Abstract
The ends of spontaneously occurring double-strand breaks (DSBs) may contain various lengths of single-stranded DNA, blocking lesions, and gaps and flaps generated by end annealing. To investigate the processing of such structures, we developed an assay in which annealed oligonucleotides are ligated onto the ends of a linearized plasmid which is then transformed into Saccharomyces cerevisiae. Reconstitution of a marker occurs only when the oligonucleotides are incorporated and repair is in frame, permitting rapid analysis of complex DSB ends. Here, we created DSBs with compatible overhangs of various lengths and asked which pathways are required for their precise repair. Three mechanisms of rejoining were observed, regardless of overhang polarity: nonhomologous end joining (NHEJ), a Rad52-dependent single-strand annealing-like pathway, and a third mechanism independent of the first two mechanisms. DSBs with overhangs of less than 4 bases were mainly repaired by NHEJ. Repair became less dependent on NHEJ when the overhangs were longer or had a higher GC content. Repair of overhangs greater than 8 nucleotides was as much as 150-fold more efficient, impaired 10-fold by rad52 mutation, and highly accurate. Reducing the microhomology extent between long overhangs reduced their repair dramatically, to less than NHEJ of comparable short overhangs. These data support a model in which annealing energy is a primary determinant of the rejoining efficiency and mechanism.
In the absence of a homologous template, double-strand breaks (DSBs) in eukaryotes are primarily repaired by direct rejoining of the ends (14). This rejoining is typically initiated by binding of the Ku heterodimer (Yku70 and Yku80 in Saccharomyces cerevisiae) to the DSB termini. Bridging of the ends is thought to be mediated by the Mre11/Rad50/Xrs2 complex in yeast and DNA-PK in mammals (6, 10). A complex of DNA ligase IV (Dnl4) and Xrcc4 (Lif1) ligates the break (36). The term nonhomologous end joining (NHEJ) is synonymous with this Ku- and DNA ligase IV-dependent rejoining mechanism. Ends that are not compatible or contain damaged bases require processing before they can be ligated. Artemis is thought to provide nuclease activity in mammalian NHEJ (21), but the end-processing nuclease(s) in yeast has not yet been identified. The yeast DNA polymerase Pol4 fills gaps in some end configurations (37), and the related enzymes Polμ and/or Polλ are thought to play this role in mammalian cells (19, 22). In the absence of NHEJ, an error-prone mechanism of DSB rejoining has been described previously (3, 20, 35, 39). DSBs situated between two direct repeats can also be repaired by single-strand annealing (SSA), a recombinational mechanism in which the sequence between the repeats is deleted (30).
Previous work on DSB repair has focused on DSBs induced by two methods: treating cells with DNA-damaging agents, such as ionizing radiation and radiomimetic chemicals, and inducing breaks with restriction and homing endonucleases. DNA-damaging agents can produce a wide variety of DNA lesions but do not allow for introduction of site-specific breaks, which limits the power of genetic readouts for monitoring repair. The use of endonucleases allows for greater control of DSB induction, but the scope of DSB ends that can be produced is severely limited. Homing endonucleases have been used to study repair of chromosomal DSBs with compatible 4-base 3′ overhangs (15, 18, 23). Transformation of plasmids cut with restriction enzymes has allowed the study of compatible and incompatible ends of both polarities (37, 38), but not the study of most blocking lesions or overhangs longer than 4 nucleotides.
We sought to develop a technique that allows for systematic testing of DSBs with a wide variety of end structures. Previously, others ligated oligonucleotides onto the ends of linearized plasmids (7, 32). To avoid extensive purification steps, we combined a similar approach with genetic selection using the yeast ADE2 gene, which ensures that only oligonucleotide-modified plasmids contribute to the output of the assay. This technique allows us to efficiently create and analyze many types of DSBs with complex end structures, limited only by the synthetic chemistries used to create the oligonucleotides. Understanding the repair of such DSBs is critical, because ends created by DNA-damaging agents in vivo are likely to contain overhangs of various lengths with lesions that block ligation, including 3′ phosphates, 3′ phosphoaldehydes, and 5′ deoxyribose phosphates. Proper handling of these overhangs and lesions is important to restorative repair. Gaps and flaps may also be generated by annealing of ends that are partially complementary, which is most relevant to inappropriate joining as seen in chromosomal rearrangements.
Here, we focus mainly on precise repair of DSBs with complementary overhangs, because this is most critical for preventing a loss of genetic information after DNA damage. Specifically, we used oligonucleotide-modified plasmids to investigate the repair of DSBs with overhangs of various lengths and to mimic restorative repair over the range of naturally occurring DSB configurations. We found that these DSBs can be repaired by three mechanisms: NHEJ, an SSA-like pathway that is catalyzed by Rad52, and a third mechanism that requires neither the NHEJ proteins nor Rad52. Pathway choice and repair efficiency are governed by the base pairing potential of the overhangs, and DSBs with longer compatible overhangs are repaired precisely more often than those with shorter overhangs. In contrast, DSBs with partially compatible long overhangs are repaired inefficiently and imprecisely. In addition, we present evidence that 5′ overhangs are not converted to 3′ overhangs during repair.
MATERIALS AND METHODS
Yeast strains.
S. cerevisiae strains used in this study are isogenic derivatives of the previously described wild-type strain YW465 (34). Strains YW450 (rad52Δ::HIS3), YW459 (yku70Δ::HIS3), YW507 (dnl4Δ::MET15), YW539 (rad52Δ::HIS3 yku70Δ::MET15), and YW878 (apn1Δ::HIS3 apn2Δ::kanMX4 tpp1Δ::MET15) were created using a PCR-mediated one-step gene replacement technique (4). All disruptions were confirmed by PCR. Strain YW1444 (apn1Δ::HIS3 apn2Δ::kanMX4 tpp1Δ::MET15 yku70Δ) was created by deleting yku70 from YW878 with URA3, then subsequently deleting URA3 and selecting for 5-fluoroorotic acid resistance.
Creation of plasmids.
Plasmid pTW423 is a derivative of the CEN plasmid pES16, which was described previously (36). pTW423 was constructed by first cutting pES16 with XhoI and SalI and ligating the ends together, thereby destroying the XhoI and SalI restriction sites in the polylinker. Next, a PCR product was prepared from pES16 using the T3 primer and primer OW758(5′-GCGAGATCTTAACTAGCTGACCTCGAGTTCCGGACTCCGTTC [theBglII and XhoI sites are in bold type and stop codons are underlined]). This product was digested with BglII and KpnI (in the polylinker) and ligated into the modified pES16 plasmid cut with the same enzymes. The resulting plasmid, pTW423, has a silent A-to-C change at ADE2 nucleotide 679, which introduces a unique XhoI site into the ADE2 coding sequence. The construction simultaneously deleted 78 essential ADE2 nucleotides (positions 598 to 676) and replaced them with a polyterminator.
Ligation of oligonucleotides onto plasmid restriction sites.
pTW423 was digested with BglII and XhoI (New England Biolabs [NEB]), producing 18-bp and 7,334-bp fragments. The 7,334-bp fragment was purified from the 18-bp fragment with the GeneClean kit (QBioGene). Complementary oligonucleotide pairs were designed to restore the ADE2 coding sequence upon repair of the DSB (see Fig. S1 in the supplemental material). Oligonucleotides were purchased from Invitrogen. 3′ and 5′ phosphates were incorporated during synthesis by the vendor as needed. To ensure that the plasmid was not recircularized during ligation, oligonucleotides were synthesized with 5′ phosphates at the plasmid ligation ends and 5′ hydroxyl groups at the DSB ends. Oligonucleotide pairs were heated to 95°C for 5 min and annealed by slow cooling to 25°C. Annealed oligonucleotides (200 pmol) were ligated onto 4 pmol of pTW423 digested with BglII and XhoI with T4 DNA ligase (Roche Molecular Biochemicals) at 16°C for 16 h. Plasmids were then purified from unligated oligonucleotides with the GeneClean kit (QBioGene). DNA concentration was quantified by UV spectrometry, and product size, linearity, and integrity were verified by agarose gel electrophoresis.
Addition of 5′ phosphates to DSB ends after ligation.
Ten micrograms of oligonucleotide-modified plasmid DNA was gel purified and incubated with 1 U of T4 polynucleotide kinase (NEB) in a 50-μl reaction mixture containing 1× T4 DNA ligase buffer (NEB) (which includes ATP) for 30 min at 37°C. The resulting plasmid was gel purified and transformed into yeast in parallel with plasmids treated similarly but without T4 polynucleotide kinase.
Primer extension.
Oligonucleotide primers OW1377 (5′-TTTGGAAGTACTGAAGGATCGTCC) and OW1378 (5′-TACCACAACCGGGAAAAGATTTG)were 5′ end labeled with [γ-32P]ATP with T4 polynucleotide kinase (Roche Molecular Biochemicals). Reaction mixtures (20 μl) containing 0.1 pmol of primer, 0.01 pmol of plasmid, 0.2 mM concentrations of the deoxynucleoside triphosphates (dNTPs) (Invitrogen), and 2.5 U of AmpliTAQ polymerase (Applied Biosystems) were incubated at 94°C for 3 min, 53°C for 1 min, and 72°C for 3 min. Twenty microliters of formamide loading dye was added, and 15 μl was run on an 8% acrylamide gel. Bands were detected and quantified with a Typhoon phosphorimager using ImageQuant version 2003.02 software.
Yeast transformation.
Plasmids were transformed into yeast using a high-efficiency lithium acetate (LiAc) method. One hundred nanograms of ligation-modified pTW423 (marked with URA3) was cotransformed with 10 ng of supercoiled pRS315 (marked with LEU2) (4). Cells were grown overnight in 2 ml of YPAD (1% yeast extract, 2% Bacto Peptone, 40 μg of adenine per ml, and 2% dextrose) and then diluted back into 25 ml of YPAD and grown for 3.5 h to an optical density at 600 nm of 0.5 to 0.8. Cells were spun down and washed with 0.1 M LiAc, resuspended to 7.5 × 108 cells/ml in 0.1 M LiAc, and incubated for 10 min at 25°C. One hundred microliters of the cell suspension was added to the DNA to be transformed with 10 μl of freshly boiled single-stranded salmon sperm DNA (10 mg/ml) (Sigma) and 700 μl of transformation solution (40% polyethylene glycol, 1× Tris-EDTA buffer, 0.1 M LiAc). Cells were then incubated at 30°C for 30 min, 85 μl of DMSO was added, and the cells were heat shocked in a 42°C water bath for 7 min. Cells were spun down, washed with 1 ml of sterile double-distilled water (ddH2O), resuspended in 1 ml of sterile ddH2O, plated in parallel on medium containing glucose (Glu) and lacking either uracil (Ura) or leucine (Leu), and grown at 30°C for 3 or 4 days. Relative repair efficiency was measured as the ratio of Ade+ (white) colonies on plates containing Glu but lacking Ura to colonies on plates containing Glu but lacking Leu.
Sequencing.
Ade+ colonies were grown overnight in medium containing Glu but lacking Ura at 30°C, and plasmid DNA was extracted using the glass bead lysis method (1). Primers OW602 (5′-CCAACTTTTGAGCGACAGAG) and OW750 (5′-GTGATTGACTCTTGCTGAC) were used to amplify a 1-kb region containing the break, and OW1377 (5′-TTTGGAAGTACTGAAGGATCGTCC) was used to sequence across the break site. Sequencing was performed by the University of Michigan sequencing core.
Filling of 5′ ends.
To analyze the effect of prefilling a 5′ overhang on NHEJ, 5 μg of BglII-digested pES16 (36) was incubated at room temperature for 20 min in the presence of 5 U of Klenow fragment and buffer (NEB) and a 50 μM concentration of the appropriate mixtures of dNTPs (either G, G plus A, G plus A plus T, or G plus A plus T plus C). Unfilled overhangs were incubated in the same buffer but without enzyme or dNTPs. After incubation, all DNAs were purified by phenol-chloroform extraction and ethanol precipitation prior to transformation. Agarose gel electrophoresis confirmed that all filled plasmids were intact and at equal concentrations.
RESULTS
Oligonucleotide-modified plasmids for studying DSB repair.
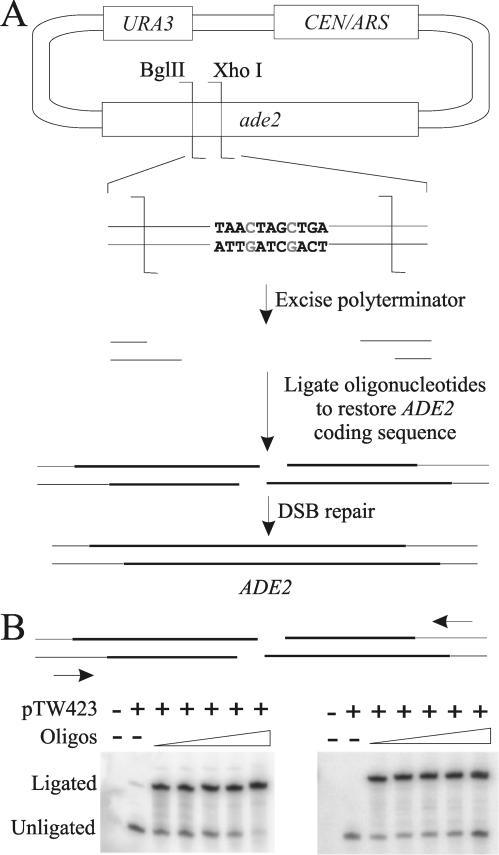
Plasmid recircularization assays have long been used to study DSB repair in vivo in yeast (2, 3, 37). We sought to improve this technique by ligating oligonucleotides onto plasmid ends. Others have ligated oligonucleotides onto plasmids for use in vitro (7, 32), but laborious purification of the modified product was essential to remove unmodified plasmids. We eliminated this requirement by using genetic selection. Our plasmid, pTW423, contains a nonfunctioning ade2 allele in which essential sequence between an endogenous BglII site and an engineered XhoI site is replaced with a polyterminator sequence containing a stop codon in each reading frame (Fig. 1A). Unmodified or single-cut plasmid can thus never give rise to an Ade+ colony. After removal of the polyterminator by digestion with BglII and XhoI, annealed oligonucleotide pairs that will reconstitute the wild-type ADE2 sequence upon DSB repair are ligated onto the BglII and XhoI ends (Fig. 1A; also see Fig. S1 in the supplemental material). To prevent recircularization of the plasmid during ligation, the termini of the oligonucleotides that will form the DSB contain 5′ hydroxyl groups. Previous results establish that damage-induced DSBs often contain 5′ hydroxyls (9) and that such DSBs can be rejoined in yeast even though the organism lacks a 5′ kinase (34). The oligonucleotide-modified plasmid is then transformed into yeast, with cotransformation of a supercoiled LEU2-marked plasmid used as a transformation control. Since ADE2 was deleted in the strains used and the cotransformed plasmid does not contain ADE2 sequence, the DSB cannot be repaired to Ade+ by homologous recombination. Therefore, Ade+ colonies can be generated only by direct in-frame rejoining of the DSB and only when both oligonucleotide pairs are successfully ligated to provide the complete ADE2 sequence. Imprecise repair will most often lead to frameshifts or deletions in ADE2 that result in Ade− colonies. Such colonies are phenotypically indistinguishable from those resulting from incomplete oligonucleotide ligation, so this system is not optimized for the quantitative study of imprecise repair. However, some information about imprecise joining can be revealed by sequencing (see below).
FIG. 1.
Oligonucleotide-modified plasmid assay. (A) Plasmid modification scheme. pTW423 is digested with BglII and XhoI and purified, removing the polyterminator. Annealed oligonucleotides are then ligated onto the BglII and XhoI ends, restoring the ADE2 coding sequence. Precise in-frame repair of the break yields Ade+ colonies. (B) Primer extension assay to determine the oligonucleotide ligation efficiency. Annealed oligonucleotides (Oligos) were added to the ligation reaction mixture at concentrations of 50, 100, 500, 1,000, and 5,000-fold molar excess over the concentration of the linearized plasmid (indicated by the thickness of the triangle over the lanes). Primer extension was performed after ligation as described in Materials and Methods.
To verify the above predictions, we first ligated each oligonucleotide pair for pTW423-34 (see Fig. S1 in the supplemental material) in the absence of the other pair and transformed the resulting half-modified plasmids into yeast. As expected, only Ade− colonies arose (data not shown). We also sequenced many joined plasmids during the course of the experiments described below (see Fig. S2 in the supplemental material). Again, as expected, Ade+ colonies always contained both ligated oligonucleotide pairs. Moreover, Ade+ colonies represented reconstitution of the wild-type ADE2 sequence in the large majority of instances. Imprecise repair did occasionally result in functional ADE2 when the reading frame was restored and five codons or fewer were lost. Thus, ADE2 selection largely, but not completely, selects for accurate rejoining. We finally note the occasional occurrence of single-base errors in sequenced plasmids, typically confined to the span of the ligated oligonucleotides but outside the repair joint itself. We attribute these errors to failed or misincorporated bases during oligonucleotide synthesis.
Primer extension was used to measure the efficiency of oligonucleotide ligation onto each modified plasmid (Fig. 1B). In this assay, ligation-modified and unmodified ends give extension products of different lengths, allowing independent assessment of the ligation extent at each end. The ligation frequency was typically in the range of 70 to 90% (see Fig. S1 in the supplemental material). Importantly, while differences in the ligation efficiency will not affect comparisons between strains for a given plasmid, they might affect comparison of one plasmid to another. For most plasmids, the ligation frequency varied within a range of approximately 20%. Therefore, interplasmid differences of 20% or less observed in the transformation assay could be accounted for by differences in ligation frequency rather than repair of the DSB, and we did not consider these differences to be meaningful. As seen below, in many cases repair rates of different plasmids varied across many orders of magnitude, however, which clearly cannot be accounted for by the level of variation observed in ligation frequencies.
Overhang length is positively correlated with repair rate.
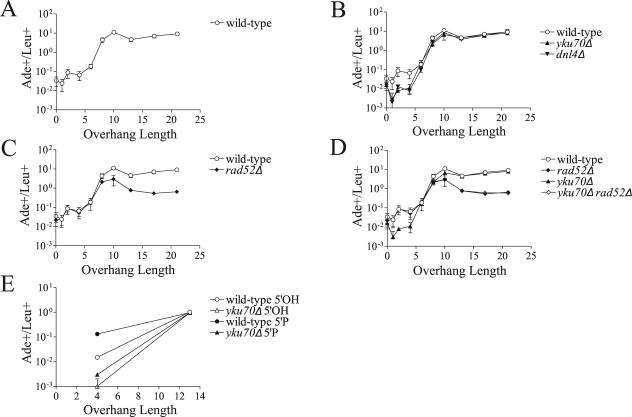
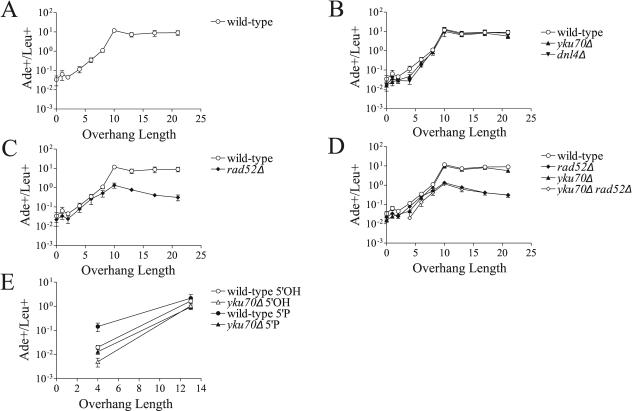
The first question we have addressed with oligonucleotide-modified plasmids is how the overhang length affects the repair of a DSB. Previous studies using restriction endonucleases have been unable to test DSBs with overhangs longer than 4 bases. Because overhangs of different lengths will vary in their ability to anneal and because, upon annealing, the distance between the ligation points will vary, these DSBs may be repaired by different mechanisms. We generated a series of modified plasmids with DSBs containing overhangs ranging from 0 (blunt ends) to 22 nucleotides in both 3′ and 5′ orientations (see Fig. S1 in the supplemental material) and transformed them into wild-type yeast. This panel of plasmids mimics the variation expected to occur at naturally occurring DSBs that arise from single chromosome breaks, where overhangs are necessarily complementary. Moreover, our design targets the precise rejoining that is most desirable at such DSBs. Strikingly, the rate of Ade+ colony formation increased approximately 100-fold as overhang lengths increased from 4 to 10 nucleotides for both 3′ and 5′ overhangs (Fig. 2A and 3A). No additional increase in repair rate was observed when overhang lengths were increased to 13, 17, and 22 bases (Fig. 2A and 3A).
FIG. 2.
Rejoining of DSBs with 3′ overhangs. (A) DSBs with 3′ overhangs are repaired more efficiently as the single-stranded overhang length increases. To control for transformation efficiency, repair is expressed as the ratio of Ade+ colonies arising from accurate repair of the linearized plasmid over Leu+ colonies resulting from uptake of a cotransformed supercoiled plasmid. This ratio can exceed 1 in part because 10-fold-more linear plasmid than circular plasmid is transformed. Each point represents the mean ± standard deviation (error bar) from four independent transformations. (B) DSB repair is independent of Yku70 and Dnl4 when overhangs are 6 nucleotides and longer. (C) Repair of DSBs with overhangs 8 nucleotides and longer is partially compromised in a rad52Δ strain. (D) In the absence of Rad52, DSBs with overhangs of >8 nucleotides are not repaired by Yku70-dependent NHEJ. Due to the decreased transformation efficiency of the yku70Δ rad52Δ strain, we were able to reliably assay only DSBs with overhangs of 6 nucleotides or longer at the plasmid concentration used. (E) Adding 5′ phosphates to DSBs enhances the repair of DSBs with 4- but not 13-base overhangs. Different carrier DNA and plasmid preparations were used in the experiment shown panel E than those used in the experiments shown in panels A to D, resulting in a different range in the linear to supercoiled plasmid transformation ratio. Variability in this ratio is common when changing reagents in yeast transformation experiments, as carrier DNA has nonequivalent effects on the uptake of linear and supercoiled DNA. This does not affect the interpretation of the data within an experiment, and similar trends are observed in panel E versus panels A to D.
FIG. 3.
Rejoining of DSBs with 5′ overhangs. The experiments are analogous to those shown in Fig. 2. Data are expressed as the ratio of Ade+ to Leu+ colonies as in Fig. 2.
Repair of DSBs with overhangs of 6 nucleotides or longer is independent of NHEJ.
In the absence of a homologous donor, repair of DSBs with overhangs of 4 nucleotides or fewer is known to be dependent on NHEJ (2, 3, 15). To determine whether repair of DSBs with longer overhangs is also dependent on NHEJ, we transformed our oligonucleotide-modified plasmids into yku70Δ and dnl4Δ strains. Repair of DSBs with 3′ overhangs of 1, 2, and 4 nucleotides decreased by approximately 10-fold in the NHEJ mutants (Fig. 2B), similar to previous results with the plasmid recircularization assay (37). Sequencing the break junctions revealed that the residual repair in yku70Δ yeast indeed represented precise rejoining in all 20 Ade+ colonies arising from 4-base 3′ overhangs (see Fig. S2 in the supplemental material). NHEJ dependence decreased dramatically at longer overhangs, with the yku70Δ mutant approaching the wild-type level in the 6- to 8-base range (Fig. 2B). A similar pattern was observed with 5′ overhangs, although 1- to 4-base 5′ overhangs were less dependent on NHEJ than 3′ overhangs were (Fig. 3B). Repair of blunt overhangs was reduced to about 50% of wild-type in the NHEJ mutants (Fig. 2B and 3B, 0-base overhang point). This observation was not unexpected, because blunt ends are known to be joined imprecisely in wild-type yeast (3, 28). Since Ade+ colonies are mainly generated by precise repair, this effectively reduces the defect seen in the NHEJ mutants.
To determine whether the pattern of NHEJ dependence we observed could be due to the use of 5′ hydroxyl termini, we used T4 polynucleotide kinase to add 5′ phosphates to the ends of the 4- and 13-base overhang DSBs and repeated transformations into wild-type and yku70Δ strains. Indeed, adding 5′ phosphates increased the repair rate of DSBs with 4-base overhangs seven- to ninefold in wild-type yeast but did not significantly affect rejoining in yku70Δ yeast, resulting in a net increase in the NHEJ defect to 50-fold for 3′ overhangs and 11-fold for 5′ overhangs (Fig. 2E and 3E). In contrast, repair of DSBs with 13-base overhangs was independent of the status of the 5′ termini and was still 7- to 15-fold greater than 4-base overhangs with 5′ phosphates (Fig. 2E and 3E). Thus, the NHEJ-independent increase in rejoining observed when overhangs increase from 4 to 13 bases is not an artifact of 5′ hydroxyl usage.
These data indicate that complementary DSB ends can be repaired precisely in the absence of both NHEJ and a homologous donor and that this process occurs at a markedly higher frequency as the overhang lengths increase. Notably, repair rates were nearly identical in yku70Δ and dnl4Δ strains at all overhang lengths (Fig. 2B and 3B). This supports the idea that the Dnl4/Lif1 complex and the Yku70/Yku80 heterodimer can promote repair only when both complexes are present.
Repair of overhangs 8 nucleotides or longer is partially dependent on Rad52.
Since DSBs with longer overhangs were not dependent on NHEJ, we hypothesized that their joining may be mechanistically similar to the annealing of exposed repeats during SSA. SSA repairs DSBs between two direct repeats, resulting in deletion of one repeat and the intervening sequence (13). SSA is initiated by resection of the 5′ ends of the DSB, which reveals the region of homology and creates 3′ single-stranded tails (30). The homologous strands are then annealed in a Rad52-dependent process (26), nonhomologous flaps are cleaved by Rad1/Rad10 (11), and the strands are ligated. Repair efficiency increases when the repeats are moved closer to the DSB and when their length is increased (31). In the absence of a competing recombination donor, SSA can repair a DSB between terminal repeats as short as 10 bases, but SSA is not active on 4-base repeats (15). In parallel with this, repair of DSBs with 3′ and 5′ overhangs of 6 bases or less was not significantly compromised in rad52Δ cells (Fig. 2C and 3C). When overhangs reached 8 nucleotides, however, repair efficiency was decreased by about 50% in the rad52Δ strain (Fig. 2C and 3C). DSBs with overhangs of 10 bases or longer showed an approximately 10-fold decrease in Ade+ colony formation in the absence of Rad52, indicating that Rad52 participates in the repair of these breaks. Importantly, however, Rad52 was not absolutely required for repair of DSBs with overhangs greater than 8 bases, as their repair rate in the absence of Rad52 was still about 10-fold higher than that of DSBs with 1- to 4-base overhangs in wild-type cells (Fig. 2C and 3C). Sequencing again showed that all 20 sequenced Ade+ colonies generated by the rad52Δ strain with 3′ 13-base overhang DSBs represented precise repair (see Fig. S2 in the supplemental material). There was no additional decrease in formation of Ade+ colonies when yku70 was deleted in addition to rad52 (Fig. 2D and 3D), indicating that repair of DSBs with longer overhangs does not require NHEJ even in the absence of Rad52.
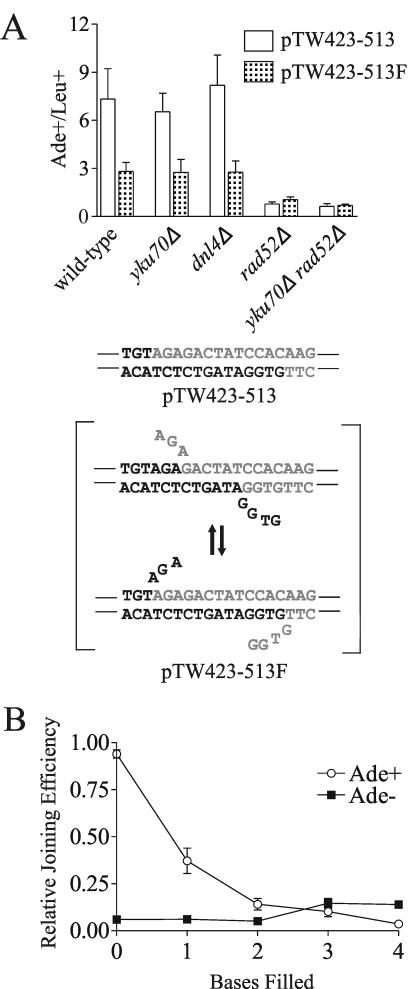
5′ ends are not converted to 3′ overhangs during Rad52-dependent repair or NHEJ.
Rad52 has single-stranded DNA annealing activity in vitro (24) and is thought to anneal 3′ ends in the synthesis-dependent strand annealing (25) and SSA pathways of recombinational repair (26). Whether Rad52 can anneal 5′ overhangs in vitro or in vivo has not yet been tested, however. Our finding that mutation of rad52 impaired joining of both 3′ and 5′ overhangs implies that Rad52 can anneal both types of ends. However, it is also possible that 5′ single-stranded tails are converted to 3′ tails via filling by a polymerase and subsequent 5′ resection. To address this possibility, we constructed a DSB with 13-base 5′ overhangs similar to the sequence of pTW423-513, except that the ends are partially filled on either side, which we named pTW423-513F (Fig. 4A). If 5′ ends are indeed converted to 3′ ends by filling and resection, partial filling of the ends should not affect the repair efficiency. Alternatively, if 5′ ends are directly annealed, partially filling the ends will create a flap structure that would be expected to inhibit repair. Because the flaps are homologous, these ends have the potential to form a duplex with 6 to 13 annealed bases depending on where the flaps form (Fig. 4A). Indeed, pTW423-513F was repaired at a lower frequency than pTW423-513 was (Fig. 4A). This reduction in repair efficiency was strictly dependent on Rad52 and was independent of NHEJ (Fig. 4A). These data support the idea that longer 5′ overhangs can be annealed by Rad52.
FIG. 4.
Partially filling 5′ overhangs inhibits their repair by both the Rad52-dependent and NHEJ pathways. (A) The oligonucleotide ligation technique was used to construct DSBs with partially filled 5′ 13-base overhangs. Substrate pTW423-513F is expected to equilibrate between two different annealing states. Data are expressed as the ratio of Ade+ to Leu+ colonies as in Fig. 2. Each point represents the mean ± standard deviation (error bar) from four independent transformations. (B) 4-Base 5′ overhangs were filled one base at a time with Klenow polymerase in vitro (see Materials and Methods). Data are expressed as colony counts from linearized pES16 normalized to colony counts from parallel transformations with supercoiled pES16. For comparison, these ratios were further normalized so that the total repair rate of the unfilled overhangs equaled 100%. Each point represents the mean ± standard deviation (error bar) from three independent transformations. Accurate repair yields white Ade+ colonies, whereas imprecise repair results in red Ade− colonies.
We also asked whether 4-base 5′ overhangs, which are repaired primarily by NHEJ, are joined directly or are filled and subsequently resected to form 3′ overhangs. To address this question, we used the BglII site in ADE2. Plasmid pES16, which contains wild-type ADE2, was cut with BglII, and Klenow polymerase was used to fill in 1, 2, 3, or 4 bases of the overhang. Repair efficiency decreased markedly as more bases were filled (Fig. 4B). The greatest decrease was evident when two bases of the overhangs were filled, which is the point at which all base pairing potential in the overhangs is eliminated. This supports the idea that short 5′ overhangs are also joined directly by overhang-to-overhang base pairing.
Repair of DSBs with overhangs that had a higher GC content is less dependent on NHEJ.
It is likely that the increase in repair efficiency and the loss of NHEJ dependence that we observed with increasing overhang length is due to increased stability of the annealed DSB. To further test this hypothesis, we created DSBs with compatible 4-base 3′ overhangs containing 1, 2, or 3 GC base pairs to vary the annealing energy (Fig. 5). Although the repair efficiency of each of these three DSBs was comparable in wild-type yeast, the requirement for NHEJ was inversely correlated with the GC content (Fig. 5). Together with the data presented in Fig. 2, this indicates that NHEJ is most required for accurate repair of DSBs when annealing of the overhangs is not energetically favorable.
FIG. 5.
Increasing the GC content of overhangs relaxes the requirement for NHEJ. Oligonucleotide-modified plasmids were designed to contain DSBs with 3′ 4-base overhangs with one, two, or three GC base pairs. Data are expressed as the ratio of Ade+ to Leu+ colonies as in Fig. 2. Each point represents the mean ± standard deviation (error bar) from four independent transformations.
Repair of DSBs with long overhangs is partially dependent on the single-strand break repair machinery.
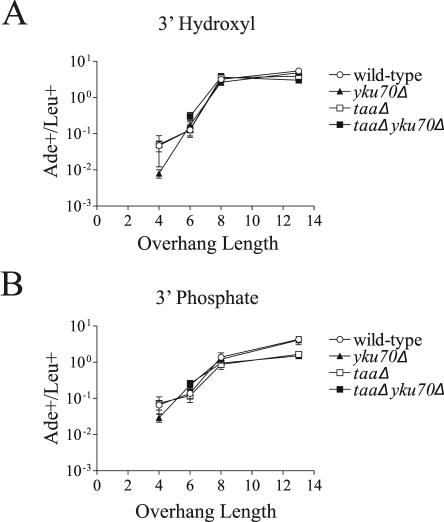
We hypothesized that the Rad52- and NHEJ-independent repair of DSBs with overhangs of 6 or more bases was mediated by spontaneous annealing of the overhangs followed by nick ligation via the base excision repair/single-strand break repair (BER/SSBR) pathway. To explore this question, we constructed DSBs containing 3′ phosphate blocking lesions on both strands. In BER/SSBR, 3′ phosphates are removed by the apurinic endonucleases Apn1 and Apn2 or the DNA 3′ phosphatase Tpp1 (33). Therefore, we can ask whether a DSB is repaired via annealing followed by BER/SSBR by transforming 3′ phosphorylated plasmids into an apn1Δ apn2Δ tpp1Δ strain. The apn1Δ apn2Δ tpp1Δ strain repaired 3′ phosphate-containing DSBs with 3′ overhangs of 8 and 13 with an approximately twofold-reduced efficiency (Fig. 6B). Although small, this effect was reproducible and dependent on the presence of the 3′ phosphate (Fig. 6A). In contrast, 3′ phosphate-containing DSBs with 4- or 6-base overhangs showed no reduction in the apn1Δ apn2Δ tpp1Δ strain (Fig. 6B). This likely indicates that these breaks can be repaired by NHEJ, which can remove the 3′ phosphate via nuclease action. To ask whether the residual repair in the apn1Δ apn2Δ tpp1Δ strain is catalyzed by NHEJ, we transformed these same plasmids into an apn1Δ apn2Δ tpp1Δ yku70Δ strain. Longer overhangs showed no additional defect upon deletion of yku70 (Fig. 6B). We were unable to directly test whether the residual repair in the apn1Δ apn2Δ tpp1Δ strain depends on Rad52 or Rad1/Rad10, because these mutations are lethal in combination with apn1Δ apn2Δ tpp1Δ.
FIG. 6.
Rejoining of DSBs with 3′ phosphate termini. Oligonucleotide ligation was used to generate plasmids containing DSBs with 3′ overhangs of 4, 6, 8, and 13 bases terminating in 3′ hydroxyls (A) or 3′ phosphates (B). The tpp1Δ apn1Δ apn2Δ genotype is represented as taaΔ. Data are expressed as the ratio of Ade+ to Leu+ colonies as in Fig. 2.
The rejoining of long overhangs is very accurate.
The above analysis focuses on accurate repair, but context-dependent imprecise repair of DSBs is also important in both spontaneous and targeted genome rearrangements. We therefore wanted to determine whether there are differences in the accuracy of repair between DSBs with short and long overhangs. We sequenced break junctions in both Ade+ and Ade− colonies from DSBs with 4- and 13-base 3′ overhangs in both wild-type and mutant strains (see Fig. S2 in the supplemental material). With 4-base overhangs, 2 of 40 Ade+ colonies obtained from wild-type yeast showed evidence of in-frame but imprecise joining. Among 20 Ade− colonies, 5 showed small deletions generated by imprecise repair, 12 could not be amplified or sequenced (presumably due to deletion of a primer annealing site), 2 were the unmodified parent plasmid, and one had a 1-base deletion attributed to an oligonucleotide synthesis error. Thus, while precise repair is the predominant outcome with 4-base overhangs, imprecise repair does occur at a detectable level. With 13-base overhang DSBs, all 20 sequenced Ade+ colonies represented precise rejoining. Among Ade− colonies, 3 of 20 colonies could not be sequenced, but the other 17 were all apparently Ade− because of oligonucleotide synthesis errors and not due to imprecise joining. Therefore, all the colonies that we were able to sequence from 13-base overhangs showed evidence of precise repair, even those that were Ade−. Combining this sequencing data with the fraction of Ade+ and Ade− colonies obtained during transformation provides an estimate of the fraction of colonies that were rejoined precisely, which is notably lower for shorter overhangs (Table 1).
TABLE 1.
Accuracy of DSB rejoining
| Plasmida | Ade+ colonies
|
Ade− colonies
|
Fraction precise joiningd | |||||
|---|---|---|---|---|---|---|---|---|
| Fraction of total coloniesb | Sequencing resultsc
|
Fraction of total coloniesb | Sequencing resultsc
|
|||||
| Precise | Imprecise | Precise | Imprecise | Other | ||||
| pTW423-34 | 0.59 ± 0.07 | 38/40 | 2/40 | 0.41 ± 0.07 | 1/20 | 5/20 | 14/20 | 0.81 ± 0.07 |
| pTW423-313 | 0.84 ± 0.04 | 20/20 | 0/20 | 0.16 ± 0.04 | 15/20 | 0/20 | 5/20 | 1.00 ± 0.04 |
| pTW423-313/4 | 0.06 ± 0.04 | 11/15 | 4/15 | 0.94 ± 0.04 | 0/24 | 4/24 | 20/24 | 0.20 ± 0.03 |
| pTW423-313/9 | 0 | 1.00 | 0/24 | 3/24 | 21/24 | 0 | ||
See Fig. 7 (also see Fig. S1 in the supplemental material) for plasmid configurations.
For each plasmid, the mean fraction of Ade+ and Ade− colonies ± 1 standard deviation was calculated from four transformations.
Joints were amplified, sequenced, and categorized as “precise,” “imprecise,” or “other.” “Precise” indicates either wild-type ADE2 sequence or, in the case of Ade− colonies, joints that showed correct overhang alignment but had mutations outside the overhangs introduced during oligonucleotide synthesis. In the case of pTW423-313/4 and pTW423-313/9, precise alignment refers to pairing of the available 4-base microhomology. “Imprecise” indicates sequences in which oligonucleotide ligation could be verified but overhangs were misaligned or recessed microhomologies were utilized. “Other” indicates parent plasmids, sequences in which one oligonucleotide pair was missing, or joints that could not be amplified or sequenced due to deletion of primer sites. The data are shown as the number of colonies with each sequencing result relative to the total number of colonies sequenced.
The fraction of precise rejoining was calculated from the combined data from Ade+ and Ade− colonies, but excluding the “other” category. This category was excluded, because it could not be determined that the plasmids responsible for these events had been successfully ligated to both oligonucleotide pairs. The number thus reflects the fraction (not frequency) of precise joints among all recovered transformants known to result from plasmids bearing both oligonucleotide pairs.
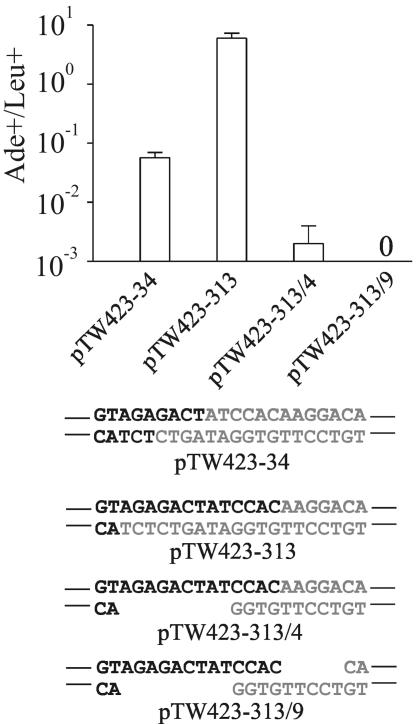
DSBs with long overhangs but short regions of homology are not repaired efficiently.
Long overhangs might be substrates for Rad52 binding regardless of whether they are complementary. To determine whether Rad52 engagement itself is sufficient to promote efficient repair, we generated two DSB configurations with long overhangs but short regions of homology (Fig. 7). Such ends might arise by limited resection of fully complementary overhangs or as a microhomology fortuitously available between noncomplementary overhangs. Thus, these ends present a model for comparing joining at compatible versus extensively processed ends. The first of these DSBs, pTW423-313/4, pairs a 13-base overhang with a 4-base overhang, yielding a single 9-base gap upon annealing (Fig. 7). This break was repaired at a much lower rate than a DSB with complementary 4-base overhangs, yielding only 15 Ade+ colonies over the course of four transformations into wild-type yeast (Fig. 7). Sequencing showed that 11 of these events occurred by annealing of the 4-base microhomology (although one also had a 3-base inversion in the overhang formed by an unknown mechanism [see Fig. S2 in the supplemental material]). The remaining four joints utilized recessed microhomologies that generated deletions but restored the ADE2 reading frame.
FIG. 7.
Inefficient repair of DSBs with long overhangs but short regions of homology. DSBs with long overhangs but limited base-pairing potential, which form gap structures upon annealing, were created by oligonucleotide ligation. Data are expressed as the ratio of Ade+ to Leu+ colonies as in Fig. 2. No Ade+ colonies were recovered for substrate pTW423-313/9.
Among Ade− colonies, 4 of 24 also showed microhomology-dependent imprecise joining, with the remainder a mixture of large deletions, parent plasmid, and a captured DNA of unknown origin (see Fig. S2 in the supplemental material). This DSB was therefore predominantly repaired imprecisely, with the 4-base microhomology sequence in the overhangs utilized in a minority of joints (Table 1). Because the repair of this DSB was so inefficient, we were unable to quantitatively assess its pathway dependence. Four transformations into a yku70Δ strain yielded nine Ade+ colonies, however, which all corresponded to annealing of the 4-base microhomology (see Fig. S2 in the supplemental material). Therefore, this event does not strictly require NHEJ.
We also created pTW423-313/9, which maintains the 4-base microhomology of pTW423-313/4 but increases the overhang length on the right side to 9 bases, such that annealing generates gaps on both sides of the break (Fig. 7). Transformation of this plasmid into wild-type yeast did not yield any Ade+ colonies. Sequencing of Ade− colonies showed a pattern similar to that seen with pTW423-313/4 (see Fig. S2 in the supplemental material). Thus, microhomologies within long overhangs support a rate of repair many times lower than that of NHEJ of comparable short overhangs and vastly lower than that of fully compatible long overhangs. Therefore, long regions of homology, not simply long overhangs, are required for the overhang length-dependent increase in DSB rejoining.
DISCUSSION
DSB rejoining mechanism is a function of overhang length.
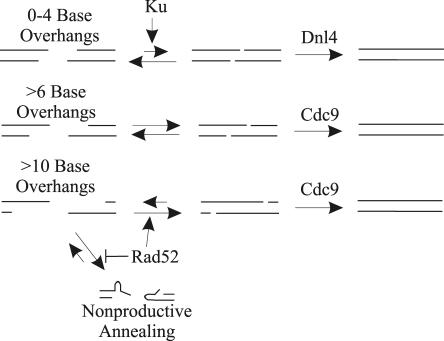
The model that underlies these studies is that many naturally occurring DSBs are little more than two single-strand breaks that have occurred close enough to each other that the duplex is unstable. Therefore, DSB overhangs are expected to be complementary, although the length of these overhangs will be variable. Even when a terminal nucleotide is damaged, restorative repair in the absence of a donor template entails the rejoining of overhangs that were originally base paired in the parent duplex. We describe three mechanisms for such rejoining (Fig. 8): NHEJ, a Rad52-dependent SSA-like pathway, and a third pathway independent of both Rad52 and NHEJ. The relative contribution of each mechanism is determined by the overhang length, creating three distinct phases in Fig. 2 and 3. DSBs with overhangs of 0 to 4 bases are primarily repaired by NHEJ. They can also be repaired less efficiently by the NHEJ- and Rad52-independent mechanism. DSBs with 6- to 8-base overhangs comprise the second phase. They show only slight reductions in repair efficiency in NHEJ-deficient strains and little or no dependence on Rad52. Finally, repair of DSBs with overhangs of 10 nucleotides or longer is completely independent of NHEJ. These DSBs can be repaired by the third mechanism, but they are principally substrates for the SSA-like pathway.
FIG. 8.
A model for rejoining of DSBs as a function of overhang length. Three pathways can rejoin DSBs. NHEJ, catalyzed by the Ku and DNA ligase IV complexes, is mainly used when joint stabilization is energetically unfavorable, such as when overhangs are less than 4 bases. DSBs with overhangs of 6 or more bases can anneal and be rejoined without the need for Ku and DNA ligase IV. Longer overhangs (>10 bases) can also function as substrates for Rad52, which promotes rejoining through an SSA-like pathway. See text for further discussion.
Implications regarding the mechanism of NHEJ.
We had initially hoped to determine the overhang length that could no longer be productively engaged by the NHEJ machinery. However, the fact that NHEJ was not required once overhangs reached 6 to 8 bases precluded this assessment. Indeed, the rate of NHEJ-independent repair at even 6 bases was greater than that of NHEJ at 4 bases (Fig. 2 and 3). It initially seemed possible that DSBs with 6- and 8-base overhangs were being joined by NHEJ, i.e., by DNA ligase IV, even though they were Ku independent. In this model, the greater annealing strength of longer overhangs would bypass the need for Ku. This would predict a greater defect in a dnl4Δ mutant than a yku70Δ mutant at these intermediate overhangs lengths, however, and this was not observed (Fig. 2B and 3B). A corollary is that the very efficient joining of longer overhangs must be mediated by DNA ligase I (Cdc9), the only other DNA ligase in yeast. However, we do note the slightly greater defect of dnl4Δ than yku70Δ yeast in NHEJ of GC-rich overhangs (Fig. 5). Still, our combined data more strongly support the notion that the NHEJ machinery works as a whole during precise repair of breaks with complementary overhangs.
We also found that DSBs with 3′ 4-base overhangs containing 3′ phosphate termini can be efficiently repaired by NHEJ in the absence of Apn1, Apn2, and Tpp1, the enzymes required for removing this lesion during BER (Fig. 6) (33). This was surprising, given that PNKP, the human homolog of Tpp1, interacts with XRCC4 (the Lif1 homolog), and has been implicated in NHEJ on the basis of in vitro and in vivo assay results (5, 17). It is possible that Tpp1 is exclusively associated with BER in yeast, with the NHEJ function of mammalian PNKP having evolved later. Indeed, the XRCC4-PNKP interaction is mediated by the PNKP FHA domain, which is not present in Tpp1 (17). Therefore, it appears that Tpp1 can be efficiently compensated for by unidentified NHEJ nucleases.
An SSA-like mechanism of DSB rejoining.
We have identified a Rad52-dependent pathway that enhances the repair of DSBs with overhangs 8 nucleotides or longer (Fig. 2C and 3C), presumably by a mechanism similar to SSA. Rad52 most likely functions to eliminate secondary structure that would inhibit annealing of complementary single-stranded DNAs while helping to anneal the overhangs (Fig. 8), as previously proposed (15, 31). Loss of this pathway has no effect on repair of DSBs with overhangs of 6 bases or shorter, probably because these overhangs are not long enough to function as substrates for Rad52, although this has not yet been tested in vitro. Extending the overhangs beyond 10 bases did not induce a further increase in the repair rate (Fig. 2A and 3A). These findings correlate well with previous observations that Rad52 can stimulate repair by SSA when a DSB is introduced between 10-base, but not 4-base, terminal direct repeats and that such repair reaches its maximum in the range of 20 to 30 nucleotides (15). That the maximum repair rate was reached at a shorter length in the present study is likely due to the lack of a requirement for resection to expose the complementary sequence. Finally, a longer annealing surface, not just longer overhangs, is required for the Rad52-dependent rejoining mechanism to function (Fig. 7).
DSBs can be repaired precisely in the absence of NHEJ and SSA.
Our data clearly show that DSBs can be repaired precisely in the absence of both NHEJ and the SSA-like pathway (Fig. 2D and 3D). This mechanism of repair likely represents annealing of the overhangs followed by single-strand break repair (Fig. 8). Although not definitively established by our data, this notion is supported by the observation that a 3′ phosphatase-deficient strain is partially compromised at repairing DSBs with longer overhangs and 3′ phosphates (Fig. 6). The residual repair of these DSBs in this strain likely represents removal of the 3′ phosphate by Rad1/Rad10, which has been shown to remove nonhomologous flaps in SSA (11) and to participate in the processing of 3′ phosphates (16, 33). We could not test this directly, because rad1Δ apn1Δ apn2Δ tpp1Δ yeast cells are inviable.
The strong positive correlation between overhang length and the efficiency of NHEJ- and Rad52-independent repair supports the idea that the efficiency of this mechanism is determined by the likelihood of overhang annealing. The overhang melting temperature (Tm) at the midpoint of the length-dependence curve, about 7 nucleotides, should thus correspond to 30°C, the temperature at which yeast is grown. Indeed, the Tms of our 6- and 8-base overhangs are 24 and 32°C, respectively. Might plasmids with longer overhangs have thus simply entered the cell in an annealed state, such that there never was a DSB? We first note that the yeast cells were heat shocked at 42°C prior to outgrowth as the standard final step of transformation, which should drive apart overhangs with a lower Tm. Second, the fact that Rad52 enhances the repair of long overhangs by about 10-fold demonstrates that a large fraction of the plasmids must enter the cell in a nonannealed state. Finally, the Tm of an overhang pair is not a static representation of its annealing state. Instead, there is an equilibrium such that even longer overhangs will have a finite transition rate to the DSB state. Likewise, even shorter overhangs will anneal transiently, so that some NHEJ-independent repair would be expected. Indeed, we and others have observed residual precise rejoining of 4-base overhangs in the absence of NHEJ (see Fig. S2 in the supplemental material) (12, 15).
The importance of NHEJ-independent rejoining mechanisms is highlighted by the fact that NHEJ-deficient mice paradoxically develop cancer due to accumulation of translocations bearing short regions of microhomology at the junctions (29). Ku- and Rad52-independent translocations with 7- to 22-bp regions of homology at the breakpoints have also been observed in yeast (39). We suggest that the NHEJ-independent repair mechanisms revealed here are substantively related to the backup NHEJ and microhomology-mediated end-joining mechanisms recently reported (3, 20, 35, 39). However, these mechanisms often result in nucleotide loss. In our system, imprecise joining of DSBs with 4-base overhangs was detected, but inaccurate repair of DSBs with longer compatible overhangs is rare enough that such events were below our detection limits (Table 1). This difference in accuracy might be accounted for by the fact that long regions of homology would typically not be present at incompatible overhangs. In agreement with this, DSBs that model imprecise repair of long overhangs, by reducing the region of microhomology, were repaired inefficiently (Fig. 7) and inaccurately (Table 1). Low colony yields made it impossible to quantitatively determine the pathway dependence of the joints that did form; chromosomal assays such as those reported by Ma et al. (20) have been more informative in this regard.
Overcoming the energetic barrier of annealing short overhangs.
Extending the thermodynamic analysis, our data indicate that the main function of NHEJ is to catalyze the synapsis of short overhangs, using protein-protein contacts to supplement the limited energy provided by base pairing (Fig. 8). This model is supported by our observation that overhangs that had a higher GC content and therefore have greater base-pairing stability are less dependent on NHEJ (Fig. 5), in direct agreement with recent in vitro results using Xenopus egg extracts (27). We were unable to repeat our experiments in chromosomes, but previous experiments comparing the repair of plasmid and chromosomal DSBs have shown strong correlation (15, 37). NHEJ-independent precise repair of DSBs with 3′ 4-base overhangs has been observed with chromosomal DSBs, but at rates about 100-fold lower than that of wild-type, as opposed to the 10- to 50-fold difference we observed using plasmid-based assays (15). The ends of plasmid DSBs are tethered by the plasmid backbone, which could promote annealing by decreasing entropy. On the other hand, chromosomal DSB ends are likely be held in proximity by chromatin elements and/or the Mre11/Rad50/Xrs2 complex (8). In total, we argue that the general principles observed here will also apply to chromosomal DSBs but that the curves may be shifted toward longer overhang lengths.
5′ overhangs are joined directly.
Rad52 catalyzes the annealing of complementary single-stranded DNAs in vitro (24), but whether overhang polarity affects this process has not yet been tested. The known roles of Rad52 in recombination involve only 3′ overhangs (25), but our results raise the possibility that Rad52 can anneal both 3′ and 5′ overhangs. The impairment of Rad52-dependent DSB joining by partial filling of 5′ overhangs supports this possibility (Fig. 4A). The same pattern was observed for NHEJ at short overhangs (Fig. 4B). Together, these data indicate that 5′ overhangs are not filled in during repair.
Supplementary Material
Acknowledgments
We thank the members of the Wilson laboratory for their support, ideas, and helpful comments on the manuscript and John Vance for construction of pTW423.
This work was supported in part by the Pew Scholars Program in the Biomedical Sciences of the Pew Charitable Trusts and Public Health Service grants R01CA90911 (T.E.W.) and T32GM007315 (J.M.D.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. E. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulton, S. J., and S. P. Jackson. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15:5093-5103. [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Chappell, C., L. A. Hanakahi, F. Karimi-Busheri, M. Weinfeld, and S. C. West. 2002. Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 21:2827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S., K. V. Inamdar, P. Pfeiffer, E. Feldmann, M. F. Hannah, Y. Yu, J. W. Lee, T. Zhou, S. P. Lees-Miller, and L. F. Povirk. 2001. Accurate in vitro end joining of a DNA double strand break with partially cohesive 3′-overhangs and 3′-phosphoglycolate termini: effect of Ku on repair fidelity. J. Biol. Chem. 276:24323-24330. [DOI] [PubMed] [Google Scholar]
- 8.Connelly, J. C., and D. R. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27:410-418. [DOI] [PubMed] [Google Scholar]
- 9.Coquerelle, T., A. Bopp, B. Kessler, and U. Hagen. 1973. Strand breaks and K′ end-groups in DNA of irradiated thymocytes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 24:397-404. [DOI] [PubMed] [Google Scholar]
- 10.DeFazio, L. G., R. M. Stansel, J. D. Griffith, and G. Chu. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 12.Guirouilh-Barbat, J., S. Huck, P. Bertrand, L. Pirzio, C. Desmaze, L. Sabatier, and B. S. Lopez. 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14:611-623. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov, E. L., N. Sugawara, J. Fishman-Lobell, and J. E. Haber. 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, S. P. 2002. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687-696. [DOI] [PubMed] [Google Scholar]
- 15.Karathanasis, E., and T. E. Wilson. 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161:1015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karumbati, A. S., R. A. Deshpande, A. Jilani, J. R. Vance, D. Ramotar, and T. E. Wilson. 2003. The role of yeast DNA 3′-phosphatase Tpp1 and rad1/Rad10 endonuclease in processing spontaneous and induced base lesions. J. Biol. Chem. 278:31434-31443. [DOI] [PubMed] [Google Scholar]
- 17.Koch, C. A., R. Agyei, S. Galicia, P. Metalnikov, P. O'Donnell, A. Starostine, M. Weinfeld, and D. Durocher. 2004. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 23:3874-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer, K. M., J. A. Brock, K. Bloom, J. K. Moore, and J. E. Haber. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. W., L. Blanco, T. Zhou, M. Garcia-Diaz, K. Bebenek, T. A. Kunkel, Z. Wang, and L. F. Povirk. 2004. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 279:805-811. [DOI] [PubMed] [Google Scholar]
- 20.Ma, J.-L., E. M. Kim, J. E. Haber, and S. E. Lee. 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23:8820-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 22.Mahajan, K. N., S. A. N. McElhinny, B. S. Mitchell, and D. A. Ramsden. 2002. Association of DNA polymerase μ (pol μ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell. Biol. 22:5194-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prado, F., F. Cortes-Ledesma, P. Huertas, and A. Aguilera. 2003. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42:185-198. [DOI] [PubMed] [Google Scholar]
- 26.Rudin, N., and J. E. Haber. 1988. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol. Cell. Biol. 8:3918-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandoval, A., and P. Labhart. 2004. High G/C content of cohesive overhangs renders DNA end joining Ku-independent. DNA Repair 3:13-21. [DOI] [PubMed] [Google Scholar]
- 28.Schar, P., G. Herrmann, G. Daly, and T. Lindahl. 1997. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11:1912-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharpless, N. E., D. O. Ferguson, R. C. O'Hagan, D. H. Castrillon, C. Lee, P. A. Farazi, S. Alson, J. Fleming, C. C. Morton, K. Frank, L. Chin, F. W. Alt, and R. A. DePinho. 2001. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol. Cell 8:1187-1196. [DOI] [PubMed] [Google Scholar]
- 30.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugawara, N., G. Ira, and J. E. Haber. 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomer, G., and Z. Livneh. 1999. Analysis of unassisted translesion replication by the DNA polymerase III holoenzyme. Biochemistry 38:5948-5958. [DOI] [PubMed] [Google Scholar]
- 33.Vance, J. R., and T. E. Wilson. 2001. Repair of DNA strand breaks by the overlapping functions of lesion-specific and non-lesion-specific DNA 3′ phosphatases. Mol. Cell. Biol. 21:7191-7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vance, J. R., and T. E. Wilson. 2001. Uncoupling of 3′-phosphatase and 5′-kinase functions in budding yeast. Characterization of Saccharomyces cerevisiae DNA 3′-phosphatase (TPP1). J. Biol. Chem. 276:15073-15081. [DOI] [PubMed] [Google Scholar]
- 35.Wang, H., A. R. Perrault, Y. Takeda, W. Qin, and G. Iliakis. 2003. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 31:5377-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, T. E., and M. R. Lieber. 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 274:23599-23609. [DOI] [PubMed] [Google Scholar]
- 38.Wu, X., T. E. Wilson, and M. R. Lieber. 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96:1303-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, X., and A. Gabriel. 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163:843-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.