Abstract
D cyclins (D1, D2, and D3) are components of the core cell cycle machinery in mammalian cells. It is unclear whether each of the D cyclins performs unique, tissue-specific functions or the three proteins have virtually identical functions and differ mainly in their pattern of expression. We previously generated mice lacking cyclin D1, and we observed that these animals displayed hypoplastic retinas and underdeveloped mammary glands and a presented developmental neurological abnormality. We now asked whether the specific requirement for cyclin D1 in these tissues reflected a unique pattern of D cyclin expression or the presence of specialized functions for cyclin D1 in cyclin D1-dependent compartments. We generated a knock-in strain of mice expressing cyclin D2 in place of D1. Cyclin D2 was able to drive nearly normal development of retinas and mammary glands, and it partially replaced cyclin D1's function in neurological development. We conclude that the differences between these two D cyclins lie mostly in the tissue-specific pattern of their expression. However, we propose that subtle differences between the two D cyclins do exist and they may allow D cyclins to function in a highly optimized fashion. We reason that the acquisition of multiple D cyclins may allow mammalian cells to drive optimal proliferation of a diverse array of cell types.
The progression of mammalian cells through the G1 phase of the cell cycle is driven by the D-type and E-type cyclins (43). These cyclins bind, activate, and provide substrate specificity for their associated cyclin-dependent kinases (CDKs). In contrast to other cyclins, which are induced periodically during cell cycle progression, the expression of D cyclins is controlled largely by the extracellular environment. For this reason, D cyclins are regarded as links between the external mitogenic milieu and the core cell cycle machinery (39, 45).
Three D-type cyclins, D1, D2 and D3, have been enumerated in mammalian cells (21, 33, 34, 37, 38, 57). These three proteins are encoded by separate genes located on different chromosomes, but they show significant amino acid similarity, suggesting that they arose from a common primordial ancestor gene (19, 58). On average, D cyclins show 50 to 60% identity throughout the entire coding sequence and 75 to 78% identity within the most conserved cyclin box domain (19, 58). All three D cyclins associate with CDK4 or CDK6, yielding six different combinations of cyclin D-CDK holoenzymes (2, 10, 20, 31, 32, 36).
An important issue is whether each of the D cyclins performs unique, possibly cell type-specific functions or the three proteins represent tissue-specific isoforms with virtually identical functions. At a biochemical level, all three D cyclins were shown to physically associate with CDK4 and CDK6 and to drive phosphorylation of the retinoblastoma protein, pRB, and pRB-related “pocket” proteins p107 and p130 (3, 28, 32, 36, 54, 56). The phosphorylation of these pocket proteins may represent the major function for cyclin D-CDK complexes in cell cycle progression, as shown by the observations that cells lacking pRB or p107 and p130 no longer require D cyclins for proliferation (1, 4, 16, 23, 29, 35, 40, 53).
However, biochemical differences between the three D cyclins were noted. Thus, cyclins D2 and D3 can form active complexes with CDK2, while cyclin D1 was reported to lack this ability (10, 17). Moreover, in addition to their well-established CDK-dependent functions, D cyclins were shown to interact with tissue-specific transcription factors, such as estrogen receptor, androgen receptor, thyroid receptor, and retinoic acid receptor alpha, C/EBP binding protein β, DMP1, and others (8, 27). In some cases, this interaction was uniquely ascribed to a particular D-type cyclin (9, 60).
To address the functions of the D-type cyclins in development, we and others generated mice lacking cyclin D1, D2, or D3 and characterized their phenotypes (12, 46-48). We found that mice lacking individual D cyclins were viable and displayed narrow, tissue-specific abnormalities. For instance, cyclin D1-deficient mice showed underdeveloped, hypoplastic retinas and presented a developmental neurological abnormality. Moreover, cyclin D1-deficient females displayed a normal mammary epithelial tree at the end of sexual maturation, but they failed to undergo full lobuloalveolar development during pregnancy (12, 48). Importantly, all these compartments developed normally in cyclin D2- or D3-deficient animals (46, 47), revealing a unique requirement for cyclin D1 in vivo in selected tissues.
In the present study, we asked whether the requirement for cyclin D1 function in these compartments was caused by tissue-specific pattern of D cyclin expression or alternatively reflected the presence of specialized tissue-specific functions for cyclin D1. To address this question by genetic means, we generated a knock-in strain of mice expressing cyclin D2 in place of cyclin D1. We next asked whether cyclin D2 could drive the normal development of cyclin D1-dependent tissues.
MATERIALS AND METHODS
Generation of cyclin D2→D1 knock-in mice.
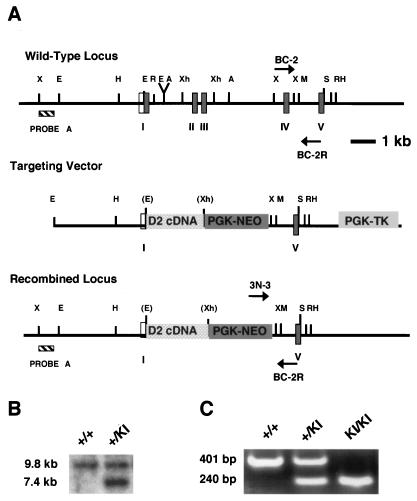
We modified the original cyclin D1 gene-targeting vector, which deleted the 5.4-kb EagI-to-XbaI fragment containing exons I to III (48) by additionally removing the 0.6-kb XbaI-XbaI fragment encompassing exon IV of the cyclin D1 gene. The 1.2-kb full-length cyclin D2 cDNA was inserted 5′ to the neomycin resistance gene into the modified cyclin D1 gene-targeting vector via EagI-XhoI sites (Fig. 1A). The 5′ UTR from the cyclin D2 gene was included in the cDNA sequence.
FIG. 1.
Generation of cyclin D2→D1 mice. (A) Cyclin D2→D1 gene-targeting strategy. Shown are the wild-type cyclin D1 allele, the cyclin D2→D1 gene-targeting construct, and the predicted product of the homologous recombination at the cyclin D1 locus. Filled boxes denote coding exons of the cyclin D1 gene, which are numbered, and an open box represents a noncoding portion of the first exon. Probe A, used for Southern blot analyses of the recombinant ES cell clones, is also indicated. D2 cDNA, cyclin D2 cDNA; PGK-NEO, neomycin resistance gene driven by the phosphoglycerokinase (PGK) promoter; PGK-TK, thymidine kinase gene driven by the PGK promoter. Restriction enzyme abbreviations: X, XbaI; E, EagI; H, Hind III; R, EcoRI; A, Asp718; Xh, XhoI; M, Mfe I; S, SalI. (B) Southern blot analysis of ES cell clones. DNA was digested with XbaI endonuclease, resolved on a gel, blotted, and probed with the probe A depicted in panel A. The sizes of wild-type and recombinant alleles are shown. The genotypes of ES cell clones are indicated above the lanes. (C) PCR genotyping of animals. Tail DNA was PCR amplified as described in Materials and Methods and resolved on a 2% agarose gel. The sizes of bands deriving from wild-type and mutant alleles are indicated. The genotypes of animals are shown above the lanes.
The cyclin D2→D1 gene-targeting construct was electroporated into J1 embryonic stem (ES) cells, and ES cell clones that underwent homologous recombination at the cyclin D1 locus were identified by Southern blotting of XbaI-digested genomic DNA, with probe A (Fig. 1A and B) (48). The probe detects a wild-type allele of 9.8 kb or a recombinant allele of 7.4 kb. Three of 330 ES cell clones screened underwent homologous recombination at the cyclin D1 locus. Subsequently, heterozygous cyclin D1+/D2→D1 ES cells were injected into C56BL/6 blastocysts, and homozygous cyclin D1D2→D1/D2→D1 mice were obtained by standard procedures (48).
PCR genotyping.
Genotype screening was done by a triple primer strategy based on a forward primer from the cyclin D1 gene (BC-2) (5′ GTCATCAAGTGTGACCCG 3′), a forward primer from the Neo cassette (3N-3) (5′ GATCTCTCGTGGGATCATTG 3′), and a reverse primer from the D1 gene sequence (BC-2R) (5′ GCACAGTCTGCCTGATGC 3′) (Fig. 1A). Samples were denatured at 95°C for 2 min, followed by 35 cycles of amplification, where each cycle consisted of the following steps: 95°C for 1 min, 60°C for 1 min, 72°C for 2 min. There was a final extension step at 72°C for 5 min. Wild-type PCR products were 401 bp and knock-in PCR products were 240 bp in size.
Histopathologic and whole-mount analyses.
Organs were dissected, fixed in Bouin's fixative (Sigma), and embedded in paraffin. Sections (each, 5 μm) were cut and stained with hematoxylin and eosin. For mammary gland whole mounts, inguinal mammary glands were removed from mice at 1 day postpartum and spread onto charged glass slides (Fisher). Glands were fixed in a 1:3 mixture of glacial acetic acid-ethanol for 24 h. The mounts were hydrated and stained overnight in 0.2% carmine red (Sigma). The slides were dehydrated in 70, 95, and 100% ethanol before clearance in toluene and storage in methyl salicylate.
Electroretinographic testing.
Wild-type homozygous cyclin D1D2→D1/D2→D1 littermates, 8 to 12 weeks old, were used. After overnight dark adaptation, mice were anesthetized with sodium pentobarbital injected intraperitoneally, and their pupils were dilated. Full-field electroretinograms (ERGs) were elicited with 10- μs flashes of white light (4.6 log ft L) presented in a Ganzfeld dome. Flashes were presented at 60-s intervals and monitored with a chlorided silver wire loop placed on the cornea. A saline cotton wick was placed in the mouth as a reference electrode and a subdermal electrode in the neck served as a ground. Responses were differentially amplified at a gain of 5,000 (−3 db at 2 and 300 Hz) and digitized at a sampling rate of 1,302 Hz. ERGs were quantified on a computer with respect to amplitude from baseline to the peak of the cornea-negative deflection (a-wave) and from the latter (or baseline, if absent) to the peak of the b-wave. Data from cyclin D1−/− animal responses were obtained from earlier experiments (48).
In situ hybridization.
In situ hybridization was done on retinas of 1-day-old mice, as described previously (48). Briefly, eyes were fixed in 4% paraformaldehyde for 48 h and dehydrated in 70 to 100% ethanol before being embedded in Paraplast. Sections (each, 6 μm) were cut and mounted before slides were dewaxed in preparation for hybridization with 35S-UTP-labeled riboprobes. Mouse cyclin D1 and D2 probes were generated with T3 and T7 polymerase, respectively, with the Superscript Reverse Transcriptase kit (Stratagene). Hybridization was done at 60°C overnight with 8 × 106 cpm of probe in 100 μl of hybridization mixture per slide. The in situ hybridization mixture contained 50% deionized formamide, 20 mM Tris-HCl (pH 8.0), 0.3 M NaCl, 10% dextran sulfate, 0.02% Ficoll 400, 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin, 0.5 mg of tRNA/ml, and 10 mM dithiothreitol.
Western blotting and immunoprecipitations.
Retinas were microdissected from 1-day-old pups. Organs were harvested from 21-day-old mice. Tissues were lysed in either ELB buffer containing 250 mM NaCl, 50 mM HEPES (pH 7.0), 5 mM EDTA, 0.5 mM dithiothreitol, 0.1% NP-40, and 1× protease inhibitor cocktail (Roche) or in immunoprecipitation buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween 20, 10% glycerol, 10 mM β-glycerophosphate, 1 mM NaF, 0.1 mM Na3VO4, and 2 mM sodium pyrophosphate. A total of 50 to 100 μg of proteins was separated on sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis gels and transferred to PVDF-Plus membranes (Osmonics). The immunoblots were probed with the following antibodies from Santa Cruz, except where indicated: anti-cyclin D1 (Ab-3; Neomarkers), anti-cyclin D2 (M-20), anti-cyclin D3 (C-16), anti-cyclin E (M-20), anti-CDK2 (M2) anti-CDK4 (H-22), anti-CDK6 (C-21), anti-p27 (C-19), and antiactin (Ms-X; Chemicon). Peroxidase-conjugated immunoglobulin G (Bio-Rad) was used as a secondary antibody, followed by enhanced chemiluminescence detection (Amersham). For immunoprecipitations, 200 to 300 μg of protein lysates was incubated with antibodies against cyclin D1 (Ab-3; Neomarkers), cyclin D2 (M-20), CDK4 (H-22), CDK6 (C-21), or p27 (C-19), all from Santa Cruz except as indicated and with Sepharose A (Amersham).
RESULTS
Generation of cyclin D2→D1 knock-in mice.
We assembled the cyclin D2→D1 gene-targeting construct by deleting exons I to IV of the cyclin D1 gene and by replacing them with mouse cyclin D2 cDNA, encompassing the entire coding sequence of cyclin D2 (Fig. 1A). Such a cyclin D2→D1 gene-targeting construct was introduced into wild-type ES cells, and heterozygous cyclin D1+/D2→D1 ES cells were obtained through homologous recombination (Fig. 1B). Subsequently, cyclin D1+/D2→D1 mice were generated by standard procedures (48). These cyclin D1+/D2→D1 animals displayed no obvious gross or histopathological abnormalities, were fertile, and had normal life spans. We next interbred the heterozygotes, which yielded homozygous cyclin D1D2→D1/D2→D1 mice; these mice will be henceforth referred to as cyclin D2→D1 knock-in mice. The genotype of knock-in mice was determined by PCR (Fig. 1C).
Molecular analyses of cyclin D2→D1 tissues.
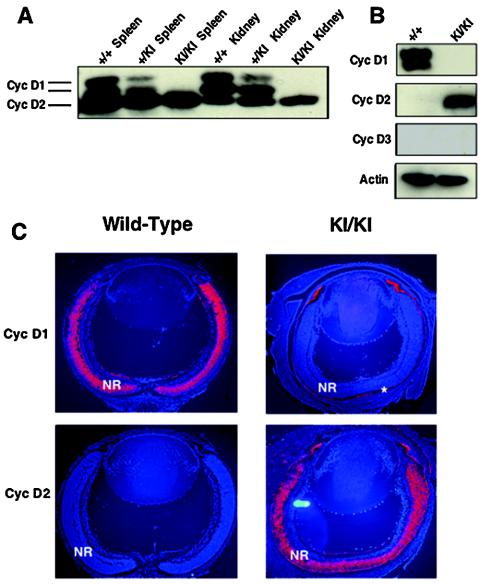
We started our analyses by verifying that cyclin D2→D1 mice indeed lacked cyclin D1 protein. To this end, we prepared lysates from different organs of knock-in and control wild-type mice and probed them with anti-cyclin D1 antibodies. As expected, we found that the tissues of cyclin D2→D1 animals lacked cyclin D1 (Fig. 2A and data not shown), confirming the genetic ablation of cyclin D1 in the mutant animals.
FIG. 2.
Molecular analyses of cyclin D2→D1 tissues. (A) Western blot analysis of indicated organs from wild-type (+/+), heterozygous (+/KI), or homozygous cyclin D2→D1 mice (KI/KI) probed with an antibody against cyclins D1 and D2. (B) Western blot analyses of retinas dissected from 1-day-old pups, probed with antibodies against cyclin D1, D2 or D3 or actin (loading control). (C) In situ hybridization analyses of retinas dissected from 1-day-old wild-type or cyclin D2→D1 (KI/KI) mice. Sections were hybridized with cyclin D1 or D2 cDNA probes. Red coloring represents the positive hybridization signal. Blue coloring represents counterstaining of cell nuclei with Hoechst stain. NR, neuroretina. The positive staining of the pigment cell layer (asterisk) represents an artifact, as this layer frequently stains with all probes.
In the cyclin D2→D1 gene-targeting strategy, the expression of knock-in cyclin D2 was placed under control of the cyclin D1 promoter. Therefore, we anticipated that in the tissues of knock-in mice, ectopic cyclin D2 would be targeted to cellular compartments normally expressing cyclin D1. Importantly, in our gene-targeting strategy cyclin D1 was replaced with wild-type, untagged version cyclin D2; hence, we were unable to distinguish between endogenous cyclin D2 versus ectopically expressed cyclin D2. Therefore, for the verification of the knock-in expression and for additional molecular analyses, we focused on the retinal tissue, as this compartment normally does not express cyclin D2 (14, 15, 55).
We first compared the expression of the D-type cyclins in the developing wild-type and knock-in retinas by Western blotting. As reported previously (14, 15, 55), wild-type retinas expressed high levels of cyclin D1 but essentially no cyclin D2 and very low levels of cyclin D3 (Fig. 2B). In contrast, the expression of the D cyclins was completely switched in cyclin D2→D1 tissues. We found that knock-in retinas lacked cyclin D1 but instead expressed cyclin D2 (Fig. 2B).
We next asked whether the expression pattern of cyclin D2 in the retinas of knock-in mice faithfully mimicked that of cyclin D1 in wild-type retinas. We prepared sections from wild-type and knock-in eyes and hybridized them with probes specific for cyclin D1 or D2. Again, we observed that wild-type retinas expressed high levels of cyclin D1 but no cyclin D2 (Fig. 2C). In contrast, the expression of the D cyclin transcripts was reversed in cyclin D2→D1 tissues. Thus, knock-in retinas lacked cyclin D1 mRNA but expressed cyclin D2 transcripts in its place. Importantly, the expression of cyclin D2 was confined to the outer, proliferating layer of the neuroretinas, precisely mirroring the expression of cyclin D1 in wild-type tissues (Fig. 2C).
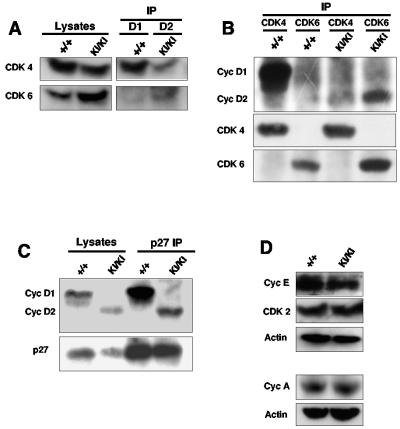
D cyclins are believed to drive cell cycle progression mainly through their association with CDK4 and CDK6 (45). To compare the composition of cyclin D-CDK complexes in wild-type versus knock-in retinas, we immunoprecipitated D cyclins from retinal lysates and probed the immunoblots with antibodies against CDK4 and CDK6. In the wild-type lysates, we detected association of cyclin D1 with CDK4 and (to a much lesser extent) with CDK6. Analyses of knock-in retinas revealed that cyclin D2 associated primarily with CDK6 and to a lesser extent with CDK4 (Fig. 3A and data not shown). We further confirmed this result by immunoprecipitating CDK4 or CDK6, followed by immunoblotting with anti-cyclin D antibodies. Again, we observed that ectopically expressed cyclin D2 interacted with significant amounts of CDK6, unlike cyclin D1 in wild-type retinas, which preferentially associated with CDK4 (Fig. 3B and data not shown). As we reported previously (14), we also detected very low levels of CDK2 associated with cyclin D1 in wild-type retinal lysates. Analyses of knock-in retinas revealed equally low levels of CDK2 associating with cyclin D2; importantly, this association was not increased in the mutant retinas (data not shown). Collectively, these findings suggest that within the retinal tissue, CDK6 might be the preferred partner of cyclin D2, while cyclin D1 associates mostly with CDK4.
FIG. 3.
Molecular analyses of cyclin D2→D1 retinas. (A) Protein lysates isolated from postnatal day 1 retinas were immunoprecipitated with an anti-cyclin D1 (+/+, wild-type samples) or anti-cyclin D2 antibodies (KI/KI, cyclin D2→D1 samples), followed by immunoblotting with anti-CDK4 or anti-CDK6 antibodies. The left panel (Lysates) shows immunoblots of straight lysates probed with antibodies against CDK4 or CDK6. (B) Retinal lysates were immunoprecipitated with antibodies against CDK4 or CDK6, followed by immunoblotting with an antibody recognizing cyclins D1 and D2 or with anti-CDK4 or anti-CDK6 antibodies. (C) Retinal lysates were immunoprecipitated with an antibody against p27Kip1. Immunoblots were probed with an antibody recognizing cyclins D1 and D2. The left panel (Lysates) shows immunoblots of straight lysates probed with antibodies against cyclin D1 and D2 or against p27Kip1. (D) Western blot analyses of retinal lysates probed with the indicated antibodies. Antiactin antibodies were used to ensure equal loading.
An important function of cyclin D-CDK4 and D-CDK6 complexes is to titrate cell cycle inhibitors p27Kip1 and p21Cip1 from cyclin E-CDK2 and cyclin A-CDK2 to cyclin D-CDK molecules, thereby controlling CDK2-associated kinase activity (6, 25, 42). While retinas do not express p21Cip1, they contain abundant p27Kip1 levels (14, 55, 59). To verify that ectopic cyclin D2-CDK6 complexes present in the retinas of cyclin D2→D1 mice were able to bind p27Kip1, we immunoprecipitated p27Kip1 from wild-type and cyclin D2→D1 retinas, and we probed immunoblots with antibodies against D cyclins. As expected, p27Kip1 in wild-type retinas was found to associate with cyclin D1. In the mutant retinas, p27Kip1 interacted with cyclin D2 (Fig. 3C), confirming an ability of ectopic cyclin D2-CDK6 complexes to bind p27Kip1. Lastly, analyses of the steady-state levels of cyclins E, A, CDK2, CDK4, and CDK6 revealed comparable levels of these proteins in the retinas of wild-type and cyclin D2→D1 mice, with the exception of slight but reproducible elevations of CDK6 levels in cyclin D2→D1 retinas (Fig. 3A and D). Collectively, these analyses indicate that in the retinas of cyclin D2→D1 mice, cyclin D2 is expressed in a pattern mirroring that of cyclin D1, and it replaces cyclin D1 in cyclin D-CDK4/CDK6 complexes.
Analyses of the retinal phenotype.
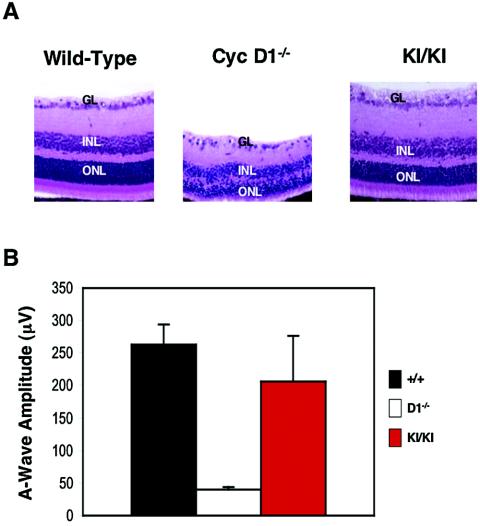
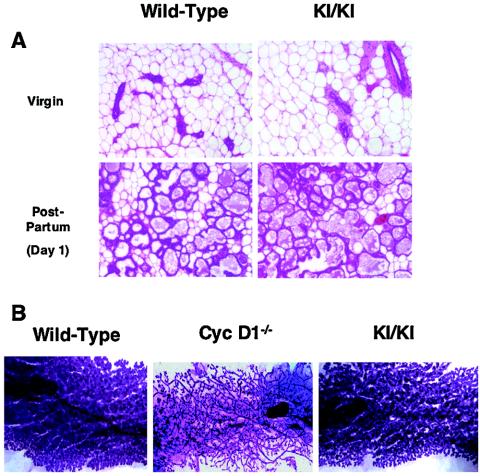
As we and others reported previously, cyclin D1-deficient mice displayed hypoplastic retinas, with all cell layers (outer nuclear, inner nuclear, and ganglion cell layers) being affected (Fig. 4A) (12, 48). In contrast, analyses of retinas collected from cyclin D2→D1 mice revealed essentially normal retinal development in knock-in animals. Indeed, at this level of resolution, cyclin D2→D1 retinas were virtually indistinguishable from wild-type tissues (Fig. 4A). This in turn, indicated that cyclin D2, when targeted to the retinal tissue in place of cyclin D1, can afford normal or nearly normal retinal development.
FIG. 4.
Rescue of the retinal abnormalities. (A) Hematoxylin and eosin-stained sections of retinas collected from 3-week-old wild-type, cyclin D1−/−, or cyclin D2→D1 (KI/KI) animals. GL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Magnification, ×40. (B) Mean amplitudes of a-waves generated in the retinas in response to a pulse of light (ERG testing). Error bars indicate standard deviations.
To obtain a functional, quantitative measure of the observed rescue of the cyclin D1−/− retinal phenotype by knock-in cyclin D2, we subjected our mice to ERG testing. In this procedure, animals are subjected to a short pulse of light, and electrophysiologic potentials generated within the retinas are recorded. Two waves of ERG potentials are generated by this procedures: a-waves, arising mainly from the photoreceptors, and b-waves, generated mostly by bipolar and Muller cells (22, 51, 52). Analyses of the amplitudes of the resulting retinal waves provide functional measure of this organ.
As we reported before (48), ERG testing of cyclin D1-deficient mice revealed reduced amplitudes of the a- and b-waves, with a-waves corresponding to 15% of those seen in wild-type animals (Fig. 4B and data not shown). In contrast, cyclin D2→D1 mice displayed a-waves with amplitudes that were 78% of normal values (Fig. 4B and data not shown); the mean amplitudes of the a-waves were 263.2 ± 30.5 μV for the wild type, 40.0 ± 4.1 μV for cyclin D1−/− mice, and 206.1 ± 71.2 μV for cyclin D2→D1 animals. Hence, ectopic expression of cyclin D2 in place of cyclin D1 can largely rescue the phenotypic manifestations of cyclin D1-deficiency and can afford nearly normal retinal function.
Analyses of mammary gland phenotypes.
Next, we turned our attention to mammary gland development. As we and others described previously, cyclin D1-deficient females developed a normal mammary epithelial tree during sexual maturation. However, these cyclin D1−/− animals failed to undergo normal pregnancy-driven lobuloalveolar development, a rapid burst of mammary epithelial expansion that takes place during late pregnancy (11, 12, 48, 49). As a consequence, cyclin D1-deficient females displayed underdeveloped mammary epithelial tree at the end of pregnancy (Fig. 5B), and they were unable to breast-feed their pups. Indeed, pups born to cyclin D1-deficient females had to be fostered to avoid starvation (11, 48).
FIG. 5.
Rescue of mammary epithelial abnormalities. (A) Histological sections of mammary glands collected from 2-month-old nulliparous females (Virgin) or from females 1 day after the delivery of pups [Post-Partum (Day 1)], wild type or cyclin D2→D1 (KI/KI), stained with hematoxylin and eosin. Magnification, ×80. (B) Whole-mount appearance of mammary glands collected 1 day after the delivery of pups from wild-type, cyclin D1−/−, or cyclin D2→D1 (KI/KI) females. The epithelium was stained with carmine red. Magnification, ×5.
In contrast, analyses of cyclin D2→D1 females revealed that knock-in animals presented extensively developed mammary epithelial lobuloalveolar structures at the end of pregnancy (Fig. 5B). Detailed histological analyses of cyclin D2→D1 mammary glands revealed abundant, milk-containing alveoli filling the entire fat pads (Fig. 5A), a feature not observed in cyclin D1-deficient females. Consequently, cyclin D2→D1 mothers were able to breast-feed their pups (data not shown). We concluded that cyclin D2, when expressed in place of cyclin D1, can drive normal or nearly normal mammary epithelial proliferation. This in turn indicates that the specific requirement for cyclin D1 in breast and retinal development is mainly caused by a tissue-specific pattern of cyclin D expression, rather then by the presence of specialized functions for cyclin D1 in those compartments.
Analyses of the developmental neurological abnormalities.
Cyclin D1-deficient mice displayed a triad of pathological features: the so-called leg-clasping reflex, growth retardation, and premature mortality (11, 48) that we interpreted as an indication of a developmental neurological abnormality. Therefore, we carefully tested for the presence of these phenotypes in cyclin D2→D1 mice.
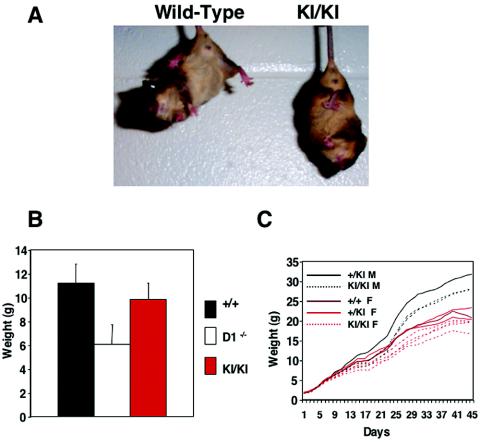
As reported previously, cyclin D1-deficient mice displayed a pathological leg-clasping reflex. Thus, when lifted by their tails, cyclin D1−/− animals responded by rapidly retracting their limbs, in contrast to wild-type mice, which extended their limbs. Analyses of cyclin D2→D1 mice revealed that knock-in animals retained the leg-clasping reflex (Fig. 6A), indicating that ectopic expression of cyclin D2 did not correct this abnormality. However, the histopathological lesion underlying this reflex in cyclin D1−/− mice remains unknown, thereby preventing us from further analyzing the molecular basis of the persistence of this abnormality in knock-in animals.
FIG. 6.
Partial rescue of neurological abnormalities by cyclin D2. (A) The leg-clasping reflex in cyclin D2→D1 mice (KI/KI). Shown are 3-week-old wild-type and mutant littermates. (B) Comparison of the mean body weight in 3-week-old wild-type (n = 15), cyclin D1−/− (n = 17), and cyclin D2→D1 (KI/KI) (n = 9) males. Error bars denote standard deviations. This time point was chosen, since at 3 weeks of age the difference between wild-type and cyclin D1−/− mice is most pronounced. (C) A growth curve of a litter born to heterozygous cyclin D1+/D2→D1 parents. Animals were weighed every other day for the first 6 weeks of life. The genotypes of the animals are indicated. M, males; F, females.
Cyclin D1−/− mice displayed severe growth retardation; by the third week of life, mutant mice were approximately one-half of the size of wild-type animals (Fig. 6B). Analyses of cyclin D2→D1 mice revealed that ectopic expression of cyclin D2 significantly rescued this growth retardation defect. We found that cyclin D2→D1 animals displayed average weights corresponding to 87% of those of wild-type littermates, compared to 51 to 59% in cyclin D1−/− mice (Fig. 6B and C).
As we and others reported before, the most affected cyclin D1−/− mice failed to thrive, and approximately 25% of the mutants died within the first 3 weeks of life (11, 48). In contrast, we did not observe any premature mortality in cyclin D2→D1 mice, and these animals displayed survival rates similar to those of wild-type littermates (data not shown). Hence, this manifestation of the neurological abnormality was fully rescued in cyclin D2→D1 mice.
Collectively, we interpreted these findings as an indication for the partial rescue of the neurological phenotypes seen in cyclin D1-deficient mice by cyclin D2.
Additional analyses.
Throughout the entire observation period, cyclin D2→D1 mice appeared healthy, were fertile, and did not display any obvious abnormalities. In addition to the studies of cyclin D1-dependent compartments, described above, we subjected organs collected from cyclin D2→D1 animals to detailed histopathological analyses. These studies revealed normal morphogenesis in all organs studied (data not shown). Hence, ectopic expression of cyclin D2 in place of cyclin D1 did not result in any appreciable abnormalities.
DISCUSSION
The three mammalian D-type cyclins are expressed in all proliferating tissues, with each cell type displaying a specific pattern of D-cyclin expression (53). In the past, we and others studied the in vivo functions of D cyclins by generating and analyzing mice lacking D cyclin genes. These studies revealed that genetic ablation of individual D cyclins led to very specific and circumscribed abnormalities. Thus, cyclin D1-deficient mice displayed neurological abnormalities, as well as retinal and mammary epithelial hypoplasia (12, 48). Cyclin D2−/− females were sterile due to the inability of ovarian granulosa cells to proliferate in response to the follicle-stimulating hormone, while cyclin D2-deficient males had underdeveloped testes (47). In addition, cyclin D2-deficient mice displayed impaired proliferation of B-lymphocytes (26, 50), as well as cerebellar abnormalities (18). Lastly, cyclin D3-deficient mice showed impaired development of immature T cells (46).
Analyses of the expression patterns of the D cyclins in wild-type mice provided a very good correlation with observed phenotypes. For instance, we noted that the developing retinas normally expressed high levels of cyclin D1 but virtually no cyclin D2 and very little cyclin D3 (14, 48). Likewise, the mammary glands of pregnant wild-type females expressed cyclin D1 (14, 49). It was also observed that ovarian granulosa cells—which are underdeveloped in cyclin D2−/− mice—normally expressed mainly cyclin D2 (44, 47), while immature T cells, a cyclin D3-dependent compartment, normally contained cyclin D3 (46). However, these results did not allow us to conclusively resolve whether the requirement for a given D cyclin in a certain tissue can be solely ascribed to the pattern of D cyclin expression or it indicates the presence of specific functions for a given D cyclin in a particular tissue. For instance, cyclin D1 might play a unique role in driving retinal or mammary epithelial development through its differential interaction with mammary- or retinal-specific interactors. Alternatively, cyclin D1-CDK, but not cyclin D2-CDK or cyclin D3-CDK complexes, might be endowed with the ability to phosphorylate mammary- or retinal-specific targets.
Two sets of past experiments strongly argued against the presence of unique, tissue-specific functions for the D cyclins. First, we previously observed that ectopic expression of cyclin E in place of cyclin D1 rescued all phenotypic manifestations of cyclin D1 deficiency (14). Importantly, these cyclin E→D1 knock-in mice displayed nearly normal retinal and breast development, suggesting that the major rate-limiting function of cyclin D1 in these tissues is to activate cyclin E (14). Since all three D cyclins would be predicted to activate cyclin E equally well, the observed rescue of the cyclin D1 deficiency by ectopic cyclin D2, reported here, is fully consistent with these earlier findings.
Moreover, we recently generated mice lacking all three D-type cyclins. We found that cyclin D1−/− D2−/− D3−/− embryos survived till the mid-gestation phase, revealing that the vast majority of embryonal cell types can proliferate in a cyclin D-independent fashion (24). D cyclins are critically required for the proliferation of the hematopoietic stem cells and cells of the myocardium, and consequently the absence of all three D cyclins led to lethality at embryonic day 16.5 (24). In contrast, double-knockout mice lacking cyclins D1 and D2 (but retaining cyclin D3) survived up to 3 weeks postnatally, and they displayed combined phenotypes of cyclin D1-null and cyclin D2-null animals but no additional obvious abnormalities. In the tissues of these cyclin D1−/− D2−/− mice, the remaining intact D cyclin (cyclin D3) became ubiquitously expressed and afforded nearly normal or normal development of many organs (7). These observations strongly supported the notion that the functions of the D cyclins are largely exchangeable, at least during this period of development. However, compartments affected in cyclin D1-null as well as in cyclin D2-null animals remained severely hypoplastic in cyclin D1−/− D2−/− animals. For instance, the appearance of cyclin D1−/− D2−/− retinas closely resembled the phenotype of cyclin D1-deficient mice; the same was true for the retinas of cyclin D1−/− D3−/− animals (7). Since no significant upregulation of the remaining, intact D cyclin was observed in these D1-dependent compartments (7), we could not conclude whether a D cyclin that is normally not expressed there could drive proliferation of cyclin D1-dependent tissues.
In the present study, we decided to directly compare the ability of cyclins D1 and D2 to drive the proliferation of cyclin D1-dependent compartments by genetic means, namely, by creating a knock-in strain of mice expressing cyclin D2 in place of cyclin D1.
Our analyses revealed that ectopically expressed cyclin D2 was able to nearly completely substitute for cyclin D1 in driving mammary and retinal development. Within the developing nervous system, the rescue of cyclin D1-null phenotype was incomplete, as one of the three neurological abnormalities, namely the leg-clasping reflex, persisted in cyclin D2→D1 animals. However, the histopathologic basis for the observed neurological abnormality remains unknown, and hence we could not study it further.
Altogether, we interpret our results as an indication that cyclin D2 can replace cyclin D1's functions to a significant degree and drive the proliferation of cyclin D1-dependent compartments. Although we did not test other combinations of D cyclins (D3→D1, D1→D2, etc.), we hypothesize that all three D cyclins can perform similar functions when expressed in an appropriate tissue.
An important distinction that needs to be made is that the demonstration that the two cyclins can perform the same function does not mean that they actually function in an identical fashion when coexpressed within the same cell. For instance, the work of Chen and Pollard (5) elegantly demonstrated that in uterine epithelial cells, treatment with 17β-estradiol causes translocation of cyclin D1-CDK4 complexes from the cytoplasm to the nucleus. However, in cyclin D1−/− uterine epithelial cells, cyclin D2 acquires cyclin D1's ability to shuttle between cytoplasm and the nucleus, thereby activating CDK4 kinase activity, but it does so slightly less efficiently then cyclin D1 (5). Hence, while the two cyclins can perform the same function in uterine epithelial cells, there is a preferred order in which these functions are carried out by particular D cyclins, with cyclin D1 being more efficient than cyclin D2.
We note that with the exception of mammary glands, where cyclin D2 seemed to fully or nearly fully correct cyclin D1−/− abnormalities, the rescue of most other phenotypes was very significant but not complete. For instance, ERG testing revealed that while cyclin D1−/− retinas responded to a light stimulus at a level of 15% of that seen in wild-type mouse retinas, ectopic expression of cyclin D2 restored this response to 78% of normal levels (Fig. 4B). Likewise, the neurologic phenotype was not completely rescued, and cyclin D2→D1 mice displayed lower weights than those seen in wild-type mice (Fig. 6B and C). One interpretation of these findings is that while the two cyclins can perform similar functions in most cyclin D1-dependent compartments, cyclin D2 is less efficient then cyclin D1 in some of them. Thus, the subtle differences between the two D cyclins may allow D cyclins to function in a highly optimized fashion. We propose that the acquisition of multiple D cyclins during evolution may have allowed mammalian cells to drive optimal proliferation of the diverse array of cell types.
We are aware of the possibility that the incomplete rescue of the cyclin D1−/− phenotype by knock-in cyclin D2 might be caused by suboptimal processing of the knock-in mRNA or protein in cyclin D2→D1 tissues. For instance, we replaced the cyclin D1 gene with cyclin D2 cDNA, and this might have led to suboptimal expression of the ectopically expressed cyclin D2 in knock-in tissues. Along these lines, we did not test whether knock-in of cyclin D1 cDNA into the cyclin D1 locus would lead to 100% correction of cyclin D1-null phenotypes. We note, however, that like D cyclins, several mammalian proteins such as Myc, Fos, and Jun exist as members of multigene families. It was previously shown that genetic replacement of the Jun gene with the gene encoding another member of the Jun family, namely, JunB, resulted in very significant but not full restoration of Jun's functions, with selected Jun−/− phenotypes not rescued at all (41). Likewise, replacement of the c-Fos gene with the Fos-related Fra-1 gene (13) or replacement of c-myc with N-myc (30) resulted in very strong but incomplete correction of the respective functions. We propose that these multigene families of proteins, including the D-type cyclins studied here, serve to perform similar and largely exchangeable functions, but they evolved to act in a most efficient manner in particular cell types or at particular developmental stages. Thus, these multigene families may allow perfect fine-tuning of the development of diverse cell types that together give rise to a mammalian organism.
Acknowledgments
We thank Sean Armour for help with collecting mutant retinas, and the members of the Sicinski lab for help and advice.
B.C.C. was supported by the UNCF-Merck Fellowship and a National Research Service Award (F31CA84293) from the National Cancer Institute. Y.G. is partially supported by the breast cancer SPORE for DFCI/Harvard. This work was supported by the DAMD17-99-1-9164 Idea Award from the Department of Defense (to P.S.).
The research on animals has complied will all relevant federal guidelines and institutional policies.
REFERENCES
- 1.Bartkova, J., J. Lukas, H. Muller, D. Lutzhoft, M. Strauss, and J. Bartek. 1994. Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer 57:353-361. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., L. Bonetta, D. MacAllan, D. Parry, A. Holder, C. Dickson, and G. Peters. 1994. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9:71-79. [PubMed] [Google Scholar]
- 3.Beijersbergen, R. L., L. Carlee, R. M. Kerkhoven, and R. Bernards. 1995. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 9:1340-1353. [DOI] [PubMed] [Google Scholar]
- 4.Bruce, J. L., R. K. Hurford, Jr., M. Classon, J. Koh, and N. Dyson. 2000. Requirements for cell cycle arrest by p16INK4a. Mol. Cell 6:737-742. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B., and J. W. Pollard. 2003. Cyclin D2 compensates for the loss of cyclin D1 in estrogen-induced mouse uterine epithelial cell proliferation. Mol. Endocrinol. 17:1368-1381. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, M., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21Cip1 and p27Kip1 CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciemerych, M. A., A. M. Kenney, E. Sicinska, I. Kalaszczynska, R. T. Bronson, D. H. Rowitch, H. Gardner, and P. Sicinski. 2002. Development of mice expressing a single D-type cyclin. Genes Dev. 16:3277-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coqueret, O. 2002. Linking cyclins to transcriptional control. Gene 299:35-55. [DOI] [PubMed] [Google Scholar]
- 9.Despouy, G., J. N. Bastie, S. Deshaies, N. Balitrand, A. Mazharian, C. Rochette-Egly, C. Chomienne, and L. Delva. 2003. Cyclin D3 is a cofactor of retinoic acid receptors, modulating their activity in the presence of cellular retinoic acid-binding protein II. J. Biol. Chem. 278:6355-6362. [DOI] [PubMed] [Google Scholar]
- 10.Ewen, M. E., H. K. Sluss, C. J. Sherr, H. Matsushime, J. Kato, and D. M. Livingston. 1993. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73:487-497. [DOI] [PubMed] [Google Scholar]
- 11.Fantl, V., P. A. Edwards, J. H. Steel, B. K. Vonderhaar, and C. Dickson. 1999. Impaired mammary gland development in Cyl-1(-/-) mice during pregnancy and lactation is epithelial cell autonomous. Dev. Biol. 212:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Fantl, V., G. Stamp, A. Andrews, I. Rosewell, and C. Dickson. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 9:2364-2372. [DOI] [PubMed] [Google Scholar]
- 13.Fleischmann, A., F. Hafezi, C. Elliott, C. E. Reme, U. Ruther, and E. F. Wagner. 2000. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 14:2695-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 15.Geng, Y., Q. Yu, E. Sicinska, M. Das, R. T. Bronson, and P. Sicinski. 2001. Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc. Natl. Acad. Sci. USA 98:194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, K. L., C. W. Jenkins, Y. Li, M. A. Nichols, X. Wu, C. L. O'Keefe, A. G. Matera, and Y. Xiong. 1994. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 8:2939-2952. [DOI] [PubMed] [Google Scholar]
- 17.Higashi, H., I. Suzuki-Takahashi, S. Saitoh, K. Segawa, Y. Taya, A. Okuyama, S. Nishimura, and M. Kitagawa. 1996. Cyclin-dependent kinase-2 (Cdk2) forms an inactive complex with cyclin D1 since Cdk2 associated with cyclin D1 is not phosphorylated by Cdk7-cyclin-H. Eur. J. Biochem. 237:460-467. [DOI] [PubMed] [Google Scholar]
- 18.Huard, J. M., C. C. Forster, M. L. Carter, P. Sicinski, and M. E. Ross. 1999. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development 126:1927-1935. [DOI] [PubMed] [Google Scholar]
- 19.Inaba, T., H. Matsushime, M. Valentine, M. F. Roussel, C. J. Sherr, and A. T. Look. 1992. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics 13:565-574. [DOI] [PubMed] [Google Scholar]
- 20.Kato, J., H. Matsushime, S. W. Hiebert, M. E. Ewen, and C. J. Sherr. 1993. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 7:331-342. [DOI] [PubMed] [Google Scholar]
- 21.Kiyokawa, H., X. Busquets, C. T. Powell, L. Ngo, R. A. Rifkind, and P. A. Marks. 1992. Cloning of a D-type cyclin from murine erythroleukemia cells. Proc. Natl. Acad. Sci. USA 89:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp, A. G., and P. H. Schiller. 1984. The contribution of on-bipolar cells to the electroretinogram of rabbits and monkeys. A study using 2-amino-4-phosphonobutyrate (APB). Vision Res. 24:1841-1846. [DOI] [PubMed] [Google Scholar]
- 23.Koh, J., G. H. Enders, B. D. Dynlacht, and E. Harlow. 1995. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 375:506-510. [DOI] [PubMed] [Google Scholar]
- 24.Kozar, K., M. A. Ciemerych, V. I. Rebel, H. Shigematsu, A. Zagozdzon, E. Sicinska, Y. Geng, Q. Yu, S. Bhattacharya, R. T. Bronson, K. Akashi, and P. Sicinski. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118:477-491. [DOI] [PubMed] [Google Scholar]
- 25.La Baer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 26.Lam, E. W., J. Glassford, L. Banerji, N. S. Thomas, P. Sicinski, and G. G. Klaus. 2000. Cyclin D3 compensates for loss of cyclin D2 in mouse B-lymphocytes activated via the antigen receptor and CD40. J. Biol. Chem. 275:3479-3484. [DOI] [PubMed] [Google Scholar]
- 27.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 28.Leng, X., M. Noble, P. D. Adams, J. Qin, and J. W. Harper. 2002. Reversal of growth suppression by p107 via direct phosphorylation by cyclin D1/cyclin-dependent kinase 4. Mol. Cell. Biol. 22:2242-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukas, J., D. Parry, L. Aagaard, D. J. Mann, J. Bartkova, M. Strauss, G. Peters, and J. Bartek. 1995. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503-506. [DOI] [PubMed] [Google Scholar]
- 30.Malynn, B. A., I. M. de Alboran, R. C. O'Hagan, R. Bronson, L. Davidson, R. A. DePinho, and F. W. Alt. 2000. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev 14:1390-1399. [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushime, H., M. E. Ewen, D. K. Strom, J. Y. Kato, S. K. Hanks, M. F. Roussel, and C. J. Sherr. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71:323-334. [DOI] [PubMed] [Google Scholar]
- 32.Matsushime, H., D. E. Quelle, S. A. Shurtleff, M. Shibuya, C. J. Sherr, and J. Y. Kato. 1994. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 14:2066-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsushime, H., M. F. Roussel, R. A. Ashmun, and C. J. Sherr. 1991. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65:701-713. [DOI] [PubMed] [Google Scholar]
- 34.Matsushime, H., M. F. Roussel, and C. J. Sherr. 1991. Novel mammalian cyclins (CYL genes) expressed during G1. Cold Spring Harb. Symp. Quant. Biol. 56:69-74. [DOI] [PubMed] [Google Scholar]
- 35.Medema, R. H., R. E. Herrera, F. Lam, and R. A. Weinberg. 1995. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl. Acad. Sci. USA 92:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyerson, M., and E. Harlow. 1994. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14:2077-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motokura, T., T. Bloom, H. G. Kim, H. Juppner, J. V. Ruderman, H. M. Kronenberg, and A. Arnold. 1991. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature 350:512-515. [DOI] [PubMed] [Google Scholar]
- 38.Motokura, T., K. Keyomarsi, H. M. Kronenberg, and A. Arnold. 1992. Cloning and characterization of human cyclin D3, a cDNA closely related in sequence to the PRAD1/cyclin D1 proto-oncogene. J. Biol. Chem. 267:20412-20415. [PubMed] [Google Scholar]
- 39.Murray, A. W. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221-234. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto, A., D. J. Demetrick, E. A. Spillare, K. Hagiwara, S. P. Hussain, W. P. Bennett, K. Forrester, B. Gerwin, M. Serrano, D. H. Beach, et al. 1994. Mutations and altered expression of p16INK4 in human cancer. Proc. Natl. Acad. Sci. USA 91:11045-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passegue, E., W. Jochum, A. Behrens, R. Ricci, and E. F. Wagner. 2002. JunB can substitute for Jun in mouse development and cell proliferation. Nat. Genet. 30:158-166. [DOI] [PubMed] [Google Scholar]
- 42.Polyak, K., J. Y. Kato, M. J. Solomon, C. J. Sherr, J. Massague, J. M. Roberts, and A. Koff. 1994. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, J. M. 1999. Evolving ideas about cyclins. Cell 98:129-132. [DOI] [PubMed] [Google Scholar]
- 44.Robker, R. L., and J. S. Richards. 1998. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 12:924-940. [DOI] [PubMed] [Google Scholar]
- 45.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 46.Sicinska, E., I. Aifantis, L. Le Cam, W. Swat, C. Borowski, Q. Yu, A. A. Ferrando, S. D. Levin, Y. Geng, H. von Boehmer, and P. Sicinski. 2003. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell 4:451-461. [DOI] [PubMed] [Google Scholar]
- 47.Sicinski, P., J. L. Donaher, Y. Geng, S. B. Parker, H. Gardner, M. Y. Park, R. L. Robker, J. S. Richards, L. K. McGinnis, J. D. Biggers, J. J. Eppig, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1996. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384:470-474. [DOI] [PubMed] [Google Scholar]
- 48.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 49.Sicinski, P., and R. A. Weinberg. 1997. A specific role for cyclin D1 in mammary gland development. J. Mammary Gland Biol. Neoplasia 2:335-342. [DOI] [PubMed] [Google Scholar]
- 50.Solvason, N., W. W. Wu, D. Parry, D. Mahony, E. W. Lam, J. Glassford, G. G. Klaus, P. Sicinski, R. Weinberg, Y. J. Liu, M. Howard, and E. Lees. 2000. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. Int. Immunol. 12:631-638. [DOI] [PubMed] [Google Scholar]
- 51.Steinberg, R. H., L. J. Frishman, and P. A. Sieving. 1991. Nagative components of the electroretinogram from proximal retina and photoreceptor, p. 121-160. In N. N. Osborne and G. J. Chader (ed.), Progress in retinal research. Pergamon, Oxford, England.
- 52.Stockton, R. A., and M. M. Slaughter. 1989. B-wave of the electroretinogram. A reflection of ON bipolar cell activity. J. Gen. Physiol. 93:101-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tam, S. W., A. M. Theodoras, J. W. Shay, G. F. Draetta, and M. Pagano. 1994. Differential expression and regulation of cyclin D1 protein in normal and tumor human cells: association with Cdk4 is required for cyclin D1 function in G1 progression. Oncogene 9:2663-2674. [PubMed] [Google Scholar]
- 54.Tedesco, D., J. Lukas, and S. I. Reed. 2002. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 16:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong, W., and J. W. Pollard. 2001. Genetic evidence for the interactions of cyclin D1 and p27Kip1 in mice. Mol. Cell. Biol. 21:1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, Z. X., D. Ginsberg, M. Ewen, and D. M. Livingston. 1996. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc. Natl. Acad. Sci. USA 93:4633-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong, Y., T. Connolly, B. Futcher, and D. Beach. 1991. Human D-type cyclin. Cell 65:691-699. [DOI] [PubMed] [Google Scholar]
- 58.Xiong, Y., J. Menninger, D. Beach, and D. C. Ward. 1992. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics 13:575-584. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, P., C. Wong, R. A. DePinho, J. W. Harper, and S. J. Elledge. 1998. Cooperation between the Cdk inhibitors p27Kip1 and p57Kip2 in the control of tissue growth and development. Genes Dev. 12:3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwijsen, R. M., E. Wientjens, R. Klompmaker, J. van der Sman, R. Bernards, and R. J. Michalides. 1997. CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405-415. [DOI] [PubMed] [Google Scholar]