Abstract
Mammalian telomeric DNA contains duplex TTAGGG repeats and single-stranded overhangs. POT1 (protection of telomeres 1) is a telomere-specific single-stranded DNA-binding protein, highly conserved in eukaryotes. The biological function of human POT1 is not well understood. In the present study, we demonstrate that POT1 plays a key role in telomeric end protection. The reduction of POT1 by RNA interference led to the loss of telomeric single-stranded overhangs and induced apoptosis, chromosomal instability, and senescence in cells. POT1 and TRF2 interacted with each other to form a complex with telomeric DNA. A dominant negative TRF2, TRF2ΔBΔM, bound to POT1 and prevented it from binding to telomeres. POT1 overexpression protected against TRF2ΔBΔM-induced loss of telomeric single-stranded overhangs, chromosomal instability, and senescence. These results demonstrate that POT1 and TRF2 share in part in the same pathway for telomere capping and suggest that POT1 binds to the telomeric single-stranded DNA in the D-loop and cooperates with TRF2 in t-loop maintenance.
Human telomeres are specialized chromosomal terminal elements, containing tandem repetitive sequences and specific proteins. Telomeric DNA is mostly composed of 2 to 30 kb of double-stranded TTAGGG repeats, which are necessary for telomeric function in somatic cells (5, 19, 24, 43). The termini of human telomeres carry an overhang (∼300 nucleotides) of single-stranded 3′ DNA (38, 41, 56). In many eukaryotes, telomere length is maintained by telomerase, a reverse transcriptase that adds TTAGGG repeats onto the 3′ ends of telomeres (5, 8, 39, 43), which can counteract the loss of terminal sequences during DNA replication. This end replication problem of human telomeres has received particular attention for its implications in ageing and cancer (4, 7, 16, 18, 30, 43, 44, 55). Maintenance of the telomeric TTAGGG repeats at human chromosome ends, either by telomerase (13) or by an alternative mechanism (9), is essential for immortalized cells in vitro to escape from the normal limitations of the proliferation capacity.
Telomeres and capping proteins allow cells to distinguish natural chromosome ends from damaged DNA. The disruption of telomeric function can trigger a DNA damage response, including p53-dependent apoptosis (28). The overloading of DNA repair activities can also threaten the integrity of chromosome ends, thereby leading to extensive genome instability (4, 15, 20, 27). Telomeric proteins stabilize the telomeres by protecting the single-stranded overhang from degradation or by remodeling the telomeres into a t-loop structure (5, 6, 15, 23). Invasion of the single-stranded overhang into the double-stranded telomeric tract forms the t-loop structure. In vitro, t-loop assembly involves the binding of telomere repeat-binding factor 2 (TRF2), near the 3′ telomeric overhang (48). A dominant-negative mutant of TRF2, TRF2ΔBΔM, effectively strips TRF2 and its interacting factors off the telomeres and causes a loss of telomeric overhangs, apoptosis, senescence, and chromosome abnormalities (15, 28, 29, 45, 48, 52).
The telomeric single-stranded overhangs have been implicated as a critical component of the telomeric structure that is required for proper telomeric function (4, 5, 15, 49). Telomeres are protected by a capping protein(s) that binds to the single-stranded DNA commonly found at the ends of chromosomes. The Oxytricha telomeric overhang is tightly bound by a single-stranded DNA-binding protein, TEBP (telomere end-binding protein) composed of α and β subunits (21). TEBP forms an extensive interface along the overhang and buries the telomeric end in a deep hydrophobic pocket to provide an effective way to hide chromosome ends from DNA repair enzymes (25). Telomeres of budding yeast are capped by a sequence-specific single-stranded DNA-binding protein, Cdc13. Unlike TEBP, Cdc13 is a recruitment factor that brings Stn1p to the telomere (42). Stn1p binds a second capping protein, Ten1p (22), to protect the telomere.
POT1 (protection of telomeres 1) is the best known candidate for a human functional homologue of TEBP and Cdc13. It binds the single-strand telomeric overhangs with exceptionally high sequence specificity (2, 32, 33, 37, 53). The protein shares weak sequence similarity with the N-terminal DNA-binding domain of TEBP from ciliated protozoa. Crystal structures show that the POT1 protein adopts an oligonucleotide-oligosaccharide-binding fold with two loops that protrude to form a clamp for single-stranded DNA binding (33). In the context of the POT1 protein, DNA self-recognition involving base stacking and unusual G-T base pairs compacts the DNA. This structure explains the sequence specificity of binding. POT1 family members are identified from diverse organisms, suggesting that all eukaryotes use a single-stranded DNA-binding protein to cap the telomere. The deletion of the fission yeast POT1 gene has an immediate effect on chromosomal stability, causing a rapid loss of telomeric DNA and chromosome circularization (2). The function of human POT1 is uncertain. Recent reports show that POT1 can act as a telomerase-dependent, positive regulator of telomere length (12), colocalize with TRF1 and TRF2 (3), interact with the TRF1 complex, and act as a terminal transducer of TRF1 telomere length control (36). In this report, we identified a key role for POT1 in the maintenance of telomeric single-stranded tails as well as phenotypic protective functions.
MATERIALS AND METHODS
Cell culture, Western blot analyses, immunoprecipitation, and TUNEL assay.
IMR90, normal human fibroblasts, and HT1080, 293, and MCF7 tumor cell lines were obtained from the American Type Culture Collection. Western blotting analyses and immunoprecipitation were essentially as described previously (47). Briefly, cell extracts were prepared in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 120 mM NaCl, and 1% NP-40 with protease inhibitors. Nuclear extracts were prepared by using the NE-PER nuclear and cytoplasmic extraction kit (Pierce, Rockford, Ill.). Cellular protein (2 mg) or nuclear extracts (0.5 mg) were immunoprecipitated with the appropriate antibodies and protein A/G beads. Proteins were separated, followed by Western blotting analyses. The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed with the TUNEL assay kit from Roche (Indianapolis, Ind.), following the manufacturer's directions.
siRNA and shRNA, PCR, expression vectors, proteins, and antibodies.
siRNA POT1-A at POT1 cDNA (NM_015450), positions 395 to 414; POT1-B at positions 195 to 214; POT1-C at positions 751 to 770; and small interfering RNA (siRNA)-TRF2 at TRF2 cDNA (AF002999) at position 1465 were designed by Oligoengine RNAi software and synthesized by Dharmacon (Lafayette, Colo.). Cells were incubated with siRNA in Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.). POT1-siRNA was repeated, with transfection carried out every 3 days until day 16 in the senescence study. Lentivirus POT1-short hairpin RNAs (shRNAs) were prepared by BioMarkers (Rockville, Md.). Lentivirus POT1-sh1 locates at POT1 cDNA position 395, and sh2 locates at position 1016. Lentivirus infection and blasticidin selection were performed as described in the manufacturer's directions. Real-time PCR was performed with ABI Sequencer 7700 with POT1 primers, and the Taqman probe was synthesized by ABI (Foster City, Calif.). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers and the probe were from ABI stock.
pLPC-Myc, pLPC-Myc-POT1, pLPC-Myc-TRF2, and pLPC-Myc-TRF2ΔBΔM retroviral constructs were gifts from Titia de Lange. RPA1 cDNA was cloned into the pLPC-Myc vector. Retroviral infection and puromycin selection were performed and modified as previously described (29). Briefly, retroviral constructs were transfected in AmphoPack 293 cells (American Type Culture Collection). After 2 days, supernatant was collected, and the virus titer was determined. Then, specific type cells were infected with 4 μg of polybrene/ml for 6 h with shaking. After 2 days of infection, the cells were selected with 2 μg of puromycin/ml. For coexpression of Myc-POT1 and Myc-TRF2ΔBΔM, the same titer viral stocks were used for infecting the cells. To maintain the same protein level, the expression levels of Myc-POT1 and Myc-TRF2ΔBΔM were determined by Western blotting analysis. pcDNA-POT1-V5 was a gift from Peter Baumann. Glutathione S-transferase (GST)-TRF2 and GST-TRF2ΔBΔM were generated by subcloning cDNA of TRF2 and TRF2ΔBΔM into pGEM-6P vector (Amersham Pharmacia Biotech, Piscataway, N.J.). Recombinant POT1 protein was purified as previously described (2) and had a purity of 95% or better. Recombinant TRF2 protein (95% purity) was a gift from Jack Griffith. POT1 peptide at amino acids 355 to 375 was synthesized (SynPep Corporation, Dublin, Calif.), conjugated to keyhole limpet hemocyanin, and used to immunize rabbits (Harlan, Indianapolis, Ind.). Anti-TRF2 was from Upstate (Waltham, Mass.) and Oncogene Research Products (Boston, Mass.). Anti-RAD50 and anti-NBS1 were from Oncogene Research Products. Anti-V5 and anti-Myc antibodies were obtained from Invitrogen. Anti-TRF1 antibody was from Santa Cruz (Santa Cruz, Calif.) and was a gift from Titia de Lange.
Telomere oligonucleotide ligation assay and in-gel hybridization assay.
A telomere oligonucleotide ligation assay (T-OLA) was carried out as previously described (11, 49). Genomic DNA (5 μg) was hybridized with a 0.5 pmol of 32P-labeled probe at 50°C overnight, followed by ligation with 20 U of Taq DNA ligase (New England Biolab, Beverly, Mass.) at 50°C for 5 h. Then, the samples were precipitated, resuspended, denatured, and separated on 6% acrylamide-6 M urea gels. T-OLA product (10 ng) was used for the quantitative PCR with the GAPDH primers. In-gel hybridization to measure telomeric overhang was carried out as previously described (17).
In vitro protein interaction, electrophoretic mobility gel shift assay (EMSA), and POT1-TRF2-TTAGGG-binding assay.
GST fusion TRF2 and TRF2ΔBΔM proteins were produced in Escherichia coli and purified according to the manufacturer's instructions (Amersham Pharmacia Biotech). POT1 protein was prepared with the TNT Quick-Coupled Transcription/Translation system (Promega, Madison, Wis.) in the presence of [35S]methionine. An in vitro binding assay was done in an immunoprecipitation buffer with rotation at room temperature for 1 h. After being washed, the samples were loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and separated by electrophoresis.
EMSA reaction mixtures (each, 20 μl) contained 25 mM HEPES-NaOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, 5% glycerol, herring sperm DNA (50 μg/ml), 200 pM of 32P-labeled [TTAGGG]4, and protein concentrations as indicated in the figures. Reaction mixtures were incubated at room temperature for 15 min, and the products were separated by 4% nondenaturing polyacrylamide gels at 4°C for 1.5 h and visualized by a PhosphorImager or film autoradiography. POT1-TRF2 and biotin-TTAGGG-binding assays were performed as previously described (60).
Indirect immunofluorescence.
Experiments were performed as previously described (57). Briefly, confocal fluorescent images were collected with a Bio-Rad MRC 1024 confocal scan head mounted on a Nikon Optiphot microscope with a 60× Zeiss LSM410 lens. Z sections were captured at 0.5-μm intervals for each cell with LaserSharp software (Bio-Rad, Hercules, Calif.). Confocal Assistant software (Bio-Rad) was used to analyze the images. For quantitation of signal overlap, the Z sections of five random cells, scored as containing foci, were analyzed and averaged with the Bio-Rad LaserSharp software. For conventional immunofluorescence microscopy, the cells were examined with a Zeiss Axioskop fluorescence microscope, equipped with a charge-coupled device camera. Images were captured, pseudocolored, and merged with IPLab image analysis software. At least 100 cells were analyzed for each experiment. The experiments were repeated at least three times. Quantitative data of colocalization represent the percentage of the proteins that colocalized with each other.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed with the ChIP kit according to the manufacturer's directions (Upstate, Waltham, Mass.). Briefly, cells were harvested and fixed in 1% formaldehyde at 37°C for 30 min. After being washed in phosphate-buffered saline (PBS), the cells were lysed in 1% sodium dodecyl sulfate, 50 mM Tris-HCI (pH 8.0), and 10 mM EDTA (107 cells ml−1) for 10 min, followed by sonication. DNA was sheared to an average size of 0.5 to 1 kb. The 200-μl extract was diluted, immunoprecipitated with indicated antibodies for 12 h at 4°C, and supplemented with 60 μl of protein A beads for 1 h at 4°C. The immunocomplexes were washed with the serial washing buffers, eluted, and heated at 65°C for 4 h to reverse the cross-links. The immunoprecipitated DNA was precipitated, denatured, and dot blotted onto Hybond membranes. A [CCCTAA]16 probe with [TTAGGG]3 primers or an Alu probe was labeled with the random primer-labeling kit from Amersham Pharmacia Biotech. The blot was prehybridized in Church's buffer for 1 h at 55°C. A heated-denatured DNA probe was added and incubated at 55°C overnight. The membranes were washed and exposed to a PhosphorImager screen. The quantification of the percent precipitated DNA was done with the ImageQuant software.
Chromosome preparation and FISH analysis.
To arrest the cells at metaphase, 0.1 μg of Colcemid/ml was added to the culture 2 h before the harvest. The cells were trypsinized and treated in hypotonic solution (0.06 M KCl) at 37°C for 8 min. Then, the cells were fixed in a fixative (3 parts of methanol with 1 part of acetic acid) three times. The cells were dropped onto clean microscopic slides, air dried, and stained with 4% Gurr's Giemsa solution (BDH Laboratory Supplies, Dorset, England). One hundred well-spread metaphase cells from four independent experiments were examined under a microscope (Nikon E400) with a 100× objective for each group. Specific chromosomal abnormalities (multicentric chromosomes, which may result from complete chromosome end fusion, and chromosome end joining and breaks) were recorded. A multicentric chromosome is a chromosome with two or more centromeres, and a chromosome end joining is a partial chromosome end fusion (only one of the two sister chromatids joined). Criteria for a chromosome break were a discontinuity of a single chromatid, in which the distance of discontinuity region was wider than the diameter of the chromatid or there was a clear misalignment of one of the chromatids. The total number of chromosomal abnormalities were divided by the number of the cells examined, and the mean number of chromosomal abnormalities per cell was recorded for statistical analysis. The telomeric peptide-nucleic acid fluorescent in situ hybridization (FISH) assay was performed with the Telomere PNA FISH kit/Cy3 from Dako (Glostrup, Denmark), following the manufacturer's directions.
SA-β-Gal assay.
IMR90 cells (passage 22) were infected by indicated constructs. The senescence-associated β-galactosidase (SA-β-Gal) assay was performed 2 days after 105 cells per well were seeded in a 6-well dish on day 14 after infection. Cells were fixed for 5 min in 2% formaldehyde-0.2% glutaraldehyde solution in PBS, washed in PBS, and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (1 mg/ml) in 150 mM NaCl, 2 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 40 mM NaPi (pH 6.0) for 12 h at 37°C.
RESULTS
POT1-RNAi induces the loss of telomeric overhangs.
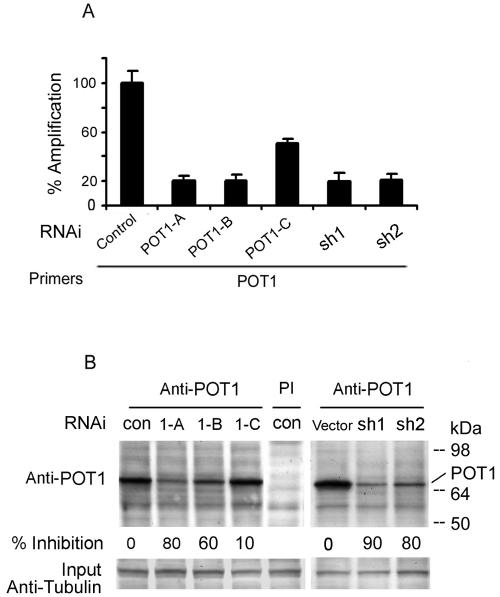
To examine the role of POT1 in human telomeres, three POT1-siRNAs and two lentivirus shRNAs were designed. Primary human fibroblasts (IMR90) were transduced with POT1-RNA interference (RNAi). After 48 h, RNA was prepared, and real-time reverse transcription-PCR was performed with POT1 primers and probe. POT1-A and POT1-B siRNAs and shRNAs inhibited the mRNA level of POT1 by approximately 80% when compared with controls (Fig. 1A); POT1-C inhibited by approximately 45%. Similar results were detected with POT1-RNAi in HT1080, 293, and MCF7 cells (data not shown). Consistent with the inhibition of the POT1 mRNA levels by expression of POT1-RNAi, the endogenous POT1 protein level was also decreased after 2 days (Fig. 1B).
FIG. 1.
Reduction of POT1 expression by POT1-RNAi. (A) IMR90 cells were transduced with three POT1-siRNAs (POT1-A, -B, and -C) or two lentivirus shRNAs (sh1 and sh2); a random sequence siRNA was used as a control. After 48 h, RNA was prepared, and real-time reverse transcription-PCR was performed with the POT1 primers and probe. (B) POT1 expression was decreased by expression of POT1-RNAi. After expression for 2 days, IMR90 cell extracts were subjected to immunoprecipitation with the anti-POT1 antibody, followed by Western blotting with the anti-POT1 antibody. PI, preimmune serum. Fifty micrograms of cell extract was loaded as input and analyzed by anti-γ-tubulin antibody.
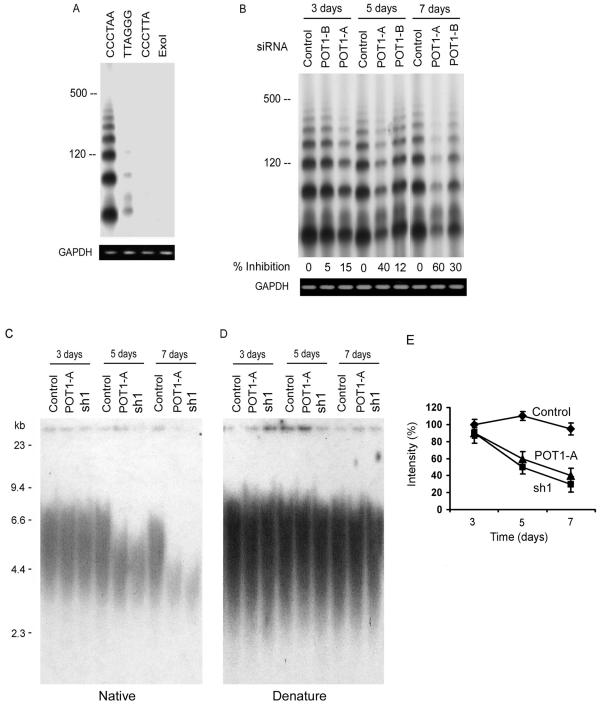
Recent observations suggest that the telomeric single-stranded overhang is a central constituent of telomere capping (5, 15, 34, 49). Because POT1 is an overhang-specific binding protein, we examined whether changes in the telomeric overhangs occurred by POT1 inhibition. To assess the length of the telomeric overhangs, a recently developed method, T-OLA, was used (11, 49). In this technique, labeled [CCCTAA]4 oligonucleotides were annealed to nondenatured genomic DNA and ligated. The length of the ligation products and signal intensity provide an indication of the overhang's length. Using the [CCCTAA]4 probe on genomic DNA derived from the IMR90 cells, the overhangs at the ends of wild-type telomeres were readily detected (Fig. 2A). In validation of the method, an oligonucleotide ([TTAGGG]4) complementary to the C-rich telomeric strand produced only a faint signal. An oligonucleotide carrying a mismatch telomeric sequence ([CCCTTA]4 or [CCCTAA]4) treated with ExoI, a single-strand-specific exonuclease, did not yield a T-OLA signal.
FIG. 2.
The expression of POT1-RNAi caused the loss of telomeric overhang signals. (A) T-OLAs were performed with oligonucleotides complementary to the G-rich strand [CCCTAA]4, the C-rich strand [TTAGGG]4, or the mismatch sequence [CCCTTA]4. (B) T-OLA analysis was used on DNA derived from IMR90 cells expressing POT1-siRNA and controls at 3, 5, and 7 days after transfection. Quantitative PCR was performed using primers specific for genomic GAPDH for equal amounts of genomic DNA in each sample (bottom). (C) The in-gel hybridization assay was performed with a native gel and probed with [CCCTTA]4. DNA was derived from IMR90 cells expressing POT1-RNAi. (D) The DNA was denatured in the gels and rehybridized with the same probes. (E) Quantitative data of the loss of telomeric overhangs were derived from three independent experiments similar to those shown in panel C, and the average value (error bars represent standard deviations) was plotted.
IMR90 cells expressing the POT1-siRNA displayed a consistent reduction in the amount of detectable telomeric overhangs (Fig. 2B). The quantification of the data showed that expression of the POT1-siRNAs for 5 to 7 days resulted in a 30 to 60% decline in the total single-strand TTAGGG repeat signal at telomeres. In contrast, no alteration in the signal was noted in the cells expressing a random control siRNA. To confirm the results, telomeric overhang signals were examined by in-gel hybridization assay of IMR90 cells treated with POT1-RNAi (Fig. 2C and D). A reduction in G-overhang signals in POT1-RNAi-treated cells showed a similar level as the T-OLA (Fig. 2E).
Inhibition of POT1 induces apoptosis, chromosomal abnormalities, and senescence.
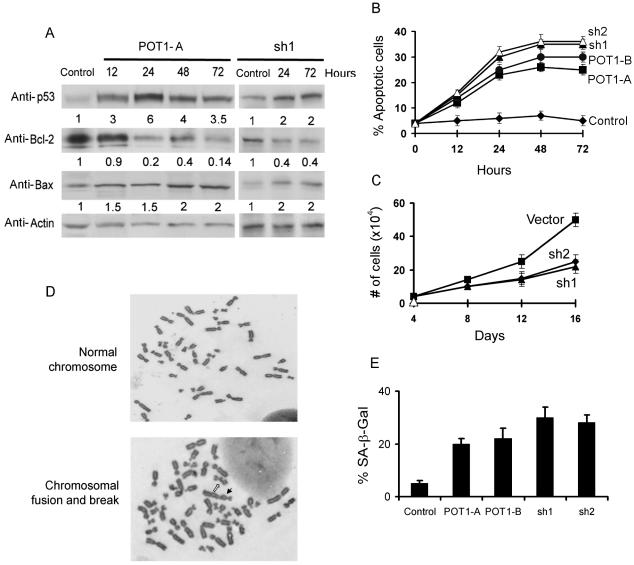
Telomere dysfunction can induce DNA damage response pathways, apoptosis, and senescence (15, 28, 45, 50). Therefore, we examined whether the erosion of the telomeric overhang by POT1-RNAi was associated with these cellular responses. Western blotting analysis of the cell lysates expressing POT1-RNAi revealed that p53 increased concurrently with a decreased expression of antiapoptotic Bcl2 and an increased expression of proapoptotic Bax (Fig. 3A) in MCF7 cells, which showed high levels of apoptosis induced by telomere dysfunction (28). Consequently, an increase in apoptosis was observed in cells expressing POT1-RNAi, as assessed by TUNEL-labeling analysis (Fig. 3B). Consistent with these results, continued cell proliferation was inhibited by infected lentivirus POT1-shRNAs (Fig. 3C).
FIG. 3.
POT1-RNAi induced the DNA damage pathway, apoptosis, chromosomal abnormalities, and the senescent phenotype. (A) The time course of Western blotting analysis in MCF-7 cells expressing POT1-siRNA (POT1-A) or lentivirus POT1-shRNA (sh1). A random sequence siRNA was used as a control. (B) TUNEL assay of MCF7 cells after expression with POT1-siRNAs or shRNAs. A random sequence siRNA was used as a control. (C) Growth curve of IMR90 cells expressing lentivirus POT1-shRNAs and vector control cells. (D) The induction of chromosomal abnormalities. IMR90 cells were transduced with indicated siRNA or shRNA for 7 days, and the chromosome was prepared and analyzed as described in Materials and Methods. Open arrow, chromosome break; solid arrow, dicentric chromosome. (E) POT1-RNAi induced the senescent phenotype. IMR90 cells were stained for β-Gal activity at pH 6.0. Graphs show the effect of expression of POT1-siRNAs and shRNAs.
To determine the association between POT1 expression levels and chromosomal abnormalities, IMR90 cells were transduced with POT1-RNAi for 7 days, and a significantly high frequency of chromosomal abnormalities (chromosome fusion and breaks) was found in the cells expressing POT1-RNAi (Fig. 3D and Table 1). We did not observe chromosome fusion in control cells. The frequency of chromosome breaks was also low in control cells (Table 1).
TABLE 1.
Induction of chromosomal abnormalities in IMR90 cells
| Expressed siRNA or genes | Mean abnormalities/ cell (SD)a
|
% of cells with abnormalities (SD) | P valuec | |
|---|---|---|---|---|
| Fusionb | Breaks | |||
| Control | 0 | 0.03 (0.05) | 3 (5) | |
| siPOT1-A | 0.04 (0.06) | 0.29 (0.18) | 25 (16) | 0.06 |
| siPOT1-B | 0.02 (0.02) | 0.20 (0.1) | 20 (9) | 0.03 |
| POT1-sh1 | 0.10 (0.08) | 0.30 (0.1) | 30 (12) | 0.03 |
| POT1-sh2 | 0.08 (0.06) | 0.30 (0.1) | 28 (9) | 0.03 |
| TRF2ΔBΔM | 0.40 (0.15) | 0.62 (0.66) | 43 (31)d | 0.02 |
| TRF2ΔBΔM+POT1 | 0 | 0.17 (0.07) | 17 (7) | 0.05 |
| POT1 | 0 | 0.04 (0.01) | 4 (1) | 1.0 |
The numbers indicated average abnormalities per metaphase cell from four independent experiments; a total of 100 cells were scored.
Chromosome fusion contained multicentric chromosomes and chromosome end joining.
Frequencies of percent of cells with abnormalities were compared with the control group.
Frequency was higher than the TRF2ΔBΔM + POT1 group with a P value of 0.08.
The expression of POT1-RNAi caused the loss of telomeric overhang signals. Loss of telomeric overhang can induce a premature senescent phenotype in primary fibroblasts (45, 52). These cells display all of the morphological and molecular signs of senescence, including a large and flat cell shape, frequent occurrence of multiple nuclei, a 2N or 4N DNA content, and positive staining with the senescence-associated marker, SA-β-Gal. A premature senescent phenotype was observed in IMR90 cells expressing POT1-RNAi, as assayed by SA-β-Gal staining (Fig. 3E). Approximately 20 to 30% of the cells displayed positive SA-β-Gal staining with senescent morphological signs after the expression of POT1-RNAi for 16 days when compared with only 4% in the control cells.
POT1 interacts with TRF2 to form a complex on telomeric DNA.
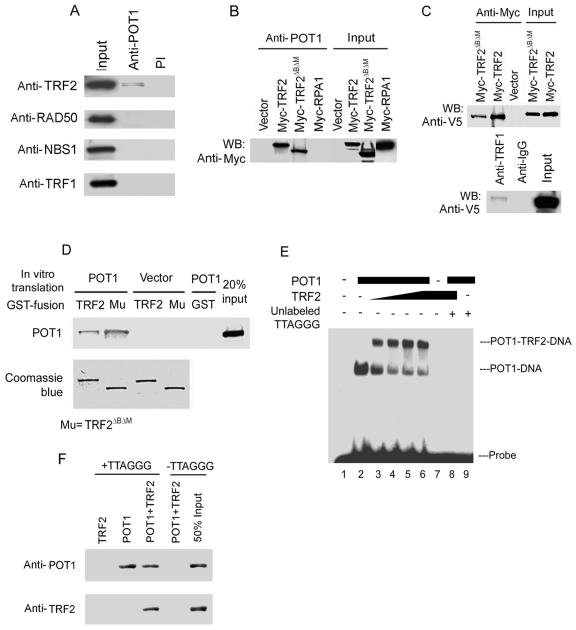
TRF2 maintains telomere integrity. The inhibition of TRF2 results in the loss of telomeric overhangs from termini (15, 52) and the induction of DNA damage response pathways (28, 45), which are the same molecular and cellular consequences as the loss of POT1. TRF2 binds to duplex telomeric DNA near the telomeric single-stranded overhangs and remodels telomeric DNA into t-loops (48), while POT1 directly binds to telomeric single-stranded DNA. These results suggest that both proteins may interact with each other to act in the same protection pathway. Therefore, we explored the possibility that POT1 interacts with TRF2 by coimmunoprecipitation. Endogenous POT1 immunoprecipitated with endogenous TRF2 (Fig. 4A). RAD50 and NBS1 (TRF2-associated proteins) and TRF1 were not detected in the POT1 complex. Moreover, the POT1 immunocomplex contained TRF2 and TRF2ΔBΔM, a dominant negative mutant of TRF2, when these proteins were coexpressed after transfection (Fig. 4B), confirming the association of POT1 with TRF2. Myc-RPA1 was not detected in the POT1 immunocomplex, excluding the possibility that the coimmunoprecipitations were nonspecific. To further investigate the association of POT1 with TRF2, the immunoprecipitation of Myc-TRF2 and TRF2ΔBΔM brought down the POT1-V5 (V5 is 14 amino acid peptides from the V5 epitope of paramyxovirus) fusion protein from nuclear extracts of 293 cells cotransfecting these expression constructs but not in the negative control (empty vectors, pLPC-Myc, and pcDNA-V5) (Fig. 4C, top). The POT1-V5 fusion protein was found in the TRF1 immunoprecipitates (Fig. 4C, bottom). Consistently, GST-TRF2 and GST-TRF2ΔBΔM bound to the POT1 protein generated by in vitro transcription-translation with [35S]methionine (Fig. 4D).
FIG. 4.
POT1 interacted with TRF2. (A) Endogenous TRF2 bound to POT1 in IMR90 fibroblasts by coimmunoprecipitation. Nuclear extracts (0.5 mg) were subjected to immunoprecipitation with the anti-POT1 antibody, followed by Western blotting with anti-TRF2, anti-RAD50, anti-NBS1, or anti-TRF1 antibodies. Preimmune serum (PI) was used as a control. Fifty micrograms of nuclear extract was loaded as input. (B) TRF2 and TRF2ΔBΔM bound to POT1 in 293 cells. Myc-TRF2 and TRF2ΔBΔM were transfected into 293 cells. Myc-TRF2 and TRF2ΔBΔM were coimmunoprecipitated with an anti-POT1 antibody from 0.5 mg of nuclear extract. Immunoprecipitated proteins were visualized by Western blotting analyses with antibodies against the Myc peptide. Myc-RPA1 was used as a control. (C) Myc-TRF2 and Myc-TRF2ΔBΔM brought down POT1-V5 (top) coexpressing POT1-V5 and Myc-TRF2 or Myc-TRF2ΔBΔM in 293 cells. TRF1 brought down POT1-V5 in 293 cells (bottom) expressing the POT1-V5 fusion protein. (D) POT1 interacted with TRF2 and TRF2ΔBΔM in vitro. Two micrograms of GST-TRF2 and TRF2ΔBΔM fusion proteins was incubated with 5 μl in vitro-translated POT1 protein labeled with [35S]methionine, and the proteins were brought down by GST beads. A 20% input of POT1 protein was included in this lane. GST-TRF2 and TRF2ΔBΔM protein inputs were verified by Coomassie blue staining. (E) POT1 and TRF2 formed a complex with telomeric single-stranded overhang DNA (TTAGGG probe, 200 pM). EMSA assay with increasing amounts of TRF2 (lanes 3 to 6, threefold steps up to 80 pM) with 20 pM POT1. Unlabeled [TTAGGG]4, 10-fold excess. (F) POT1 and TRF2 bound on [TTAGGG]4 oligomers. The biotinylated [TTAGGG]4 (10 nM) was attached to streptavidin beads. POT1 (40 pM) and/or TRF2 (40 pM) was added and incubated with the [TTAGGG]4-coated beads. POT1 and TRF2 associated with DNA were detected by immunoblotting.
To further examine the POT1 and TRF2 complex on telomeric DNA, EMSA was performed. Consistent with previous reports (2, 37), recombinant POT1 protein specifically bound to telomeric [TTAGGG]4 (Fig. 4E). When baculovirus-expressed TRF2 was added to the reaction mixture, we observed an additional complex that migrated above the POT1-DNA complex. The appearance of this new band was strictly dependent on the addition of TRF2. The shifted bands were competed effectively by the unlabeled [TTAGGG]4, suggesting that these bands were specific. To verify the gel shift complex-containing proteins, purified POT1 and/or TRF2 proteins were incubated with biotinylated single-stranded [TTAGGG]4 DNA. In the presence of POT1, TRF2 was associated with the POT1-DNA complex retrieved by streptavidin beads (Fig. 4F). TRF2 alone bound on double-stranded telomeric DNA but not on single-stranded [TTAGGG]4 (Fig. 4F and data not shown).
TRF2ΔBΔM and TRF2-siRNA remove POT1 from telomeres.
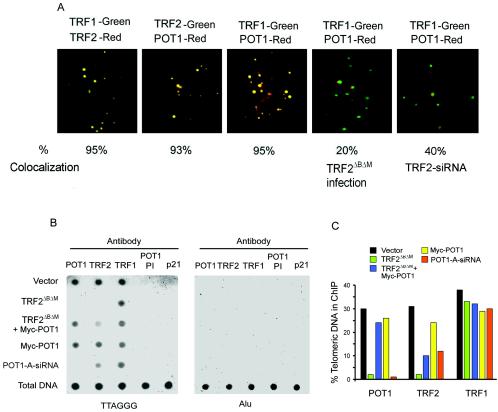
Using indirect immunofluorescence with TRF1, which is a specific marker for interphase telomeres (10, 46, 51), we showed that POT1 colocalized with TRF1 and TRF2 in the nuclei of IMR90 cells (Fig. 5A). This is in agreement with a previous report (3). To demonstrate the specificity of POT1 foci, the cells were treated with POT1-siRNA (see Fig. S1 in the supplemental material). The number of foci per cell decreased significantly with much weaker immunofluorescence signals in the positively stained cells. The number of the positively stained cells also decreased to 50% compared with the control. To determine POT1 localization on telomeres, IMR90 cells were expressed by a retroviral construct expressing TRF2ΔBΔM or TRF2-siRNA. Foci of colocalized POT1 and TRF1 significantly decreased after expression of TRF2ΔBΔM or TRF2-siRNA. Approximately 20 to 40% of POT1 foci colocalized with TRF1. In contrast, approximately 95% of the POT1 foci in control cells were sites of TRF1 accumulation, suggesting that TRF2ΔBΔM binds to POT1 and removes it from the telomeres.
FIG. 5.
(A) POT1 colocalized with endogenous TRF1 and TRF2 in IMR90 fibroblasts by immunostaining with anti-TRF1, anti-TRF2, and anti-POT1 antibodies. POT1 reduced the colocalization with TRF1 2 days after expressing TRF2ΔBΔM or TRF2-siRNA. (B) POT1 associated with telomeric DNA. ChIP of HT1080 cells expressing pLPC-Myc, Myc-POT1, Myc-TRF2ΔBΔM, or POT1-A-siRNA with indicated antibodies or preimmune serum (PI) of POT1 antibody for 2 days after infection. Duplicate dot blots were probed for telomeric or Alu repeats. (C) The quantification of the data in panel B representing the percentage of TTAGGG DNA recovered in each sample. Averaged signals obtained with total DNA samples were used as 100% value for the quantification.
To study the association of POT1 and TRF2 with telomeres, we performed a ChIP assay, using anti-POT1 and anti-TRF2 antibodies. Whole cells were treated with the cross-linking reagent formaldehyde, and DNA dot blots were probed with a CCCTAA probe to determine relative levels of protein-associated telomeric DNA. Alu sequences, preimmune POT1 antibody serum, and the p21 antibody were used as negative controls in each experiment. We observed an enrichment of telomeric DNA coimmunoprecipitated with POT1, TRF2, and TRF1 antibodies in control cells (empty vector) (Fig. 5B and C). As previously seen (36), expressing TRF2ΔBΔM resulted in a significant reduction in the binding of TRF2 to telomeres as assessed by ChIP. Consistent with immunofluorescence data, POT1 was not detected at telomeric DNA in cells expressing TRF2ΔBΔM. Furthermore, POT1-siRNA reduced POT1 from telomeric DNA, and TRF2 also showed a decreased association with telomeric DNA in the sample expressing POT1-siRNA (Fig. 5B and C). As a control, neither TRF2ΔBΔM nor POT1-siRNA affected the binding of TRF1 on telomeric DNA.
POT1 blocks the effects of TRF2ΔBΔM.
Because TRF2ΔBΔM interacts with POT1 and removes it from telomeres, we then investigated whether the overexpression of exogenous POT1 could block the biological alterations induced by TRF2ΔBΔM. Using ChIP assays, POT1-associated telomeric DNA could be recovered in cells coexpressing Myc-POT1 and Myc-TRF2ΔBΔM (Fig. 5B and C). In this group, ChIP with TRF2 antibody resulted in a faint signal, suggesting that the TRF2-associated telomeric DNA was partially recovered. In agreement with previous reports, the overexpression of Myc-POT1 did not affect the amount of POT1 at telomeres by ChIP assay with POT1 antibody (36).
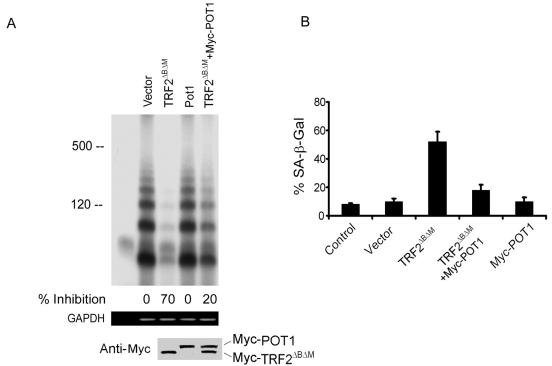
Using the T-OLA, we observed that TRF2ΔBΔM induces a 70% loss of telomeric overhangs (Fig. 6A), similar to the results of a previously published report (52). In contrast, the T-OLA signal only slightly decreased by coexpressing exogenous Myc-POT1 and Myc-TRF2ΔBΔM, compared with controls (empty vector). Thus, POT1 overexpression can block the erosive effect of TRF2ΔBΔM on telomeric overhangs. As a control, Myc-POT1 alone did not alter the telomeric overhang signal.
FIG. 6.
(A) The expression of POT1 decreased the loss of telomeric overhang induced by TRF2ΔBΔM. (Top) The T-OLA was used on genomic DNA derived from the IMR90 cells expressing the indicated constructs for 5 days after infection. Lane 1, no genomic DNA. (Middle) Quantitative PCR was performed, using primers specific for genomic GAPDH for equal amounts of genomic DNA in each sample. (Bottom) Myc-POT1- and Myc-TRF2ΔBΔM-expressing levels were determined by immunoblotting. (B) POT1 blocked the senescent phenotype induced by TRF2ΔBΔM. IMR90 cells were stained for β-Gal activity at pH 6.0. Graphs showed the effect of infection of Myc-TRF2ΔBΔM (P < 0.01), Myc-TRF2ΔBΔM plus Myc-POT1 (P > 0.5), Myc-POT1 (P > 0.5), empty vector (P > 0.5), and the untreated control for 16 days. The data show the results of three experiments; the error bars represent standard deviations. P values were determined by Student's t test representing each indicated group and untreated control group. The P value between Myc-TRF2ΔBΔM and Myc-TRF2ΔBΔM plus Myc-POT1 was <0.05.
Telomere dysfunction induced by TRF2ΔBΔM can initiate genomic instability (52). The chromosomes in cells expressing TRF2ΔBΔM show abnormally high occurrences of mitotic defects, including lagging chromosomes, anaphase bridges, and associations between chromosome ends (52). We examined chromosomal abnormalities in IMR90 cells expressing TRF2ΔBΔM for 7 days and found a significantly high frequency of chromosomal abnormalities (chromosome fusion and breaks) in cells expressing TRF2ΔBΔM (Table 1). Chromosome fusion (multicentric chromosomes and chromosome end joining) was not observed in vector control cells. Overexpressing POT1 (coexpressing Myc-POT1 and TRF2ΔBΔM) prevented the specific chromosomal abnormalities induced by TRF2ΔBΔM (Table 1). We did not observe multicentric chromosomes and chromosome end joining in cells expressing Myc-POT1. These results indicated that POT1 can block specific chromosomal abnormalities induced by TRF2ΔBΔM.
TRF2ΔBΔM can induce a premature senescent phenotype in primary fibroblasts (45, 52). Consistent with previous reports, we found that Myc-TRF2ΔBΔM induced premature senescence 16 days after infection in IMR90 cells (Fig. 6B). Coexpressing Myc-POT1 significantly decreased the senescent phenotype induced by Myc-TRF2ΔBΔM. The cells expressing POT1 alone did not display this phenotype, as assayed by SA-β-Gal staining.
DISCUSSION
Our data indicate that POT1 is required to maintain the normal structure at telomeric single-stranded overhangs, protect against apoptosis, and prevent chromosomal instability and senescence. POT1 interacts with TRF2 to form a complex on telomeres that appears to be involved in the primary functions ascribed to telomeres in normal human cells.
Telomere protection by POT1.
A striking consequence of the loss of POT1 function is the erosion of telomeric single-stranded overhangs. The telomeric single-stranded tails play a key role for telomere protection. Linear chromosomes confer both benefits and burdens on eukaryotic cells (26). The chromosome end must be capped for its maintenance. The current evidence suggests that all eukaryotes use a single-stranded DNA-binding protein to cap the telomeres (14). The erosion of the single-stranded overhangs immediately cause telomere dysfunction. Yet, while the telomeric double-stranded TTAGGG repeats persisted, the telomeres failed to protect chromosome ends (15, 48, 52). The loss of telomeric single-stranded overhangs consequently induces cellular responses and chromosomal instability.
Loss of TRF2 from telomeres, caused by expression of the dominant-negative TRF2ΔBΔM mutation, resulted in a reduction in the single-stranded overhangs (52). But the mechanism by which TRF2 governs single-stranded overhang structure is unclear. Our data reveal a crucial role for POT1 in the maintenance of telomeric overhangs. Loss of POT1 from telomeres, caused by POT1-RNAi expression or the dominant-negative TRF2ΔBΔM mutation, resulted in an approximately 60 to 70% reduction in the telomeric overhang signal. These results are consistent with a recent report that the inhibition of POT1 by antisense oligonucleotides decreased telomeric overhang signals (31). POT1-RNAi directly induces the loss of the overhangs, supporting the model that protein binding is necessary for capping. Apparently, the chromosome end is rapidly altered when POT1 is reduced, and this alteration leads to the activation of the DNA damage response pathway. This outcome induces the erosion of telomeric overhangs and causes the loss of chromosomal integrity. Complete loss of telomeric DNA at some chromosomal ends upon POT1 depletion will also reduce the G overhang signals, even if reduction of the G overhang was not the primary event. To address this possibility, a telomere FISH assay was performed, and no detectable alteration of telomeric signals was observed (see Fig. S2 in the supplemental material). TRF2ΔBΔM removes POT1 from telomeres and also induces the erosion of the telomeric overhangs, the same consequences as the inhibition of POT1 by POT1-RNAi; this suggests that TRF2 and POT1 act in the same pathway on capping telomeres.
The loss of POT1 from the telomeres by TRF2ΔBΔM expression is correlated with the loss of telomeric overhangs. We observed the removal of POT1 binding to telomeres at day 2 after TRF2ΔBΔM expression by both immunostaining and ChIP assays. At this time point, telomeric single-stranded overhangs still persisted in cells expressing TRF2ΔBΔM, as determined by the T-OLA (data not shown), suggesting that TRF2ΔBΔM-induced reduction of POT1 from telomeres is a direct result from the interaction of both proteins.
We observed TRF2ΔBΔM induction of specific chromosomal abnormalities and the senescent phenotype, which is in agreement with previous reports (45, 52). Interestingly, POT1 effectively blocked the effect of TRF2ΔBΔM. This novel finding supports the hypothesis that chromosomal abnormalities and cellular senescence are the consequences of the loss of the protective mechanism on telomeric single-stranded overhangs by POT1 and TRF2. The frequency of chromosome breaks induced by expression of POT1-RNAi and TRF2ΔBΔM was significantly increased. These results are consistent with the hypothesis that the uncapping of telomeres by removing POT1 and TRF2 may induce telomere erosion to the critical short stage, trigger DNA damage response pathways, and then induce chromosome breaks.
POT1 and TRF2 are the telomeric proteins associated with the maintenance of the correct DNA configuration of the telomeric single-stranded overhangs. It was previously shown that telomerase is not required for the maintenance of telomeric single-stranded tails in yeast and mammals (17, 38, 41, 54, 56), and none of the other telomeric proteins identified in eukaryotes are known to affect telomere synthesis. However, according to one recent analysis (39), the disruption of telomerase activity in normal human cells alters the maintenance of the telomeric single-stranded overhangs without changing the rate of overall telomere shortening. Rescue of an hTERT mutant defective in telomere elongation by fusion with POT1 suggests that POT1 may recruit hTERT on telomeres (1). Further studies are needed to define the exact relationship between POT1 and telomerase.
Recent reports show that POT1 interacts with TRF1 as a terminal transducer of TRF1 telomere length control (36). POT1-interacting protein PTOP is responsible for recruitment of POT1 to the TRF1-TRF1-interacting protein 2 (TIN2) complex on telomeres (35, 58, 59). When these proteins were overexpressed, we confirmed the association of POT1 and TRF1. Thus, POT1 may have multiple functions. It binds to the telomeric single-stranded overhangs, interacts with TRF2 to protect them, and forms a complex with TRF1 to modulate telomere elongation.
Interaction of POT1 and TRF2.
We found that POT1 interacted with TRF2 in vitro and in vivo. POT1 bound to endogenous TRF2, as determined by immunoprecipitation. TRF2 and TRF2ΔBΔM overexpression was associated with POT1, confirming the POT1 and TRF2 interactions. TRF2ΔBΔM lacks the DNA-binding domain but still bound to POT1, suggesting that recovery of POT1 in the TRF2 immunocomplex is not dependent on the DNA-binding activity of TRF2. POT1 and TRF2 directly interacted in vitro, supporting an interaction between POT1 and TRF2 and arguing against DNA tethering as the basis for their coimmunoprecipitation. Functional studies were consistent with POT1 and TRF2 complex formation. TRF2ΔBΔM removed POT1 from the telomeres, as found by a colocalization study and a ChIP assay. Moreover, exogenous POT1 formed a complex with TRF2ΔBΔM to block its interaction with POT1 and TRF2. Thus, one possibility is that telomeric single-stranded overhangs are protected by POT1 and TRF2 and telomere-capping functions are recovered. To detect the binding of endogenous POT1 and TRF2, we used three anti-POT1 antibodies for the coimmunoprecipitation assay. Only one POT1 antibody could bring down TRF2. This is a possible reason that low enrichment of POT1-TRF2 complex in cells can be difficult to detect when a different affinity antibody is used (36). We also did not detect RAD50 and NBS1 in the POT1-immunoprecipitated complex, suggesting that TRF2 may form different complexes on the telomeres: TRF2-TIN2-repressor-activator protein 1 (RAP1) in a double-stranded TTAGGG repeat and TRF2-POT1 in a single-stranded overhang. It is possible that other proteins are also present in the latter complex, such as TIN2. A recent report shows by dynamic studies of protein binding to telomeres in living cells that TRF2 resides at telomeres in two distinct fractions (40). The fast fraction has binding dynamics similar to those of TRF1, and the slow one binds with dynamics similar to those of POT1. The results support the model that TRF2 has a dual role in the maintenance of telomeric length and integrity and that POT1 and TRF2 cooperate to maintain telomeric integrity.
Mechanisms of telomeric structure and function.
In a telomere-capping model, t-loops can provide cells with an architectural solution to the telomere protection problem in mammals (15, 23). t-loops are large duplex telomeric loops that appear to be formed by the invasion of the single-stranded overhang into the duplex part. TRF2, when given the right substrate, can form t-loop-like structures in vitro (48). The exact structure at the base of the t-loops is not known, but it is clear that there is a short segment of single-stranded DNA, likely representing a D-loop of TTAGGG repeats that are displaced by the invading single-stranded overhang. The invasion of the single-stranded overhang may simply sequester the telomere terminus from being mistaken for a site of damage (15, 23).
TRF2 and POT1 could also form a complex that can bind to the base of t-loops. The current model suggests that telomeric single-stranded binding proteins and single-stranded DNA overhangs are required for t-loop formation (23). E. coli single-stranded binding protein (SSB) can form a protein-DNA complex at the t-loop-tail junction, when HeLa t-loops are incubated with SSB (23). This suggests that a single-stranded binding protein might be located at the base of t-loops. As a specifically telomeric single-stranded DNA-binding protein, POT1 could bind to the telomeric tails in the D-loop and stabilize t-loop configuration. TRF2 is a duplex TTAGGG repeat-binding protein and does not interact with a single-stranded DNA overhang (15, 52), making it unlikely that TRF2 directly protects the single-stranded DNA end. TRF2 binds to the junction of double- and single-stranded telomeric DNA (15, 23, 48), very near the POT1-binding site. POT1 interacted with TRF2 to form a protein-DNA complex with telomeric single-stranded DNA. These data suggest that POT1 may be a component of the base of t-loops. POT1-RNAi and TRF2ΔBΔM cause the same consequence of the loss of telomeric overhangs, implying that TRF2ΔBΔM induces the loss of telomeric overhangs by both disrupting TRF2-mediated t-loops and removing POT1 from telomeres. Furthermore, POT1 can correct the specific chromosomal abnormalities and senescent phenotypes induced by TRF2ΔBΔM, which support this model.
At the ends of human chromosomes, three interconvertible states may exist: t-loops, POT1 capping, and engagement with telomerase (2). These different states could correlate with particular stages of the cell cycle. POT1, as a telomeric-specific single-stranded DNA-binding protein, may be involved in telomere capping and t-loop maintenance. It will be of interest to determine if POT1 contributes to these various regulated states during the cell cycle.
Supplementary Material
Acknowledgments
We thank Xin-Wei Wang and John Bradsher for their advice and assistance and Titia de Lange for providing the Myc-TRF2, Myc-TRF2ΔBΔM, and Myc-POT1 constructs and the anti-TRF1 antibody. We thank Peter Baumann for providing the pcDNA-POT1-V5 construct and his expert advice, Jack Griffith for providing the TRF2 protein, and Susan Garfield for confocal image assistance. We also thank Dorothea Dudek and the NCI CCR Fellows Editorial Board for their editorial assistance.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Armbruster, B. N., C. M. Linardic, T. Veldman, N. P. Bansal, D. L. Downie, and C. M. Counter. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol. Cell. Biol. 24:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292:1171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, P., E. Podell, and T. R. Cech. 2002. Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell. Biol. 22:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 2000. Telomere states and cell fates. Nature 408:53-56. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, E. H., S. Chan, J. Chang, T. B. Fulton, A. Krauskopf, M. McEachern, J. Prescott, J. Roy, C. Smith, and H. Wang. 2000. Molecular manifestations and molecular determinants of telomere capping. Cold Spring Harb. Symp. Quant. Biol. 65:253-263. [DOI] [PubMed] [Google Scholar]
- 7.Blasco, M. A. 2003. Mammalian telomeres and telomerase: why they matter for cancer and aging. Eur. J. Cell Biol. 82:441-446. [DOI] [PubMed] [Google Scholar]
- 8.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 10.Chong, L., B. van Steensel, D. Broccoli, H. Erdjument-Bromage, J. Hanish, P. Tempst, and T. de Lange. 1995. A human telomeric protein. Science 270:1663-1667. [DOI] [PubMed] [Google Scholar]
- 11.Cimino-Reale, G., E. Pascale, E. Battiloro, G. Starace, R. Verna, and E. D'Ambrosio. 2001. The length of telomeric G-rich strand 3′-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 29:E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colgin, L. M., K. Baran, P. Baumann, T. R. Cech, and R. R. Reddel. 2003. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 13:942-946. [DOI] [PubMed] [Google Scholar]
- 13.Counter, C. M., A. A. Avilion, C. E. LeFeuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lange, T. 2001. Cell biology. Telomere capping—one strand fits all. Science 292:1075-1076. [DOI] [PubMed] [Google Scholar]
- 15.de Lange, T. 2004. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5:323-329. [DOI] [PubMed] [Google Scholar]
- 16.DePinho, R. A. 2000. The age of cancer. Nature 408:248-254. [DOI] [PubMed] [Google Scholar]
- 17.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djojosubroto, M. W., Y. S. Choi, H. W. Lee, and K. L. Rudolph. 2003. Telomeres and telomerase in aging, regeneration and cancer. Mol. Cells 15:164-175. [PubMed] [Google Scholar]
- 19.Farr, C., J. Fantes, P. Goodfellow, and H. Cooke. 1991. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88:7006-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldser, D. M., J. A. Hackett, and C. W. Greider. 2003. Telomere dysfunction and the initiation of genome instability. Nat. Rev. Cancer 3:623-627. [DOI] [PubMed] [Google Scholar]
- 21.Gottschling, D. E., and T. R. Cech. 1984. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell 38:501-510. [DOI] [PubMed] [Google Scholar]
- 22.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 24.Hanish, J. P., J. L. Yanowitz, and T. de Lange. 1994. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 91:8861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath, M. P., V. L. Schweiker, J. M. Bevilacqua, J. A. Ruggles, and S. C. Schultz. 1998. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell 95:963-974. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa, F., and T. Naito. 1999. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat. Res. 434:99-107. [DOI] [PubMed] [Google Scholar]
- 27.Karlseder, J. 2003. Telomere repeat binding factors: keeping the ends in check. Cancer Lett. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 28.Karlseder, J., D. Broccoli, Y. Dai, S. Hardy, and T. de Lange. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283:1321-1325. [DOI] [PubMed] [Google Scholar]
- 29.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295:2446-2449. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S.-h., P. Kaminker, and J. Campisi. 2002. Telomeres, aging and cancer: in search of a happy ending. Oncogene 21:503-511. [DOI] [PubMed] [Google Scholar]
- 31.Kondo, T., N. Oue, K. Yoshida, Y. Mitani, K. Naka, H. Nakayama, and W. Yasui. 2004. Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer Res. 64:523-529. [DOI] [PubMed] [Google Scholar]
- 32.Lei, M., P. Baumann, and T. R. Cech. 2002. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry 41:14560-14568. [DOI] [PubMed] [Google Scholar]
- 33.Lei, M., E. R. Podell, P. Baumann, and T. R. Cech. 2003. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426:198-203. [DOI] [PubMed] [Google Scholar]
- 34.Li, G. Z., M. S. Eller, R. Firoozabadi, and B. A. Gilchrest. 2003. Evidence that exposure of the telomere 3′ overhang sequence induces senescence. Proc. Natl. Acad. Sci. USA 100:527-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, D., A. Safari, M. S. O'Connor, D. W. Chan, A. Laegeler, J. Qin, and Z. Songyang. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6:673-680. [DOI] [PubMed] [Google Scholar]
- 36.Loayza, D., and T. de Lange. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 424:1013-1018. [DOI] [PubMed] [Google Scholar]
- 37.Loayza, D., H. Parsons, J. Donigian, K. Hoke, and T. de Lange. 2004. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 279:13241-13248. [DOI] [PubMed] [Google Scholar]
- 38.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657-666. [DOI] [PubMed] [Google Scholar]
- 39.Masutomi, K., E. Y. Yu, S. Khurts, I. Ben Porath, J. L. Currier, G. B. Metz, M. W. Brooks, S. Kaneko, S. Murakami, J. A. DeCaprio, R. A. Weinberg, S. A. Stewart, and W. C. Hahn. 2003. Telomerase maintains telomere structure in normal human cells. Cell 114:241-253. [DOI] [PubMed] [Google Scholar]
- 40.Mattern, K. A., S. J. Swiggers, A. L. Nigg, B. Lowenberg, A. B. Houtsmuller, and J. M. Zijlmans. 2004. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 24:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 16:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 43.Reddel, R. R. 2003. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 194:155-162. [DOI] [PubMed] [Google Scholar]
- 44.Shay, J. W., and W. E. Wright. 2002. Telomerase: a target for cancer therapeutics. Cancer Cell 2:257-265. [DOI] [PubMed] [Google Scholar]
- 45.Smogorzewska, A., and T. de Lange. 2002. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21:4338-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smogorzewska, A., B. van Steensel, A. Bianchi, S. Oelmann, M. R. Schaefer, G. Schnapp, and T. de Lange. 2000. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 20:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spillare, E. A., A. I. Robles, X. W. Wang, J. C. Shen, G. D. Schellenberg, and C. C. Harris. 1999. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 13:1355-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stansel, R. M., T. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart, S. A., I. Ben Porath, V. J. Carey, B. F. O'Connor, W. C. Hahn, and R. A. Weinberg. 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33:492-496. [DOI] [PubMed] [Google Scholar]
- 50.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 13:1549-1556. [DOI] [PubMed] [Google Scholar]
- 51.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 52.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 53.Wei, C., and C. M. Price. 2004. Cell cycle localization, dimerization, and binding domain architecture of the telomere protein cPot1. Mol. Cell. Biol. 24:2091-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright, W. E., M. A. Piatszek, W. E. Rainey, W. Byrd, and J. W. Shay. 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18:173-179. [DOI] [PubMed] [Google Scholar]
- 55.Wright, W. E., and J. W. Shay. 2002. Historical claims and current interpretations of replicative aging. Nat. Biotechnol. 20:682-688. [DOI] [PubMed] [Google Scholar]
- 56.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 11:2801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, L., S. L. Davies, P. S. North, H. Goulaouic, J. F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 58.Ye, J. Z.-S., J. R. Donigian, M. van Overbeek, D. Loayza, Y. Luo, A. N. Krutchinsky, B. T. Chait, and T. de Lange. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279:47264-47271. [DOI] [PubMed] [Google Scholar]
- 59.Ye, J. Z., D. Hockemeyer, A. N. Krutchinsky, D. Loayza, S. M. Hooper, B. T. Chait, and T. de Lange. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.