Abstract
The NF-κB family of transcription factors is activated by a wide variety of signals to regulate a spectrum of cellular processes. The proper regulation of NF-κB activity is critical, since abnormal NF-κB signaling is associated with a number of human illnesses, such as chronic inflammatory diseases and cancer. We report here that PIAS1 (protein inhibitor of activated STAT1) is an important negative regulator of NF-κB. Upon cytokine stimulation, the p65 subunit of NF-κB translocates into the nucleus, where it interacts with PIAS1. The binding of PIAS1 to p65 inhibits cytokine-induced NF-κB-dependent gene activation. PIAS1 blocks the DNA binding activity of p65 both in vitro and in vivo. Consistently, chromatin immunoprecipitation assays indicate that the binding of p65 to the promoters of NF-κB-regulated genes is significantly enhanced in Pias1−/− cells. Microarray analysis indicates that the removal of PIAS1 results in an increased expression of a subset of NF-κB-mediated genes in response to tumor necrosis factor alpha and lipopolysaccharide. Consistently, Pias1 null mice showed elevated proinflammatory cytokines. Our results identify PIAS1 as a novel negative regulator of NF-κB.
A large variety of signals, such as proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-1 [IL-1]) and bacterial lipopolysaccharide (LPS), activate the NF-κB signaling pathway. NF-κB is a family of dimeric transcription factors composed of members of the Rel family of DNA binding proteins, including NF-κB1 (p50 and its precursor p105), NF-κB2 (p52 and its precursor p100), c-Rel, RelA (p65), and RelB (11, 18). Upon stimulation, NF-κB translocates into the nucleus, where it binds to specific DNA sequences and regulates transcription. NF-κB is involved in mediating a wide spectrum of cellular responses, including infections, inflammation, and apoptosis (2, 27). Inappropriate regulation of NF-κB is involved in a wide range of human diseases, including cancer, neurodegenerative disorders, arthritis, asthma, and chronic inflammation (3, 4, 10, 12). The NF-κB signaling pathway is tightly modulated at various levels by distinct regulatory proteins. For example, the binding of the IκB family of proteins prevents the nuclear translocation of NF-κB (16). However, a protein factor that can regulate the DNA binding activity of NF-κB has not been documented.
The PIAS (protein inhibitor of activated STAT) family of proteins consists of four members: PIAS1, PIAS3, PIASx, and PIASy (33). Members of the PIAS family have been suggested to regulate STAT-mediated transcription. Upon cytokine stimulation, PIAS binds to STAT and inhibits STAT-mediated gene activation (1, 8, 21, 22). Among the PIAS family, PIAS1 and PIASy have been shown to inhibit STAT1-dependent transcription through distinct mechanisms. PIAS1 inhibits the transcriptional activity of STAT1 by blocking the DNA binding activity of STAT1. In contrast, PIASy does not affect the DNA binding activity of STAT1. It has been suggested that PIASy may act as a transcriptional corepressor of STAT1. The PIAS family of proteins has also been suggested to regulate a number of other transcription factors, including nuclear hormone receptors (13, 28, 36, 37), LEF1 (31), and p53 (15, 26, 32).
To understand the physiological role of PIAS1, we have recently generated Pias1 null mice (23). Detailed gene activation analysis indicates that PIAS1 selectively regulates a subset of interferon (IFN)-inducible genes. The antiviral activity of IFNs is significantly enhanced by Pias1 disruption. In addition, Pias1 null mice show enhanced protection against pathogenic infection. These results support a physiological role of PIAS1 in the negative regulation of IFN-activated STAT1-mediated gene activation and demonstrate an important role of PIAS1 in innate immune responses.
Since STAT1 and the Rel family of proteins share structural similarity in their DNA binding domains (6), we explored the possible involvement of PIAS1 in the regulation of NF-κB. Here we report that PIAS1 interacts with the p65 subunit of NF-κB and represses its transcriptional activity. In vitro and in vivo studies indicate that PIAS1 inhibits the DNA binding activity of p65. Microarray analysis indicates that the disruption of Pias1 results in elevated expression of a subset of NF-κB-dependent genes. Our results demonstrate a critical role of PIAS1 in the negative regulation of the NF-κB signaling pathway.
MATERIALS AND METHODS
Materials.
Flag-PIAS1, glutathione S-transferase (GST)-PIAS1, and Gal4-p65 plasmids have been described (20, 22). Flag-p65, Flag-p65(1-313), Flag-p65(299-551), Flag-PIAS1(1-415), Flag-PIAS1(416-650), Flag-PIAS1(1-344), Flag-PIAS1(89-344), Myc-PIAS1, and Myc-p65 were cloned by PCR amplification of the corresponding coding regions followed by subcloning into pCMV-Flag or pMyc expression vectors. The following antibodies were utilized in the coimmunoprecipitation and Western blotting analyses: anti-p65 (C-20; Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-p50 (H-119; Santa Cruz Biotechnology), anti-E2F-1 (C-20; Santa Cruz Biotechnology), anti-IκBα (C-20; Santa Cruz Biotechnology), antiactin (C-11; Santa Cruz Biotechnology), anti-Flag (M2; Sigma), anti-Myc (Cell Signaling), and antitubulin (Sigma). The anti-PIAS1 antibody was raised against a GST fusion protein containing the C-terminal region of human PIAS1 (amino acids 551 to 650).
Coimmunoprecipitation assays.
Coimmunoprecipitation assays were performed as described previously (8). Briefly, whole-cell lysates were prepared 30 h post-transient transfection in a lysis buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 1% Brij, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin/ml, and 3 μg of aprotinin/ml. The mixture was incubated on ice for 30 min and centrifuged at 13,000 × g for 5 min. The supernatant was used in coimmunoprecipitation assays with various antibodies.
Transient transfection and luciferase assays.
Human 293T cells were transfected by a calcium-phosphate procedure as described previously (34), and cell lysates were collected for luciferase assays (Promega) 30 h posttransfection. The relative luciferase units were corrected for relative expression of β-galactosidase. Human A549 cells were transfected with Lipofectamine reagent (Invitrogen) and assayed for luciferase activities with a dual-luciferase system (Promega) using the cotransfected pRLTK to correct for the differences in transfection efficiency.
Northern blot analysis.
Northern blot analysis was performed essentially as described previously (24).
Microarray analysis.
Microarray analysis was performed essentially following the manufacturer's instructions (Affymetrix). Briefly, bone marrow-derived macrophages (BMMs) from wild-type or Pias1−/− littermates were either untreated or treated with TNF-α (20 ng/ml) for 30 min or LPS (10 ng/ml) for 1 h. Total RNA was prepared with RNA-STAT60 (TEL-TEST) and purified with the RNeasy kit (QIAGEN). Double-stranded cDNA was synthesized from 20 μg of total RNA according to Affymetrix’s methodology and purified with Phase Lock gels (Eppendorf). Biotin-labeled RNA was synthesized with the BioArray High Yield RNA transcript labeling kit (Enzo). Samples were cleaned, fragmentated, and hybridized to murine genome (MGU74Av2) GeneChips (Affymetrix) as instructed. GeneChips were stained with phycoerythrin-streptavidin (Molecular Probes) and scanned with a GeneChip scanner (Affymetrix).
Bone marrow-derived macrophages.
BMMs were differentiated from marrow cells from 4- to 8-week-old Pias1 null mice and their wild-type littermates as described previously (7). BMMs were maintained in 1× Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 30% L929-conditioned medium containing macrophage colony-stimulating factor for 7 days before they were either untreated or treated with TNF-α (20 ng/ml) or LPS (10 ng/ml) for various times. Total RNA was prepared and subjected to real-time PCR analyses.
Immunofluorescence.
Immunofluorescence analysis was performed as described previously (21). A mouse monoclonal anti-p65 (1:100; F-6; Santa Cruz Biotechnology) and a rabbit polyclonal anti-PIAS1 (1:400) were added to the cells simultaneously as primary antibodies. A mixture of anti-rabbit immunoglobulin G (IgG) Fluor 488 (1:200; Molecular Probes), anti-mouse IgG Cy3 (1:200, Jackson Labs), and Hoechst (1 μg/ml; Sigma) was added to the cells during the secondary antibody incubation.
Electrophoretic mobility shift assay.
The electrophoretic mobility shift assay (EMSA) was performed as described previously (8). The sequence of the NF-κB oligonucleotide is 5′-GATCCGAGAGGGGATTCCCCGATCG-3′. The sequence of the SP-1 oligonucleotide is 5′-ATTCGATCGGGGCGGGGCGAG-3′.
Quantitative real-time PCR.
Quantitative real-time PCR (Q-PCR) was performed as described previously (9). Briefly, first-strand cDNA was produced by reverse transcription of 3 μg of total RNA using SuperScript II (Invitrogen). Q-PCR was carried out using the iCycler thermocycler (Bio-Rad) in a final volume of 25 μl containing the following: Taq polymerase, 1× Taq buffer (Stratagene), 125 μM deoxynucleoside triphosphates, SYBR Green I (Molecular Probes), and fluorescein (Bio-Rad). Amplification conditions were 95°C (3 min) and 40 cycles of 95°C (30 s), 55°C (30 s), and 72°C (30 s). Actin was used to standardize the levels of cDNA. The specific primers used in Q-PCR analyses are given in Table S2 of the supplemental material.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed using the ChIP assay kit (Upstate Biotech) as instructed by the manufacturer. Wild-type or Pias1−/− BMMs (107) were either untreated or treated with LPS (10 ng/ml) for 20 min or 1 h. Cell extracts were prepared, and chromatin was sheared by sonication (six 10-s pulses at 30% of the maximum strength). ChIP assays were performed with an anti-p65 antibody or rabbit IgG as a negative control. Bound DNA was quantified by Q-PCR and normalized with the input DNA. Approximately 10% of the immunoprecipitated samples were analyzed by Western blotting with anti-p65 to reveal that similar amounts of NF-κB p65 were present in each sample. Similar ChIP assays were performed with the tetracycline (TET)-off PIAS1 cell line, except that cells grown in the presence or absence of doxycycline (DOX) for 12 h were either untreated or treated with TNF-α (20 ng/ml) for 20 min or 1 h. The sequences of the primers used are given in Table S2 of the supplemental material.
Flow cytometric analysis.
Flow cytometric analysis was carried out as described previously (19, 25). Briefly, single-cell suspensions from spleens or thymuses were depleted of red blood cells by hypotonic lysis and stained with combinations of the following antibodies (Pharmingen): anti-B220-phycoerythrin (PE), anti-IgM-fluorescein isothiocyanate, anti-CD4-fluorescein isothiocyanate, anti-CD8-PE, and anti-Gr-1-PE. Data were acquired on a FACScan (Becton Dickinson) and analyzed with CellQuest software.
Measurement of serum cytokines.
Serum samples were collected from 4- to 15-week-old Pias1 null mice and their wild-type littermates, and serum cytokine levels (TNF-α, IL-1β, IFN-γ, and IL-4) were measured by enzyme-linked immunosorbent assay (ELISA) as instructed by the manufacturer (Biosources, Camarillo, Calif.).
RESULTS
PIAS1 specifically interacts with the p65 subunit of NF-κB in vivo.
STAT and NF-κB are two important families of transcription factors activated by cytokines. Upon ligand stimulation, both STAT and NF-κB translocate from the cytoplasm into the nucleus, where they bind DNA and activate transcription of specific genes. We explored the possible involvement of PIAS1 in the regulation of NF-κB signaling.
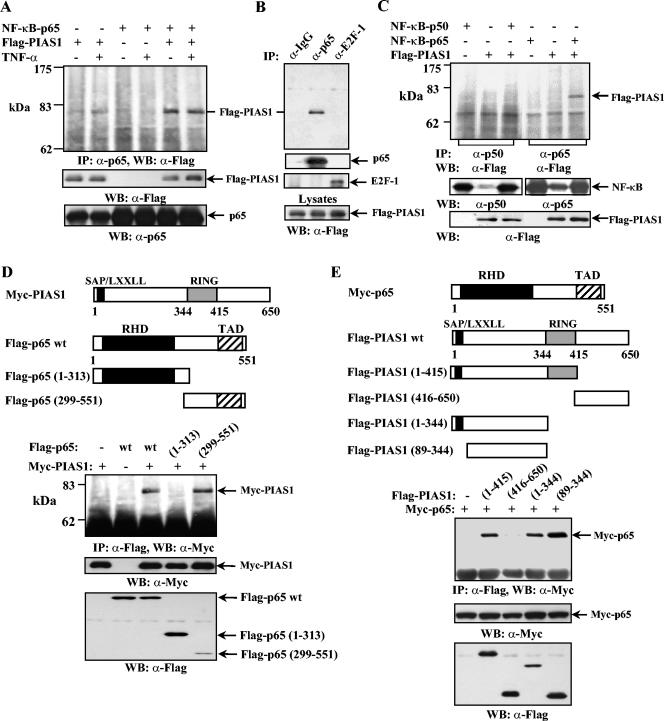
To test whether PIAS1 can interact with NF-κB in vivo, human 293T cells were transiently transfected with expression constructs encoding Flag-PIAS1 and the p65 subunit of NF-κB, alone or together. Thirty hours posttransfection, cells were either left untreated or treated with TNF-α for 15 min, and whole-cell lysates were utilized in coimmunoprecipitation assays using an anti-p65 antibody. After extensive washing, the immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting using an anti-Flag antibody. When both p65 and Flag-PIAS1 were overexpressed in 293T cells, PIAS1 was coimmunoprecipitated by anti-p65, indicating that PIAS1 and p65 interact in vivo (Fig. 1A, top panel, lane 5). This interaction was not affected by TNF-α treatment. The proper expression of Flag-PIAS1 and p65 was confirmed by Western blot analysis of the same lysates (Fig. 1A, middle and bottom panels). To validate the specific p65-PIAS1 interaction, coimmunoprecipitation assays were carried out with 293T lysates overexpressing both Flag-PIAS1 and p65, using rabbit IgG, anti-p65, or an antibody against E2F-1, an irrelevant transcription factor, as a negative control. As shown in Fig. 1B, Flag-PIAS1 was immunoprecipitated only by anti-p65 but not by anti-E2F-1 or rabbit IgG. These results indicate that PIAS1 interacts with NF-κB p65 in vivo.
FIG. 1.
PIAS1 specifically interacts with the p65 subunit of NF-κB in vivo. (A, top panel) Human 293T cells were transiently transfected with either Flag-PIAS1 or p65 alone or together, as indicated. Thirty hours posttransfection, cells were either untreated or treated with TNF-α for 15 min. The whole-cell lysates were subjected to coimmunoprecipitation (IP) with anti-p65 (Santa Cruz Biotechnology) followed by Western blotting (WB) with anti-Flag (Sigma). (Middle and bottom panels) The same lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-Flag or anti-p65 as indicated. (B) The same procedure described for panel A was performed except that 293T cells were transiently transfected with both Flag-PIAS1 and NF-κB p65, and coimmunoprecipitation assays were performed with rabbit IgG, anti-p65, or anti-E2F-1 (Santa Cruz Biotechnology) followed by Western blotting with anti-Flag (top panel). The same filter was reprobed with anti-p65 or anti-E2F-1 (middle two panels). The same lysates were also analyzed by Western blotting with anti-Flag to show equal amounts of Flag-PIAS1 present in each sample (bottom). (C) The same procedure described for panel A was performed except that 293T cells were transfected with either Flag-PIAS1, p65, or p50 alone or together, as indicated, and coimmunoprecipitation assays were performed with anti-p50 (lanes 1 to 3) or anti-p65 (lanes 4 to 6) (top). The same lysates were probed with anti-p50, anti-p65, or anti-Flag, as indicated (middle and bottom panels). (D) The same procedure described for panel A was performed except that Myc-PIAS1 and Flag-p65 wt, Flag-p65(1-313), or Flag-p65(299-551) was used as indicated. (E) The same procedure described for panel A was performed except that Myc-p65 and Flag-PIAS1 wt or different mutants were used as indicated.
To test whether PIAS1 interacts with other subunits of NF-κB, 293T cells were transiently transfected with Flag-PIAS1, NF-κB p65, or NF-κB p50 alone or Flag-PIAS1 together with one of the NF-κB subunits. Coimmunoprecipitation assays were performed as described above using the antibodies specifically recognizing p65 or p50. PIAS1 interacts with p65 but not the p50 subunit of NF-κB (Fig. 1C, top panel, lanes 3 and 6). The proper expression of each component was confirmed by Western blot analysis (Fig. 1C, middle and bottom panels). These results revealed that PIAS1 specifically interacts with the p65 subunit of NF-κB in vivo.
p65 contains a transcriptional activation domain and a Rel homology domain. To examine the PIAS1 interaction region of p65, 293T cells were transiently transfected with Myc-PIAS1 together with Flag-p65(1-313) or Flag-p65(299-551) followed by coimmunoprecipitation analysis with anti-Flag. PIAS1 was found to interact with Flag-p65(299-551) but not Flag-p65(1-313). Thus, the C-terminal region of p65 containing the transcriptional activation domain is responsible for binding to PIAS1 (Fig. 1D).
Similar coimmunoprecipitation analysis was performed to identify the p65 interaction region of PIAS1. 293T cells were transiently transfected with Myc-p65 together with various deletion mutants of PIAS1. The region encoding amino acid residues 89 to 344, which is located between the PIAS1 SAP (SAF-A, Acinus, PIAS) domain and the RING domain, was found to be sufficient for interacting with p65 (Fig. 1E).
PIAS1 inhibits NF-κB-mediated gene activation.
To study the effect of PIAS1 on NF-κB-mediated gene activation, luciferase reporter assays were carried out in human 293T cells. Cells were transiently transfected with a luciferase reporter construct containing two copies of the NF-κB binding site together with increasing amounts of Flag-PIAS1. Twenty-four hours posttransfection, cells were either left untreated or treated with TNF-α for 6 h, followed by luciferase assays. As shown in Fig. 2A, TNF-α treatment leads to an approximately 100-fold increase in the endogenous NF-κB-mediated gene activation, which was inhibited by PIAS1 in a dose-dependent manner (Fig. 2A, left panel). Similar results were observed in human A549 lung cancer cells (Fig. 2A, right panel). The proper expression of Flag-PIAS1 in both cell lines was confirmed by Western blot analysis (Fig. 2A, bottom panels). These results suggest that PIAS1 is an inhibitor of the NF-κB signaling pathway.
FIG. 2.
PIAS1 inhibits TNF-α-induced NF-κB-dependent gene activation. (A) Luciferase (Luc.) reporter assays. Human 293T cells (left) or lung cancer A549 cells (right) were transiently transfected with luciferase reporter construct 2×NF-κB together with increasing amounts of Flag-PIAS1. Twenty-four hours posttransfection, cells were either untreated (gray bar) or treated with TNF-α for 6 h (black bar), and cell lysates were subjected to luciferase assays. Shown is the average of results from three independent experiments. (B) Inducible expression of Flag-PIAS1 upon removal of DOX in Utf-PIAS1 cells. Lysates (10 μg of protein/lane) from Utf-PIAS1 cells grown in the presence or absence of DOX for 12 h were subjected to Western blot analysis using anti-Flag and antitubulin antibodies. (C) PIAS1 blocks the endogenous NF-κB-inducible genes in response to TNF-α stimulation. Utf and Utf-PIAS1 cells growing in the presence or absence of DOX for 12 h were either untreated or treated with TNF-α for 1 or 3 h. Total RNA (10 μg/lane) was subjected to Northern analysis using a cDNA probe of Bfl1, IκBα, or GAPDH. Shown is a representative of three independent experiments.
To further confirm the role of PIAS1 in the regulation of the NF-κB signaling pathway, we examined the effect of PIAS1 on the activation of the endogenous NF-κB downstream genes in response to TNF-α stimulation. We established a TET-off PIAS1 cell line (Utf-PIAS1) from human osteosarcoma U2OS cells, where the expression of PIAS1 is induced in the absence of DOX, an analog of TET (24) (Fig. 2B). To examine the effect of PIAS1 on the transcriptional activation of endogenous NF-κB-dependent genes, Utf-PIAS1 cells growing in the presence or absence of DOX for 12 h were either left untreated or treated with TNF-α for 1 or 3 h. Total RNA was collected and subjected to Northern blot analysis using a cDNA probe of Bfl-1 or IκBα, two known NF-κB downstream genes (17, 20). Quantitative analysis indicated that the induction of PIAS1 by DOX removal in Utf-PIAS1 cells inhibited the transcriptional activation of Bfl-1 by approximately 60% in response to 3 h of TNF-α treatment, while DOX treatment had no significant effect on Bfl-1 induction in Utf control cells (Fig. 2C). Similarly, the TNF-α-mediated transcriptional activation of IκBα was significantly repressed by PIAS1. These results are consistent with those of the luciferase reporter assays and suggest that PIAS1 acts as an inhibitor of NF-κB-mediated gene activation.
Enhanced NF-κB-mediated gene activation in Pias1 null cells.
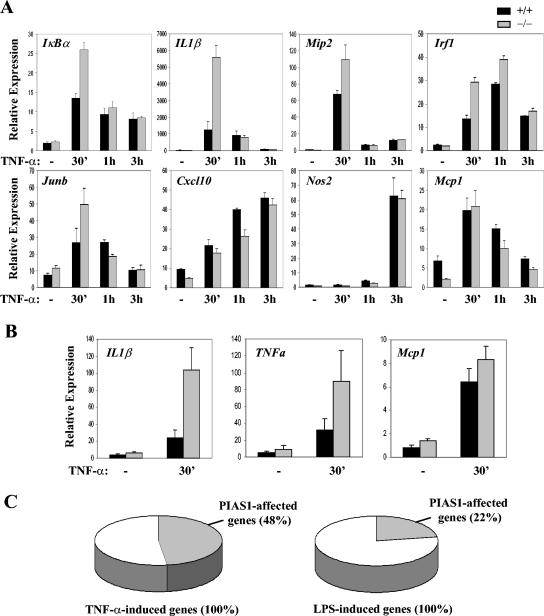
To validate a role of PIAS1 in the regulation of NF-κB, we examined whether NF-κB-mediated gene activation is altered in Pias1 null cells. BMMs from Pias1 null mice and their wild-type littermates were untreated or treated with TNF-α for various time periods (0 h, 30 min, 1 h, and 3 h). Total RNA isolated from these cells was used for the analysis of transcriptional activation of several known NF-κB-dependent genes by Q-PCR (38). The induction of IκBα, IL-1β, Mip2 (macrophage inflammatory protein 2), Irf1 (interferon regulatory factor 1), and Junb by TNF-α was significantly increased in Pias1 null cells compared with wild-type controls (Fig. 3A). Interestingly, the induction of other NF-κB target genes, including Cxcl10, Nos2, and Mcp1, was not significantly affected in the absence of PIAS1 (Fig. 3A). These data suggest that PIAS1 displays specificity in the regulation of NF-κB target genes. Similarly, the TNF-α-induced activation of the IL-1β or TNF-α genes, but not Mcp1, was significantly enhanced in Pias1 null primary embryonic fibroblasts (Fig. 3B).
FIG. 3.
Enhanced expression of NF-κB-regulated genes in response to TNF-α and LPS in Pias1 null cells. (A) Q-PCR analyses of BMMs from wild-type (black bars, +/+) or Pias1 null (gray bars, −/−) littermates, untreated or treated with TNF-α for various times. Genes are indicated at the top left of each panel. Shown is a representative of results from three independent experiments. (B) The procedure described for panel A was carried out except that wild-type and Pias1 null primary MEF cells were used. Shown is the average of results from three independent pairs of MEF cells. (C) Microarray analyses of genes affected by PIAS1. Total RNA from wild-type or Pias1 null BMMs untreated or treated with TNF-α for 30 min (left) or LPS for 1 h (right) was subjected to microarray analyses. TNF- or LPS-induced genes are defined as having at least twofold induction over untreated samples. PIAS1-affected genes are defined as having at least 1.3-fold higher induction in Pias1 null cells than in wild-type cells.
To further examine the specificity of PIAS1 in the regulation of NF-κB signaling, BMMs from Pias1 null cells and their wild-type control cells were either untreated or treated with TNF-α for 30 min or LPS for 1 h. Total RNA isolated from these cells was subjected to microarray analysis. Under these conditions, 65 genes were induced at least twofold after TNF-α stimulation for 30 min, 48% of which showed at least 1.3-fold more induction in Pias1 null cells than wild-type cells. Similarly, 98 genes were induced at least twofold after 1 h of LPS stimulation, but only 22% of them were affected by PIAS1 (Fig. 3C and Table S1 in the supplemental material). These data indicate that PIAS1 selectively affects the induction of a subgroup of TNF-α or LPS-induced genes.
Elevated proinflammatory cytokine production in Pias1 null mice.
The induction of IL-1β and TNF-α genes by TNF-α treatment was significantly enhanced in Pias1 null cells (Fig. 3), consistent with a known important role of NF-κB in the regulation of proinflammatory cytokine gene expression. To directly determine the cytokine levels in mice under physiological conditions, serum samples were prepared from Pias1 null mice and their wild-type littermates and subjected to ELISAs. The levels of two proinflammatory cytokines, IL-1β and TNF-α, were elevated in Pias1 null mice 6-fold and 3.5-fold, respectively (Fig. 4). In contrast, serum levels of IFN-γ and IL-4, two cytokines not directly regulated by the NF-κB pathway, were not significantly altered in Pias1 null mice.
FIG. 4.
Elevated serum cytokine levels in Pias1 null mice compared to their wild-type littermates. Serum samples were collected from 4- to 15-week-old Pias1 null mice (open circles) and their wild-type littermates (filled circles). Serum IL-1β (n = 9), TNF-α (n = 6), IFN-γ (n = 6), and IL-4 (n = 5) levels were determined by ELISA (Biosources). P values were determined by paired t test. The average of cytokine levels in each group is indicated by a dash.
Normal T- and B-lymphocyte development and enhanced granulopoiesis in Pias1 null mice.
Given a potential role of PIAS1 in cytokine-activated signaling pathways, we analyzed whether Pias1 null mice have defects in lymphocyte development. PIAS1 protein is normally expressed in the spleen and thymus. Flow cytometric analyses were carried out with cells from spleens and thymuses of 4- to 8-week-old Pias1 null mice and their wild-type littermates. The CD4/CD8 profiles of the thymocytes and the B220/IgM profiles of the splenocytes were similar for these mice (Fig. 5A), suggesting that both T and B lymphocytes developed normally in the absence of PIAS1.
FIG. 5.
Flow cytometric analysis of the hematopoietic cells from Pias1 null mice and their wild-type littermates. (A) Normal T- and B-lymphoid development in Pias1 null mice. Cells from the thymus and spleen of 6-week-old mice were stained with anti-CD4 and anti-CD8 (thymus) or anti-B220 and anti-IgM (spleen) and analyzed by flow cytometry. The percentages of cells within defined regions are indicated. Data are representative of three separate analyses. (B) Enhanced granulopoiesis in Pias1 null mice. The same splenocytes as in panel A were stained with the granulocyte-specific marker Gr-1 followed by flow cytometric analysis. Data are representative of three separate analyses. (C) Increased expression of G-CSF in Pias1 null thymus. Total RNA from the thymuses of Pias1 null mice and their wild-type littermates (n = 6) was subjected to Q-PCR analyses.
The NF-κB pathway regulates the proliferation and differentiation of granulocytes. In fact, IκBα null mice exhibited enhanced granulopoiesis (5). When splenocytes were stained with the granulocyte-specific marker Gr-1, an increased population of mature granulocytes was reproducibly observed in Pias1 null mice compared to that in the wild-type controls (Fig. 5B). It has been implicated that granulocyte colony-stimulating factor (G-CSF), a key cytokine regulated by the NF-κB pathway, plays an important role in the production of granulocytes (5). Therefore, we determined the RNA levels of G-CSF in wild-type and Pias1 null mice. Total RNA isolated from the thymus of the wild-type and Pias1 null mice was subjected to Q-PCR analysis using specific primers for murine G-CSF. As shown in Fig. 5C, the relative expression of G-CSF was increased in Pias1 null thymus compared to that of the wild-type littermates. In contrast, the RNA levels of GM-CSF were not altered in Pias1 null thymus. Similar observations have been described for IκBα null mice (5). These results further support the role of PIAS1 in the negative regulation of NF-κB in vivo.
PIAS1 has no effect on the activation and nuclear translocation of NF-κB.
To understand how PIAS1 regulates NF-κB signaling, we examined whether PIAS1 affects the activation and nuclear translocation of NF-κB p65. The localization of the endogenous PIAS1 and p65 was examined by immunofluorescence analyses. Wild-type (6+/+) and Pias1 null (7−/−) mouse embryo fibroblasts (MEFs) were either untreated or treated with TNF-α for 15 min and then stained with anti-p65 and anti-PIAS1 antibodies simultaneously. In wild-type MEFs, NF-κB p65 resided in the cytoplasm of untreated cells and translocated into the nucleus upon TNF-α stimulation, which is consistent with published results. In contrast, PIAS1 remained in the nucleus with or without TNF-α stimulation. The colocalization of p65 and PIAS1 was observed in the nucleus after TNF-α stimulation (Fig. 6A). In Pias1 null MEFs, anti-PIAS1 did not reveal specific nuclear staining as observed in Pias1+/+ cells, which validates the specificity of the anti-PIAS1 antibody (Fig. 6A). Most importantly, the nuclear translocation of p65 appeared normal in Pias1 null MEFs, suggesting that PIAS1 does not affect the nuclear translocation of NF-κB p65 in response to TNF-α. Similar results were also obtained in wild-type and Pias1 null BMMs untreated or treated with LPS (data not shown).
FIG. 6.
PIAS1 does not affect the activation and nuclear translocation of NF-κB. (A) Immunofluorescence analysis of PIAS1 and NF-κB p65 in wild-type (6+/+) and Pias1 null (7−/−) MEFs. Cells were untreated or treated with TNF-α for 30 min followed by costaining with anti-p65 and anti-PIAS1 antibodies. The nucleus is visualized by the Hoechst DNA dye. (B) IκBα degradation and p65 translocation were not altered in Pias1 null cells. BMMs from Pias1+/+ and Pias1−/− mice were either untreated or treated with TNF-α (20 ng/ml) or LPS (10 ng/ml) for various times. Cytoplasmic and nuclear fractions were prepared and subjected to Western blot analyses using anti-IκBα or anti-p65 as indicated. The same filters were probed with antiactin to reveal equal loadings in each lane.
To further examine the effect of PIAS1 on the activation and nuclear translocation of NF-κB, cytoplasmic and nuclear extracts of Pias1+/+ and Pias1−/− BMMs untreated or treated with TNF-α or LPS for various times were prepared, followed by Western blot analysis using anti-IκBα or anti-p65. In Pias1+/+ BMMs, IκBα was degraded in the cytoplasm with the concurrent translocation of p65 into the nucleus upon stimulation (Fig. 6B). In Pias1−/− BMMs, IκBα degradation and p65 translocation were normal compared to the wild-type controls (Fig. 6B, compare lanes 2 to 6 and 8 to 12). Thus, PIAS1 does not affect the signaling events leading to the nuclear translocation of p65.
PIAS1 blocks the DNA binding activity of NF-κB.
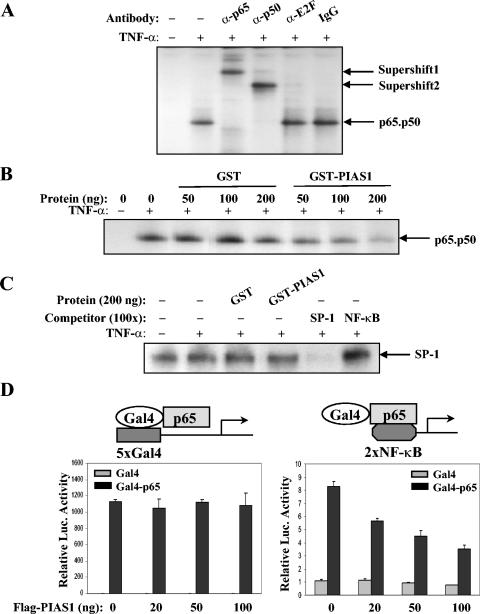
We examined the effect of PIAS1 on the DNA binding activity of NF-κB in vitro by EMSA. Nuclear extracts from MCF-7 cells untreated or treated with TNF-α were analyzed by EMSA using an NF-κB binding site as the probe. TNF-α treatment induced the formation of a specific shift band, which represents the NF-κB p50-p65 heterodimer, since it was specifically supershifted by anti-p50 or anti-p65 antibody but not by anti-E2F or rabbit IgG (Fig. 7A). To test the effect of PIAS1 on the DNA binding activity of NF-κB, GST-PIAS1 protein was prepared and used in EMSA. The addition of purified GST-PIAS1 protein inhibited the DNA binding activity of NF-κB p50-p65 in a dose-dependent manner (Fig. 7B, lanes 6 to 8). As a control, the addition of the same amounts of GST protein had no effect on the DNA binding activity of NF-κB (Fig. 7B, lanes 3 to 5). Under similar conditions, GST-PIAS1 does not affect the DNA binding activity of SP-1 (Fig. 7C), supporting the conclusion that the effect of GST-PIAS1 on the DNA binding of NF-κB is specific.
FIG. 7.
PIAS1 can block the DNA binding activity of NF-κB. (A) Nuclear extracts from MCF-7 cells untreated or treated with TNF-α for 15 min were analyzed by EMSA using an oligonucleotide containing the NF-κB binding site as a probe. The identity of the shift band was examined by incubating with anti-p65, anti-p50, anti-E2F, or rabbit IgG as indicated. (B) The same extracts from panel A were mixed with increasing amounts of purified GST (lanes 3 to 5) or GST-PIAS1 proteins (lanes 6 to 8) followed by EMSA analysis. (C) The same procedure as described for panel B was carried out except that an SP-1 oligonucleotide was used as a probe. (D) Luciferase assays were performed with 293T cells with an expression construct encoding Gal4 (gray bar) or a Gal4-p65 fusion protein (black bar), increasing amounts of Flag-PIAS1, and either 5×Gal4 reporter (left) or 2×NF-κB reporter (right). Shown is the average of results from three independent experiments.
To test the hypothesis that PIAS1 inhibits the NF-κB-mediated gene activation by blocking the DNA binding activity of p65 in vivo, we used a Gal4-p65 fusion protein, which can activate the luciferase reporter constructs carrying either the Gal4-binding site or the NF-κB binding site. When human 293T cells were transiently transfected with the Gal4-p65 expression construct, the 5×Gal4 reporter containing five copies of the Gal4 binding site and increasing amounts of PIAS1, PIAS1 showed no inhibition on Gal4-p65-mediated gene activation (Fig. 7D). In contrast, when the 2×NF-κB reporter construct was used in the luciferase assays, PIAS1 inhibited the transcriptional activity of the Gal4-p65 fusion protein (Fig. 7D). These results support the conclusion that PIAS1 inhibits NF-κB-mediated transcription by blocking the DNA binding activity of NF-κB p65.
To further examine whether PIAS1 has the ability to regulate the DNA binding activity of p65 in vivo, we performed ChIP assays to examine the binding of p65 to the promoters of endogenous NF-κB-regulated genes. Protein extracts from wild-type and Pias1 null BMMs untreated or treated with LPS for 20 min or 1 h were immunoprecipitated with anti-p65 or anti-IgG. The bound DNA was quantified by Q-PCR analysis using specific primers (Fig. 8A). The binding of p65 to the endogenous IκBα promoter upon LPS stimulation was significantly enhanced in Pias1 null cells. Similar analysis was also performed with protein extracts prepared from the TET-off PIAS1 cell line (Utf-PIAS1). The induction of PIAS1 in the absence of DOX significantly repressed the binding of p65 to the promoter of IκBα in response to TNF-α treatment (Fig. 8B). These results further support the conclusion that PIAS1 inhibits the activity of NF-κB by interfering with the recruitment of p65 to the promoters of NF-κB-regulated genes.
FIG. 8.
PIAS1 affects the binding of p65 to the endogenous promoters revealed by ChIP assays. (A) The binding of p65 to the IκBα promoter is increased in Pias1 null cells. ChIP assays were performed with BMMs from wild-type (+/+) or Pias1 null (−/−) littermates untreated or treated with LPS (10 ng/ml). p65-coprecipitated DNA was analyzed by Q-PCR with specific primers against the κB site in the murine IκBα promoter (mIκBα). Rabbit IgG was used as a negative control. (B) The procedure described for panel A was performed except that extracts used were from PIAS1 TET-off cells in the presence or absence of DOX for 12 h. Q-PCR was performed with specific primers against the κB site in the human IκBα promoter (hIκBα).
DISCUSSION
In this paper, we examined the role of PIAS1 in the regulation of NF-κB signaling. We showed that PIAS1 interacts with the p65 subunit of NF-κB by coimmunoprecipitation assays. We demonstrated that the ectopic expression of PIAS1 inhibits NF-κB-mediated gene activation. Consistently, the induction of a subset of NF-κB-mediated genes by TNF-α and LPS is significantly enhanced in Pias1−/− cells. Taking together the biochemical and genetic results, we suggest that PIAS1 is a novel negative regulator of NF-κB. We further explored the molecular mechanism of PIAS1-mediated inhibition on NF-κB. We showed that PIAS1, which is expressed mainly in the nucleus, does not affect the activation or the nuclear translocation of NF-κB p65. PIAS1 can block the DNA binding activity of p65 both in vitro and in vivo. Through analysis of the Gal4-p65 fusion protein, we showed that the inhibition by PIAS1 of the transcriptional activity of Gal4-p65 specifically occurs on the NF-κB binding site but not the Gal4-binding site. Consistently, ChIP assays indicate that the recruitment of p65 to the promoters of NF-κB-regulated genes is significantly enhanced in Pias1−/− cells but repressed in PIAS1-overexpressing cells. These data suggest that PIAS1 is a physiologically important negative regulator of NF-κB. PIAS1 inhibits the transcriptional activity of NF-κB by blocking the DNA binding activity of NF-κB p65.
In addition to regulating the DNA binding activity of a transcription factor, PIAS proteins have also been suggested to regulate transcription through other molecular mechanisms (33). For example, PIAS proteins possess SUMO E3 ligase activity. It has been suggested that PIASy may repress the transcriptional activity of LEF1 by targeting LEF1 to nuclear bodies (31). Experiments described in this paper indicate that the lack of PIAS1 has no effect on the cellular localization of p65. Similarly, the nuclear translocation of STAT1 in response to IFNs is not affected in the absence of PIAS1 (23). The physiological significance of PIAS1 SUMO E3 ligase activity in the regulation of NF-κB or STAT1 remains to be determined.
NF-κB plays an important role in immune and inflammatory responses. The dysregulation of NF-κB activity is associated with a number of human diseases (3, 4, 10, 12). Thus, the activity of NF-κB must be properly regulated. In the cytoplasm, the binding of the IκB family of proteins prevents the nuclear translocation of NF-κB (16). In the nucleus, it has been shown that Twist-2 can directly bind and repress the transcriptional activity of NF-κB (35). Interestingly, both IκB and Twist proteins are cytokine inducible, suggesting that they act as negative feedback loops of NF-κB signaling. In this paper, we showed that the activity of NF-κB is tightly regulated by PIAS1: either the increased or the decreased expression of PIAS1 results in abnormal NF-κB activity. PIAS1 is constitutively expressed in the nucleus. It is possible that PIAS1 may act as a threshold to control the strength of NF-κB signaling. Alternatively, the activity of PIAS1 may be regulated by TNF-α through an as-yet-unidentified mechanism.
Our results suggest that the endogenous PIAS1 and p65 reside in different compartments in unstimulated cells, and they become colocalized to the nucleus upon stimulation. However, we have not been able to detect the endogenous PIAS1-p65 interaction by coimmunoprecipitation analysis. It is possible that the PIAS1-p65 complex is not stable under the coimmunoprecipitation conditions used. Alternatively, only a small portion of p65 may interact with PIAS1 in vivo. Ligand-independent interaction between PIAS1 and p65 was observed when PIAS1 and p65 were coexpressed in 293T cells. This ligand-independent PIAS1-p65 interaction may result from the constitutive nuclear localization of p65 when it is overexpressed. Under physiological conditions, ligand stimulation is required to cause the translocation of p65 into the nucleus, where it interacts with PIAS1. Our results indicate that PIAS1 does not affect the signaling events leading to the nuclear translocation of p65. Instead, PIAS1 acts in the nucleus to repress the DNA binding activity of p65. The effect of PIAS1 on the DNA binding activity of p65 in Pias1 null cells was validated by in vivo ChIP assays. However, an enhanced DNA binding activity of p65 was not observed in Pias1 null cell extracts when examined by in vitro EMSA (unpublished observation). It is possible that under physiological conditions, the local concentration of PIAS1 and/or the native chromatin structure of the promoter regions may affect the inhibitory activity of PIAS1 on NF-κB.
Studies using PIAS1-deficient cells indicate that PIAS1 affects only a subset of TNF-α-mediated gene expression. The selective effect of PIAS1 on NF-κB-dependent gene expression is similar to its specific role in STAT1 signaling (23). Interestingly, the transcriptional activation of certain genes requires the participation of both NF-κB and STAT1. For example, STAT1 is involved in the induction of Irf1 and Cxcl10 by IFN-γ, while the induction of Irf1 and Cxcl10 by TNF-α requires NF-κB (29, 30). However, Irf1 is a PIAS1-insensitive gene in response to IFNs (23), whereas the induction of Irf1 by TNF-α is significantly enhanced in the absence of PIAS1. In contrast, the induction of Cxcl10 by IFNs, but not by TNF-α, is PIAS1 sensitive (23). These results support the conclusion that PIAS1 can independently regulate the transcriptional activity of STAT1 and NF-κB. The selective effect of PIAS1 on the regulation of TNF-α-responsive genes is not fully understood yet. One possible explanation is the redundant role of other PIAS family members in NF-κB signaling. Interestingly, a potential role of PIAS3 in regulating NF-κB activity has been proposed (14). Further studies are required to understand whether PIAS3 or other PIAS members are indeed involved in the transcriptional regulation of endogenous NF-κB-dependent genes.
Supplementary Material
Acknowledgments
We thank J. Black and W. Xie for help on generating constructs.
This work was supported by grants from the NIH (M.A.T., G.C., H.W., and K.S.) and the Howard Hughes Medical Institute (H.W.). B.L. is a special fellow of the Leukemia and Lymphoma Society. K.A.W. was supported by a Dr. Norman Sprague, Jr., Fellowship and U.S. Public Health Service National Research Service award GM07185.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arora, T., B. Liu, H. He, J. Kim, T. L. Murphy, K. M. Murphy, R. L. Modlin, and K. Shuai. 2003. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J. Biol. Chem. 278:21327-21330. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle, P. A., and V. R. Baichwal. 1997. NF-kappa B as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv. Immunol. 65:111-137. [PubMed] [Google Scholar]
- 3.Barnes, P. J., and I. M. Adcock. 1997. NF-kappa B: a pivotal role in asthma and a new target for therapy. Trends Pharmacol. Sci. 18:46-50. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, P. J., and M. Karin. 1997. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336:1066-1071. [DOI] [PubMed] [Google Scholar]
- 5.Beg, A. A., W. C. Sha, R. T. Bronson, and D. Baltimore. 1995. Constitutive NF-kappa B activation, enhanced granulopoiesis, and neonatal lethality in I kappa B alpha-deficient mice. Genes Dev. 9:2736-2746. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X., U. Vinkemeier, Y. Zhao, D. Jeruzalmi, J. E. Darnell, Jr., and J. Kuriyan. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827-839. [DOI] [PubMed] [Google Scholar]
- 7.Chin, A. I., P. W. Dempsey, K. Bruhn, J. F. Miller, Y. Xu, and G. Cheng. 2002. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416:190-194. [DOI] [PubMed] [Google Scholar]
- 8.Chung, C. D., J. Liao, B. Liu, X. Rao, P. Jay, P. Berta, and K. Shuai. 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803-1805. [DOI] [PubMed] [Google Scholar]
- 9.Doyle, S., S. Vaidya, R. O'Connell, H. Dadgostar, P. Dempsey, T. Wu, G. Rao, R. Sun, M. Haberland, R. Modlin, and G. Cheng. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17:251-263. [DOI] [PubMed] [Google Scholar]
- 10.Foxwell, B., K. Browne, J. Bondeson, C. Clarke, R. de Martin, F. Brennan, and M. Feldmann. 1998. Efficient adenoviral infection with IkappaB alpha reveals that macrophage tumor necrosis factor alpha production in rheumatoid arthritis is NF-kappaB dependent. Proc. Natl. Acad. Sci. USA 95:8211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore, T. D., M. Koedood, K. A. Piffat, and D. W. White. 1996. Rel/NF-kappaB/IkappaB proteins and cancer. Oncogene 13:1367-1378. [PubMed] [Google Scholar]
- 13.Gross, M., B. Liu, J. Tan, F. S. French, M. Carey, and K. Shuai. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 20:3880-3887. [DOI] [PubMed] [Google Scholar]
- 14.Jang, H. D., K. Yoon, Y. J. Shin, J. Kim, and S. Y. Lee. 2004. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J. Biol. Chem. 279:24873-24880. [DOI] [PubMed] [Google Scholar]
- 15.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 16.Karin, M. 1999. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 17.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 18.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 19.Le, L. Q., J. H. Kabarowski, Z. Weng, A. B. Satterthwaite, E. T. Harvill, E. R. Jensen, J. F. Miller, and O. N. Witte. 2001. Mice lacking the orphan G protein-coupled receptor G2A develop a late-onset autoimmune syndrome. Immunity 14:561-571. [DOI] [PubMed] [Google Scholar]
- 20.Lee, H. H., H. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-kappaB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA 96:9136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, B., M. Gross, J. ten Hoeve, and K. Shuai. 2001. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 98:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 95:10626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, B., S. Mink, K. A. Wong, N. Stein, C. Getman, P. W. Dempsey, H. Wu, and K. Shuai. 2004. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat. Immunol. 5:891-898. [DOI] [PubMed] [Google Scholar]
- 24.Liu, B., and K. Shuai. 2001. Induction of apoptosis by protein inhibitor of activated Stat1 through c-Jun NH2-terminal kinase activation. J. Biol. Chem. 276:36624-36631. [DOI] [PubMed] [Google Scholar]
- 25.Majeti, R., Z. Xu, T. G. Parslow, J. L. Olson, D. I. Daikh, N. Killeen, and A. Weiss. 2000. An inactivating point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell 103:1059-1070. [DOI] [PubMed] [Google Scholar]
- 26.Megidish, T., J. H. Xu, and C. W. Xu. 2002. Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 277:8255-8259. [DOI] [PubMed] [Google Scholar]
- 27.Mercurio, F., and A. M. Manning. 1999. NF-kappaB as a primary regulator of the stress response. Oncogene 18:6163-6171. [DOI] [PubMed] [Google Scholar]
- 28.Moilanen, A. M., U. Karvonen, H. Poukka, W. Yan, J. Toppari, O. A. Jeanne, and J. J. Palvimo. 1999. A testis-specific androgen receptor coregulator that belongs to a novel family of nuclear proteins. J. Biol. Chem. 274:3700-3704. [DOI] [PubMed] [Google Scholar]
- 29.Nazar, A. S., G. Cheng, H. S. Shin, P. N. Brothers, S. Dhib-Jalbut, M. L. Shin, and P. Vanguri. 1997. Induction of IP-10 chemokine promoter by measles virus: comparison with interferon-gamma shows the use of the same response element but with differential DNA-protein binding profiles. J. Neuroimmunol. 77:116-127. [DOI] [PubMed] [Google Scholar]
- 30.Pine, R. 1997. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is mediated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 25:4346-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuai, K., and B. Liu. 2003. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3:900-911. [DOI] [PubMed] [Google Scholar]
- 34.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 35.Sosic, D., J. A. Richardson, K. Yu, D. M. Ornitz, and E. N. Olson. 2003. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell 112:169-180. [DOI] [PubMed] [Google Scholar]
- 36.Tan, J., S. H. Hall, K. G. Hamil, G. Grossman, P. Petrusz, J. Liao, K. Shuai, and F. S. French. 2000. Protein inhibitor of activated Stat-1 is a nuclear receptor co-regulator expressed in human testis. Mol. Endocrinol. 14:14-26. [DOI] [PubMed] [Google Scholar]
- 37.Tan, J. A., S. H. Hall, K. G. Hamil, G. Grossman, P. Petrusz, and F. S. French. 2002. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J. Biol. Chem. 277:16993-17001. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, A., S. Scoggin, R. B. Gaynor, and N. S. Williams. 2003. Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22:2054-2064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.