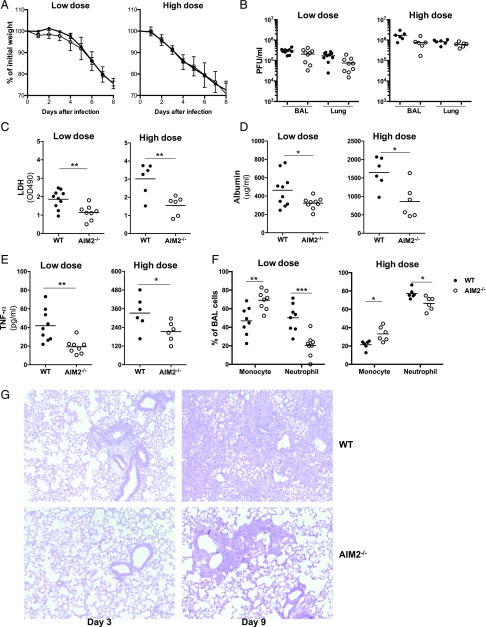
To further determine the role of AIM2 in influenza pathogenesis, we challenged WT and AIM2−/− mice with low (0.5LD50) and high doses (50LD50) of PR8 virus and evaluated the weight loss, viral replication, and lung damage (Fig. 4). Both WT and AIM2−/− mice showed significant weight loss and infectious virus release with viral infections, but no difference was detected between WT and AIM2−/− groups (Fig. 4A, 4B). To test whether AIM2 deficiency affects influenza virus-induced acute lung injury, we examined the lung barrier function and inflammation at 3 dpi (Fig. 4C–F). BAL samples from PR8-infected AIM2−/− mice exhibited a significantly lower amount of LDH (Fig. 4C) and albumin (Fig. 4D), decreased TNF-α (Fig. 4E), and reduced proportion of neutrophils and increased proportion of macrophages (Fig. 4F) indicating attenuated lung injury and inflammation when compared with the samples from WT mice. As expected, high-dose infection caused more weight loss, higher viral burden, and more lung injury and inflammation than low-dose infection (Fig. 4A–F). Despite the significant weight loss, viral burden, and obvious indication of lung injury (Fig. 4A–F), PR8-infected mice did not show significant tissue damage as represented in H&E staining of lung sections from WT or AIM2-deficient mice at low dose of infection (data not shown). At early stage of high-dose infection (day 3) mild inflammatory cell infiltration around bronchus and edema around blood vessels were observed in WT, but not in AIM2−/− mice (Fig. 4G). At 9 dpi, when the viral burden had been almost cleared (data not shown), infected WT mouse lungs displayed significant lung destruction and inflammatory cell infiltration in parenchyma, whereas the cell infiltration was mainly limited in parabronchi in AIM2-deficient mice (Fig. 4G). These results indicate that AIM2 deficiency attenuates influenza-induced lung damage.
FIGURE 4.
AIM2-deficient mice display less lung injury and inflammation, but no difference in viral burden and weight loss compared with WT mice. Sex- and age-matched WT and AIM2−/− mice were challenged with low (0.5LD50) or high (50LD50) dose of PR8 virus intranasally. Mice weight (A) was monitored daily (n = 10 for low-dose infection and n = 6 for high-dose infection), and viral replication (B) lung tissue and BALF were collected at 3 dpi for assays of viral burden (B), LDH (C), albumin (D), release of TNF-α by ELISA (E), and BAL cell distribution by Hema-3 staining of BAL cytospin slides (F). Mouse pathology (G) was examined by H&E staining of fixed lung sections at 3 and 9 dpi. Representative images of H&E slides (original magnification ×100) are shown. *p < 0.05, **p < 0.01, ***p < 0.001, between WT and AIM2−/− groups.
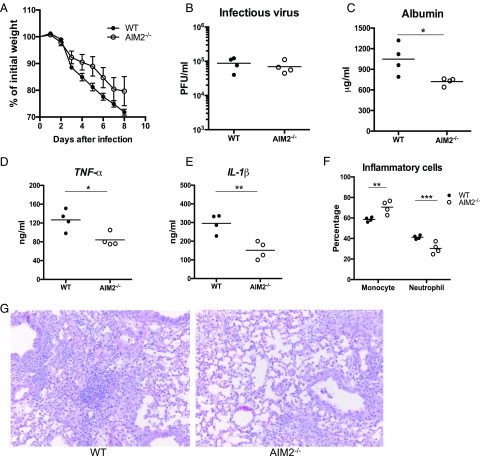
To further validate the findings with contemporary IAV, we examined viral infections in WT and AIM2−/− mice with pandemic influenza CA07 (Figs. 5, 6). Consistent with the findings from PR8 infection, CA07 infection led to inflammasome activation, and AIM2 deficiency significantly reduced cleavage of Casp1 and processing of IL-1β (Fig. 5), but did not show significant changes in weight loss or viral replication when compared with WT mice (Fig. 6A, 6B). Also, there was less albumin (Fig. 6C), TNF-α and IL-1β (Fig. 6D, 6E), and neutrophils (Fig. 6F) present in the BAL of infected AIM2−/− mice than WT mice at 3 dpi. H&E staining of fixed AIM2−/− lung sections exhibited significantly less inflam-matory cell infiltration and tissue damage than WT lungs at 9 dpi (Fig. 6G). These data support the results observed with PR8 infection that AIM2 deficiency attenuates inflammasome activation and lung injury and inflammation after influenza A infection, suggesting this is not a viral strain-specific phenotype.
FIGURE 5.
AIM2 deficiency attenuates contemporary IAV-induced inflammasome activation. Sex- and age-matched WT and AIM2−/− mice were challenged with influenza CA07 (50LD50) or DMEM (Ctrl) intranasally. Mice were harvested at 3 dpi for evaluation of the abundance of Casp1 and IL-1β by Western blotting. (A) Lung homogenate. (B) BAL. Representative images of Western blotting (left panel). Quantification of Western blotting signal intensity (right panels). n = 4. *p < 0.05, **p < 0.01, compared with the Ctrl group; #p < 0.05, ##p < 0.01, between AIM2−/− and WT groups.
FIGURE 6.
AIM2 deficiency reduces contemporary IAV-induced lung injury and inflammation but does not alter viral burden and weight loss. Sex- and age-matched WT and AIM2−/− mice were challenged with CA07 (50LD50) intranasally. Mice weight (A) was monitored daily (n = 8 per group), and lung tissue and BAL fluid were collected at 3 dpi for assays of viral burden (B), albumin (C), release of TNF-α (D) and IL-1β (E), and BAL cell distribution (F). Mouse pathology (G) was examined at 9 dpi. Representative images of H&E-stained lung sections (original magnification ×200) are shown. This experiment was repeated once. *p < 0.05, **p < 0.01, ***p < 0.001, between WT and AIM2−/− groups.
To examine whether AIM2 deficiency alters adaptive immune response against IAV infection, we evaluated the flu-specific IgG level in serum samples of survivors from low-dose PR8 infection by ELISA (Supplemental Fig. 2). All samples were positive for influenza-specific serum IgG at 2 wk postinfection, but no difference was present between AIM2−/− and WT groups (Supplemental Fig. 2A). At 8 wk postinfection, the serum IgG titer dropped, but still no difference was shown between the two groups (Supplemental Fig. 2B). With CA07 infection, we examined the percentage of CD4+ and CD8+ T lymphocytes in infected lungs at 9 dpi by flow analysis (Supplemental Fig. 2C). AIM2-deficient mice displayed similar amounts of lung CD4+ and CD8+ cells as WT mice. These data suggest that AIM2 deficiency may not affect the adaptive immune response against IAV infections.
To further examine whether AIM2 deficiency provides protection against influenza-induced mortality, we compared mouse survival between WT and AIM2−/− groups with lethal doses of PR8 and CA07 challenge. The results in Fig. 7 show that AIM2-deficient mice display steadily improved survival compared with WT mice after different doses and strains of IAV challenge. The survival rate in AIM2−/− mice doubled with the low dose of infection (0.5LD50) and increased to 6-fold with the high dose of PR8 infection (50LD50) at the end of observation (Fig. 7A). These results have been confirmed in experiments with AIM2 littermate controls (Fig. 7B, left). Both AIM2 knockout homozygous and heterozygous mice showed significantly prolonged survival compared with littermate WT controls post PR8 infection. With CA07 infection, AIM2-deficient mice also showed significantly improved survival compared with WT mice (Fig. 7B, right). These results demonstrate that AIM2 deficiency provides protection against IAV-associated mortality.
FIGURE 7.
AIM2 deficiency conveys protection against IAV-induced mortality. (A) Sex- and age-matched WT and AIM2−/− mice were infected with PR8 intranasally. (B) AIM2−/− and littermate AIM2+/− and AIM2+/+ mice were infected with PR8 (100LD50 intratracheally) or CA07 (50LD50 intranasally). Mice survival was monitored daily. The low-dose and high-dose survival experiment (A) was repeated once. *p < 0.05, **p < 0.01, between AIM2−/− and WT groups.
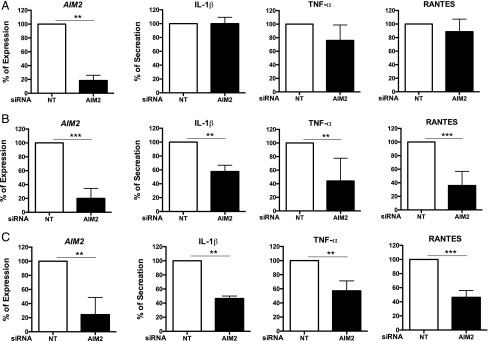
Data in Fig. 1 indicate that influenza infection increases AIM2 expression in primary human AMs and ATII cells, and AMs express a higher level of AIM2 than ATII. To test whether AMs are the primary targets for AIM2 function in the lung, we performed additional in vitro experiments in primary murine and human lung cells (Figs. 8, 9). We infected ATII cells and AMs isolated from WT and AIM2−/− groups, and compared cellular expression of IL-1β gene and release of IL-1β in culture supernatants. Representative results in Fig. 8 show that viral infection induced a similar level of IL-1β mRNA (Fig. 8A) and protein release (Fig. 8B) in both WT and AIM2-deficient ATII cells. However, IL-1β production was impaired in AMs from the AIM2-deficient group. We further validated the cell-specific function of AIM2 in human ATII cells and AMs (Fig. 9) treated with nontarget or AIM2-specific siRNA (siAIM2). Consistent with the results from AIM2-deficient mouse cells (Fig. 8A), siAIM2 did not affect the proinflammatory response in PR8-infected human ATII cells despite an 80% reduction in AIM2 expression achieved by the treatment in five tested donors (Fig. 9A). siAIM2 did not alter viral replication in these cells either (data not shown). However, siAIM2 significantly reduced PR8-stimulated IL-1β, TNF-α, and RANTES release from human AMs (Fig. 9B). Similar results were observed with CA07 infection that siAIM2 treatment significantly reduced IL-1β, TNF-α, and RANTES secretion (Fig. 9C) but did not alter IL-1β gene expression (data not shown) in human AMs. These results strongly support a macrophage-specific role for AIM2 in regulating influenza-stimulated proinflammatory response.
FIGURE 8.
Ams, but not ATII cells, from AIM2-deficient mice exhibit an impairment in IL-1β response post IAV infection. ATII cells and AMs isolated from WT and AIM2−/− mice were infected with PR8 at an moi of 1. At 24 hpi, RNA was harvested from cells for evaluation of IL-1β gene expression by real-time RT-PCR, and culture supernatants were collected for detection of IL-1β using Bio-Plex IL-1β singleplex set (BioRad) following manufacturer’s protocol. (A) ATII cells. (B) AMs. Data represent results from one of three experiments.
FIGURE 9.
siAIM2 treatment significantly decreases AIM2 expression and release of proinflammatory cytokines from AMs after IAV infections. Primary human ATII cells and AMs were treated with siAIM2 or nontarget control siRNA (NT) 2 d before infections with PR8 or CA07 (moi = 1). At 24 hpi, RNA was extracted for examining AIM2 gene expression, and culture supernatants were collected for detection of IL-1β, TNF-α, and RANTES using DuoSet ELISA kits from R&D Systems. (A) ATII cells with PR8 infection. n = 5. (B) AMs with PR8 infection. n = 5. (C) AMs with CA07 infection. n = 3. Data represent % expression or secretion to NT siRNA-treated condition. **p < 0.01, ***p < 0.001, between NT and AIM2 siRNA-treated conditions.
Discussion
AIM2 is critical for IAV-induced inflammasome activation
In this study, we have demonstrated that IAV activates the AIM2 inflammasome. Viral infection increases gene expression of AIM2 and IL-1β in both human and mouse lung primary cells (Figs. 1A, 8A) and mouse lungs (Fig. 1B), which constitutes the first signal, the “priming” signal, required for activation of the inflammasome (25, 26). Additionally, viral infection stimulates dsDNA release from infected WT and AIM2−/− mouse lungs (Fig. 1C), and the size of dsDNA fragments (Fig. 1D) meets the minimum requirement for the second signal, the “activation” signal, for the AIM2 inflammasome (3). Moreover, significantly increased cCasp1 and cIL-1β from the BAL and lung tissue of PR8- or CA07-infected mice (Figs. 2, 3, 5, Supplemental Fig. 1) provides direct evidence for inflammasome activation by IAV. AIM2-deficient mice display significantly reduced Casp1 activation and cleavage of IL-1β compared with WT mice post IAV infections (Figs. 3B, 5, Supplemental Fig. 1). These results indicate that AIM2 is critical for IAV-stimulated inflammasome activation. Interestingly, AIM2-deficient mice do not show differences in Casp1 and IL-1β transcription in the lung (Fig. 3A) and dsDNA release (Fig. 1C) from WT mice, suggesting that AIM2 inflammasome does not participate in transcriptional regulation of proinflammatory genes. There is no difference in levels of pro-Casp1 and pro–IL-1β in the lung or viral replication between WT and AIM2−/− groups. These results indicate that the significantly reduced release of cIL-1β into the BAL from the AIM2−/− group is independent of viral replication and likely the result of significantly decreased Casp1 activation and impaired processing of IL-1β precursor because of AIM2 deficiency (Figs. 3, 5, Supplemental Fig. 1).
We speculate that there are two possible mechanisms for the AIM2 inflammasome activation post influenza infection. One is that macrophages uptake the alveolar epithelial cells damaged by IAV (17, 22) and bring genomic dsDNA from epithelial cells into the cytosol of macrophages, which is recognized by intracellular AIM2. This hypothesis is supported by the fact that AMs from AIM2-deficient mice (Fig. 8B) and siAIM2-treated human AMs (Fig. 9B, 9C) produce much less IL-1β in response to IAV infection. Another possible mechanism is that IAV causes mitochondrial damage and release of mitochondrial DNA into the cytosol, which directly activates the AIM2 inflammasome. ATII cells and AMs are mitochondria-rich cells (27), and this may help to explain why influenza virus increases AIM2 expression in both primary human and mouse ATII cells and AMs (Figs. 1A, 8A). It will be interesting to determine how influenza infection causes mitochondrial damage (28, 29) in these cells and clarify the mechanism for activation of the AIM2 inflammasome in the future.
AIM2 deficiency provides protection against IAV-induced lung injury and mortality
As shown in Fig. 4, infection with PR8 causes dose-dependent weight loss, viral replication, and lung injury in mice. Infection with CA07 also leads to significant weight loss and lung injury (Figs. 6, 7). We believe that the significant lung damage and death is partly due to the inappropriate recognition of cytoplasmic self-DNA by AIM2; this promotes the cleavage of Casp1 and subsequent release of mature IL-1β (Figs. 2, 3, 5) and cytokine storm, which is often observed with severe influenza infections (30–32). IL-1β is a well-known master proinflammatory cytokine and can stimulate the secretion of many cytokines and chemokines, as well as the subsequent infiltration of immune cells into the infection site. In addition, IL-1β is a critical molecule for tissue injury and repair (33–35). Several studies have reported a detrimental role for IL-1β for severe influenza infections (36–38). We recently also reported that IL-1β treatment reduces the transepithelial electrical resistance of human primary alveolar epithelial cells and leads to epithelial barrier injury (19). In this study, accompanied by the striking reduction in inflammasome activation (Figs. 3, 5, Supplemental Fig. 1), AIM2-deficient mice exhibit decreased acute lung injury and tissue damage (Figs. 4C–G, 6C–G), and significantly improved survival post IAV infections (Fig. 7). Also, AIM2-deficient mouse and human AMs show less secretion of IL-1β, TNF-α, and RANTES stimulated by IAV (Figs. 8, 9). Together, these results demonstrate that AIM2 deficiency attenuates virus-stimulated proinflammatory response and results in prolonged survival against IAV infections. Similar to other inflammasome studies (39–41) from the literature, AIM2 deficiency apparently does not affect viral replication (Figs. 4B, 6B). AIM2 deficiency does not affect the expression of antiviral genes in the lung either (data not shown). In addition, inhibition of AIM2 expression by siAIM2 does not change viral replication in primary human ATII cells (data not shown), the main viral replication site (42, 43) in vivo, for both laboratory-adapted and contemporary IAVs. Collectively, these results indicate that AIM2 deficiency conveys protection to influenza-infected mice, and this protection likely results from attenuation of proinflammatory response through impaired inflammasome activation. Targeting AIM2 might be an effective approach to limit IAV-induced mortality or morbidity without compromising the host antiviral responses.
We notice that there is still partial release of cIL-1β and cleavage of Casp1 in PR8- or CA07-infected AIM2−/− mice (Figs. 3, 5, Supplemental Fig. 1). This might be because of the activation of other inflammasomes such as the NLRP3 inflammasome (6, 9, 23, 39–41, 44), the well-studied inflammasome in influenza infection. AIM2 deficiency did not alter the expression of NLRP3 or ASC, the common adaptor for AIM2 and NLRP3 inflammasomes, in infected lung tissue or primary cells (data not shown), suggesting that virus could still activate the NLRP3 and other inflammasomes in the absence of AIM2. We examined the survival of ASC and NLRP3 knockout mice after lethal doses of PR8 or CA07 infection (Supplemental Fig. 3). We did not evaluate Casp1 knockout mice because the commercial Casp1-deficient mice also exhibit a deficiency in Casp11 (http://www.jax.org). Consistent with the reports from other groups (23, 40), we confirmed a protective role for ASC in influenza-associated mortality. ASC-deficient mice display further reduced survival against PR8 and CA07 infections (Supplemental Fig. 3, top). However, NLRP3-deficient mice do not exhibit different survival when compared with WT mice (Supplemental Fig. 3, bottom). There are controversial reports in the literature about the role of NLRP3 in influenza infection. Some studies report a positive role for NLRP3 in host defense against influenza infection. NLRP3-deficient mice have been shown to have worsened mortality and delayed viral clearance (40, 41). Our results are consistent with the results from Iwasaki’s group (23) that NLRP3 deficiency does not alter mouse susceptibility to influenza-induced mortality. Also, a recent study using NLRP3 inhibitor reported that NLRP3 plays a protective role in the early stage of influenza infection, but a detrimental role in the late stage through increased cytokine production and lung cellular infiltrates (39). The NLRP3 inflammasome can be activated by many conditions including a variety of pathogen- and damage-associated molecular patterns and different stresses (45–47). Therefore, a difference in viral dose, virus production, course of infection, animal facility, and the sex of mice could lead to different results. The clarification of NLRP3 on influenza infection is beyond the scope of this study and requires carefully designed future experiments.
AMs are the likely target cells for AIM2 function in regulating inflammation
Consistent with findings with NLRP3 inflammasome (23, 48), AIM2 deficiency seems to be dispensable for regulating the adaptive immune response in influenza infection. AIM2-deficient mice display similar levels of influenza-specific IgG and similar amounts of CD4+ and CD8+ T cell influx into the lung (Supplemental Fig. 2) post IAV infections. These results suggest that AIM2 mainly functions in innate immunity against influenza infection. Both ATII cells and macrophages are critically important sentinel cells in lung innate immunity. In an effort to determine which target cells are critical for AIM2 function, ATII or AM, we examined the effect of AIM2 knockdown on IAV-stimulated proinflammatory response (Figs. 8, 9). Despite the fact that IAV increases AIM2 expression in both cell types (Fig. 1A), siAIM2-treated human AMs, but not ATII cells, exhibit significantly reduced release of the proinflammatory cytokine IL-1β, TNF-α, and RANTES (Fig. 9). These results are consistent with the findings with primary lung cells from AIM2-deficient mice (Fig. 8), in which AIM2-deficient AMs fail to produce IL-1β in response to IAV infection. Although in vitro inflammasome studies have shown that influenza infection can activate inflammasome in respiratory epithelial cells (23, 49, 50) and immune cells (51–53), our results reveal a central role for AMs in AIM2-related inflammasome regulation.
Interestingly, our results are partially in conflict with the recent report of an anti-inflammatory function for AIM2 (54). Although both studies have shown that PR8 infection induces dsDNA release into the BAL, the published study reported that AIM2-deficient mice displayed decreased survival compared with WT mice, and AIM2 played an anti-inflammatory role. However, our study indicates a proinflammatory role for AIM2 in influenza infection. We noticed several significant differences in the two studies. First, in our study, we used in vitro primary cell culture as well as an in vivo mouse model of IAV infections, whereas the other study was focused solely on a mouse study. Second, the two studies used a different source of PR8 virus, and viral doses seem significantly different. The published study used PR8 produced in eggs, whereas our PR8 virus was propagated from mammalian cells. Our PR8 study is mainly lethal dose infections, whereas the other study seems to be sublethal dose infection, although they used 40,000 PFU of virus (54), reflecting variable infectivity of viruses propagated from different sources. When we performed the survival experiment using the 40,000 PFU of PR8 propagated in mammalian cells, all mice died, but AIM2−/− mice survived longer than WT mice in response to PR8 or CA07 infection (p < 0.01). We believe the apparent discrepancy between the two studies is mainly due to the different doses, which may cause a different immune response. Also, the different animal facility may affect the microbiome of the animals and result in different host innate and adaptive responses against influenza (55, 56). Our primary cell system data (Figs. 8, 9) support the proinflammatory role of AIM2 in influenza infection in the absence of microbiome effects.
In summary, we have performed in vitro and in vivo studies using laboratory and contemporary strains of IAVs to determine the role of AIM2 in influenza infection. The results from this study demonstrate that IAV activates the AIM2 inflammasome, which is critical for influenza-induced acute lung injury and mortality. Our results revealed that AIM2 has the potential to serve as a therapeutic target for limiting influenza-associated mortality and morbidity.
Supplementary Material
Acknowledgments
We thank William Horne, Kevin J. McHugh, Jennifer Profozich, and Ye Liu for technical help.
This work was supported by National Institutes of Health Grants R03AI101953 and R01HL113655, startup funding from the University of Pittsburgh (to J.W.), and the Cystic Fibrosis Foundation Research Development Program.
The online version of this article contains supplemental material.
- AIM2
- absent in melanoma 2
- AM
- alveolar macrophage
- ATII
- alveolar type II
- BAL
- bronchoalveolar lavage
- BALF
- BAL fluid
- CA07
- A/California/07/09
- Casp1
- caspase-1
- cCasp1
- cleaved Casp1
- cIL-1β
- cleaved IL-1β
- dpi
- day postinfection
- hpi
- hour postinfection
- IAV
- influenza A virus
- LDH
- lactate dehydrogenase
- moi
- multiplicity of infection
- PR8
- A/PR8/34
- siAIM2
- AIM2-specific siRNA
- siRNA
- small interfering RNA
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Lamkanfi M., Dixit V. M. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 227: 95–105. [DOI] [PubMed] [Google Scholar]
- 2.Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. 2009. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L., Superti-Furga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10: 266–272. [DOI] [PubMed] [Google Scholar]
- 4.Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinon F. 2012. Dangerous liaisons: mitochondrial DNA meets the NLRP3 inflammasome. Immunity 36: 313–315. [DOI] [PubMed] [Google Scholar]
- 7.Rathinam V. A., Jiang Z., Waggoner S. N., Sharma S., Cole L. E., Waggoner L., Vanaja S. K., Monks B. G., Ganesan S., Latz E., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strittmatter G. E., Sand J., Sauter M., Seyffert M., Steigerwald R., Fraefel C., Smola S., French L. E., Beer H. D. 2016. IFN-γ primes keratinocytes for HSV-1-induced inflammasome activation. J. Invest. Dermatol. 136: 610–620. [DOI] [PubMed] [Google Scholar]
- 9.Pang I. K., Iwasaki A. 2011. Inflammasomes as mediators of immunity against influenza virus. Trends Immunol. 32: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Man S. M., Karki R., Kanneganti T. D. 2016. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 46: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathinam V. A., Fitzgerald K. A. 2010. Inflammasomes and anti-viral immunity. J. Clin. Immunol. 30: 632–637. [DOI] [PubMed] [Google Scholar]
- 12.Shieh W. J., Blau D. M., Denison A. M., Deleon-Carnes M., Adem P., Bhatnagar J., Sumner J., Liu L., Patel M., Batten B., et al. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes-Alnemri T., Yu J. W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E., et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saiga H., Kitada S., Shimada Y., Kamiyama N., Okuyama M., Makino M., Yamamoto M., Takeda K. 2012. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int. Immunol. 24: 637–644. [DOI] [PubMed] [Google Scholar]
- 15.Wonderlich E. R., Swan Z. D., Bissel S. J., Hartman A. L., Carney J. P., O’Malley K. J., Obadan A. O., Santos J., Walker R., Sturgeon T. J., et al. 2017. Widespread virus replication in alveoli drives acute respiratory distress syndrome in aerosolized H5N1 influenza infection of macaques. J. Immunol. 198: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mauad T., Hajjar L. A., Callegari G. D., da Silva L. F., Schout D., Galas F. R., Alves V. A., Malheiros D. M., Auler J. O., Jr., Ferreira A. F., et al. 2010. Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181: 72–79. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Nikrad M. P., Phang T., Gao B., Alford T., Ito Y., Edeen K., Travanty E. A., Kosmider B., Hartshorn K., Mason R. J. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 45: 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Nikrad M. P., Travanty E. A., Zhou B., Phang T., Gao B., Alford T., Ito Y., Nahreini P., Hartshorn K., et al. 2012. Innate immune response of human alveolar macrophages during influenza A infection. PLoS One 7: e29879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travanty E., Zhou B., Zhang H., Di Y. P., Alcorn J. F., Wentworth D. E., Mason R., Wang J. 2015. Differential susceptibility of human lung primary cells to H1N1 influenza viruses. J. Virol. 89: 11935–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed L. J, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497. [Google Scholar]
- 21.Hidvegi T., Stolz D. B., Alcorn J. F., Yousem S. A., Wang J., Leme A. S., Houghton A. M., Hale P., Ewing M., Cai H., et al. 2015. Enhancing autophagy with drugs or lung-directed gene therapy reverses the pathological effects of respiratory epithelial cell proteinopathy. J. Biol. Chem. 290: 29742–29757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosmider B., Messier E. M., Janssen W. J., Nahreini P., Wang J., Hartshorn K. L., Mason R. J. 2012. Nrf2 protects human alveolar epithelial cells against injury induced by influenza A virus. Respir. Res. 13: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichinohe T., Lee H. K., Ogura Y., Flavell R., Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K., McAleer J. P., Lin Y., Paterson D. L., Zheng M., Alcorn J. F., Weaver C. T., Kolls J. K. 2011. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity 35: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAuley J. L., Tate M. D., MacKenzie-Kludas C. J., Pinar A., Zeng W., Stutz A., Latz E., Brown L. E., Mansell A. 2013. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog. 9: e1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opitz B., van Laak V., Eitel J., Suttorp N. 2010. Innate immune recognition in infectious and noninfectious diseases of the lung. Am. J. Respir. Crit. Care Med. 181: 1294–1309. [DOI] [PubMed] [Google Scholar]
- 27.Miller M. L., Andringa A., Hastings L. 1995. Relationships between the nuclear membrane, nuclear pore complexes, and organelles in the type II pneumocyte. Tissue Cell 27: 613–619. [DOI] [PubMed] [Google Scholar]
- 28.Tran A. T., Cortens J. P., Du Q., Wilkins J. A., Coombs K. M. 2013. Influenza virus induces apoptosis via BAD-mediated mitochondrial dysregulation. J. Virol. 87: 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichinohe T., Yamazaki T., Koshiba T., Yanagi Y. 2013. Mitochondrial protein mitofusin 2 is required for NLRP3 inflammasome activation after RNA virus infection. Proc. Natl. Acad. Sci. USA 110: 17963–17968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beigel J. H., Farrar J., Han A. M., Hayden F. G., Hyer R., de Jong M. D., Lochindarat S., Nguyen T. K., Nguyen T. H., Tran T. H., et al. Writing Committee of the World Health Organization (WHO) Consultation on Human Influenza A/H5 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353: 1374–1385. [DOI] [PubMed] [Google Scholar]
- 31.Peiris J. S., Cheung C. Y., Leung C. Y., Nicholls J. M. 2009. Innate immune responses to influenza A H5N1: friend or foe? Trends Immunol. 30: 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Short K. R., Kroeze E. J., Fouchier R. A., Kuiken T. 2014. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 14: 57–69. [DOI] [PubMed] [Google Scholar]
- 33.Capaldo C. T., Nusrat A. 2009. Cytokine regulation of tight junctions. Biochim. Biophys. Acta 1788: 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Sadi R. M., Ma T. Y. 2007. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J. Immunol. 178: 4641–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borthwick L. A., McIlroy E. I., Gorowiec M. R., Brodlie M., Johnson G. E., Ward C., Lordan J. L., Corris P. A., Kirby J. A., Fisher A. J. 2010. Inflammation and epithelial to mesenchymal transition in lung transplant recipients: role in dysregulated epithelial wound repair. Am. J. Transplant. 10: 498–509. [DOI] [PubMed] [Google Scholar]
- 36.Palomo J., Dietrich D., Martin P., Palmer G., Gabay C. 2015. The interleukin (IL)-1 cytokine family--balance between agonists and antagonists in inflammatory diseases. Cytokine 76: 25–37. [DOI] [PubMed] [Google Scholar]
- 37.Roux J., Kawakatsu H., Gartland B., Pespeni M., Sheppard D., Matthay M. A., Canessa C. M., Pittet J. F. 2005. Interleukin-1beta decreases expression of the epithelial sodium channel alpha-subunit in alveolar epithelial cells via a p38 MAPK-dependent signaling pathway. J. Biol. Chem. 280: 18579–18589. [DOI] [PubMed] [Google Scholar]
- 38.Dolinay T., Kim Y. S., Howrylak J., Hunninghake G. M., An C. H., Fredenburgh L., Massaro A. F., Rogers A., Gazourian L., Nakahira K., et al. 2012. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am. J. Respir. Crit. Care Med. 185: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tate M. D., Ong J. D., Dowling J. K., McAuley J. L., Robertson A. B., Latz E., Drummond G. R., Cooper M. A., Hertzog P. J., Mansell A. 2016. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci. Rep. 6: 27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen I. C., Scull M. A., Moore C. B., Holl E. K., McElvania-TeKippe E., Taxman D. J., Guthrie E. H., Pickles R. J., Ting J. P. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30: 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas P. G., Dash P., Aldridge J. R., Jr., Ellebedy A. H., Reynolds C., Funk A. J., Martin W. J., Lamkanfi M., Webby R. J., Boyd K. L., et al. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Oberley-Deegan R., Wang S., Nikrad M., Funk C. J., Hartshorn K. L., Mason R. J. 2009. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J. Immunol. 182: 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W. C., Chan R. W., Wang J., Travanty E. A., Nicholls J. M., Peiris J. S., Mason R. J., Chan M. C. 2011. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J. Virol. 85: 6844–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ong J. D., Mansell A., Tate M. D. 2017. Hero turned villain: NLRP3 inflammasome-induced inflammation during influenza A virus infection. J. Leukoc. Biol. 101: 863–874. [DOI] [PubMed] [Google Scholar]
- 45.Ting J. P., Harton J. A. 2015. NLRP3 moonlights in TH2 polarization. Nat. Immunol. 16: 794–796. [DOI] [PubMed] [Google Scholar]
- 46.Kuriakose T., Kanneganti T. D. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol. Immunol. 2017 doi: 10.1016/j.molimm.2017.01.023. DOI: 10.1016/j.molimm.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo E. K., Kim J. K., Shin D. M., Sasakawa C. 2016. Molecular mechanisms regulating NLRP3 inflammasome activation. Cell. Mol. Immunol. 13: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellebedy A. H., Lupfer C., Ghoneim H. E., DeBeauchamp J., Kanneganti T. D., Webby R. J. 2011. Inflammasome-independent role of the apoptosis-associated speck-like protein containing CARD (ASC) in the adjuvant effect of MF59. [Published erratum appears in 2013 Proc. Natl. Acad. Sci. USA 110: 4429.] Proc. Natl. Acad. Sci. USA 108: 2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer R. N., Brighton L. E., Mueller L., Xiang Z., Rager J. E., Fry R. C., Peden D. B., Jaspers I. 2012. Influenza enhances caspase-1 in bronchial epithelial cells from asthmatic volunteers and is associated with pathogenesis. J Allergy Clin. Immunol. 130: 958–967.e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pothlichet J., Meunier I., Davis B. K., Ting J. P., Skamene E., von Messling V., Vidal S. M. 2013. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 9: e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tate M. D., Pickett D. L., van Rooijen N., Brooks A. G., Reading P. C. 2010. Critical role of airway macrophages in modulating disease severity during influenza virus infection of mice. J. Virol. 84: 7569–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tate M. D., Schilter H. C., Brooks A. G., Reading P. C. 2011. Responses of mouse airway epithelial cells and alveolar macrophages to virulent and avirulent strains of influenza A virus. Viral Immunol. 24: 77–88. [DOI] [PubMed] [Google Scholar]
- 53.Ioannidis L. J., Verity E. E., Crawford S., Rockman S. P., Brown L. E. 2012. Abortive replication of influenza virus in mouse dendritic cells. J. Virol. 86: 5922–5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schattgen S. A., Gao G., Kurt-Jones E. A., Fitzgerald K. A. 2016. Cutting edge: DNA in the lung microenvironment during influenza virus infection tempers inflammation by engaging the DNA sensor AIM2. J. Immunol. 196: 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichinohe T., Pang I. K., Kumamoto Y., Peaper D. R., Ho J. H., Murray T. S., Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108: 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salk H. M., Simon W. L., Lambert N. D., Kennedy R. B., Grill D. E., Kabat B. F., Poland G. A. 2016. Taxa of the nasal microbiome are associated with influenza-specific IgA response to live attenuated influenza vaccine. PLoS One 11: e0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.